Abstract
Background
Whole-exome sequencing (WES) has recently emerged as an appealing approach to systematically study coding variants. However, the requirement for a large amount of high-quality DNA poses a barrier that may limit its application in large cancer epidemiologic studies. We evaluated the performance of WES with low input amount and saliva DNA as an alternative source material.
Methods
Five breast cancer patients were randomly selected from the Pathways Study. From each patient, four samples, including 3 µg, 1 µg, and 0.2 µg blood DNA and 1 µg saliva DNA, were aliquoted for library preparation using the Agilent SureSelect kit and sequencing using Illumina HiSeq2500. Quality metrics of sequencing and variant calling, as well as concordance of variant calls from the whole exome and 21 known breast cancer genes, were assessed by input amount and DNA source.
Results
There was little difference by input amount or DNA source on the quality of sequencing and variant calling. The concordance rate was about 98% for single nucleotide variant calls and 83–86% for short insertion/deletion calls. For the 21 known breast cancer genes, WES based on low input amount and saliva DNA identified the same set variants in samples from a same patient.
Conclusions
Low DNA input amount, as well as saliva DNA, can be used to generate WES data of satisfactory quality.
Impact
Our findings support the expansion of WES applications in cancer epidemiologic studies where only low DNA amount or saliva samples are available.
Keywords: whole-exome sequencing, cancer epidemiology, breast cancer, SNV, indel
Introduction
The advent of next-generation sequencing (NGS) techniques and the reduction in cost overtime have transformed the landscape of human genetic research by offering a widely accessible tool to interrogate the genome at an unprecedented pace and scale (1). Compared to whole genome sequencing (WGS), which remains costly for population-wide applications, whole-exome sequencing (WES), which targets the approximately 1% coding sequences of the human genome, provides an appealing solution with a balanced trade-off between cost, genome coverage, functional annotation, and analytical burden (2, 3). It thus has been widely adopted to study Mendelian diseases (4, 5) and characterize cancer genomes (6), and begun to make its way into clinical practice for novel diagnosis and identification of therapy targets (7–9).
In epidemiologic research, WES is emerging as a new powerhouse in searching for coding risk variants (10–12), surpassing genome-wide genotyping microarrays that are limited to common and known variants. Several previous studies have evaluated the performance of different WES technologies and platforms (13–15). However, two practical issues remain that may impede the application of WES to large epidemiologic populations, namely the apparent need for relatively large amounts of high-quality DNA, and the current need to source this from peripheral blood. The large amount of needed genomic DNA (e.g., 3 µg) poses a practical challenge to studies where such amounts are unavailable or would deplete the resource. As saliva samples are now routinely collected in many epidemiologic studies as an inexpensive alternative source of genomic DNA using non-invasive methods, there could be broader use of WES if it were shown that saliva DNA performs comparably well to blood DNA on WES platforms.
To address these two aforementioned issues in WES, we evaluated the WES performance of the Agilent SureSelect Human All Exon kit in conjunction with the Illumina HiSeq 2500 platform, which is currently one of the few mainstream choices for WES library preparation and sequencing, respectively (16, 17). Our goal was to determine the performance of sequencing, variant calling for single nucleotide variations (SNVs) and short insertion/deletion (indels), and the accuracy in identifying coding variants in known breast cancer-related genes, using different DNA input amounts (0.2 µg, 1 µg, and 3 µg genome DNA) from peripheral blood, and different DNA sources (1 µg DNA from saliva).
Materials and Methods
Genomic DNA samples
Genomic DNA samples were obtained from the Pathways Study, a prospective cohort study that recruited recently-diagnosed breast cancer patients from the Kaiser Permanente Northern California (KPNC) health plan membership (18). At the baseline in-person interview after patient consent, blood samples were collected from 90% of participants via phlebotomy, and saliva samples were also collected from 96% of participants by the Oragene™ DNA Self-Collection kit (DNA Genotek Inc., Kanata, Ontario, Canada) as an alternative source of genomic DNA. The biospecimens were shipped to Roswell Park Cancer Institute (RPCI) for processing and storage under the auspices of the RPCI Data Bank and Biorepository (DBBR) (19). Whole blood was aliquoted for DNA extraction using the Qiagen FlexiGene kit (Valencia, CA). DNA from approximately 2 ml saliva samples was extracted using the Oragene kit. Nucleotide concentration of DNA samples was determined by both NanoDrop and PicoGreen techniques. DNA samples were stored at −80°C until analysis. For this study, we included randomly-selected samples from five women diagnosed with triple-negative breast cancer [estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and human epidermal growth factor receptor 2 (Her2)-negative] who had DNA available from both peripheral blood and saliva samples. The study was approved by the Institutional Review Boards (IRB) of RPCI and KPNC.
Library preparation and sequencing
Genomic DNA from whole blood (3 µg, 1 µg, and 0.2 µg DNA) and from saliva (1 µg DNA) was captured using the Agilent SureSelect Human All Exon v5 Kit (Santa Clara, CA). The 3 µg and 1 µg input amounts were fragmented to a size range of 150–200 bp followed by end repair, adaptor ligation, and low PCR cycle (5 cycles). The 0.2 µg input followed the same procedures, except using a higher number of PCR cycles (11 cycles). Individual libraries were barcoded, pooled (5-plex) and loaded to four lanes of a HiSeq Flow Cell, followed by 101 bp paired-end sequencing using Illumina HiSeq 2500 (San Diego, CA) according to manufacturer’s protocol. To eliminate potential batch effects, the libraries were randomly assigned to four sequencing lanes using the OSAT program to ensure that the distribution of DNA input amount and DNA source was even across lanes (20). The library preparation and sequencing was performed by the RPCI Genomics Shared Resource.
Variant calling for SNVs and Indels
The raw sequence reads were aligned to the Human Reference Genome (NCBI build 37) using the Burrows-Wheeler Aligner (21). After removing PCR duplicates using Picard (22), the GATK software version 3.0 (23) was used for local realignment, base quality recalibration, and variant calling of SNVs and small indels. In the variant-calling step, variants were first called in each sample separately, and then joint genotyping analysis was performed on the samples from the same DNA source and same DNA input amount, followed by variant recalibration to generate analysis-ready variants. Only the variants that passed the GATK quality filter (tranche sensitivity threshold 99.9%) were used in our analysis.
Benchmark rate of variant calling concordance
As the bioinformatics pipeline may have a major impact on variant calling concordance, we estimated the concordance level of our pipeline based on a reference WES dataset with high-quality variant callsets and used the concordance rate as a benchmark in our evaluation. The publicly-available WES data of a CEU trio (NA12878, NA12891, and NA12892) was downloaded from the 1000 Genomes Project. The WES data were originally generated using Agilent SureSelect All Exon v2 Kit, followed by 76 bp paired-end sequencing. Variant calls for NA12878 from our pipeline were compared with two comprehensive variant callsets compiled by the Genome in a Bottle Consortium (GIBA) for this particular individual (24). The two callsets contain high-quality variant calls in the whole exome and in the high-confidence portion of the exome, respectively. The high-confidence portion of the exome excludes simple repeats, known segmental duplications, known structural variants reported in dbVar (25) for NA12878, regions paralogous to the 1000 Genomes Project “decoy reference”, and regions in the RepeatSeq database (26). The calculated concordance rates of SNV and indel calls for the NA12878 subject were then used as guidelines for assessing the consistency of variant calling for samples of varying DNA input amount and DNA source from each patient in our study.
Results
Sequencing performance
From each exome library, we obtained 63–102 million reads, with an average sequencing depth of 67–111× and ≥94% bases covered by at least 20× (Table 1). The PCR duplicate rates in all samples ranged from 0.03–0.13, except for one outlier library generated from 1 µg blood DNA with a duplicate rate of 0.30. The mapping rate of the sequenced reads to the reference genome in each sample was 98–100%; the exome capture rate was 50% on average; and the average insert size was 200 bp. All were within the expected range, indicating overall good performance of exome sequencing.
Table 1.
Summary of whole-exome sequencing data statistics
| Sample ID | DNA source | DNA amount |
Average sequencing depth |
% bases covered by at least 20× |
Exome capture rate* |
PCR duplicate rate |
Mapping % |
Mean insert size |
|---|---|---|---|---|---|---|---|---|
| PBCTNPLT001 | whole blood | 3 µg | 111 | 97.5 | 0.55 | 0.08 | 99.7 | 215 |
| PBCTNPLT002 | whole blood | 3 µg | 93 | 96.2 | 0.57 | 0.08 | 99.7 | 206 |
| PBCTNPLT003 | whole blood | 3 µg | 92 | 95.9 | 0.57 | 0.06 | 99.7 | 207 |
| PBCTNPLT004 | whole blood | 3 µg | 70 | 94.0 | 0.55 | 0.06 | 99.7 | 216 |
| PBCTNPLT005 | whole blood | 3 µg | 77 | 95.2 | 0.59 | 0.03 | 99.7 | 208 |
| PBCTNPLT001 | whole blood | 1 µg | 74 | 94.3 | 0.42 | 0.30 | 99.6 | 207 |
| PBCTNPLT002 | whole blood | 1 µg | 92 | 96.5 | 0.54 | 0.08 | 99.7 | 214 |
| PBCTNPLT003 | whole blood | 1 µg | 88 | 95.6 | 0.55 | 0.09 | 99.7 | 212 |
| PBCTNPLT004 | whole blood | 1 µg | 79 | 95.8 | 0.56 | 0.06 | 99.7 | 217 |
| PBCTNPLT005 | whole blood | 1 µg | 111 | 98.0 | 0.56 | 0.06 | 99.7 | 209 |
| PBCTNPLT001 | whole blood | 0.2 µg | 67 | 93.8 | 0.48 | 0.12 | 99.2 | 186 |
| PBCTNPLT002 | whole blood | 0.2 µg | 75 | 94.8 | 0.51 | 0.08 | 99.0 | 185 |
| PBCTNPLT003 | whole blood | 0.2 µg | 77 | 94.9 | 0.53 | 0.07 | 99.0 | 184 |
| PBCTNPLT004 | whole blood | 0.2 µg | 74 | 94.9 | 0.51 | 0.08 | 99.0 | 182 |
| PBCTNPLT005 | whole blood | 0.2 µg | 79 | 95.5 | 0.53 | 0.09 | 98.9 | 186 |
| PBCTNPLT001 | saliva | 1 µg | 72 | 94.9 | 0.56 | 0.08 | 99.1 | 205 |
| PBCTNPLT002 | saliva | 1 µg | 77 | 95.7 | 0.54 | 0.05 | 99.4 | 214 |
| PBCTNPLT003 | saliva | 1 µg | 99 | 97.4 | 0.50 | 0.13 | 98.2 | 203 |
| PBCTNPLT004 | saliva | 1 µg | 73 | 95.0 | 0.52 | 0.10 | 97.7 | 210 |
| PBCTNPLT005 | saliva | 1 µg | 69 | 93.9 | 0.51 | 0.12 | 98.5 | 201 |
The exome capture rate is calculated as the sequenced bases in the capture regions divided by the length sum of all mapped reads.
We then examined whether the DNA input amount and DNA source affected sequencing quality. In comparisons of the two lower DNA input amounts (1 µg and 0.2 µg) with the standard 3 µg blood DNA, no significant differences were found in total sequenced reads, sequencing depth, percent bases covered by at least 20×, PCR duplicate rate, or exome capture rate. The only significant differences were the mapping rate and the mean insert size. The mapping rate from the 0.2 µg DNA input was marginally lower, and the mean insert size was shorter than the two higher input amounts (Student’s t-test p-values ≤ 0.001). Similarly, in comparisons between saliva DNA and blood DNA, the only significant differences were also observed in the mapping rate and the mean insert size (p-values<0.05). It should be noted, however, that all the mapping rates exceeded 98%. Using a multivariable linear model to relate each of the sequencing statistics in Table 1 with patient ID, DNA amount and DNA source, only the mean insert size was significantly different by input amount, with shorter insert size when using 0.2 µg DNA compared to 1µg DNA (p<0.001).
Quality of variant calls
We next investigated the performance of variant calling by DNA input amount and DNA source. For each of the five breast cancer patients, we detected 42.6–52.3 k variants including SNVs and indels. The number of indels was approximately 10.8–12.0% of the number of SNVs, consistent with that from the 1000 Genomes data (27). We investigated the overall variant calling quality on the basis of three commonly-used quality metrics: transition-transversion ratio (Ti/Tv) for SNV calls, heterozygous-homozygous ratio (Het/Homo), and percentage of overlap with known variants in dbSNP (Table 2). The Ti/Tv ratio for each sample ranged from 2.59–2.64, consistent with that commonly-observed in WES studies (13, 28, 29).
Table 2.
Quality metrics of variant calls
| Sample ID | DNA source | DNA input amount |
# variants | Transition/ transversion ratio |
Heterozygous /homozygous call ratio |
% overlapping with dbSNP |
|---|---|---|---|---|---|---|
| PBCTNPLT001 | whole blood | 3 µg | 43014 | 2.59 | 1.59 | 98.46 |
| PBCTNPLT002 | whole blood | 3 µg | 44353 | 2.63 | 1.59 | 98.45 |
| PBCTNPLT003 | whole blood | 3 µg | 44868 | 2.61 | 1.66 | 98.13 |
| PBCTNPLT004 | whole blood | 3 µg | 52352 | 2.62 | 2.03 | 98.07 |
| PBCTNPLT005 | whole blood | 3 µg | 43781 | 2.62 | 1.64 | 98.46 |
| PBCTNPLT001 | whole blood | 1 µg | 42610 | 2.60 | 1.58 | 98.61 |
| PBCTNPLT002 | whole blood | 1 µg | 44202 | 2.64 | 1.60 | 98.51 |
| PBCTNPLT003 | whole blood | 1 µg | 44645 | 2.62 | 1.66 | 98.19 |
| PBCTNPLT004 | whole blood | 1 µg | 52206 | 2.61 | 2.04 | 98.10 |
| PBCTNPLT005 | whole blood | 1 µg | 43841 | 2.62 | 1.66 | 98.40 |
| PBCTNPLT001 | whole blood | 0.2 µg | 42592 | 2.59 | 1.57 | 98.51 |
| PBCTNPLT002 | whole blood | 0.2 µg | 44060 | 2.64 | 1.60 | 98.51 |
| PBCTNPLT003 | whole blood | 0.2 µg | 44707 | 2.61 | 1.65 | 98.17 |
| PBCTNPLT004 | whole blood | 0.2 µg | 52161 | 2.61 | 2.03 | 98.08 |
| PBCTNPLT005 | whole blood | 0.2 µg | 43626 | 2.62 | 1.64 | 98.43 |
| PBCTNPLT001 | saliva | 1 µg | 42711 | 2.59 | 1.58 | 98.56 |
| PBCTNPLT002 | saliva | 1 µg | 44317 | 2.64 | 1.60 | 98.50 |
| PBCTNPLT003 | saliva | 1 µg | 44998 | 2.61 | 1.71 | 98.16 |
| PBCTNPLT004 | saliva | 1 µg | 52287 | 2.62 | 2.04 | 98.14 |
| PBCTNPLT005 | saliva | 1 µg | 43700 | 2.62 | 1.64 | 98.50 |
The Het/Homo call ratios varied notably by patients’ racial/ethnic background. Among the three patients of European descent (PBCTNPLT001, 002, 005) and one patient of Hispanic descent (PBCTNPLT003), the ratio ranged from 1.58–1.71, while the ratio was higher in all four samples from one patient of African descent (PBCTNPLT004), with values as high as 2.03–2.04. This racial/ethnic variation is consistent with the literature (16, 30). Of note, DNA from this patient also showed the highest number of variants among the five patients evaluated, probably due to high genetic diversity in African ancestry (31, 32). The overlap between the called variants and known variants from dbSNP was high, and the percentage of novel variants was below 2%, with the highest in samples from the two patients of non-European descent (PBCTNPLT003, 004). Despite these racial/ethnic variations, we noticed little difference in any of the above three quality metrics of variant calls by DNA input amount or DNA source.
Concordance of variant calls by DNA input amount and DNA source
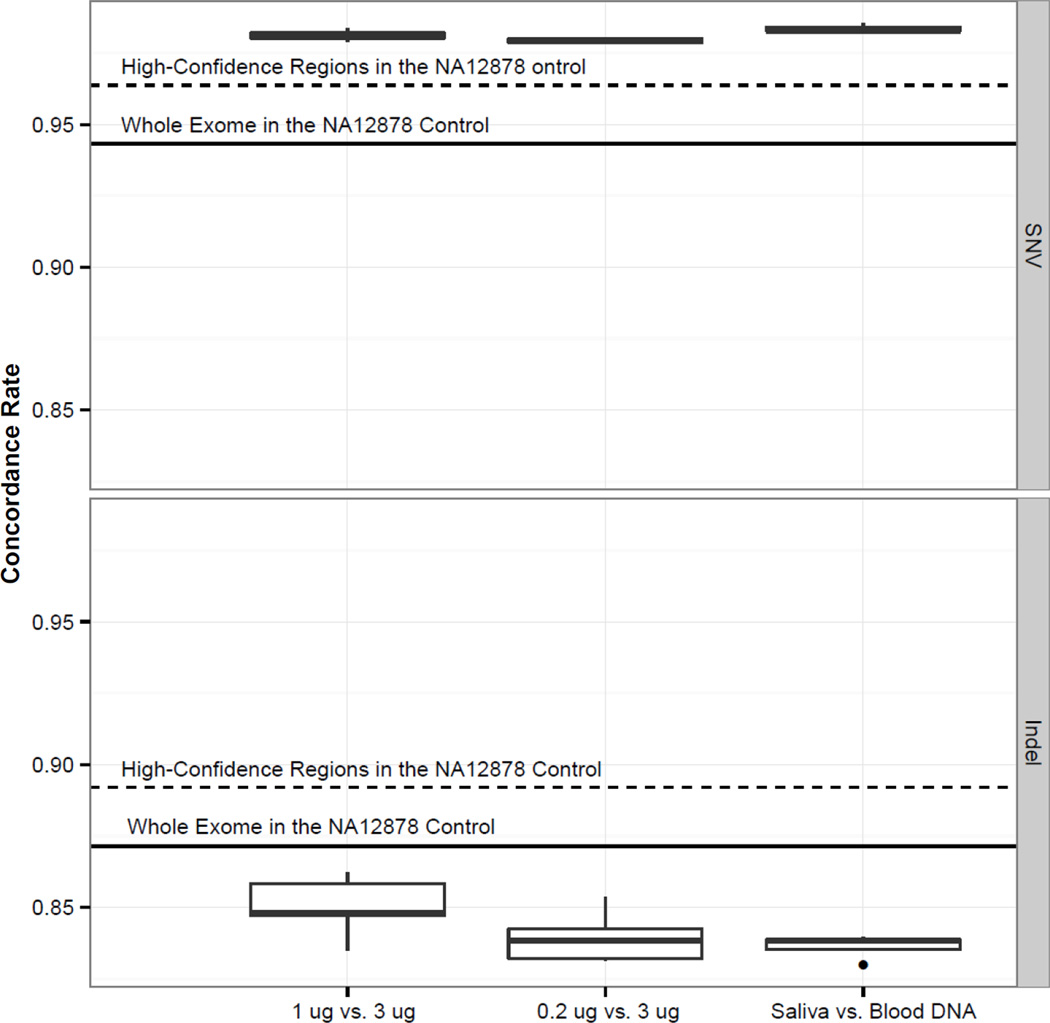
We next assessed the concordance of variant calls within each patient between each of the two lower DNA input amounts and the 3 µg input amount, as well as between saliva DNA and blood DNA. In all comparisons, the concordance rate was close to or exceeded 98% for SNVs, and for indels the rate ranged from 83–86% (Figure 1 and Supplementary Table 1s). When comparing the concordance rate of our samples with the benchmark rates estimated for our pipeline based on the NA12878 data (see Patients and Methods), we found the average SNV concordance rate for each of the five breast cancer patients was higher than the reference concordance rates calculated for the NA12878 subject in whole-exome and high-confidence exome regions, respectively (94.3% and 96.4), while the concordance rate of indel calls was only slightly lower than the reference concordance rate of NA12878 in whole-exome (87.1%) (Figure 1). When compared to the 3 µg DNA input, the 0.2 µg DNA input amount had a marginally-lower concordance rate than the 1 µg DNA input, particularly for indel calls (83.9% vs. 85%). For saliva DNA, the SNV concordance remained at a high level (98.3%) but the indel concordance was the lowest among all comparisons (83.6%), which could be due to shorter DNA fragments from saliva than those from blood DNA. Nevertheless, the slightly inferior indel concordance is still in an acceptable range (33).
Figure 1.
Concordance of single nucleotide variant (SNV) calls (upper panel) and short insertion/deletion (indel) calls (lower panel). Boxplots of concordance rates between each pair of samples from the same patient are displayed: 1 µg vs. 3 µg DNA; 0.2 µg vs. 3 µg DNA; and 1 µg saliva DNA vs. 1 µg blood DNA. The top and bottom of the box correspond to the 3rd and 1st quartiles, respectively, and the band inside the box corresponds to the median. The ends of the whiskers represent the most extreme data points within 1.5 times the interquartile range from the box, and the dots indicate outliers that are beyond 1.5 times the interquartile range from the box.
We further investigated the quality metrics of the discordant variant calls by DNA input amount and DNA source (Supplementary Figures 1 and 2). We found these variants were enriched with potential false positives, as characterized by lower quality scores, higher novel variant percentage, indel length, and Het/Homo call ratio. These findings suggest that we might underestimate the actual variant concordance concerning only bona fide variants after excluding false variant calls.
Detection of coding variants in known breast cancer genes
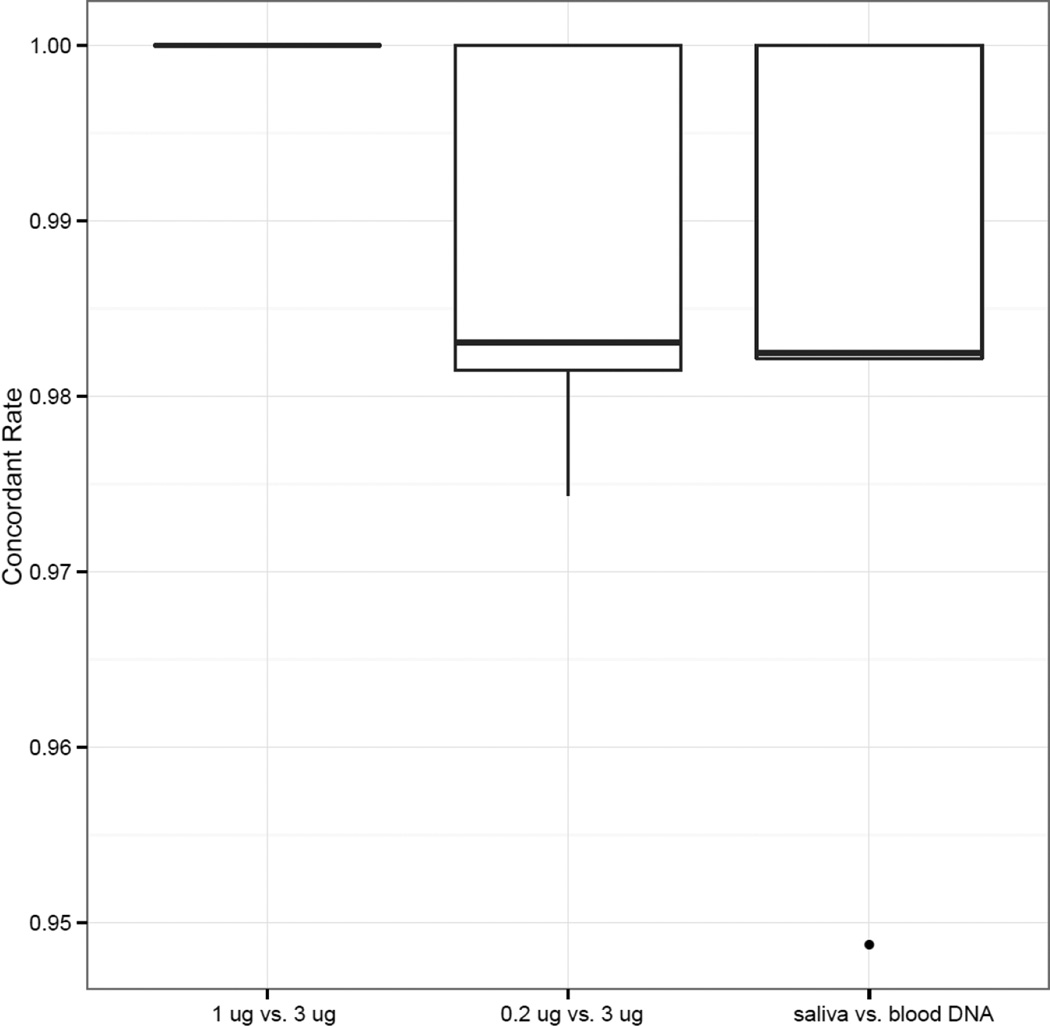
Lastly, as all samples evaluated in our study were collected from women diagnosed with triple-negative breast cancer, we examined whether the use of a lower DNA input amount or saliva samples had any impact on the detection of coding variants that may be underlying breast cancer etiology. We compiled a list of 21 breast cancer-related genes from the Cancer Gene Census (34) (Supplementary Table 2s) and assessed the concordance of variants within these genes among the four samples from each patient. As shown in Figure 2 and Supplementary Table 3s, compared to the coding variants detected from the 3 µg blood DNA input amount (39–59 per sample including both SNVs and indels), the number of variants detected from the two lower DNA input amounts differed slightly by 0 to 2, and for DNA sourced from saliva by −1 to 2. The concordance rate was 100%, with the 1 µg blood DNA input amount, 97.4–100% with the 0.2 µg DNA input amount, and 94.9–100% with the saliva DNA. All discordant calls came from one SNV and four indels (Supplementary Table 4s). After manual review of the sequence alignment files, we concluded that these discordant calls were either false Indel calls introduced by homopolymer (35), or the variants reside in regions where sequencing coverage was too low to make reliable calls. Therefore, the true variant concordance rate can reach 100% with respect to true variants.
Figure 2.
Concordance of variant calls in known breast cancer genes. Boxplots of concordance rates between each pair of samples from the same patient are displayed: 1 µg vs. 3 µg DNA; 0.2 µg vs. 3 µg DNA; and 1 µg saliva DNA vs. 1 µg blood DNA. The top and bottom of the box corresponds to the 3rd and 1st quartiles, respectively, and the band inside the box corresponds to the median. The ends of the whiskers represent the most extreme data points within 1.5 times the interquartile range from the box, and the dots indicate outliers that are beyond 1.5 times the interquartile range from the box.
Discussion
Our results demonstrate that lower DNA input amounts and DNA from saliva have relatively small effects on WES quality and variant-calling consistency. To the best of our knowledge, this is the first comprehensive evaluation of the impact of lower DNA input amount and DNA source on the performance of WES with potential applications for cancer epidemiology. We further demonstrated that lower DNA input amount and saliva DNA can reliably detect variants in breast cancer-related genes, which supports their use in epidemiologic studies searching for coding risk variants, when sample requirements according to a manufacturer’s standard protocol cannot be readily met.
Among various commonly-used sequencing and variant-calling quality metrics evaluated, we found that the data generated from 1 µg blood DNA was essentially the same as the 3 µg blood DNA, and that there was little impact on most quality metrics when using DNA input amounts as low as 0.2 µg. The only differences were shorter insert size and lower mapping rates when using 0.2 µg DNA. The shorter insert size may result from extra fragmentation in the DNA shearing step due to lower DNA amount and high cycle number of PCR (n=11) performed. The slightly lower mapping rate could also result from more random errors introduced by increased PCR cycles. Nevertheless, the shorter insert size or slightly lower mapping rate has little effect on the rate of PCR duplication, sequencing depth, or downstream variant calling.
We demonstrated that the WES performance relevant to sequencing and variant calling qualities for saliva DNA samples was similar to that of blood DNA samples. The mapping rate to the reference genome and the insert size of saliva samples were only slightly lower than that of blood samples (98–99% vs. 100% for mapping rate; 201–214 vs. 207–219 for insert size), indicating very low bacterial DNA contamination and shorter DNA fragments in the saliva samples. As the saliva samples used in our study were collected and processed without any special optimization for NGS applications, we expect this finding has wide generalizability to saliva samples collected routinely in many epidemiologic studies.
Regarding variant calling concordance according to input amount and source of DNA, we did observe inferior indel concordance to that of SNV calls, especially in the lower 0.2 µg DNA input amount and in saliva DNA. This could be due to the more complex structure of indel variants themselves, which make their calling from short-read data more challenging than SNVs. In addition, the lower insert size associated with lower DNA input amount and saliva DNA had a greater impact on indel calling. Nonetheless, the magnitude of the difference was small and negligible in most applications.
Although it is possible to infer copy number variations (CNVs) from WES data and several algorithms have been developed for this purpose, previous studies evaluating the performance of these algorithms concluded that the sensitivity, accuracy, and power were still limited (36, 37). We thus did not evaluate the impact of DNA input amount and saliva DAN on CNV detection in our study.
The motivation of our study is to test whether we can reliably detect rare variants related to breast cancer etiology using low DNA input and saliva DNA. Therefore, we designed the study with a sequencing depth typically used for detecting rare variants. We expect the concordance rate would be lower at a substantially lower sequencing depth, particularly when DNA input is low or saliva DNA is used. Future studies are warranted to assess the impact of varying sequencing depth on the concordance rate.
In summary, we provide compelling evidence that when the standard DNA requirement of a manufacturer’s WES protocol cannot be satisfied, lower DNA input amounts (down to 0.2 µg) or using saliva as an alternative DNA source can generate comparable results. These findings may allow the expansion of WES applications in epidemiologic studies in which DNA specimens may be a finite resource or only low DNA amounts or saliva samples are available. However, caution should taken for indel calls, as we found a larger impact of low DNA input and saliva DNA on indels than on CNVs. Currently, there are two exome capture platforms that require less than 0.2 µg input DNA: the Ion AmpliSeq™ Exome Kit, which can only be run on the Ion Proton™ Sequencer, and the Illumina Nextera Exome Kit. Both kits use as little as 50 ng DNA as the starting material. It will be interesting to investigate comprehensively the performance of WES data generated using such low input amounts, and to compare the performance among different exome-capture platforms with such low DNA input. Our study did show larger impact on calling indels than SNVs when lowering DNA input amount or using saliva DNA. We may anticipate that the performance difference will be even larger when using 50 ng input DNA. In addition, such difference may become stronger when using other exome capture platforms, as the Agilent platform was reported to have increased sensitivity for indels than other platforms (16).
Supplementary Material
Acknowledgments
Financial Support:
The Pathways Study is supported by NIH R01 CA105274 (L.H. Kushi). The RPCI Bioinformatics Shared Resource, Biostatistics Shared Resource, Data Bank and BioRepository, and Genomics Shared Resource are CCSG Shared Resources supported by NIH grant P30 CA016056.
References
- 1.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 2.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teer JK, Mullikin JC. Exome sequencing: the sweet spot before whole genomes. Hum Mol Genet. 2010;19:R145–R151. doi: 10.1093/hmg/ddq333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabbani B, Tekin M, Mahdieh N. The promise of whole-exome sequencing in medical genetics. J Hum Genet. 2014;59:5–15. doi: 10.1038/jhg.2013.114. [DOI] [PubMed] [Google Scholar]
- 8.Johansen Taber KA, Dickinson BD, Wilson M. The promise and challenges of next-generation genome sequencing for clinical care. JAMA Intern Med. 2014;174:275–280. doi: 10.1001/jamainternmed.2013.12048. [DOI] [PubMed] [Google Scholar]
- 9.Garraway LA. Genomics-driven oncology: framework for an emerging paradigm. J Clin Oncol. 2013;31:1806–1814. doi: 10.1200/JCO.2012.46.8934. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald LM, Kumar A, Boyle EA, Zhang Y, McIntosh LM, Kolb S, et al. Germline missense variants in the BTNL2 gene are associated with prostate cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 2013;22:1520–1528. doi: 10.1158/1055-9965.EPI-13-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gracia-Aznarez FJ, Fernandez V, Pita G, Peterlongo P, Dominguez O, de la Hoya M, et al. Whole exome sequencing suggests much of non-BRCA1/BRCA2 familial breast cancer is due to moderate and low penetrance susceptibility alleles. PLoS One. 2013;8:e55681. doi: 10.1371/journal.pone.0055681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteban-Jurado C, Vila-Casadesus M, Garre P, Lozano JJ, Pristoupilova A, Beltran S, et al. Whole-exome sequencing identifies rare pathogenic variants in new predisposition genes for familial colorectal cancer. Genet Med. 2014 doi: 10.1038/gim.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark MJ, Chen R, Lam HY, Karczewski KJ, Chen R, Euskirchen G, et al. Performance comparison of exome DNA sequencing technologies. Nat Biotechnol. 2011;29:908–914. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sulonen AM, Ellonen P, Almusa H, Lepisto M, Eldfors S, Hannula S, et al. Comparison of solution-based exome capture methods for next generation sequencing. Genome Biol. 2011;12:R94. doi: 10.1186/gb-2011-12-9-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asan, Xu Y, Jiang H, Tyler-Smith C, Xue Y, Jiang T, et al. Comprehensive comparison of three commercial human whole-exome capture platforms. Genome Biol. 2011;12:R95. doi: 10.1186/gb-2011-12-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark MJ, Chen R, Lam HYK, Karczewski KJ, Chen R, Euskirchen G, et al. Performance comparison of exome DNA sequencing technologies. Nat Biotech. 2011;29:908–914. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asan, Xu Y, Jiang H, Tyler-Smith C, Xue Y, Jiang T, et al. Comprehensive comparison of three commercial human whole-exome capture platforms. Genome Biology. 2011;12:R95. doi: 10.1186/gb-2011-12-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwan ML, Ambrosone CB, Lee MM, Barlow J, Krathwohl SE, Ergas IJ, et al. The Pathways Study: a prospective study of breast cancer survivorship within Kaiser Permanente Northern California. Cancer Causes Control. 2008;19:1065–1076. doi: 10.1007/s10552-008-9170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambrosone CB, Nesline MK, Davis W. Establishing a cancer center data bank and biorepository for multidisciplinary research. Cancer Epidemiol Biomarkers Prev. 2006;15:1575–1577. doi: 10.1158/1055-9965.EPI-06-0628. [DOI] [PubMed] [Google Scholar]
- 20.Yan L, Ma C, Wang D, Hu Q, Qin M, Conroy JM, et al. OSAT: a tool for sample-to-batch allocations in genomics experiments. BMC Genomics. 2012;13:689. doi: 10.1186/1471-2164-13-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zook JM, Chapman B, Wang J, Mittelman D, Hofmann O, Hide W, et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotech. 2014;32:246–251. doi: 10.1038/nbt.2835. [DOI] [PubMed] [Google Scholar]
- 25.Lappalainen I, Lopez J, Skipper L, Hefferon T, Spalding JD, Garner J, et al. DbVar and DGVa: public archives for genomic structural variation. Nucleic Acids Res. 2013;41:D936–D941. doi: 10.1093/nar/gks1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Highnam G, Franck C, Martin A, Stephens C, Puthige A, Mittelman D. Accurate human microsatellite genotypes from high-throughput resequencing data using informed error profiles. Nucleic Acids Res. 2013;41:e32. doi: 10.1093/nar/gks981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y, Tsung EF, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19:1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Li B, Li C, Cai Q, Zheng W, Long J. Improved variant calling accuracy by merging replicates in whole-exome sequencing studies. Biomed Res Int. 2014;2014:319534. doi: 10.1155/2014/319534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kidd Jeffrey M, Gravel S, Byrnes J, Moreno-Estrada A, Musharoff S, Bryc K, et al. Population Genetic Inference from Personal Genome Data: Impact of Ancestry and Admixture on Human Genomic Variation. The American Journal of Human Genetics. 2012;91:660–671. doi: 10.1016/j.ajhg.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ionita-Laza I, Lange C, N ML. Estimating the number of unseen variants in the human genome. Proc Natl Acad Sci U S A. 2009;106:5008–5013. doi: 10.1073/pnas.0807815106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montgomery SB, Goode DL, Kvikstad E, Albers CA, Zhang ZD, Mu XJ, et al. The origin, evolution, and functional impact of short insertion-deletion variants identified in 179 human genomes. Genome Res. 2013;23:749–761. doi: 10.1101/gr.148718.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang H, Wu Y, Narzisi G, O'Rawe JA, Barron LT, Rosenbaum J, et al. Reducing INDEL calling errors in whole genome and exome sequencing data. Genome Med. 2014;6:89. doi: 10.1186/s13073-014-0089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samarakoon PS, Sorte HS, Kristiansen BE, Skodje T, Sheng Y, Tjonnfjord GE, et al. Identification of copy number variants from exome sequence data. BMC Genomics. 2014;15:661. doi: 10.1186/1471-2164-15-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan R, Wang Y, Kleinstein SE, Liu Y, Zhu X, Guo H, et al. An evaluation of copy number variation detection tools from whole-exome sequencing data. Hum Mutat. 2014;35:899–907. doi: 10.1002/humu.22537. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.