Abstract
Recently it was discovered that a transient activation of transcription factor NF-κB can give cells properties essential for invasiveness and cancer initiating potential. In contrast, most oncogenes to date were characterized on the basis of mutations or by their constitutive overexpression. Study of NF-κB actually leads to a far more dynamic perspective on cancer: tumors caused by diverse oncogenes apparently evolve into cancer after loss of feedback regulation for NF-κB. This event alters the cellular phenotype and the expression of hormonal mediators, modifying signals between diverse cell types in a tissue. The result is a disruption of stem cell hierarchy in the tissue, and pervasive changes in the microenvironment and immune response to the malignant cells.
Keywords: Immunity, Inflammation, Homeostasis, Cancer, Nuclear Factor kappa B
1. Introduction
The role of transcription factor Nuclear Factor kappa B (“NF-κB”) in cell physiology has been reviewed extensively, and excellent articles describe mutations on genes that encode for NF-κB regulators in lymphoid malignancy [1]. Such mutations are relatively rare in solid tumors [2]. Lineages that give rise to solid tumors normally restrict their growth to generate solid tissue – this restriction can be overcome by NF-κB in carcinogenesis [3]. However, in recent years, study models for adenocarcinoma show oncogenes acting through NF-κB to cause cancer [1,4,5]. We selected a few of these models to present key changes in cell signaling to highlight the common theme. Lastly, we use leukemia as a model for metabolic homeostasis. Cell lineages giving rise to leukemia differ from adenocarcinoma in that they possess a natural capacity to initiate rapid clonal growth and migration.
NF-κB is a dimer of proteins with Rel homology domain at the N-terminus (e.g. RelA/p65, RelB, NF-κB1/p50, NF-κB2/p52), which forms a complex the Ikappa B protein (IκB) that restricts NF-κB from entering the nucleus (Table 1). IκB can be inducibly or constitutively degraded, depending on the signaling context [1]. In many cell types a dimer of RelA with p50 regulates NF-κB target genes, including other Rel domain proteins. We focus on RelA as a paradigm for NF-κB effects in study systems discussed here unless otherwise specified.
Table 1.
Identities of representative isoforms of the proteins referred herein, according to the National Center of Biotechnology Information (NCBI) and the Online Mendelian Inheritance in Man catalog (OMIM) that outlines the current consensus for the biological role of each entry.
| Protein name | NCBI (Entrez Gene ID) | OMIM entry |
|---|---|---|
| AKT1 | 207 | 164730 |
| AMPK | 5562 | 602739 |
| AR | 367 | 313700 |
| Bcl-2 | 596 | 151430 |
| Bfl1 | 597 | 601056 |
| Calpain1 / mu I | 823 | 114220 |
| Calpain2 / m II | 824 | 114230 |
| CCL2 | 6347 | 158105 |
| CCL20 | 6364 | 601960 |
| CCL5 | 6352 | 187011 |
| ccnd1 (cyclin D1) | 595 | 168461 |
| CD11b | 3684 | 120980 |
| CD8 | 925 | 186910 |
| CD44 | 960 | 107269 |
| COX2 | 5743 | 600262 |
| CSF3 | 1440 | 138970 |
| CXCL1 | 2919 | 155730 |
| CXCL1O | 3627 | 147310 |
| ER | 2099 | 133430 |
| Fascin-1 | 6624 | 602689 |
| Foxp3 | 50943 | 300292 |
| GATA3 | 2625 | 131320 |
| GRα | 2908 | 138040 |
| Hexokinase 2 | 3099 | 601125 |
| HIF1 | 3091 | 603348 |
| HMG-CoA reductase | 3156 | 142910 |
| IFNα | 3439 | 147660 |
| IFNβ | 3456 | 147640 |
| IFNγ | 3458 | 147570 |
| IkBα | 4792 | 164008 |
| IKK1 | 1147 | 600664 |
| IKK2 | 3551 | 603258 |
| IL-10 | 3586 | 124092 |
| IL-12A | 3592 | 161560 |
| IL-12B | 3593 | 161561 |
| IL-1β | 3553 | 147720 |
| IL-2 | 3558 | 147680 |
| IL-23A | 51561 | 605580 |
| IL-6 | 3569 | 147620 |
| IL-8 | 3576 | 146930 |
| JAK2 | 3717 | 147796 |
| Mcl-1 | 4170 | 159552 |
| MMP2 | 4313 | 120360 |
| MMP9 | 4318 | 120361 |
| Myc | 4609 | 190080 |
| Nanog | 79923 | 607937 |
| NFkB1 (p50) | 4790 | 164011 |
| NFkB2 | 4791 | 164012 |
| P53 | 7157 | 191170 |
| PFKB3 | 5209 | 605319 |
| Proteasome subunit A1 | 5682 | 602854 |
| Proteasome subunit C5 | 5705 | 601681 |
| Ras (KRAS1) | 3845 | 190070 |
| Rb (Rb1) | 5925 | 614041 |
| Rel | 5966 | 164910 |
| RelA | 5970 | 164014 |
| RelB | 5971 | 604758 |
| Src | 6714 | 190090 |
| STAT3 | 6774 | 102582 |
| Tert (telomerase) | 7015 | 187270 |
| TGFβ | 7040 | 190180 |
| TLR3 | 7098 | 603029 |
| TNF | 7124 | 191160 |
| VEGF | 7422 | 192240 |
NF-κB activation can proceed through the canonical pathway, or the noncanonical pathway [6]. In canonical signaling, IκB protein restricts the Rel dimer. The protein kinase complex IKK, which interacts with a variety of proteins (Supporting Table S1) can phosphorylate IκB; after phosphorylation, the proteasome degrades IκB [7,8], enabling a rapid nuclear entry of Rel proteins, where, depending upon their posttranslational modifications, they activate or repress specific groups of target genes [9]. “Noncanonical” signaling takes place when the restricting protein is p100. p100 processing gives rise to the protein p52, which forms a dimer with RelB [6]. During cell stress, other proteins, such as tumor suppressor p53, can restrict RelA from entering the mitochondria [10]. Multiple proteins thereby ensure a tight regulation of NF-κB activity. Under conditions of high expression of the RelA protein, or mutations of enzymes that modify RelA function, some cell types escape feedback regulation of NF-κB activity, as we discuss in section 5.2. We focus on proteins that show why feedback control of NF-κB activity is critical in shaping the microenvironment in malignancy, including cell phenotypes, immune response, and material exchange within a niche.
2. NF-κB subunit RelA is modified to control multiple signal transduction pathways
NF-κB regulates cell differentiation and its inflammatory responses [1]. What makes NF-κB unique is the fact that it is activated in response to diverse changes in the host tissue, and has the capacity to alter the state of the host tissue and of multiple components of the immune system profoundly. Signals that modulate RelA (Fig. 1A) activity affect stability, tertiary structure, and specific combination of charged and hydrophobic residues exposed on RelA, and determine: 1. whether RelA associates specifically with nuclear, mitochondrial, or cytoplasmic proteins; 2. the gene promoters or enhancers RelA associates, modulating transcriptional activity.
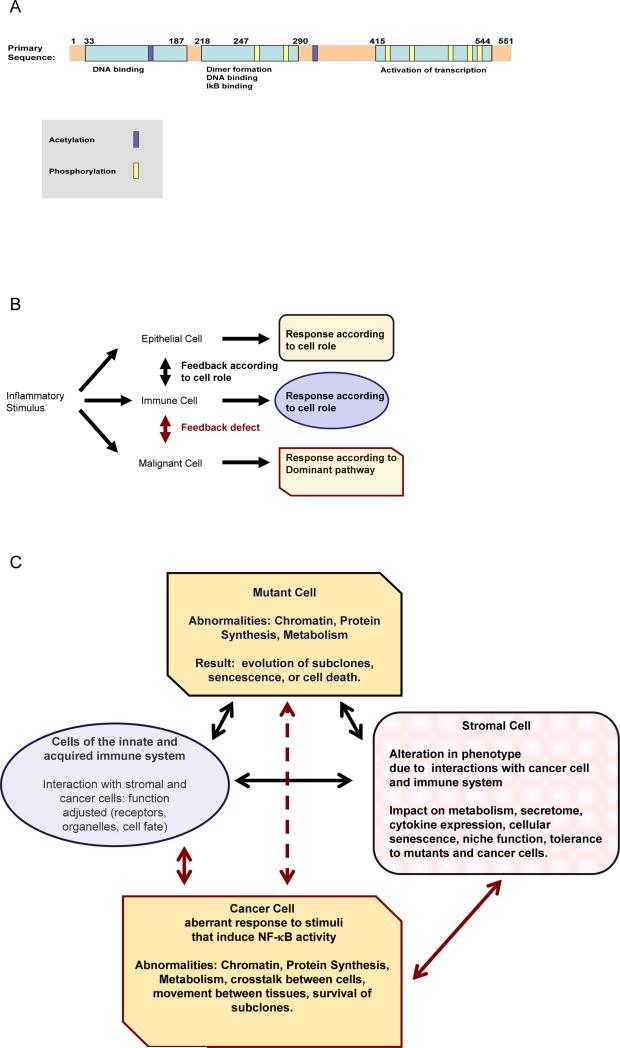
Fig.1.
A: depiction of the primary structure of RelA. A few representative examples of posttranslational modifications are shown. A schematic outline of relevant pathways is given in the Kyoto Encyclopedia of Genes and Genomes resource at http://www.kegg.jp/pathway/hsa04064
RelA interacts with a number of key regulatory proteins, such as nuclear hormone receptors, either by direct physical association, or through competition for coactivators and corepressors [11]. In this way steroid hormones and inflammatory cytokines regulate one another. Several growth factors, or cytokines, binding to their transmembrane receptors, as well as cell stress, elicit intracellular signal cascades that activate distinct Rel proteins, depending on the cell type [12,13]. Recipient cells, in turn, respond by integrating those signals and expressing adhesion molecules, enzymes, and mediators that coordinate cellular function within their microenvironment [13,14].
Metabolism and oxidant stress can impact RelA transcriptional activity [12,14]. As an example, in human pancreatic ductal adenocarcinoma cells, abnormally high enzymatic activities of the hexosamine biosynthetic pathway modify RelA and upstream kinases IKKα and IKKβ; inducing RelA phosphorylation on serine 536, nuclear translocation, NF-κB transcriptional activity, target gene expression, and thereby facilitating anchorage-independent growth [15].
In contrast RelA can also turn into an activator of apoptosis, by phosphorylation on Threonine 505, which can be induced by cisplatin in susceptible cells [16]. Depending, therefore, on the cellular assortment of proteins that interact with RelA, its activation can either kill or reprogram the cell. In contrast to non-malignant cells, where regulation of the activity of Rel-modifying enzymes makes cell survival depend on tissue integrity, in a cancer cell enzymes that modify RelA operate according to mechanisms overriding tissue-imposed control (Fig 1B). This uncouples cell survival from tissue integrity and normal function (Fig 1C).
It is very important to note that RelA controls expression of genes encoding several of its’own regulators, including the inhibitor IκBα and activating kinases IKKα and IKKβ [17] As these kinases interact with diverse signal mediators (Supplementary Table S1), many effects of transiently induced RelA on regulation of cell physiology may escape attention. In contrast, it is already known that synthesis of non-canonical pathway proteins RelB and p52, is controlled by canonical signaling [18]. Hence, aberrant function of the canonical pathway can have indirect effects on cellular function downstream of the non-canonical pathway, with a severe impact on tissue integrity.
3. Interactions between diverse cell types shape tissue function through NF-κB
RelA on one hand mediates the expression of many inflammatory genes, and on the other it can activate survival genes in both normal and cancer cells [19]. A central theme in RelA activity is that it opposes growth restrictions on the cell [20], and prepares the cell for developmental change.
Almost any type of cell stress or inflammatory mediator can induce NF-κB activity [14]. Infection, injury, or toxic organ damage, can cause cell death: reactive oxygen species (ROS), and varied sources of genotoxic and metabolic stress can be elevated above the threshold that a cell tolerates [21]. Under cytotoxic conditions, certain cell types are needed for survival of the organism. In this case NF-κB, by increasing expression of several cohorts of metabolic enzymes and key hormones, can balance stress signals, and thereby protects essential cells [11,14,22,23]. NF-κB target genes consequently include cytokines, adhesion molecules, and specialized functional units of immune cells, as well as antiapoptotic proteins like Bcl-2 and Bfl-1, [13,24-26]. Cytokines and adhesion molecules mediate communication between an individual cell and its surrounding tissue, as well as the rest of the organism. Which are their effects on tissue?
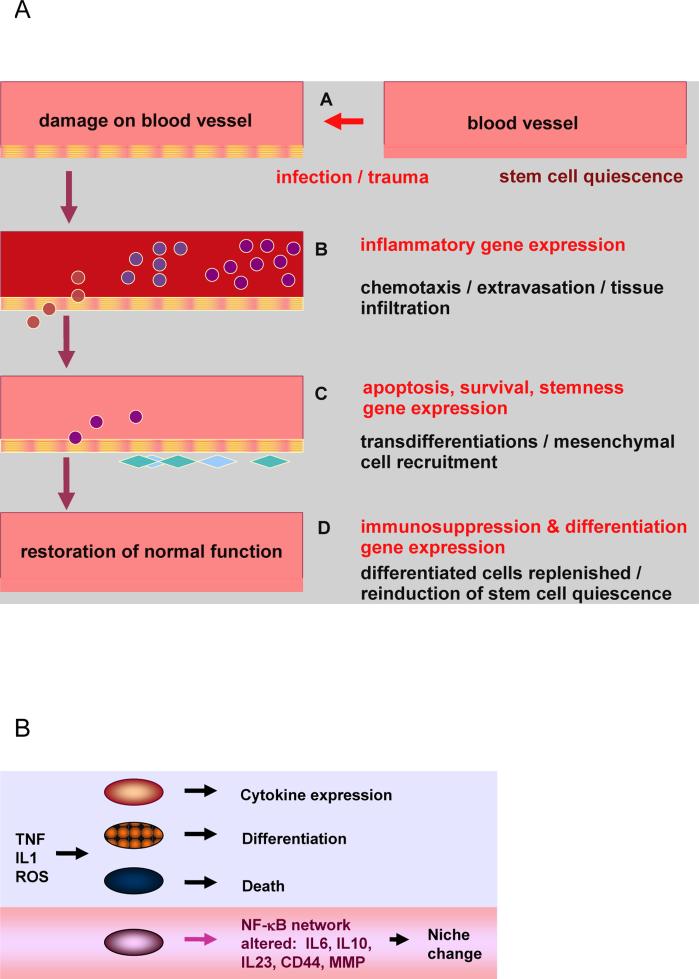
Inflammation lasts from the phase of initiation to the phase of resolution, with restoration of normal function [27]. It activates changes in numerous cell types in the microenvironment: complementary groups of leukocytes, epithelial, mesenchymal, and endothelial cells interact through adhesion molecules and cytokines, to sequentially activate the processes of immune cell recruitment, extravasation (passing through blood vessel walls into tissues), tissue infiltration, ROS release, induction of antioxidant systems, cell reprogramming and transdifferentiation, matrix component deposition, and immune system suppression (Fig. 2A) [28-30]. Therefore, to protect host tissue, activity of NF-κB is tightly regulated:
-
a)
Certain cell types that function in immune response need to persist as long as the response lasts. For example, innate immune response will ideally last for a restricted time in order to protect the host tissue [31]. The lymphocytes that facilitate adaptive immunity, however, need to live on and retain essential properties: they undergo lasting changes in their chromosomes. NF-κB target genes like Bfl-1 help such cells survive, essentially protecting cells with genomic rearrangements - a property shared with cancer cells [13,32].
-
b)
Cells necessary to fill in developing or damaged tissues, are either formed by mesenchymal cells, adult tissue stem cells, or by reversal of differentiation of cells that form local tissue [33]. Cytokines can increase the pool of available cell precursors, both through differentiation of stem cells [34], and by reprogramming of tissue resident cells [35]. Other cells, instead of forming replacement units, are induced to supply essential molecules to the stem cells, forming “feeder” niches [36,37].
Fig. 2.
A: During inflammation, a sequential activation of different types of leukocytes takes place, induced by cytokines, chemokines, and adhesion factors. Malignant cells express subsets of genes that are normally induced in cell subtypes that are essential for survival of the organism.
Through these processes NF-κB -dependent genes coordinate interaction between tissues, with drastic effects on cell function. This cross-talk is disrupted in cancer (Fig 1C): in a normal cell, enzymes that mediate activation of NF-κB are regulated according to the role of the cell in development and inflammation by inducing signal pathways that lead to resolution of the initial trigger [27]. In contrast, in tumor cells, under certain conditions, NF-κB mediates propagation of cell clones that have lost a key feedback inhibiting mechanism (Fig 2B); the result is cancer [14,15,38]. Malignant cells may respond to a lethal stimulus through a signal pathway that is disproportionately activated (Fig 1B), and has a critical influence on downstream enzymes [14,22], as we discuss in section (6).
4. Control of NF-κB impacts cell differentiation
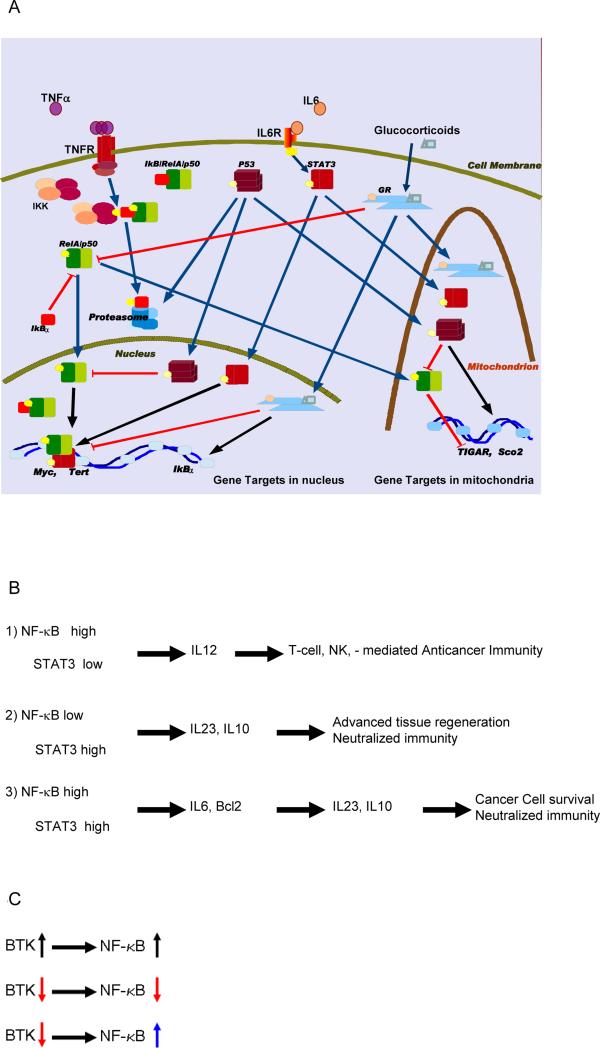
RelA is rather stable and abundant in most cell types [1,10]. Its main natural inhibitor, IκBα, on the contrary, is susceptible to proteolysis by proteasome, lysosome, or calpain, and may differ greatly between different cell types, different states of growth, and diverse signals [7,8]. Within tumors, both proteasome and calpain activity can be increased [39]. As RelA activation is downstream of receptor-initiated signals, and subject to amplification cascades, its activity can become independent from cell surface receptors (Fig. 3) [1,11].
Fig.3.
A: A simplified, schematic depiction of three representative signal pathways with proven potential to interact with intracellular signal relay mechanisms regulated by NF-κB. B: STAT3 and NF-κB induce unique combinations of gene cohorts with an activity profile that can distinguish subgroups of malignant cells. C: A model drug (red arrow) that inhibits a tyrosine kinase involved in NF-κB activation (here Bruton tyrosine kinase “BTK”), will fail to work when NF-κB is constitutively active (blue arrow), and can thereby saturate regulatory sequences (promoters and enhancers) of its oncogenic targets.
Interference of RelA with cellular factors linked to differentiation is critical for its role in epithelial neoplasia. Components of mechanisms that guide differentiation and migration of epithelial lineage cells are disrupted in neoplastic development of many solid tumors [33,34]. Estrogen-regulated pathways via the estrogen receptor (ER) are often involved in breast cancer, and androgen via the androgen receptor (AR) in prostate cancer [11,40]. RelA plays an interesting role in each of these processes: it may enhance ER or AR - mediated signal pathways that activate cell growth and proliferation, while inhibiting potently the ER- or AR- mediated signals that lead to cell differentiation [11,23].
Additionally, embryonal stem cells regulate STAT3 (signal transducer and activator of transcription 3), Nanog, and noncanonical NF-κB signaling in balance with canonical signaling to maintain pluripotency [18, 41-43], which implies that loss of control for canonical signaling in neoplasia could give rise to a subpopulation of malignant cells with properties overlapping stem cells.
In innate immune system cells, including microglial cells of the central nervous system, RelA nuclear translocation is inducible (as in most mammalian cells), and leads to inflammation; because IκBα gene expression is induced by RelA, innate immune responses are normally self-limiting (reversible [29,35]).
Cells that mediate adaptive immunity tend to increase basal levels of NF-κB activity. Rapidly induced RelA activates c-Rel gene expression, facilitating survival of activated B- and T-lymphocytes [13], while cells that function in memory storage for the immune system, maintain a balanced level of Rel activity [1,13,44]. Similarly, during dendritic cell maturation, rapidly activated dimers (e.g., p50/RelA) bound to a subset of target promoters are gradually replaced by slowly activated dimers (e.g., p52/RelB); this prolongs expression of specific NF-κB target subsets in dendritic cells [45]. Consequently, regulation of cytokine expression by dendritic cells, reflected in the proportions between secreted cytokines IL10, IL23, and IL-12, depends on the cell maturation stage [46]. Tumors that express cyclooxygenase (COX)-2, and secrete its’ product prostaglandin E2, inhibit RelB activity in dendritic cells; result is increased IL-10, decreased IL-12 expression, and impaired antigen presentation by the dendritic cells [47]. At the molecular level dendritic cell maturation and function in antitumor immunity, including T-cell priming, are impaired by induction of the transcription factor STAT3 [48].
In the organism, however, the ability to limit RelA activity is important to prevent chronic inflammation and myeloid malignancy: knockout of the miR-146a gene (a Rel target and feedback regulator) in C57BL/6 mice, led to increased transcription of NF-κB-regulated genes, and development of myeloid malignancies with high content of nuclear RelA. This is an example of loss of feedback control on RelA [49].
5. Constitutive NF-κB activity and feedback changes affect cancer
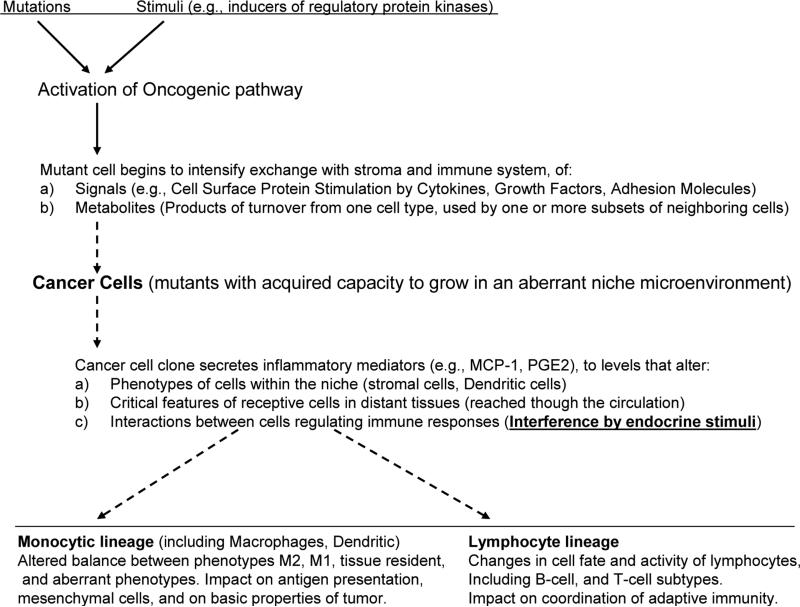
In diverse types of cancer, constitutive NF-κB activity enables a malignant cell to survive oncogene activation, tumor suppressors, radiation, drug treatments, extensive genetic alterations and the surveillance of both innate and adaptive immune cells [19,50,51]. As an example of biochemical challenge, oxidant stress can impact several distinct stages of NF-κB dependent signals, inducing different subsets of genes in different cell types. Depending on the cellular assortment of coregulators, NF-κB can mediate either positive or negative feedback to oxidant stress causing the propagation or termination of inflammatory cascades [29]. The cell exposed to oxidant stress may survive or die, depending on the posttranslational modifications of Rel proteins and the rest of the cellular proteome that interacts with them, resulting in the expression of different sets of target genes [9,19]. The gene products themselves interact with the cell's genome and proteome, and thereby modulate further: gene expression, DNA repair, cell cycle control, mitochondrial function, vesicle transport and contents, oxidant-neutralizing enzymes, salvage metabolism, and competition with surrounding cells for nutrients and tissue space. Consequently, the ability of the organism to kill this cell by inducing biochemical stress is diminished (Figs 2B, 3B).
In a tissue, through disrupted regulation of NF-κB activity and ensuing cytokine expression, tumors may perpetuate inflammation by concomitant expression of factors that in normal cells are expressed inducibly [24], and by blending features that belong to cells of different types [52,53], and different stages of maturation (Fig. 3B) [1,12,54]. Increased levels of inflammatory mediators that activate cell movement across tissues facilitate cancer cell passage through organs, and ultimately, metastasis [39,55].
5.1. Sequential expression of inflammatory mediators is altered in cancer
The function of NF-κB -dependent hormonal mediators in orchestrating interaction between innate and adaptive immunity with host tissue is pivotal in cancer development. Through these mediators cancer cells shape their microenvironment, which is shown by their effects on cell differentiation and phenotypic adjustments, changes in material turnover, and cross-talk with a range of immune cell types. We limit discussion of this subject on the interactions between a few selected feedback regulators of NF-κB that control key aspects of its' impact on host tissue, which provides the targets for design of subtype-specific intervention strategies. As we discuss in detail below, loss of NF-κB control in malignant cells changes expression of inflammatory mediators. Result is that signals for tissue regeneration can coexist with propagation of inflammation, even though in a normal tissue, regeneration signals suppress inflammation. This affects multiple cell types and is essential feature of cancer. In particular, characterization of protein families of transcription factors NF-κB, AP-1 (activating protein-1) and STAT3, which have gene targets that include inflammatory mediators, adhesion molecules and antiapoptotic proteins [24,50,55] provided a molecular basis for the role of inflammation in cancer. Many inflammatory mediators influence transendothelial migration of inflammatory cells and vascular permeability. Their presence in cancer, however, is not ubiquitous.
Notably, the fact that cancer can remain undetected for years until disease has progressed [56], means that it can develop without overt inflammation [26]. Disease progression, in contrast, is often marked by systemic increase of inflammatory cytokines [24]. And furthermore, an inflammatory response may also lead to resolution of the tumor: for example, experimental NF-κB hyperactivation in tumor tissues, by combined stimulation with IFNα with poly-I:C (a TLR3 ligand) can allow reprogramming of the chemokine microenvironment to enhance the recruitment of cytolytic T effector cells [57]. When applied individually each chemokine modulator generated a heterogeneous response for different tumors, while the response was uniform for the combination of IFNα and poly-I:C, and was enhanced by inhibitors of prostaglandin E2 production [57]. The clinical significance of the proportions between secreted products of NF-κB target genes can be appraised by the discovery that the ratio of inflammation-inducing to inflammation resolving cytokines can be reverse between a) pathological conditions involving overt inflammation [25] and b) cancer [58].
As is often evident by the display of different sets of markers by a single fraction of a specific malignant tumor [59], the cancer cell can become a mosaic of gene expression patterns and phenotypic markers that are normally encountered in cells from different tissues and different developmental stages (stem cells, precursor cells, and mature, differentiated cells) [33], [55], [60], [61], [62]. Somatic tumors do not originate from a single source or developmental phase. For example, different stages of B-cell maturation can give rise to lymphomas, and NF-κB is involved in many cases [1]. Mantle cell lymphoma is an aggressive malignancy supported by aberrant B cell receptor (BCR) signaling, which is targeted by inhibitors ibrutinib and sotrastaurin. While either drug can inhibit BCR-induced canonical NF-κB signaling, in many cases malignant B-cells can survive through the alternative NF-κB pathway [63]. Oncogenesis does not follow a strict pattern of signal relay, and is not confined to a single cell source, even if gene expression follows certain lineage-dependent restrictions. However, analysis of gene expression gives information on the NF-κB target gene signature that is essential to identify critical downstream pharmacological therapeutic targets [64]. When combined with the study of cultured cells, genetic analysis enables a personalized approach to tumors unforeseen by standard therapeutic protocols [65]. Apart from personalized treatment, a clinical approach for mantle cell lymphoma, is to interfere with malignant cell metabolism by inhibiting mammalian target of rapamycin (mTOR) threonine kinase, which is a target of IKKβ and acts in synergy with NF-κB [66],[67].
A large number of genes that are activated by NF-κB and associated with oncogenesis and chronic inflammation contain STAT3 DNA-binding sites. In contrast, many genes associated with antitumor immunity lack STAT3 DNA-binding sites and can only be activated by NF-κB when STAT3 is inhibited in tumors [68]. STAT3 facilitates NF-κB binding to genes that are important for tumor growth while inhibiting its binding to Th-1 (T-helper cell type 1) stimulatory genes in growing tumors, including tumor-infiltrating immune cells (Fig. 3B). The result is that the tumor cell is not attacked by the immune system. In contrast, in normal T-cells STAT3 limits NF-κB activation, IL-2 production, and proliferation [69].
The change of NF-κB targets brought about by STAT3 in a malignant cell [68], allows the cell to reach high NF-κB activity in its nucleus, by overcoming negative feedback mechanisms (such as IκBα activation by NF-κB) [1]. Tumor cells that exhibit this type of constitutive NF-κB/STAT3 activity become competitive for niche occupation, and can induce cells from adjacent tissues to secrete biomolecules and metabolites, and possibly to undergo autophagy [19,36,37]. The resulting secreted products aid malignant cell propagation, specifically favoring cells that have neutralized tumor suppressors. STAT3 physically interacts with NF-κB to activate the catalytic subunit of telomerase (tert) in human breast cancer stem cells [70]. What is interesting is that once NF-κB overcomes the restraints posed by p53 activity, it is no longer subject to inhibition by glucocorticoid receptor (“GR”) [71]. NF-κB then can perturb the homeostatic effects of STAT3, GR, and p53 in mitochondria (schematic illustration in Fig. 3) [10,72].
STAT3 and p53, in normal cells limit NF-κB-driven immune responses to protect tissue integrity; this task is performed in coordination with GR [71,72]. Malignant cells that have lost the gene that encodes p53, exhibit decreased response to glucocorticoids [71], increased expression of NF-κB target genes [10], and increased ratio of incoming/secreted exosomes [73]. Similar effects can be expected when genes that mediate control of NF-κB by p53 and STAT3 are epigenetically repressed.
Cytokines regulated by transcription factors, NF-κB and STAT3, found to participate in cancer-related inflammation include IL-1β, IL-6, IL-23, and TNF-α [14,24] (Fig. 3 B). They can facilitate tumor growth through activation of other cytokines such as IL-8, and several types of adhesion molecules [12,36] culminating in: 1). 1) induction of expression for survival proteins ( Mcl-1, Bcl-2, etc.,) on tumor cells [74], 2) growth arrest, senescence, autophagy, secretory phenotype [36], and apoptosis in cohorts of surrounding cells (due to the presence of intact p53), 3) proliferation or quiescence in different cohorts of neoplastic cells, 4) decrease in the efficiency of immune response against the tumor, and 5) metastasis [24]. It is very important to emphasize that through the restrictions on NF-κB activity, an organism links tissue function and system homeostasis to the control of cell survival (Figure 4). As a specific example, abnormally high levels of the cytokine IL-6 can confer multiple carcinogenic properties to immortalized cells, as we discuss in the next section.
Fig. 4.
Immune response is impaired in the vicinity of cells secreting mediators as IL-10, or prostaglandin E2, while other tissue sites, upon a local increase in e.g., IL-12, or TNFα have increased risk of cellular infiltrations and organ damage.
5.2. NF-κB dysregulation permits oncogenes like myc to cause cancer
Many different types of oncogenes cause cancer that depends on NF-κB; here we address a few representative studies of adenocarcinoma models. Inflammatory breast cancer, a particularly lethal disease, is characterized by NF-κB activity [75]. In cell culture, transient activation of Src oncoprotein (Rous sarcoma virus proto-oncogene tyrosine-protein kinase homolog) can mediate an epigenetic switch from immortalized breast cells to a stably transformed line that forms self-renewing mammospheres that contain cancer stem cells [76]. This switch is possible because Src activates NF-κB, inducing thereby Lin28 expression, thus decreasing levels of let7 miRNA; the result is activation of the cytokine IL-6, which induces STAT3 expression. Result is further increase in NF-κB activity. Normal breast epithelial cells express at least one negative regulator of NF-κB activity, namely miR-146b, which is a direct STAT3 target gene [77]. However, in cancer cells, in spite of high STAT3 activity, miR-146b levels may remain low, especially when the miR-146b gene promoter is methylated. Breast cancer patient samples that express IL-6 and show STAT3 activity correlate with a negative prognosis when miR-146b levels are low [77]. Thereby in breast cancer cells, a shift in RelA target genes, caused by transcription factor STAT3, could play a role at least in some phases of carcinogenesis. The higher activity of STAT3 in some tumors could result from activation of NF-κB by an “above-threshold” event such as transient activation of Src [76]. Therefore, a positive feedback loop that works in a tumor - such as above-threshold activation of NF-κB or of its inflammatory gene targets (Fig. 3B) - may constitute an identifiable drug target. Such a therapeutic intervention may become safer when specific tissues or cells are targeted [11].
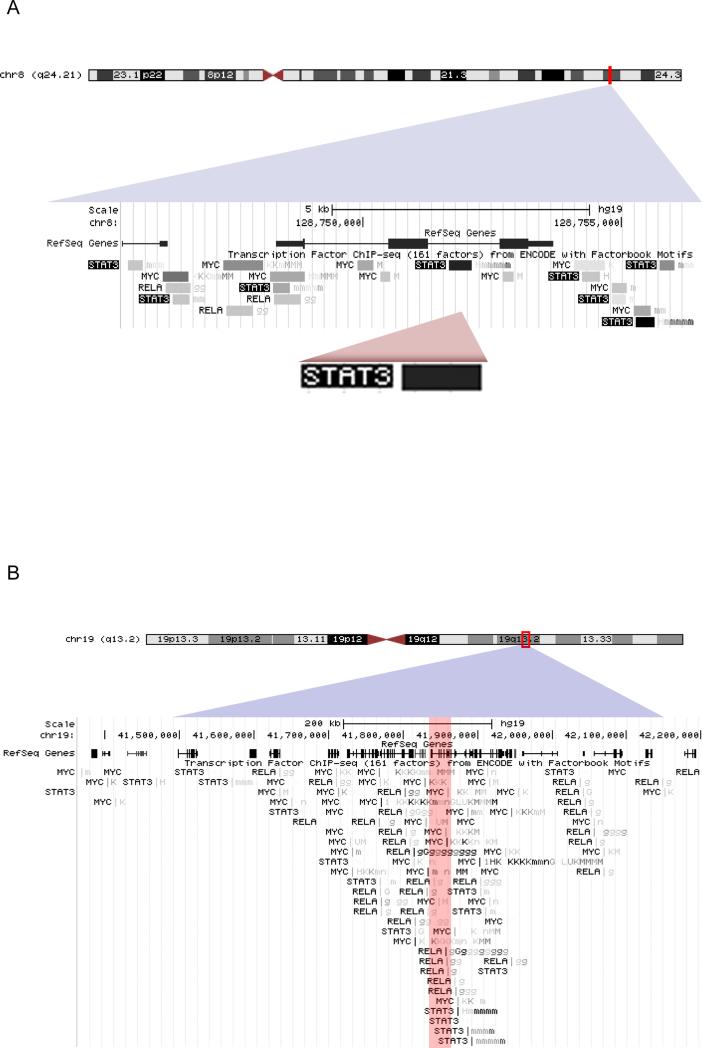
The NF-κB/STAT3 positive feedback loop noted [76], had an interesting aspect: myc mRNA levels increased at a pace that followed stat3 mRNA. Interestingly, in the transcription factor chromatin immunoprecipitation results obtained with the ENCODE project on the human genome [78], binding sites for STAT3 on the myc gene are particularly dense (Fig. 5A). Analysis of p53 transcriptional function via genome-wide chromatin occupancy and gene expression identified STAT3 as antagonist for p53 on the myc gene [79]. Inhibition of STAT3 activity can suppress myc expression in cultured cells [80]. In conclusion, the NF-κB-STAT3 interaction can affect cell fate by activating expression of myc [81]. In human T lymphocytes, for example Protein Kinase C theta-activated NF-κB signaling regulates the expression of telomerase (tert) via c-Myc, which inhibits cellular senescence. More important, is probably the fact that c-Myc activates the coordinated expression of genes that modulate cellular metabolism, growth, and proliferation; when expressed at high levels, is also capable of mediating resistance to cell cycle arrest at the G1 phase by rapamycin [82], or to metabolic effects of AMPK activators [83]. By increasing the metabolic and oxidant burden, c-Myc increases cellular dependence on the NF-κB functions that decrease growth restrictions:
-
a)
NF-κB competes with p53 both in the nucleus and in mitochondria. Many tumor cells have defects either in the tp53 gene itself, or in p53-linked pathways [84], or have Serine 536-phosphorylated RelA, which is an effective inhibitor of p53 activity [85,86]. With Ser-536-phosphorylated RelA, tumors may override IκBα and p53-imposed restrictions on metabolism and growth [87]. This enables tumors to use a higher glycolytic rate that is consistent with the Warburg effect [88], and thereby compete with cells that have wildtype p53.
-
b)
NF-κB activates myc expression, and at the same time, expression of survival proteins that can quench apoptotic signals from Myc [21]. In three-dimensional cell cultures, incorporation of a cell in its natural niche restricts several oncogenes including myc, ccnd1, akt1, from completing cancer cell development [89]. This means that oncogenes have increased need for cooperating events within a natural niche. Importantly, NF-κB can cooperate with Myc for telomerase gene expression [90]. Higher telomerase activity allows a tumor cell to gain a “competitive edge” over stromal cells, and utilize them as a feeder stromal layer:
Fig. 5.
Chromatin immunoprecipitation data of the ENCODE project, for binding of transcription factors NF-κB RelA, STAT3, and c-Myc. A: on human chromosome 8q24.21 locus of the myc gene . B: On the chromosome 19q13.2, locus of the tgfb1 gene (red zone).
Indeed, while human and mouse fibroblasts can become tumorigenic upon lentiviral transduction with an SV40 large T antigen (to sequester p53 and Rb tumor suppressors) and oncogenic Ras, the naked mole-rat fibroblast proved resistant. This resistance to oncogene-induced tumorigenesis was reversed by transduction with human telomerase. Naked mole-rat fibroblasts then formed tumors that grew rapidly in immunodeficient mice [91]. It is important to note that they have high constitutive levels of p53 protein [92], which is apparently overridden by increased telomerase expression. Therefore disruption of the negative feedback between factors such as STAT3 and RelA, can augment the capacity of transformed cells for malignant growth by amplifying expression of target genes like myc, and thereby fitting subclones of tumor cells with biochemical properties that are essential for malignant growth. One of the simplest approaches, therefore, to restore function of tumor suppressors in cancer is to inhibit key components of the resulting inflammatory response, regardless of whether it is a direct or an indirect consequence of cancer itself [93]. This approach is also supported by the fact that drugs such as metformin, as we will discuss next, inhibit synergistic NF-κB activity with STAT3 at concentrations that do not kill normal cells.
6. NF-κB effects on tissue: shaping the microenvironment
In spite of enormous genetic heterogeneity between cancers, common themes exist. One of them is a higher mutation rate in the later stages [94]; this is consistent with dysregulation of the p53 network that normally links cell survival to genetic integrity [95]. As p53 interacts with RelA [71] dysregulation of the p53 network leads to the question if RelA targets are overexpressed in cancer [96], and which molecular mechanisms mediate their effects on the microenvironment [97]. We know that malignant cells secrete signaling molecules such as cytokines and metabolites to redirect the phenotype of cells in surrounding tissue [37,98,99]. and that diverse types of cancer can secrete signaling molecules that disrupt the immune responses [51,68]; interaction between cancer and stroma may allow a tumor escape rejection, even after being recognized by adaptive immunity [100]. Carcinoma cells through secretion of IL-6, IL-8, CXCL10, and CCL5, (their expression depending on RelA) overcome the control of Hypoxia-Induced Factors (HIF) and induce CD11b+Gr-1+ myeloid cells to promote tumor growth and angiogenesis [101]. Loss of the control of cytokine expression by the NF-κB /p53 interaction network, enables immunosuppression and disruption of tissue integrity, in spite of signals for regeneration. Pancreatic adenocarcinoma recently emerged as a model for the effects of aberrant NF-κB feedback by its interaction partners, especially p53: multiple cytokines and chemokines including IL-6, IL-23, CXCL1, CCL20, and CSF3, were expressed by premalignant pancreatic ductal epithelial cells in a RelA-dependent manner to promote development of metastatic cancer [102]. Thereby loss of feedback restriction on RelA results to aberrant coordination of secretion of the mediators for inflammation and regeneration, preventing the restoration of tissue integrity, and the physiological function of the immune response (indications exist that this role can be generalized in neoplasia: eg., for a variety of pediatric malignancies, IL-6 is a predictor of severe infections [103] ). In pancreatic cancer IL6 is known to promote an immunosuppressive microenvironment [104]. Systemic inflammation with activity of NF-κB, IL-6, and STAT3 not only did not clear mice from pancreatic cancer xenografts, but instead blocked chemotherapy and gemcitabine from clearing the tumors [105]. In confirmation of the role of NF-κB, blocking of gemcitabine-induced RelA nuclear translocation, decreases levels of MMP (metalloprotease)-2, MMP-9, VEGF, and IL-8, inhibiting angiogenesis and invasion [106].
How could loss of feedback control of NF-κB disrupt the components of the microenvironment? Two target molecules that can contribute to this effect are fascin and TGFβ.
In most normal or transformed cells, as we have seen, NF-κB has the capacity to interfere with transcription factors important in cell phenotype and fate. The main reason for lifting phenotypic restrictions for most cells and reversing properties of cellular differentiation is that during inflammation the organism needs to concentrate tissue-resident cells on the challenge (e.g. infection, or necrotic tissue), and after cessation of the challenge, to prime resident cells for terminating the inflammatory response and restoring function. One gene essential in cell migration is fascin [107]. In metastatic breast cancer cells, IL6-induced STAT3 cooperates with TNF-induced NF-κB to activate fascin expression [108]. In the process of normal mammary gland morphogenesis or luminal differentiation, transcription factor GATA3 would limit fascin gene expression [109]. Moreover, glucocorticoids can further reduce fascin expression, and thereby allow the formation of tight junctions, which are essential for building epithelial barriers [110]. Thus normal tissue function relies on negative feedback regulation of inflammatory mediators. This negative feedback regulation results in an on-off cycle for NF-κB in normal cells and tissues [111], which is lost in adenocarcinoma, as indicated by the effects of deregulated expression of its downstream targets that we discuss next.
In adenocarcinoma TGFβ can suppress antigen presentation, cytotoxic T cells, natural killer (NK) cells, and induce FoxP3+ regulatory T cells that inhibit the antitumor immune response [112,113]. It is very interesting to note the convergence of binding sites for NF-κB, STAT3 and c-Myc toward the gene encoding for TGFβ on the human chromosome 19 (Fig 5 B). TGFβ on the one hand regulates epithelial-mesenchymal transition [114], and on the other hand suppresses systemic immunity, hence facilitating metastasis [115]. In fact, impaired control of signaling between NF-κB, TGFβ, and HIF1 is implicated in cancer metastasis and organ failure [116,117]. Importantly, however, disruption of the NF-κB feedback control by TGFβ, demonstrates the potential to compromise tissue regeneration and integrity: elevated TGFβ levels in the tumor microenvironment can contribute to fascin overexpression [118,119]. It is interesting to note that TGFβ can use NF-κB as a mediator for gene expression in epithelial-mesenchymal transition [120], and therefore is a key node for positive feedback to NF-κB during cell transformation by oncogene ras. In the presence of oncogenic Ras, inflammatory stimuli trigger NF-κB-mediated positive feedback that amplifies Ras activity to pathological levels, generating pancreatic adenocarcinoma in mice [4].
The same positive feedback is likely to occur also in colon, lung, and other types of adenocarcinoma [4]. In a normal cell TGFβ poses certain growth restrictions, however in subpopulations of eg. [121], pancreatic cancer cells NF-κB can substitute for TGFβ signaling in epithelial-mesenchymal transition [120] and thus increase metastatic tumor burden. Furthermore, by inducing overexpression of myc as we discussed in the previous chapter, dysregulated NF-κB activity can overcome restrictions on angiogenesis and neovascularisation, which are normally imposed by TGFβ [122]. Thereby TGFβ ceases to function as a tumor suppressor in the adenocarcinoma cell. Hence on the one hand cancer cells can grow unrestricted by the presence of TGFβ, and on the other hand TGFβ still affects other cells that are abundant in the tissue. Specifically, TGFβ restricts the antigen-presenting function of myeloid cells [123], and in parallel, enables myeloid cells to prevent CD8(+) T cells from blocking adenocarcinoma metastasis [115]. In adenocarcinoma cells, therefore, a loss of negative regulation of NF-κB by TGFβ, allows them to respond differently to stimuli that normally induce tumor suppression, and to amplify downstream signals, affecting surrounding tissue. Components of the metastatic niche in this way disrupt the ordered mobilization of cells involved in inflammation and regeneration, compromising the integrity of tissue function and immune response (Figure 4).
7. Niche effects and material turnover: insight enabled by study of lymphoid malignancy
Cells of lymphoid malignancies in, can survive in niches that produce autocrine or paracrine factors such as IL-6, which stimulate parts of the stem cell signal transduction apparatus [37,124]. IL6 - by activating STAT3 - inhibits cell death from cytotoxic drugs such as arsenite [125]. In the hematopoietic stem cell niche myeloma cells compete with normal hematopoietic cells for mediators and metabolites supplied by stromal cells [37,124,125].
Resistance of cancer cells to toxic agents and immune responses is augmented by a flow of secretory microvesicles and exosomes, resulting in a deterioration of the stroma in favor of the tumor. One trigger for this flow are cytokines, such as IL-6 [124]. The flow of microvesicles and exosomes between stromal and tumor cells is regulated by NF-κB [36], STAT3 [126], and p53 [73]. The exchange of contents between tumor and normal cells can therefore have a specific direction (due to partial or total inactivation of p53 networks in the former). Secretory vesicles transport a variety of lipids, proteins, and nucleic acids, including mRNA, and microRNAs: flow of microvesicles and exosomes results in redistribution of regulatory, metabolic and structural components [127]. The result is a substantial enhancement of cancer cell resistance to stress, in spite of deregulated material turnover [21]. How does homeostasis function in cancer cells? In contrast with carcinoma, leukemia originates from cell lineages with inherent capacity for clonal growth and cell migration: lower burden of complexity in carcinogenesis makes leukemia a better model for resistance of cancer by induction of homeostatic responses, as we see next.
Glucocorticoids kill lymphoblasts causing acute leukemia [128]. High-dose glucocorticoid treatment may cause a certain degree of hyperglycemia. During induction therapy, this is associated with poorer survival in children with acute lymphocytic leukemia [129]. Glycolysis has been identified as one potential pathway of resistance of leukemia cells to glucocorticoid [130], and STAT3 has been identified as a stress-responsive transcription factor, capable of interacting with glucocorticoid-regulated pathways [131]. High activity of STAT3 was observed in lymphoma cells with signatures corresponding to NF-κB activity, proliferation, and glycolysis [132]. STAT3 activated by IL-6 can enhance expression of the glycolytic enzymes hexokinase 2 and 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (PFKFB3) [133].
Cultured leukemia cells that were resistant to glucocorticoid treatment [128], could be killed by proteasome inhibition, the main signature effect being inhibition of STAT3 target genes [21]. At least a portion of glucocorticoid-resistant leukemia, therefore, can be treated by drug combinations that include inhibitors of proteasome and modulators of STAT3-dependent metabolism. Indeed, experimental evidence suggests that leukemia cells do not operate a metabolism identical to other components of the immune system, and seem to be particularly sensitive to inhibition of STAT3-regulated pathways: the hypoglycemic drug metformin, which inhibits STAT3 transcriptional activity in lymphocytes [134], elicits autophagy and apoptosis in leukemic cells (IC50 for patient samples ranged between 0.6 and 0.9mM at 96h), while not having a comparable toxicity on normal proliferating CD4(+) T-lymphocytes from healthy donors [135].
Hyperactivation of the STAT3 inducer JAK2 can make malignant cells sensitive to metabolic inhibition: 3-hydroxy-3-methyl-glutaryl coenzyme A (HMG-CoA) reductase inhibitors - widely used to treat hypercholesterolemia - induce apoptosis and inhibit JAK2-V617F-dependent neoplastic cell growth. These cells are more sensitive to statin treatment than non-JAK2-V617F-dependent cells. Importantly, statin treatment inhibited erythropoietin-independent erythroid colony formation of primary cells from myeloproliferative neoplasia patients, but had no effect on erythroid colony formation from healthy individuals [136]. A hypercholesterolemia that is followed by severe hypocholesterolemia, indicates that at least a portion of neoplastic cells utilize cholesterol and can be targeted by statin treatment [137].
8. Homeostatic challenges: impact on NF-κB activity in solid and lymphoid tumors
Subclones of cancer cells may escape death by activating intracellular protein turnover, through exchange of vesicles with stromal cells, and by selective recruitment of groups of cells of the immune system. Inflammatory mediators and metabolites also enable diverse subpopulations of cancer cells to cooperate or compete, influence their microenvironment, and ultimately develop new phenotypes. This lifts key tissue-mediated restrictions on cell fate.
The ability of tumor cells to evade chemotherapy, radiation, and targeted therapy is very often proportional to their capacity to activate salvage pathways and exploit homeostasis mechanisms that are normally heavily restricted in normal cells [14,106, 138-140]. Normal undifferentiated cells, for example, are very sensitive to changes in conditions and differentiation stimuli [10,34]. Tumor initiating cells, in contrast, may retain stem cell properties even after exposure to differentiation stimuli [141]; maintaining homeostasis is a challenge. A solution utilized by tumor cells, is to increase proteasome activity [142]. Higher activity of the proteasome in healthy cells ensures a good probability of the organism living longer, by removing misfolded proteins [143]. Aberrantly high activity of the proteasome in tumor cells ensures their selective advantage over healthy cells, and the removal of apoptosis inducers [142].
Malignant cells can respond to proteasome inhibition by activating NF-κB and restoring mechanisms for their survival by activating salvage pathways, such as lysosome acidification [8]. Cascade-initiating mechanisms are redundant in neoplasia under certain conditions: under selective pressure exerted by drugs, a malignant cell may ultimately retain only the enzymatic activities that are essential to its very existence. However, deregulated gene expression, combined with a high mutational load, generates aberrantly expressed, misfolded proteins. Removal of aberrant products is achieved by degradation through multifunctional complexes such as the proteasome, which is a therapeutic target [142]. Inhibition of the proteasome [8], or epigenetic reprogramming via a deacetylase inhibitor [144], may lead to activation of homeostatic responses and NF-κB, and rescue a malignant cell from antineoplastic treatments. One such example is the induction of lysosome activity after inhibition of the proteasome: this is not only a trigger for lysosome-dependent degradation of IκB, but it can also result in rescue of the cancer cell through autophagy, which enables it to survive by recycling many of its contents [145]. It is therefore important to target therapeutic intervention to the drug-induced metabolic pathway that rescues the malignant cell.
It is therefore evident that interactions between cancer and stroma can include exchange of microvesicles, exosomes, cytokines, growth factors, and metabolites. Drastic changes in proteins that function as critical nodes of the NF-κB network disrupt key feedback responses and essential mechanisms of oscillation for NF-κB activity. Neoplastic tissue is characterized by particularly high levels of certain metabolites and enzymatic activities, with resultant breakdown of regulatory mechanisms and establishment of abnormal ratios between critical rheostat molecules located in organelles and on chromatin. As metabolites and homeostatic mechanisms depend heavily on the host tissue, no animal model or current in vitro system can be viewed as faithful reproduction of a cancer host: key challenge in the field is to develop in vitro models that will enable characterizing changes of inflammatory mediators in specific states of cancer development.
Conclusions and outlook
It is becoming increasingly clear that many aspects of inflammation influence the course of malignant disease decisively. NF-κB driven inflammatory gene expression programs are known mediators of cancer-related mortality [146]. This article presents a few mechanisms that have drastic effects on the cancer cell microenvironment, when their impact on NF-κB activity is altered. The next decade is expected to define cancer in terms of an aberrant function of the NF-κB network. Inflammatory signal pathways and NF-κB activation in malignant tumors neither follow a normal pattern of function nor respond to modulators in a readily expected fashion [74,144]; their secreted targets, nonetheless, can be readily identified. Compounds that inhibit the activation of selected subsets of NF-κB-dependent genes are increasingly being assayed in preclinical studies [29]; metabolic intervention, however, is closer to clinical application [14]. NF-κB driven signals, through their downstream targets convey the capacity for rapid responses and systemic amplification to inflammation, being capable to change cellular function, especially in relation to host tissue. Disruption of NF-κB feedback regulation explains is why this factor from a mechanism of tumor control gains the potential to transform the clinical course of the disease, since tumors that involve diverse oncogenes evolve into cancer after dysregulation of NF-κB. There are multiple control mechanisms with partly overlapping effects on NF-κB activity. As a paradigm, regulation of NF-κB by transcription factors STAT3, GR, and p53, which mediate effects of metabolism and stress, does not function as in normal cells. Loss of feedback for those mechanisms changes key properties of a neoplastic cell and alters tissue steady state. The result is an aberrant interaction with the host tissue and the immune system, with impact on discrete cancer cell subpopulations and the resulting microenvironment. Importantly, dysregulation of NF-κB activity affects oncogenesis, because it enables a tumor cell to combine key properties of stem cells with essential adaptation features of a differentiated cell. Characterization of the modules that control NF-κB activity points at designing treatment according to the biological state of host tissue. In the near future, selective inhibition of signal pathways interacting with NF-κB could prove effective in combination with cell-targeted agents, and contribute significantly to personalized cancer treatment.
Supplementary Material
*Highlights (for review).
- During inflammation, a sequential activation of different cell types takes place, induced by cytokines, chemokines, and adhesion factors. Malignant cells can express combinations of cytokines, chemokines, and adhesion factors from different stages of the inflammatory cascade.
- Dysregulation of NF-κB activity affects oncogenesis, because it enables a tumor cell to combine selected properties of stem cells with tissue adaptation features of a differentiated cell. This dysregulation influences several gene cohorts in subsets of cancer cells; however, it has far-ranging effects on the entire organism through disruption of essential mediators of homeostasis.
- Regulation of NF-κB by transcription factors signal transducer and activator of transcription-3 (STAT3), glucocorticoid receptor (GR), and p53 protein, which mediate effects of metabolism and stress, does not function in cancer cells as in normal cells. The result is an aberrant interaction with the host tissue and the immune system, with impact on discrete cancer cell subpopulations and their microenvironment.
Acknowledgments
This work was supported by grants NIEHS R01-ES018948 and NIAID P01-AI062885 to I.B.. The authors wish to thank Dr. Moshe Yaniv for very helpful and thought-provoking comments; and Dr. D. Baltimore for encouragement. Figure 5 used depiction derived from the Human Genome Browser. The University of California at Santa Cruz project team for Human Genome Browser (assembly hg19) is as follows: Hiram Clawson, Brooke Rhead, Pauline Fujita, Ann Zweig, Katrina Learned, Donna Karolchik and Robert Kuhn (website: http://genome.ucsc.edu/goldenPath/credits.html#human_credits) Starting with the hg19 assembly, the human genome sequence is provided by the Genome Reference Consortium (GRC). Credits for GRC are given in: (http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/credits.shtml)
Biography
 Spiros Vlahopoulos. Earned Diploma in Biology, in 1993, at the University of Cologne, and PhD in Biology, in 1999, at the University of Texas. Conducted postdoctoral study at Baylor College of Medicine. Main interests: cellular signaling mechanisms elicited by homeostatic imbalance, interference between signals transduced by steroid hormone receptors and inflammation-induced transactivators. Studying effects of Nuclear Factor kappa B on malignant cells, and modulation of immune system interactions with other systems and host tissue. Member of FASEB since 2004.
Spiros Vlahopoulos. Earned Diploma in Biology, in 1993, at the University of Cologne, and PhD in Biology, in 1999, at the University of Texas. Conducted postdoctoral study at Baylor College of Medicine. Main interests: cellular signaling mechanisms elicited by homeostatic imbalance, interference between signals transduced by steroid hormone receptors and inflammation-induced transactivators. Studying effects of Nuclear Factor kappa B on malignant cells, and modulation of immune system interactions with other systems and host tissue. Member of FASEB since 2004.
 Osman Cen earned M.S. in Biology, at the Illinois Institute of Technology, Chicago, IL (1996), and Ph.D. in Microbiology & Immunology, at the University of Texas Medical Branch (2001), earning the prestigious McLaughlin predoctoral fellowship award. AAAS and ASM member since 2007. Studied cytokine receptor signaling and Src kinase activation with the yeast two-hybrid system, and characterized the role of Src kinases and SAP in the development of NKT cells. Current focus is the developmental signaling pathways in lymphoid cells and the modulation of these pathways by the Epstein-Barr virus protein LMP2A in human malignancies.
Osman Cen earned M.S. in Biology, at the Illinois Institute of Technology, Chicago, IL (1996), and Ph.D. in Microbiology & Immunology, at the University of Texas Medical Branch (2001), earning the prestigious McLaughlin predoctoral fellowship award. AAAS and ASM member since 2007. Studied cytokine receptor signaling and Src kinase activation with the yeast two-hybrid system, and characterized the role of Src kinases and SAP in the development of NKT cells. Current focus is the developmental signaling pathways in lymphoid cells and the modulation of these pathways by the Epstein-Barr virus protein LMP2A in human malignancies.
Nina Hengen, Earned M.D. at the University of Belgrade School of Medicine, Belgrade, Serbia, and Ph.D. in Pharmacology & Toxicology, at the University of Texas. Expert in Pharmacokinetics, Pathophysiology, the mechanisms underlying the response of human organism to Disease, and Integrated Pharmaceutical Care & Science. Associate Professor, Shenandoah University, Winchester, VA.
James Agan. Graduated 1996 from Southern Illinois University. Til 2002 at Baylor College of Medicine, worked in embryo cell development, modulation of differentiation of progenitor cells from diverse lienages by inflammatory mediators.
Maria Moschovi, MD, PhD. Her research interests include molecular aetiology of childhood neoplasia, cytogenetic abnormalities and angiogenesis; molecular responses and metabolic disorders that occur due to malignant disease and following treatment. Assistant Professor of Medicine in the University of Athens.
Elena Critselis completed undergraduate study at in Nutritional Sciences at Cornell University, USA, dual Master of Public Health degrees in Chronic Disease Epidemiology and Global Health at Yale University, USA, and PhD at the Department of Nutritional Sciences–Dietetics at Harokopio University. Work includes expression of gene expression polymorphisms of cytokines and correlation analysis of their effects on carcinoma, and epidemiologic analysis of pharmacological intervention.
Maria Adamaki earned a B.Sc. in Molecular Biology from the University of Hertfordshire, UK and a M.Sc. in Molecular Medicine from the Imperial College School of Medicine, UK. Her research interests include models of leukemogenesis and gene expression patterns in leukemia. Worked with Cancer Research UK on the topic of mutation pathogenesis in childhood leukemia.
Flora Bacopoulou, MD, with Pharmacy and Medical Degrees from Athens University; studied pediatrics and neonatology in University Hospitals Coventry and Warwickshire’ NHS Trust; pediatric neurology and respiratory medicine in Birmingham Children's Hospital reference center, UK; Adolescent Gynecology, earning the certification of the International Fellowship of Pediatric and Adolescent Gynecology. Interested in factors influencing pharmacologic intervention, alterations of immune system during metabolic disease, and the effects of hormonal mediators during adolescence.
John A. Copland, MS, PhD. Professor of Cancer Biology at the Mayo Clinic College of Medicine, Jacksonville, Florida, and Consultant for the Department of Cancer Biology, Mayo Clinic Cancer Center. Study section member for Department of Defense, National Institute for Health, reviewer for National Cancer Institute. Research interests include molecular mechanisms of carcinogenesis and tumor progression.
Istvan Boldogh, MD, PhD, Earned with Summa Cum Laude the title Doctor of Sciences in Medicine by the Hungarian Academy of Science, Budapest in 1986. University of Texas McLaughlin Distinguished Fellow, for 1987-1988. Professor at the School of Medicine, Graduate School of Biomedical Sciences and Department of Microbiology and Immunology, University of Texas at Galveston. Research interests include basic mechanisms by which mitochondrial reactive oxygen species is generated and are etiological agents in aging and a number of diseases.
 Michael Karin, Ph.D. recipient of the 2010 Harvey prize, is distinguished Professor of Pharmacology and Pathology at the School of Medicine, University of California, San Diego, where has been on the faculty since 1987. Dr. Karin also served as a member of the National Advisory Council for Environmental Health Sciences and has been an American Cancer Society Research Professor since 1999. Dr. Karin was elected as a member of the US National Academy of Sciences in 2005 and as an associate member of the European Molecular Biology Association in 2007. Ranked first worldwide by the Institute of Scientific Information in a listing of most-cited molecular biology and genetic research papers published in prestigious journals. In addition to establishing molecular links between obesity, inflammation and cancer, his work revealed new targets for cancer prevention and therapy.
Michael Karin, Ph.D. recipient of the 2010 Harvey prize, is distinguished Professor of Pharmacology and Pathology at the School of Medicine, University of California, San Diego, where has been on the faculty since 1987. Dr. Karin also served as a member of the National Advisory Council for Environmental Health Sciences and has been an American Cancer Society Research Professor since 1999. Dr. Karin was elected as a member of the US National Academy of Sciences in 2005 and as an associate member of the European Molecular Biology Association in 2007. Ranked first worldwide by the Institute of Scientific Information in a listing of most-cited molecular biology and genetic research papers published in prestigious journals. In addition to establishing molecular links between obesity, inflammation and cancer, his work revealed new targets for cancer prevention and therapy.
 George P. Chrousos, MD recipient of the 2014 Fred Conrad Koch Award, Professor and Chairman of the First Department of Pediatrics at the University of Athens School of Medicine, Greece; former Chief of the Pediatric and Reproductive Endocrinology Branch of the National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, Maryland. Member: American Society for Clinical Research, the Association of American Physicians, the Institute of Medicine of the National Academy of Sciences, Washington DC, USA, and the Academia Europaea, London, UK. World leading authority in the glucocorticoid signaling system of the cell, on the diseases of the hypothalamic-pituitary-adrenal axis, and on the physiological and molecular mechanisms of stress, inflammatory, autoimmune and allergic diseases. ISI highly cited in the list of Clinical Medicine, and in Biology and Biochemistry.
George P. Chrousos, MD recipient of the 2014 Fred Conrad Koch Award, Professor and Chairman of the First Department of Pediatrics at the University of Athens School of Medicine, Greece; former Chief of the Pediatric and Reproductive Endocrinology Branch of the National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, Maryland. Member: American Society for Clinical Research, the Association of American Physicians, the Institute of Medicine of the National Academy of Sciences, Washington DC, USA, and the Academia Europaea, London, UK. World leading authority in the glucocorticoid signaling system of the cell, on the diseases of the hypothalamic-pituitary-adrenal axis, and on the physiological and molecular mechanisms of stress, inflammatory, autoimmune and allergic diseases. ISI highly cited in the list of Clinical Medicine, and in Biology and Biochemistry.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
Literature Cited
- 1.Staudt LM. Oncogenic activation of NF-kappaB. Cold Spring Harb. Perspect. Biol. 2010;2:a000109. doi: 10.1101/cshperspect.a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parker M, Mohankumar KM, Punchihewa C, Weinlich R, et al. C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma. Nature. 2014;506:451–5. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin D-C, Zhang Y, Pan Q-J, Yang H, et al. PLK1 Is transcriptionally activated by NF-κB during cell detachment and enhances anoikis resistance through inhibiting β-catenin degradation in esophageal squamous cell carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011;17:4285–95. doi: 10.1158/1078-0432.CCR-10-3236. [DOI] [PubMed] [Google Scholar]
- 4.Daniluk J, Liu Y, Deng D, Chu J, et al. An NF-κB pathway-mediated positive feedback loop amplifies Ras activity to pathological levels in mice. J. Clin. Invest. 2012;122:1519–28. doi: 10.1172/JCI59743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao Y, Luo J-L, Karin M. IkappaB kinase alpha kinase activity is required for self-renewal of ErbB2/Her2-transformed mammary tumor-initiating cells. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15852–7. doi: 10.1073/pnas.0706728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Razani B, Reichardt AD, Cheng G. Non-canonical NF-κB signaling activation and regulation: principles and perspectives. Immunol. Rev. 2011;244:44–54. doi: 10.1111/j.1600-065X.2011.01059.x. [DOI] [PubMed] [Google Scholar]
- 7.Han Y, Weinman S, Boldogh I, Walker RK, et al. Tumor necrosis factor-alpha-inducible IkappaBalpha proteolysis mediated by cytosolic m-calpain. A mechanism parallel to the ubiquitin-proteasome pathway for nuclear factor-kappab activation. J. Biol. Chem. 1999;274:787–94. doi: 10.1074/jbc.274.2.787. [DOI] [PubMed] [Google Scholar]
- 8.Jia L, Gopinathan G, Sukumar JT, Gribben JG. Blocking autophagy prevents bortezomib-induced NF-κB activation by reducing I-κBα degradation in lymphoma cells. PloS One. 2012;7:e32584. doi: 10.1371/journal.pone.0032584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang B, Yang X-D, Lamb A, Chen L-F. Posttranslational modifications of NF-kappaB: another layer of regulation for NF-kappaB signaling pathway. Cell. Signal. 2010;22:1282–90. doi: 10.1016/j.cellsig.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson RF, Witzel I-I, Perkins ND. p53-dependent regulation of mitochondrial energy production by the RelA subunit of NF-κB. Cancer Res. 2011;71:5588–97. doi: 10.1158/0008-5472.CAN-10-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Copland JA, Sheffield-Moore M, Koldzic-Zivanovic N, Gentry S, et al. Sex steroid receptors in skeletal differentiation and epithelial neoplasia: is tissue-specific intervention possible? BioEssays. 2009;31:629–41. doi: 10.1002/bies.200800138. [DOI] [PubMed] [Google Scholar]
- 12.Vlahopoulos S, Boldogh I, Casola A, Brasier AR. Nuclear factor-kappaB-dependent induction of interleukin-8 gene expression by tumor necrosis factor alpha: evidence for an antioxidant sensitive activating pathway distinct from nuclear translocation. Blood. 1999;94:1878–89. [PubMed] [Google Scholar]
- 13.Gerondakis S, Siebenlist U. Roles of the NF-kappaB pathway in lymphocyte development and function. Cold Spring Harb. Perspect. Biol. 2010;2:a000182. doi: 10.1101/cshperspect.a000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tornatore L, Thotakura AK, Bennett J, Moretti M, et al. The nuclear factor kappa B signaling pathway: integrating metabolism with inflammation. Trends Cell Biol. 2012;22:557–66. doi: 10.1016/j.tcb.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Ma Z, Vocadlo DJ, Vosseller K. Hyper-O-GlcNAcylation is anti-apoptotic and maintains constitutive NF-κB activity in pancreatic cancer cells. J. Biol. Chem. 2013;288:15121–30. doi: 10.1074/jbc.M113.470047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Msaki A, Sánchez AM, Koh LF, Barré B, et al. The role of RelA (p65) threonine 505 phosphorylation in the regulation of cell growth, survival, and migration. Mol. Biol. Cell. 2011;22:3032–40. doi: 10.1091/mbc.E11-04-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Mitra A, Dojer N, Fu S, et al. A probabilistic approach to learn chromatin architecture and accurate inference of the NF-κB/RelA regulatory network using ChIP-Seq. Nucleic Acids Res. 2013;41:7240–59. doi: 10.1093/nar/gkt493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basak S, Shih VF-S, Hoffmann A. Generation and activation of multiple dimeric transcription factors within the NF-kappaB signaling system. Mol. Cell. Biol. 2008;28:3139–50. doi: 10.1128/MCB.01469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao G, Fu J. NF-κB and cancer: a paradigm of Yin-Yang. Am. J. Cancer Res. 2011;1:192–221. [PMC free article] [PubMed] [Google Scholar]
- 20.Toruner M, Fernandez-Zapico M, Sha JJ, Pham L, et al. Antianoikis effect of nuclear factor-kappaB through up-regulated expression of osteoprotegerin, BCL-2, and IAP-1. J. Biol. Chem. 2006;281:8686–96. doi: 10.1074/jbc.M512178200. [DOI] [PubMed] [Google Scholar]
- 21.Lambrou GI, Papadimitriou L, Chrousos GP, Vlahopoulos SA. Glucocorticoid and proteasome inhibitor impact on the leukemic lymphoblast: multiple, diverse signals converging on a few key downstream regulators. Mol. Cell. Endocrinol. 2012;351:142–51. doi: 10.1016/j.mce.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Wright CJ, Agboke F, Muthu M, Michaelis KA, et al. Nuclear factor-κB (NF-κB) inhibitory protein IκBβ determines apoptotic cell death following exposure to oxidative stress. J. Biol. Chem. 2012;287:6230–9. doi: 10.1074/jbc.M111.318246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKay LI, Cidlowski JA. Molecular control of immune/inflammatory responses: interactions between nuclear factor-kappa B and steroid receptor-signaling pathways. Endocr. Rev. 1999;20:435–59. doi: 10.1210/edrv.20.4.0375. [DOI] [PubMed] [Google Scholar]
- 24.Grivennikov SI, Karin M. Dangerous liaisons: STAT3 and NF-kappaB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev. 2010;21:11–9. doi: 10.1016/j.cytogfr.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang W, Shao S, Jiao Z, Guo M, et al. The Th17/Treg imbalance and cytokine environment in peripheral blood of patients with rheumatoid arthritis. Rheumatol. Int. 2012;32:887–93. doi: 10.1007/s00296-010-1710-0. [DOI] [PubMed] [Google Scholar]
- 26.De Santis E, Di Vito M, Perrone GA, Mari E, et al. Overexpression of pro-inflammatory genes and down-regulation of SOCS-1 in human PTC and in hypoxic BCPAP cells. Biomed. Pharmacother. Bioméd. Pharmacothérapie. 2013;67:7–16. doi: 10.1016/j.biopha.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Anders H-J, Romagnani P, Mantovani A. Pathomechanisms: homeostatic chemokines in health, tissue regeneration, and progressive diseases. Trends Mol. Med. 2014;20:154–65. doi: 10.1016/j.molmed.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Agelopoulos M, Thanos D. Epigenetic determination of a cell-specific gene expression program by ATF-2 and the histone variant macroH2A. EMBO J. 2006;25:4843–53. doi: 10.1038/sj.emboj.7601364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Natoli G. NF-κB and chromatin: ten years on the path from basic mechanisms to candidate drugs. Immunol. Rev. 2012;246:183–92. doi: 10.1111/j.1600-065X.2012.01103.x. [DOI] [PubMed] [Google Scholar]
- 30.Hajishengallis G, Chavakis T. Endogenous modulators of inflammatory cell recruitment. Trends Immunol. 2013;34:1–6. doi: 10.1016/j.it.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 2012;122:787–95. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garding A, Bhattacharya N, Claus R, Ruppel M, et al. Epigenetic upregulation of lncRNAs at 13q14.3 in leukemia is linked to the In Cis downregulation of a gene cluster that targets NF-kB. PLoS Genet. 2013;9:e1003373. doi: 10.1371/journal.pgen.1003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, et al. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene. 2007;26:711–24. doi: 10.1038/sj.onc.1209808. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Liu J, Yao S, Li F, et al. Nuclear factor kappa B signaling initiates early differentiation of neural stem cells. Stem Cells Dayt. Ohio. 2012;30:510–24. doi: 10.1002/stem.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J, Sayed N, Hunter A, Au KF, et al. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151:547–58. doi: 10.1016/j.cell.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salminen A, Kauppinen A, Kaarniranta K. Emerging role of NF-κB signaling in the induction of senescence-associated secretory phenotype (SASP). Cell. Signal. 2012;24:835–45. doi: 10.1016/j.cellsig.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 37.Noll JE, Williams SA, Purton LE, Zannettino ACW. Tug of war in the haematopoietic stem cell niche: do myeloma plasma cells compete for the HSC niche? Blood Cancer J. 2012;2:e91. doi: 10.1038/bcj.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonavida B. RKIP-mediated chemo-immunosensitization of resistant cancer cells via disruption of the NF-κB/Snail/YY1/RKIP resistance-driver loop. Crit. Rev. Oncog. 2014;19:431–45. doi: 10.1615/critrevoncog.2014011929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spirina LV, Kondakova IV, Choynzonov EL, Chigevskaya SY, et al. Expression of vascular endothelial growth factor and transcription factors HIF-1, NF-kB expression in squamous cell carcinoma of head and neck; association with proteasome and calpain activities. J. Cancer Res. Clin. Oncol. 2013;139:625–33. doi: 10.1007/s00432-012-1366-0. [DOI] [PubMed] [Google Scholar]
- 40.Ko YJ, Balk SP. Targeting steroid hormone receptor pathways in the treatment of hormone dependent cancers. Curr. Pharm. Biotechnol. 2004;5:459–70. doi: 10.2174/1389201043376616. [DOI] [PubMed] [Google Scholar]
- 41.Yang C, Atkinson SP, Vilella F, Lloret M, et al. Opposing putative roles for canonical and noncanonical NFκB signaling on the survival, proliferation, and differentiation potential of human embryonic stem cells. Stem Cells Dayt. Ohio. 2010;28:1970–80. doi: 10.1002/stem.528. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Hu W. NFκB signaling regulates embryonic and adult neurogenesis. Front. Biol. 2012;7 doi: 10.1007/s11515-012-1233-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takase O, Yoshikawa M, Idei M, Hirahashi J, et al. The role of NF-κB signaling in the maintenance of pluripotency of human induced pluripotent stem cells. PloS One. 2013;8:e56399. doi: 10.1371/journal.pone.0056399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gugasyan R, Horat E, Kinkel SA, Ross F, et al. The NF-κB1 transcription factor prevents the intrathymic development of CD8 T cells with memory properties. EMBO J. 2012;31:692–706. [Google Scholar]
- 45.Saccani S, Pantano S, Natoli G. Modulation of NF-kappaB activity by exchange of dimers. Mol. Cell. 2003;11:1563–74. doi: 10.1016/s1097-2765(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 46.Hayashi F, Yanagawa Y, Onoé K, Iwabuchi K. Dendritic cell differentiation with prostaglandin E results in selective attenuation of the extracellular signal-related kinase pathway and decreased interleukin-23 production. Immunology. 2010;131:67–76. doi: 10.1111/j.1365-2567.2010.03275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma S, Stolina M, Yang S-C, Baratelli F, et al. Tumor cyclooxygenase 2-dependent suppression of dendritic cell function. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2003;9:961–8. [PubMed] [Google Scholar]
- 48.Ullrich E, Ménard C, Flament C, Terme M, et al. Dendritic cells and innate defense against tumor cells. Cytokine Growth Factor Rev. 2008;19:79–92. doi: 10.1016/j.cytogfr.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 49.Zhao JL, Rao DS, Boldin MP, Taganov KD, et al. NF-kappaB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc. Natl. Acad. Sci. U. S. A. 2011;108:9184–9. doi: 10.1073/pnas.1105398108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prasad S, Ravindran J, Aggarwal BB. NF-kappaB and cancer: how intimate is this relationship. Mol. Cell. Biochem. 2010;336:25–37. doi: 10.1007/s11010-009-0267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang DJ, Ratnam NM, Byrd JC, Guttridge DC. NF-κB functions in tumor initiation by suppressing the surveillance of both innate and adaptive immune cells. Cell Rep. 2014;9:90–103. doi: 10.1016/j.celrep.2014.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aggarwal BB, Kunnumakkara AB, Harikumar KB, Gupta SR, et al. Signal transducer and activator of transcription-3, inflammation, and cancer: how intimate is the relationship? Ann. N. Y. Acad. Sci. 2009;1171:59–76. doi: 10.1111/j.1749-6632.2009.04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Helm O, Held-Feindt J, Grage-Griebenow E, Reiling N, et al. Tumor-associated macrophages exhibit pro- and anti-inflammatory properties by which they impact on pancreatic tumorigenesis. Int. J. Cancer J. Int. Cancer. 2014;135:843–61. doi: 10.1002/ijc.28736. [DOI] [PubMed] [Google Scholar]
- 54.Vlahopoulos S, Zoumpourlis VC. JNK: a key modulator of intracellular signaling. Biochem. Biokhimiia. 2004;69:844–54. doi: 10.1023/b:biry.0000040215.02460.45. [DOI] [PubMed] [Google Scholar]
- 55.Ndlovu 'Matladi N, Van Lint C, Van Wesemael K, Callebert P, et al. Hyperactivated NF-{kappa}B and AP-1 transcription factors promote highly accessible chromatin and constitutive transcription across the interleukin-6 gene promoter in metastatic breast cancer cells. Mol. Cell. Biol. 2009;29:5488–504. doi: 10.1128/MCB.01657-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kodama K, Ozasa K, Okubo T. Radiation and cancer risk in atomic-bomb survivors. J. Radiol. Prot. Off. J. Soc. Radiol. Prot. 2012;32:N51–4. doi: 10.1088/0952-4746/32/1/N51. [DOI] [PubMed] [Google Scholar]
- 57.Muthuswamy R, Berk E, Junecko BF, Zeh HJ, et al. NF-κB hyperactivation in tumor tissues allows tumor-selective reprogramming of the chemokine microenvironment to enhance the recruitment of cytolytic T effector cells. Cancer Res. 2012;72:3735–43. doi: 10.1158/0008-5472.CAN-11-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao L, Yang J, Wang H-P, Liu R-Y. Imbalance in the Th17/Treg and cytokine environment in peripheral blood of patients with adenocarcinoma and squamous cell carcinoma. Med. Oncol. Northwood Lond. Engl. 2013;30:461. doi: 10.1007/s12032-013-0461-7. [DOI] [PubMed] [Google Scholar]
- 59.Baccelli I, Trumpp A. The evolving concept of cancer and metastasis stem cells. J. Cell Biol. 2012;198:281–93. doi: 10.1083/jcb.201202014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cowell CF, Weigelt B, Sakr RA, Ng CKY, et al. Progression from ductal carcinoma in situ to invasive breast cancer: revisited. Mol. Oncol. 2013;7:859–69. doi: 10.1016/j.molonc.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malouf GG, Job S, Paradis V, Fabre M, et al. Transcriptional profiling of pure fibrolamellar hepatocellular carcinoma reveals an endocrine signature. Hepatol. Baltim. Md. 2014;59:2228–37. doi: 10.1002/hep.27018. [DOI] [PubMed] [Google Scholar]
- 62.Mia-Jan K, Munkhdelger J, Lee M-R, Ji S-Y, et al. Expression of CD133 in neuroendocrine neoplasms of the digestive tract: a detailed immunohistochemical analysis. Tohoku J. Exp. Med. 2013;229:301–9. doi: 10.1620/tjem.229.301. [DOI] [PubMed] [Google Scholar]
- 63.Rahal R, Frick M, Romero R, Korn JM, et al. Pharmacological and genomic profiling identifies NF-κB-targeted treatment strategies for mantle cell lymphoma. Nat. Med. 2014;20:87–92. doi: 10.1038/nm.3435. [DOI] [PubMed] [Google Scholar]
- 64.Peri S, Devarajan K, Yang D-H, Knudson AG, et al. Meta-analysis identifies NF-κB as a therapeutic target in renal cancer. PloS One. 2013;8:e76746. doi: 10.1371/journal.pone.0076746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Armstrong MD, Von Hoff D, Barber B, Marlow LA, et al. An effective personalized approach to a rare tumor: prolonged survival in metastatic pancreatic acinar cell carcinoma based on genetic analysis and cell line development. J. Cancer. 2011;2:142–52. doi: 10.7150/jca.2.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Renner C, Zinzani PL, Gressin R, Klingbiel D, et al. A multicenter phase II trial (SAKK 36/06) of single-agent everolimus (RAD001) in patients with relapsed or refractory mantle cell lymphoma. Haematologica. 2012;97:1085–91. doi: 10.3324/haematol.2011.053173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chaturvedi NK, Rajule RN, Shukla A, Radhakrishnan P, et al. Novel treatment for mantle cell lymphoma including therapy-resistant tumor by NF-κB and mTOR dual-targeting approach. Mol. Cancer Ther. 2013;12:2006–17. doi: 10.1158/1535-7163.MCT-13-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee H, Deng J, Xin H, Liu Y, et al. A requirement of STAT3 DNA binding precludes Th-1 immunostimulatory gene expression by NF-κB in tumors. Cancer Res. 2011;71:3772–80. doi: 10.1158/0008-5472.CAN-10-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oh H-M, Yu C-R, Golestaneh N, Amadi-Obi A, et al. STAT3 protein promotes T-cell survival and inhibits interleukin-2 production through up-regulation of Class O Forkhead transcription factors. J. Biol. Chem. 2011;286:30888–97. doi: 10.1074/jbc.M111.253500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chung SS, Aroh C, Vadgama JV. Constitutive activation of STAT3 signaling regulates hTERT and promotes stem cell-like traits in human breast cancer cells. PloS One. 2013;8:e83971. doi: 10.1371/journal.pone.0083971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Murphy SH, Suzuki K, Downes M, Welch GL, et al. Tumor suppressor protein (p)53, is a regulator of NF-kappaB repression by the glucocorticoid receptor. Proc. Natl. Acad. Sci. U. S. A. 2011;108:17117–22. doi: 10.1073/pnas.1114420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mantel C, Messina-Graham S, Moh A, Cooper S, et al. Mouse hematopoietic cell-targeted STAT3 deletion: stem/progenitor cell defects, mitochondrial dysfunction, ROS overproduction, and a rapid aging-like phenotype. Blood. 2012;120:2589–99. doi: 10.1182/blood-2012-01-404004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu X, Harris SL, Levine AJ. The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res. 2006;66:4795–801. doi: 10.1158/0008-5472.CAN-05-4579. [DOI] [PubMed] [Google Scholar]
- 74.Stam RW, Den Boer ML, Schneider P, de Boer J, et al. Association of high-level MCL-1 expression with in vitro and in vivo prednisone resistance in MLL-rearranged infant acute lymphoblastic leukemia. Blood. 2010;115:1018–25. doi: 10.1182/blood-2009-02-205963. [DOI] [PubMed] [Google Scholar]
- 75.Lerebours F, Vacher S, Andrieu C, Espie M, et al. NF-kappa B genes have a major role in inflammatory breast cancer. BMC Cancer. 2008;8:41. doi: 10.1186/1471-2407-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xiang M, Birkbak NJ, Vafaizadeh V, Walker SR, et al. STAT3 induction of miR-146b forms a feedback loop to inhibit the NF-κB to IL-6 signaling axis and STAT3-driven cancer phenotypes. Sci. Signal. 2014;7 doi: 10.1126/scisignal.2004497. ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nikulenkov F, Spinnler C, Li H, Tonelli C, et al. Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ. 2012;19:1992–2002. doi: 10.1038/cdd.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yan S, Li Z, Thiele CJ. Inhibition of STAT3 with orally active JAK inhibitor, AZD1480, decreases tumor growth in Neuroblastoma and Pediatric Sarcomas In vitro and In vivo. Oncotarget. 2013;4:433–45. doi: 10.18632/oncotarget.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Han S-S, Yun H, Son D-J, Tompkins VS, et al. NF-kappaB/STAT3/PI3K signaling crosstalk in iMyc E mu B lymphoma. Mol. Cancer. 2010;9:97. doi: 10.1186/1476-4598-9-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hosoi H, Dilling MB, Liu LN, Danks MK, et al. Studies on the mechanism of resistance to rapamycin in human cancer cells. Mol. Pharmacol. 1998;54:815–24. doi: 10.1124/mol.54.5.815. [DOI] [PubMed] [Google Scholar]
- 83.Blandino G, Valerio M, Cioce M, Mori F, et al. Metformin elicits anticancer effects through the sequential modulation of DICER and c-MYC. Nat. Commun. 2012;3:865. doi: 10.1038/ncomms1859. [DOI] [PubMed] [Google Scholar]
- 84.Son D-S, Kabir SM, Dong Y-L, Lee E, et al. Inhibitory effect of tumor suppressor p53 on proinflammatory chemokine expression in ovarian cancer cells by reducing proteasomal degradation of IκB. PloS One. 2012;7:e51116. doi: 10.1371/journal.pone.0051116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gurova KV, Hill JE, Guo C, Prokvolit A, et al. Small molecules that reactivate p53 in renal cell carcinoma reveal a NF-kappaB-dependent mechanism of p53 suppression in tumors. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17448–53. doi: 10.1073/pnas.0508888102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jeong S-J, Pise-Masison CA, Radonovich MF, Park HU, et al. A novel NF-kappaB pathway involving IKKbeta and p65/RelA Ser-536 phosphorylation results in p53 Inhibition in the absence of NF-kappaB transcriptional activity. J. Biol. Chem. 2005;280:10326–32. doi: 10.1074/jbc.M412643200. [DOI] [PubMed] [Google Scholar]
- 87.Kawauchi K, Araki K, Tobiume K, Tanaka N. Activated p53 induces NF-kappaB DNA binding but suppresses its transcriptional activation. Biochem. Biophys. Res. Commun. 2008;372:137–41. doi: 10.1016/j.bbrc.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 88.Liu X, Wang X, Zhang J, Lam EKY, et al. Warburg effect revisited: an epigenetic link between glycolysis and gastric carcinogenesis. Oncogene. 2010;29:442–50. doi: 10.1038/onc.2009.332. [DOI] [PubMed] [Google Scholar]
- 89.Leung CT. Epithelial cell translocation: new insights into mechanisms of tumor initiation. BioEssays. 2013;35:80–3. doi: 10.1002/bies.201200151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sheng W-Y, Chen Y-R, Wang T-CV. A major role of PKC theta and NFkappaB in the regulation of hTERT in human T lymphocytes. FEBS Lett. 2006;580:6819–24. doi: 10.1016/j.febslet.2006.11.044. [DOI] [PubMed] [Google Scholar]
- 91.Liang S, Mele J, Wu Y, Buffenstein R, et al. Resistance to experimental tumorigenesis in cells of a long-lived mammal, the naked mole-rat (Heterocephalus glaber). Aging Cell. 2010;9:626–35. doi: 10.1111/j.1474-9726.2010.00588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lewis KN, Mele J, Hornsby PJ, Buffenstein R. Stress resistance in the naked mole-rat: the bare essentials - a mini-review. Gerontology. 2012;58:453–62. doi: 10.1159/000335966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang L, Karin M. Roles of tumor suppressors in regulating tumor-associated inflammation. Cell Death Differ. 2014;21:1677–86. doi: 10.1038/cdd.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McGranahan N, Favero F, de Bruin EC, Birkbak NJ, et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med. 2015;7:283ra54. doi: 10.1126/scitranslmed.aaa1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Poltz R, Naumann M. Dynamics of p53 and NF-κB regulation in response to DNA damage and identification of target proteins suitable for therapeutic intervention. BMC Syst. Biol. 2012;6:125. doi: 10.1186/1752-0509-6-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Di Minin G, Bellazzo A, Dal Ferro M, Chiaruttini G, et al. Mutant p53 reprograms TNF signaling in cancer cells through interaction with the tumor suppressor DAB2IP. Mol. Cell. 2014;56:617–29. doi: 10.1016/j.molcel.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 97.Malaquin N, Carrier-Leclerc A, Dessureault M, Rodier F. DDR-mediated crosstalk between DNA-damaged cells and their microenvironment. Front. Genet. 2015;6:94. doi: 10.3389/fgene.2015.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Leibovich-Rivkin T, Buganim Y, Solomon H, Meshel T, et al. Tumor-promoting circuits that regulate a cancer-related chemokine cluster: dominance of inflammatory mediators over oncogenic alterations. Cancers. 2012;4:55–76. doi: 10.3390/cancers4010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Leibovich-Rivkin T, Liubomirski Y, Meshel T, Abashidze A, et al. The inflammatory cytokine TNFα cooperates with Ras in elevating metastasis and turns WT-Ras to a tumor-promoting entity in MCF-7 cells. BMC Cancer. 2014;14:158. doi: 10.1186/1471-2407-14-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yu P, Fu Y-X. Targeting tumors with LIGHT to generate metastasis-clearing immunity. Cytokine Growth Factor Rev. 2008;19:285–94. doi: 10.1016/j.cytogfr.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu G, Tang Y, Geng N, Zheng M, et al. HIF-α/MIF and NF-κB/IL-6 axes contribute to the recruitment of CD11b+Gr-1+ myeloid cells in hypoxic microenvironment of HNSCC. Neoplasia N. Y. N. 2014;16:168–79. doi: 10.1593/neo.132034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ying H, Elpek KG, Vinjamoori A, Zimmerman SM, et al. PTEN is a major tumor suppressor in pancreatic ductal adenocarcinoma and regulates an NF-κB-cytokine network. Cancer Discov. 2011;1:158–69. doi: 10.1158/2159-8290.CD-11-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Abrahamsson J, Påhlman M, Mellander L. Interleukin 6, but not tumour necrosis factor-alpha, is a good predictor of severe infection in febrile neutropenic and non-neutropenic children with malignancy. Acta Paediatr. Oslo Nor. 1992. 1997;86:1059–64. doi: 10.1111/j.1651-2227.1997.tb14807.x. [DOI] [PubMed] [Google Scholar]
- 104.Mace TA, Ameen Z, Collins A, Wojcik S, et al. Pancreatic cancer-associated stellate cells promote differentiation of myeloid-derived suppressor cells in a STAT3-dependent manner. Cancer Res. 2013;73:3007–18. doi: 10.1158/0008-5472.CAN-12-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Knoop RF, Sparn M, Waldmann J, Plassmeier L, et al. Chronic pancreatitis and systemic inflammatory response syndrome prevent impact of chemotherapy with gemcitabine in a genetically engineered mouse model of pancreatic cancer. Neoplasia N. Y. N. 2014;16:463–70. doi: 10.1016/j.neo.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brandi G, Tavolari S, Guarnieri T, Di Marco M, et al. Antiprotease strategy in pancreatic cancer treatment: emergence from a preclinical study. Pancreas. 2014;43:53–63. doi: 10.1097/MPA.0b013e3182a6486e. [DOI] [PubMed] [Google Scholar]
- 107.Yang S, Huang F-K, Huang J, Chen S, et al. Molecular mechanism of fascin function in filopodial formation. J. Biol. Chem. 2013;288:274–84. doi: 10.1074/jbc.M112.427971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Snyder M, Huang J, Huang X-Y, Zhang JJ. A STAT3/NFκB Complex is Necessary for the Expression of Fascin in Metastatic Breast Cancer Cells in Response to IL-6 and TNF-α. J. Biol. Chem. 2014 doi: 10.1074/jbc.M114.591719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sun J, He H, Pillai S, Xiong Y, et al. GATA3 transcription factor abrogates Smad4 transcription factor-mediated fascin overexpression, invadopodium formation, and breast cancer cell invasion. J. Biol. Chem. 2013;288:36971–82. doi: 10.1074/jbc.M113.506535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wong V, Ching D, McCrea PD, Firestone GL. Glucocorticoid down-regulation of fascin protein expression is required for the steroid-induced formation of tight junctions and cell-cell interactions in rat mammary epithelial tumor cells. J. Biol. Chem. 1999;274:5443–53. doi: 10.1074/jbc.274.9.5443. [DOI] [PubMed] [Google Scholar]
- 111.Owens MB, Hill AD, Hopkins AM. Ductal barriers in mammary epithelium. Tissue Barriers. 2013;1:e25933. doi: 10.4161/tisb.25933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yang L, Pang Y, Moses HL. TGF-beta and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 2010;31:220–7. doi: 10.1016/j.it.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ellermeier J, Wei J, Duewell P, Hoves S, et al. Therapeutic efficacy of bifunctional siRNA combining TGF-β1 silencing with RIG-I activation in pancreatic cancer. Cancer Res. 2013;73:1709–20. doi: 10.1158/0008-5472.CAN-11-3850. [DOI] [PubMed] [Google Scholar]
- 114.Balanis N, Wendt MK, Schiemann BJ, Wang Z, et al. Epithelial to mesenchymal transition promotes breast cancer progression via a fibronectin-dependent STAT3 signaling pathway. J. Biol. Chem. 2013;288:17954–67. doi: 10.1074/jbc.M113.475277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pang Y, Gara SK, Achyut BR, Li Z, et al. TGF-β signaling in myeloid cells is required for tumor metastasis. Cancer Discov. 2013;3:936–51. doi: 10.1158/2159-8290.CD-12-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.López-Novoa JM, Nieto MA. Inflammation and EMT: an alliance towards organ fibrosis and cancer progression. EMBO Mol. Med. 2009;1:303–14. doi: 10.1002/emmm.200900043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cufí S, Vazquez-Martin A, Oliveras-Ferraros C, Martin-Castillo B, et al. Metformin against TGFβ-induced epithelial-to-mesenchymal transition (EMT): from cancer stem cells to aging-associated fibrosis. Cell Cycle Georget. Tex. 2010;9:4461–8. doi: 10.4161/cc.9.22.14048. [DOI] [PubMed] [Google Scholar]
- 118.Zubeldia IG, Bleau A-M, Redrado M, Serrano D, et al. Epithelial to mesenchymal transition and cancer stem cell phenotypes leading to liver metastasis are abrogated by the novel TGFβ1-targeting peptides P17 and P144. Exp. Cell Res. 2013;319:12–22. doi: 10.1016/j.yexcr.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 119.Sun J, He H, Xiong Y, Lu S, et al. Fascin protein is critical for transforming growth factor β protein-induced invasion and filopodia formation in spindle-shaped tumor cells. J. Biol. Chem. 2011;286:38865–75. doi: 10.1074/jbc.M111.270413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Maier HJ, Wirth T, Beug H. Epithelial-mesenchymal transition in pancreatic carcinoma. Cancers. 2010;2:2058–83. doi: 10.3390/cancers2042058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim H, Choi J-A, Kim J-H. Ras promotes transforming growth factor-β (TGF-β)-induced epithelial-mesenchymal transition via a leukotriene B4 receptor-2-linked cascade in mammary epithelial cells. J. Biol. Chem. 2014;289:22151–60. doi: 10.1074/jbc.M114.556126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dews M, Tan GS, Hultine S, Raman P, et al. Masking epistasis between MYC and TGF-β pathways in antiangiogenesis-mediated colon cancer suppression. J. Natl. Cancer Inst. 2014;106:dju043. doi: 10.1093/jnci/dju043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Novitskiy SV, Pickup MW, Chytil A, Polosukhina D, et al. Deletion of TGF-β signaling in myeloid cells enhances their anti-tumorigenic properties. J. Leukoc. Biol. 2012;92:641–51. doi: 10.1189/jlb.1211639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li J, Favata M, Kelley JA, Caulder E, et al. INCB16562, a JAK1/2 selective inhibitor, is efficacious against multiple myeloma cells and reverses the protective effects of cytokine and stromal cell support. Neoplasia N. Y. N. 2010;12:28–38. doi: 10.1593/neo.91192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ivanov VN, Wen G, Hei TK. Sodium arsenite exposure inhibits AKT and Stat3 activation, suppresses self-renewal and induces apoptotic death of embryonic stem cells. Apoptosis Int. J. Program. Cell Death. 2013;18:188–200. doi: 10.1007/s10495-012-0779-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Canino C, Mori F, Cambria A, Diamantini A, et al. SASP mediates chemoresistance and tumor-initiating-activity of mesothelioma cells. Oncogene. 2012;31:3148–63. doi: 10.1038/onc.2011.485. [DOI] [PubMed] [Google Scholar]
- 127.Van Dommelen SM, Vader P, Lakhal S, Kooijmans SAA, et al. Microvesicles and exosomes: opportunities for cell-derived membrane vesicles in drug delivery. J. Control. Release Off. J. Control. Release Soc. 2012;161:635–44. doi: 10.1016/j.jconrel.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 128.Lambrou GI, Vlahopoulos S, Papathanasiou C, Papanikolaou M, et al. Prednisolone exerts late mitogenic and biphasic effects on resistant acute lymphoblastic leukemia cells: Relation to early gene expression. Leuk. Res. 2009;33:1684–95. doi: 10.1016/j.leukres.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 129.Sonabend RY, McKay SV, Okcu MF, Yan J, et al. Hyperglycemia during induction therapy is associated with poorer survival in children with acute lymphocytic leukemia. J. Pediatr. 2009;155:73–8. doi: 10.1016/j.jpeds.2009.01.072. [DOI] [PubMed] [Google Scholar]
- 130.Hulleman E, Kazemier KM, Holleman A, VanderWeele DJ, et al. Inhibition of glycolysis modulates prednisolone resistance in acute lymphoblastic leukemia cells. Blood. 2009;113:2014–21. doi: 10.1182/blood-2008-05-157842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Langlais D, Couture C, Balsalobre A, Drouin J. Regulatory network analyses reveal genome-wide potentiation of LIF signaling by glucocorticoids and define an innate cell defense response. PLoS Genet. 2008;4:e1000224. doi: 10.1371/journal.pgen.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lam LT, Wright G, Davis RE, Lenz G, et al. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-{kappa}B pathways in subtypes of diffuse large B-cell lymphoma. Blood. 2008;111:3701–13. doi: 10.1182/blood-2007-09-111948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ando M, Uehara I, Kogure K, Asano Y, et al. Interleukin 6 enhances glycolysis through expression of the glycolytic enzymes hexokinase 2 and 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3. J. Nippon Med. Sch. Nippon Ika Daigaku Zasshi. 2010;77:97–105. doi: 10.1272/jnms.77.97. [DOI] [PubMed] [Google Scholar]
- 134.Kang KY, Kim Y-K, Yi H, Kim J, et al. Metformin downregulates Th17 cells differentiation and attenuates murine autoimmune arthritis. Int. Immunopharmacol. 2013;16:85–92. doi: 10.1016/j.intimp.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 135.Grimaldi C, Chiarini F, Tabellini G, Ricci F, et al. AMP-dependent kinase/mammalian target of rapamycin complex 1 signaling in T-cell acute lymphoblastic leukemia: therapeutic implications. Leukemia. 2012;26:91–100. doi: 10.1038/leu.2011.269. [DOI] [PubMed] [Google Scholar]
- 136.Griner LN, McGraw KL, Johnson JO, List AF, et al. JAK2-V617F-mediated signalling is dependent on lipid rafts and statins inhibit JAK2-V617F-dependent cell growth. Br. J. Haematol. 2013;160:177–87. doi: 10.1111/bjh.12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pugliese L, Bernardini I, Pacifico N, Peverini M, et al. Severe hypocholesterolaemia is often neglected in haematological malignancies. Eur. J. Cancer Oxf. Engl. 1990. 2010;46:1735–43. doi: 10.1016/j.ejca.2010.03.041. [DOI] [PubMed] [Google Scholar]
- 138.Kunnumakkara AB, Guha S, Krishnan S, Diagaradjane P, et al. Curcumin potentiates antitumor activity of gemcitabine in an orthotopic model of pancreatic cancer through suppression of proliferation, angiogenesis, and inhibition of nuclear factor-kappaB-regulated gene products. Cancer Res. 2007;67:3853–61. doi: 10.1158/0008-5472.CAN-06-4257. [DOI] [PubMed] [Google Scholar]
- 139.Märten A, Zeiss N, Serba S, Mehrle S, et al. Bortezomib is ineffective in an orthotopic mouse model of pancreatic adenocarcinoma. Mol. Cancer Ther. 2008;7:3624–31. doi: 10.1158/1535-7163.MCT-08-0393. [DOI] [PubMed] [Google Scholar]
- 140.Zhou H, Liu Y, Cheung LH, Kim S, et al. Characterization and mechanistic studies of a novel melanoma-targeting construct containing IκBa for specific inhibition of nuclear factor-κB activity. Neoplasia N. Y. N. 2010;12:766–77. doi: 10.1593/neo.10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–4. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xie Y. Structure, assembly and homeostatic regulation of the 26S proteasome. J. Mol. Cell Biol. 2010;2:308–17. doi: 10.1093/jmcb/mjq030. [DOI] [PubMed] [Google Scholar]
- 143.Li Y, de Magalhães JP. Accelerated protein evolution analysis reveals genes and pathways associated with the evolution of mammalian longevity. Age Dordr. Neth. 2013;35:301–14. doi: 10.1007/s11357-011-9361-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dai Y, Chen S, Wang L, Pei X-Y, et al. Disruption of IkappaB kinase (IKK)-mediated RelA serine 536 phosphorylation sensitizes human multiple myeloma cells to histone deacetylase (HDAC) inhibitors. J. Biol. Chem. 2011;286:34036–50. doi: 10.1074/jbc.M111.284216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vlahopoulos S, Critselis E, Voutsas IF, Perez SA, et al. New Use for Old Drugs? Prospective Targets of Chloroquines in Cancer Therapy. Curr. Drug Targets. 2014;15:843–51. doi: 10.2174/1389450115666140714121514. [DOI] [PubMed] [Google Scholar]
- 146.Loberg RD, Bradley DA, Tomlins SA, Chinnaiyan AM, et al. The lethal phenotype of cancer: the molecular basis of death due to malignancy. CA. Cancer J. Clin. 2007;57:225–41. doi: 10.3322/canjclin.57.4.225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.