Abstract
Purpose
Leiomyosarcoma (LMS) is a malignant neoplasm with smooth muscle differentiation. Little is known about its molecular heterogeneity and no targeted therapy currently exists for LMS. Recognition of different molecular subtypes is necessary to evaluate novel therapeutic options. In a previous study on 51 LMS, we identified three molecular subtypes in LMS. The current study was performed to determine whether the existence of these subtypes could be confirmed in independent cohorts.
Experimental Design
99 cases of LMS were expression profiled with 3′end RNA-Sequencing (3SEQ). Consensus Clustering was conducted to determine the optimal number of subtypes.
Results
We identified 3 LMS molecular subtypes and confirmed this finding by analyzing publically available data on 82 LMS from The Cancer Genome Atlas (TCGA). We identified two new FFPE tissue-compatible diagnostic immunohistochemical markers; LMOD1 for subtype I LMS and ARL4C for subtype II LMS. An LMS tissue microarray with known clinical outcome was used to show that subtype I LMS is associated with good outcome in extrauterine LMS while subtype II LMS is associated with poor prognosis in both uterine and extrauterine LMS. The LMS subtypes showed significant differences in expression levels for genes for which novel targeted therapies are being developed, suggesting that LMS subtypes may respond differentially to these targeted therapies.
Conclusion
We confirm the existence of 3 molecular subtypes in LMS using two independent datasets and show that the different molecular subtypes are associated with distinct clinical outcomes. The findings offer an opportunity for treating LMS in a subtype-specific targeted approach.
Keywords: Leiomyosarcoma, subtypes, outcome, biomarker, sequencing
Introduction
Leiomyosarcoma is reported to represent as many as 24% of all sarcomas (1, 2). Currently no targeted therapy exists for LMS and the tumor responds poorly to conventional chemotherapy or radiation therapy. A significant proportion of LMS originates in the uterus, the remainder of LMS originates in various soft tissue sites where it is thought to often arise from smooth muscle cells in vessel walls.
The successful stratification of several tumors (including breast cancer, lung cancer and colon cancer) into molecular subtypes in the past decades has significantly improved our knowledge of these malignancies and has led to changes in the therapeutic approach to these cancers (3–12). In a previous study, our group proposed the existence of three molecular subtypes of LMS by using microarray-based gene expression profiling on 51 fresh frozen samples of LMS (13). In order to validate this finding, we analyzed 99 LMS cases collected at different institutions from the years 1991 to 2012 as an independent cohort, and performed expression profiling with 3′end RNA-sequencing (3SEQ) (14–20). In addition, we analyzed the publically available expression profiling dataset from TCGA on 82 cases. Our analysis confirms the existence of three molecular subtypes in LMS. Using two novel markers we could distinguish the three subtypes by immunohistochemistry and correlate the subtypes with clinical outcome. The recognition of these molecular subtypes in LMS may have clinical significance as the subtypes vary in their expression of a number of genes for which novel targeted therapies either already exist or are being developed.
Materials and Methods
3SEQ library construction and bioinformatics analysis
Paraffin blocks of 99 Leiomyosarcoma (LMS), 4 Myometrium, 3 Leiomyoma and 6 Undifferentiated Pleomorphic Sarcomas (UPS) from 1991 to 2012 from 9 hospitals were obtained with IRB approval and a waiver of consent due to the archival nature of the specimens. Multiple 2mm-diameter cores were re-embedded into paraffin blocks longitudinally and sectioned again to ensure the purity of tumor cells. After RNA isolation, 3SEQ libraries for next generation sequencing-based expression profiling were sequenced and analysis was performed as described previously (14–20). All gene expression profiling data used for this study have been deposited in the Gene Expression Omnibus (GEO) and are publicly accessible through GSE45510, GSE53844 and GSE54511.
After filtering data to genes with a standard deviation greater than 100, transforming the data by log2 and gene-based centering, the Consensus Clustering (R package ConsensusClusteringPlus) (21) was used to determine the number of subtypes and to assign the subtype for each LMS case. This was ran over 1000 iterations with setting “Distance – (1-Pearson correlation), a 80% sample resampling, 80% gene resampling, a maximum evaluated k of 12, and agglomerative hierarchical clustering algorithm”. Silhouette width (R package cluster) (22) was then calculated based on the assignment from ConsensusClusteringPlus to measure the accuracy of assignments from ConsensusClusteringPlus. Separately, RNASeq data of 82 LMS cases were downloaded from the TCGA database. To compare with 3SEQ data, the TCGA data were normalized into TPM and analyzed with ConsensusClusteringPlus and Silhouette analysis as performed for 3SEQ data. Subclass mapping (23) was performed to determine the common LMS subtypes identified in 3SEQ and TCGA RNASeq datasets.
In order to get subtype specific genes, SAMSeq (24) was performed on both datasets between each subtype and all other subtypes with a FDR of 0.05. Kaplan-Meier plots were used to compute the survival curves, and Log-rank (Mantel-Cox) test was used to determine the statistical significance of survival between groups by using GraphPad Prism5 software. Significance of contingency analysis between one subtype and the other subtypes was measured by two tail Fisher’s exact test and Chi-square test with GraphPad Prism5 software. Univariate and multivariate analysis by the Cox proportional hazard method was performed using the survival package in R. Analysis of gene ontology (GO) was done using DAVID Bioinformatics Resources version 6.7 (25, 26). CSF1-signature (27) positive, and CINSARC-signature (28) positive cases were identified as those cases that coordinately highly expressed these signature genes with >0.3 correlation with the centroid as performed previously (19, 27, 29, 30). Principal component analysis (PCA) was performed on square root transformed TPM with R package pcaMethods, ellipse contour of principal components (PC) was computed and drawn with R package car.
TMA construction, Immunohistochemistry staining and scoring
The tissue microarrays used in this study (TA-201 and TA-381) were constructed with Tissue Arrayer (Beecher Instruments, Sun Prairie, WI). TA-201 included the LMS cases with clinic outcome data, while TA-381 was comprised of LMS cases analyzed by 3SEQ. For Immunohistochemistry staining, sections underwent antigen retrieval and were stained with anti-ARL4C antibody (1:120, Sigma, CAT#HPA028927) and anti-LMOD1 antibody (1:20, Sigma, CAT#HPA028325) with the EnVision+system (Dako). The staining results were scored as follows: 3 (+++), strong staining (>30% positivity); 2 (++), weak staining (10–30%); 1 (+), equivocal or uninterpretable (<10%); 0 (−), absence of any staining. Hierarchical clustering of IHC results was performed with software “Cluster3” with “uncentered Correlation” and “Centroid Linkage” and a clustered heatmap was visualized with Java TreeView (31, 32).
Results
Consensus Clustering identifies three molecular subtypes of leiomyosarcoma based on gene expression data
We analyzed 99 cases of LMS by 3SEQ, a next generation sequencing based approach that quantifies the number of 3′ ends for each polyadenylated mRNA or lncRNA and that performs well on RNA isolated from FFPE material (14–20). Clinical information of the cases is shown in Table 1 and Supplementary Table S1. The median age for this cohort is 56 years, with 49 cases originating in the uterus and 49 cases in extrauterine sites. For one case the origin was unknown. A female to male ratio of 3.3 to 1 was noted in this cohort. Half of the extrauterine LMS (24 cases) were from the extremities, while the remainder were from thoracic, abdominal, or retroperitoneal regions.
Table 1.
Patient characteristics (N=99).
| No. of Patients | Percent (%) | Subtype I | Subtype II | Subtype III | Other LMSa | |
|---|---|---|---|---|---|---|
| Age(year) | ||||||
| Median | 56 | 55 | 55 | 60 | 57 | |
| Range | 2 ~ 94 | 2 ~ 84 | 41 ~ 67 | 43 ~ 80 | 38 ~ 94 | |
| Sex | ||||||
| Female | 76 | 77% | 22 | 18 | 12 | 24 |
| Male | 23 | 23% | 13 | 4 | 1 | 5 |
| Location | ||||||
| Uterine | 49 | 49% | 9 | 13 | 12 | 15 |
| TARb | 25 | 25% | 12 | 8 | 1 | 4 |
| Extremities | 24 | 24% | 13 | 1 | 0 | 10 |
| Unknown | 1 | 1% | 1 | 0 | 0 | 0 |
| Grade | ||||||
| Low | 18 | 18% | 10 | 3 | 0 | 2 |
| Intermediate | 19 | 19% | 10 | 4 | 3 | 5 |
| High | 62 | 63% | 15 | 15 | 10 | 22 |
| Relapse | ||||||
| Primary | 62 | 63% | 22 | 9 | 12 | 19 |
| Local recur | 27 | 27% | 9 | 12 | 0 | 6 |
| Metastasis | 10 | 10% | 4 | 1 | 1 | 4 |
| Therapyc | ||||||
| Yes | 11 | 11% | 4 | 4 | 1 | 2 |
| No | 86 | 87% | 29 | 18 | 12 | 27 |
| Unknown | 2 | 2% | 2 | 0 | 0 | 0 |
|
| ||||||
| Total | 99 | 35 | 22 | 13 | 29 | |
other LMS: LMS with negative silhouette value.
TAR: Thoracic/Abdominal/Retroperitoneal sites.
Any prior chemotherapy and radiotherapy.
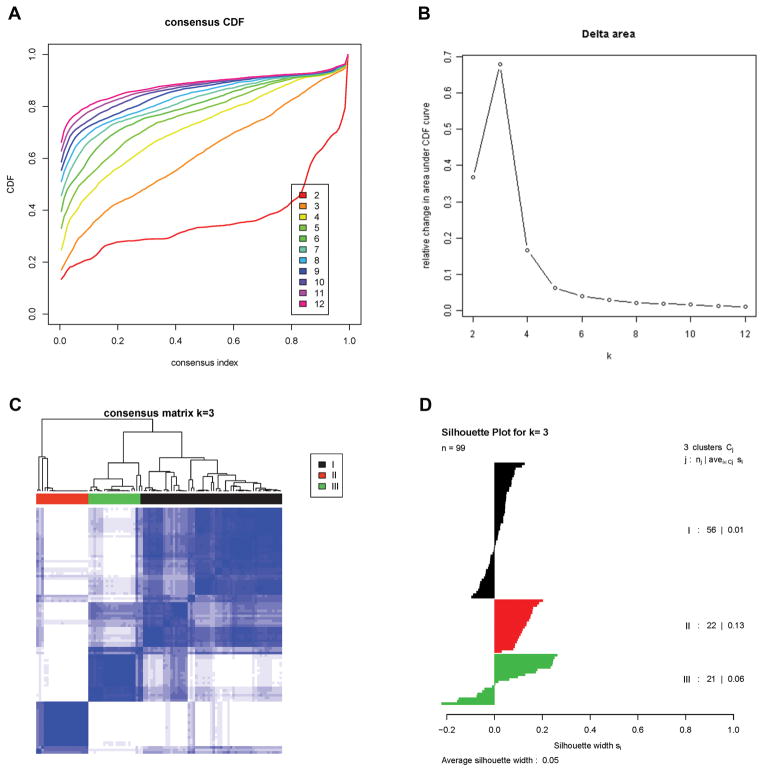
3SEQ yielded an average of 18 million total reads after quality filtering and an average of 2 million uniquely mapping reads against human transcriptome (refMrna) per sample. All reads that uniquely mapped to an individual gene (UCSC genome browser, hg19) were combined to obtain a quantification of the expression level for that gene. The genes with the highest degree of variation in expression levels across all samples were identified by filtering the dataset for genes with a standard deviation ≥100, yielding 1,300 genes. These were used to perform Consensus Clustering (21), a method that uses data-resampling followed by iterative re-clustering to estimate cluster stability and to help determine the optimal number of clusters in a dataset. As shown in Figure 1A and 1B, the greatest increase in the area under the CDF curve (empirical cumulative distribution function) is seen when 3 molecular subtypes are assumed. Assuming 4 molecular subtypes showed a lesser degree of increase in the area under the CDF curve and showed no significant increase in the ability to classify the cases (Supplementary Fig. S1). In order to keep the number of subtypes to a manageable number, we focused on the existence of three molecular subtypes of LMS for this study. A consensus matrix of the probability that 2 cases of LMS will belong to the same subtype is shown in Figure 1C. Silhouette analysis (22) was performed to measure the confidence with which an individual LMS case could be assigned to its subtype (Figure 1D). This analysis indicated that all subtype II cases had a positive silhouette value while a subset of subtype I and III LMS had negative values. Similar to studies performed by others, we used only the 70 LMS cases with positive Silhouette values as the most reliable examples (“core” LMS cases) for subsequent analyses (3,8). In the group of 70 core LMS cases, 35 belonged to subtype I, 22 to subtype II while 13 were subtype III.
Figure 1. Identification of three molecular subtypes in LMS.
A). Empirical cumulative distribution plots as a function of the number of molecular subtypes in 3SEQ cohort.
B). Relative increase in the area under the CDF curve with increasing number of molecular subtypes.
C). Consensus clustering matrix of LMS samples using 3 molecular subtypes.
D). Silhouette analysis of LMS samples based on the assignments from Consensus Clustering.
Molecular subtypes of LMS can be reproduced in an independent dataset
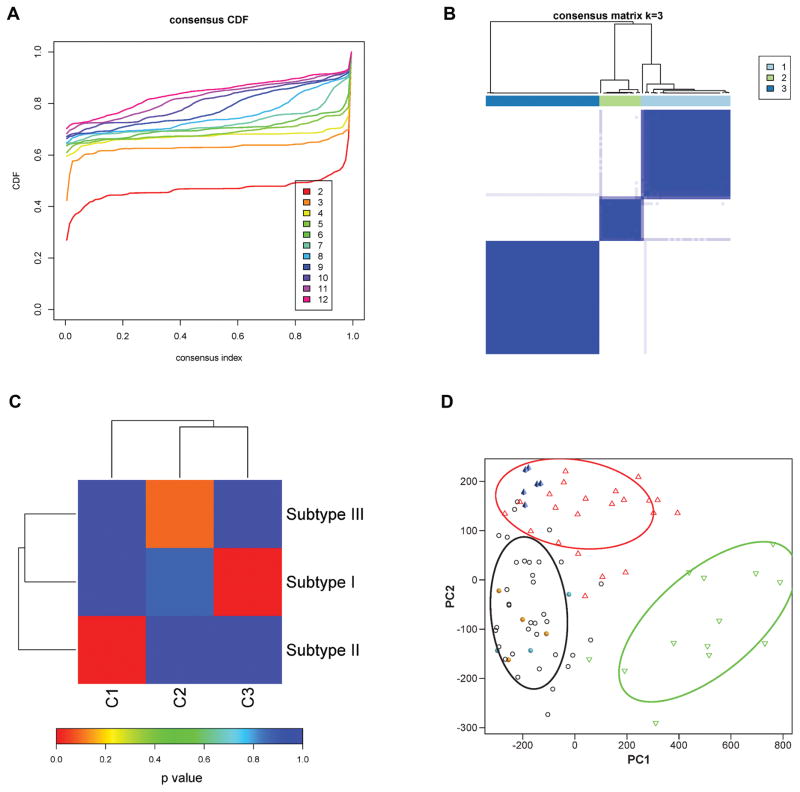
To further explore the reproducibility of the three LMS subtypes, we downloaded gene expression data for 82 LMS cases from the TCGA database. Of these samples, 80 cases were primary LMS while 2 cases were recurrences. The origin of the tumors differed from the 99 cases analysed by 3SEQ with 19 LMS originating from the uterus, 13 cases from the extremities and 32 cases from other extrauterine sites. The remaining 18 cases had an unknown primary site. In addition to the differences in the site of origin for the tumors examined there was a significant difference in the percentage of samples derived from primary lesions versus recurrences between the two cohorts. In the 3SEQ cohort 37% of the cases were recurrences or metastases while only 2% of TCGA LMS cases were recurrent and none were metastatic (Supplementary Table S2). The 2 datasets differed in two additional aspects. First, the TCGA dataset used fresh frozen material whereas our 3SEQ analysis was performed on FFPE material. Second, whole transcriptome RNASeq was used for the TCGA dataset while 3SEQ determines the gene expression level based on quantification of 3′end reads. Despite these differences, analysis of the TCGA dataset produced results very similar to those found in the 3SEQ cohort. The TCGA dataset yielded 1,174 genes when filtered for a standard deviation ≥100 (1,300 genes in the 3SEQ set). By Consensus Clustering and Silhouette analysis, the TCGA dataset also showed the greatest increase in area under the CDF curve when three subtypes were used (Figure 2A). Compared to the cases studied by 3SEQ in our cohort the homogeneity of the subtypes in the TCGA cohort was greater as determined by consensus clustering (Figure 2B), possibly due to the fact that frozen material and not FFPE material was used for the cases in the TCGA cohort. To measure the reproducibility between the 3SEQ and TCGA cohorts the cases with positive Silhouette values from both sets were analysed by Subclass mapping (23). The 3SEQ-subtype I and -subtype II types were significantly reproduced as subtype-C3 and -C1 in the TCGA cohort with FDR corrected p-values of 0.0090 for both. The 3SEQ-subtype III did not reach statistical significance when it was compared with the remaining TCGA-subtype C2 (p = 0.0959) (Figure 2C). A substantial number of genes identified by SAMSeq in the 3SEQ and TCGA datasets showed overlap between 3SEQ subtypes I and -II and the corresponding TCGA subtypes (Supplementary Table S3). Importantly, five genes for which we previously used immunohistochemical markers (13, 33) to identify subtype I LMS (CASQ2, ACTG2, MYLK, SLMAP, and CFL2) were shown to be highly expressed on the mRNA level in TCGA-subtype C3, as was the new subtype I marker LMOD1. In addition, the new marker ARL4C, which identifies 3SEQ subtype II cases, was highly expressed in the corresponding TCGA-subtype C1 (see below and Supplementary Table S4). Together, these data indicate that the existence of 3 molecular subtypes in LMS can be reproduced on the mRNA level across these different datasets.
Figure 2. Identification of molecular subtypes of LMS in TCGA data.
A). Empirical cumulative distribution plots as a function of the number of molecular subtypes in TCGA cohort.
B). Consensus clustering matrix of LMS samples using 3 molecular subtypes.
C). Subclass association (SA) matrix between 3SEQ data (subtype I, subtype II, subtype III) and TCGA data (subtype C1, subtype C2, subtype C3). p value is corrected with FDR.
D). Principal component analysis (PCA) of 70 core 3SEQ LMS cases with other diagnoses. Subtype I LMS-black circle; subtype II LMS-red triangle: subtype III LMS-green triangle; leiomyoma-cyan solid circle; myometrium-orange solid circle; UPS-blue solid pyramid. The elliptical contours represent the normal probability of 0.75.
Clinical features of LMS molecular subtypes in three datasets
In the 3SEQ dataset, approximately 26%, 59% and 92% of subtype I, subtype II and subtype III LMS were derived from the uterus with uterine LMS being significantly associated with subtype III (p = 0.0005, Chi-square test). However, when evaluating only those 34 cases arising in the uterus, we found that uterine LMS had a roughly equal chance of belonging to each of the molecular subtypes with 9, 13, and 12 cases belonging to subtype I, II, and III, respectively. In contrast, the extrauterine LMS were overrepresented in subtype I and II, with 25, 9 and 1 cases belonging to subtype I, II and III, respectively. These results indicate that a subset of uterine LMSs behave as independent molecular subtype (subtype III) while another subset of uterine LMS together with extrauterine LMS belong to subtype I and subtype II LMS. In the TCGA dataset, we observed similar demographic characteristics as in the 3SEQ cohort, with TCGA C2 like 3SEQ subtype III being significantly associated with uterine LMS (p < 0.0001), while TCGA C3 (subtype I) and TCGA C1 (subtype II) were overrepresented in extrauterine sites (Supplementary Table S2). These results correlate well with our findings on 51 LMS cases from our previous dataset analyzed by gene microarrays, where the ratio of uterine LMS (13) also was higher in subtype III (10/24) than in subtype I (1/13) and subtype II (3/12), but did not reach significance (p = 0.1178, Chi-square test).
Within the 3SEQ cohort, 18% (18) of tumors are low grade, 19% (19) are intermediate grade, and 63% (62) are high grade by histologic analysis. Low grade lesions were more frequent in subtype I LMS, with 10/35 cases showing low grade histology while 3/22 of subtype II and 0/13 of subtype III LMS cases are low grade respectively, however there was no statistically significant correlation between grade and molecular subtype (Table 1, p = 0.0989, Chi-square). The median age for subtype I, subtype II, and subtype III was 55, 55 and 60 years, respectively. One-way ANOVA analysis showed that there was no significant difference in age between the three subtypes (p = 0.3769).
Functional annotation of LMS molecular subtypes based on subtype-specific genes in the 3SEQ dataset
SAMSeq (24) was used to identify genes that were significantly over/under-expressed in each subtype of LMS analyzed by 3SEQ, again using only those cases with a positive Silhouette value. In subtype I LMS, 5900 genes are relatively overexpressed while 1900 genes are under-expressed compared to the other two subtypes (Supplementary Table S5). The overexpressed genes were enriched in Gene Ontology (GO) Biology processes that included muscle contraction, muscle system processes, and cytoskeleton organization. In addition, a number of genes (LMOD1, MYOCD, CALD1, DES, CNN1, ACTG2, SLMAP and CASQ2) known to be involved in smooth muscle function were identified in this list indicating that subtype I was most associated with normal smooth muscle function (Supplementary Table S6). Consistent with this finding, 4 samples of normal myometrium and 3 cases of benign uterine leiomyoma were grouped with subtype I LMS in principal component analysis (Figure 2D) and clustered with subtype I when analyzed by unsupervised hierarchical clustering with the 70 LMS core samples (Supplementary Fig. S2A). As stated above, the overexpression of genes associated with muscle function was also noted on the mRNA level in corresponding subtype C3 from the TCGA dataset, while the subtype I identified previously through gene microarray analysis had high levels of expression for these genes at both the mRNA and protein level (13).
In 3SEQ data on subtype II LMS, 5100 genes are significantly overexpressed and 2087 genes are expressed at lower levels as compared to the other two subtypes of LMS. These subtype II LMS specific genes are enriched in GO biological processes including translation, translational elongation, and protein localization (Supplementary Table S6). Subtype II showed significantly less muscle specific genes than subtype I. Undifferentiated pleomorphic sarcoma (UPS) is a tumor type that fails to express differentiation markers and that can be difficult to distinguish from LMS. Six cases of UPS grouped with subtype II LMS in principal component analysis (Figure 2D) and by unsupervised hierarchical clustering (Supplementary Fig. S2B). A search for the CSF1-signature, which we previously reported to be associated with poor outcome for both gynecological and non-gynecological LMS (27), in the 3SEQ data showed that the cases with a centroid correlation > 0.3 for the CSF1 signature were overwhelmingly subtype II LMS (p < 0.0001, Chi-square test).
A total of 1320 genes are significantly overexpressed and 7544 genes are under-expressed in subtype III LMS when compared to the other two subtypes. Subtype III overexpressed genes are enriched in GO biological processes including metabolic process, ion transport, and regulation of transcription (Supplementary Table S6). When considering only the genes with a mean level of expression 50 reads (TPM) or more in any subtype in the Stanford 3SEQ cohort the number of overexpressed genes identified by SAMseq will decrease from 11,125 to 3,350. The same approach will decrease the number of genes in the TCGA cohort from 11,068 to 3,845.
Identification of novel immunohistochemistry (IHC) markers for LMS subtype I and II
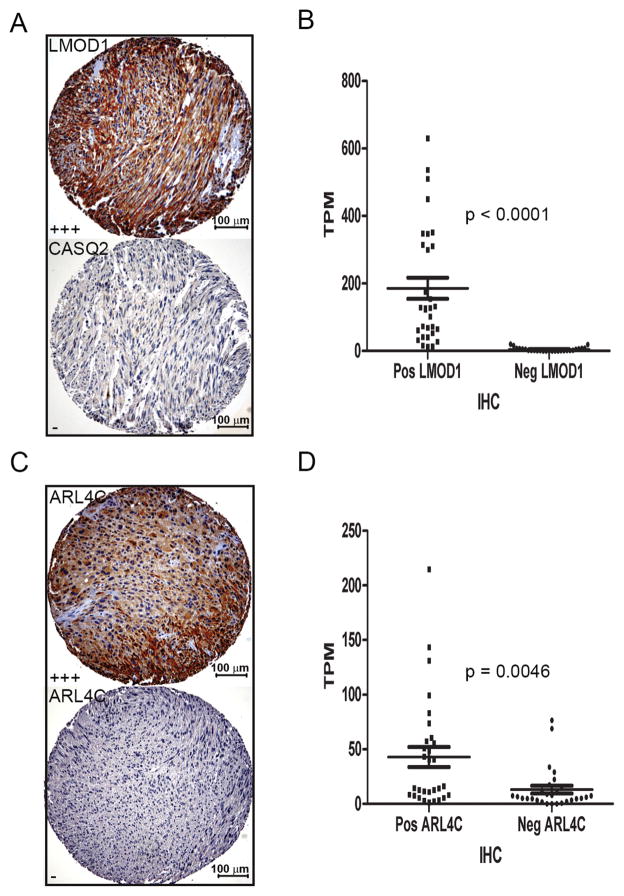
Through gene microarray analysis we previously identified 5 immunohistochemical markers (ACTG2, SLMAP, MYLK, CFL2 and CASQ2) that could identify subtype I LMS in FFPE material (13, 33). The quantitative nature of gene expression profiling by 3SEQ allowed for a more detailed search for additional immunohistochemical markers for LMS subtypes. LMOD1 mRNA was highly expressed in subtype I LMS. A tissue microarray (TA-381) was generated that contained 58 of the 70 LMS cases with positive Silhouette values. Immunohistochemistry showed strong staining of a subset LMS (Figure 3A). LMOD1 stained positive in 31 LMS cases, 19 of which were subtype I LMS as assigned by 3SEQ analysis, in contrast, 21 of 27 LMOD1 negative LMSs were subtype II or subtype III LMS. LMOD1 protein expression therefore showed a significant association with subtype I LMS (p = 0.0085, Chi-square test; r = 0.8964, p<0.0001, Spearman correlation) (Supplementary Table S7). Comparison of the LMOD1 staining results with the five previously identified subtype I biomarkers (13, 33), showed that the highest correlation of IHC staining (0.65) was obtained between genes ACTG2, SLMAP and LMOD1. CASQ2 had the lowest correlation with the 5 other genes (r = −0.22) and we refined our panel of subtype I biomarkers to include ACTG2, SLMAP, LMOD1, CFL2, and MYLK, while omitting CASQ2. Using this panel, the correlation for the new panel of markers was 0.47 (Supplementary Fig. S3). Finally, staining of TA-381 showed a significant association (p < 0.0001) between LMOD1 immunostaining and mRNA levels as determined by 3SEQ LMOD1 mRNA level (Figure 3B).
Figure 3. Immunohistochemical markers for subtype I and subtype II LMS.
A). Representative staining for LMOD1 (Score +++) and CASQ2 (Negative) for a subtype I LMS case (case # 10129, scale bar = 100μm, 10X).
B). Comparison of 3SEQ mRNA quantification and immunostaining results on TA-381 that contains 58 cases of LMS analyzed by 3SEQ; LMS cases positive for anti-LMOD1 staining have significantly higher mRNA expression levels (TPM) than LMS cases negative for anti-LMOD1 (p < 0.0001). The p value was derived from unpaired t test with Welch’s correction.
C). Positive and negative staining by ARL4C antibody for representative subtype II LMS (top, case # 10019) and subtype I LMS (bottom case # 9994, scale bar = 100μm, 10X).
D). LMS cases positive for anti-ARL4C staining have significantly higher mRNA expression levels (TPM) than LMS cases negative for anti-ARL4C (p = 0.0046).
Analysis of 3SEQ data showed high levels of mRNA expression for ARL4C in subtype II LMS. Using a FFPE compatible antibody strong staining was seen in a subset of LMS cases (Figure 3C) with positive staining in 30 LMS cases, 17 of which were subtype II LMS. Negative staining was seen in 28 LMS cases, 23 of which were subtype I or subtype III LMS. This IHC result validates ARL4C to be a subtype II LMS biomarker not only at the mRNA level (as determined by SAMSeq) but also at the protein level (p = 0.0033, Chi-square test; r = 0.5277, p < 0.0001, Spearman Correlation) (Supplementary Table S7). Comparison between 3SEQ mRNA levels and ARL4C immunohistochemistry showed a good association (p = 0.0046) (Figure 3D). Comparison between LMOD1 and ARL4C protein expression and mRNA expression levels varied as expected across the LMS subtypes as defined by 3SEQ (Supplementary Fig. S4).
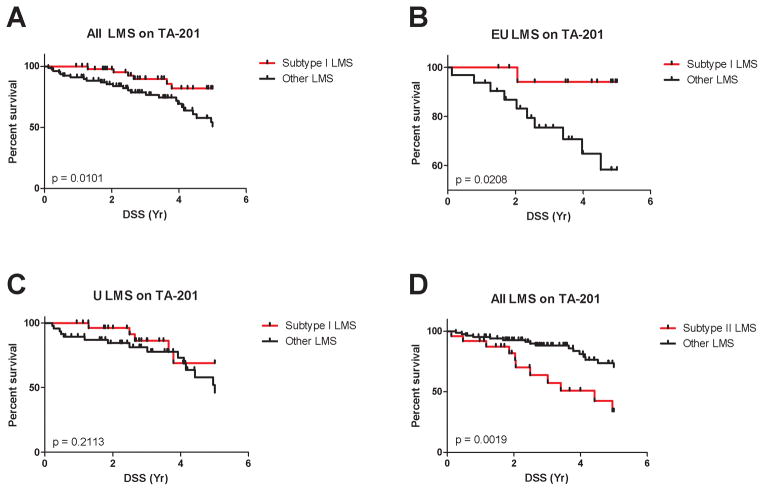
While in individual cases these markers may not definitely identify a case as belonging to a specific subtype, when combined these markers do allow us to distinguish large numbers of cases represented on a TMA in distinct molecular groups. No outcome data were available for the LMS samples used for 3SEQ analysis. To correlate the assignment of LMS subtypes with outcome we used immunostaining data on TA-201 that contains 127 cases of LMS with known clinical outcome (27, 34). In this analysis, 48 of 127 (38%) LMS cases were defined as subtype I LMS by their coordinate expression of all 5 subtype I reactive antibodies. These cases demonstrated a better disease specific survival (DSS) when uterine and extrauterine LMS were analyzed together and compared to the remaining LMS cases (p = 0.0101, Log-rank test) (Figure 4A). However, the difference in clinical outcome was mostly driven by the extrauterine LMS (p = 0.0208) (Figure 4B) and no difference in outcome was seen for subtype I LMS from uterus (p = 0.2113) (Figure 4C). The association with good outcome lost its significance in multivariate analysis when two prognostic signatures that we previously identified, ROR2 (34) and the CSF1 response protein signature (27), were included (Supplementary Table S8 and S9).
Figure 4. Immunohistochemical markers for subtype I and subtype II LMS predict different clinical outcomes.
TA-201 with 127 cases of LMS with known clinical outcome was stained for the panel of 5 subtype I LMS markers and the ARL4C marker for subtype II LMS. The p value was from Log-rank (Mantel-Cox) test.
Coordinate expression of subtype I biomarkers was associated with good outcome when analysed on all cases (panel A) but this was mainly due to an effect on extra-uterine (EU) LMS (panel B) but not uterine (U) LMS (panel C). Subtype II biomarker ARL4C predicted worse outcome in LMS (panel D), with equal effect on extra-uterine and uterine LMS (not shown).
Subtype II LMS cases were defined as those reacting for ARL4C on the outcome TMA. Staining for ARL4C was seen in 25 of 111 available LMS cases (23%) and these cases were associated with a worse DSS when all LMS were combined (p = 0.0019, Log-rank test) (Figure 4D). This finding was seen in both extrauterine (p = 0.0352) and uterine (p = 0.0461) LMS. While subtype II was associated with poor outcome in a statistically significant manner in univariate analysis (p = 0.0030, Wald test), this significance was also lost in the multivariate analysis when prognosticators ROR2 and the CSF1 response protein signature were included (27, 34) (Supplementary Table S8 and S9). For correlation with outcome in the immunohistochemistry analysis, subtype III was defined as those cases that failed to classify as either subtype -I or –II. When thus defined, subtype III had an intermediate outcome (Supplementary Fig. S5) but failed to reach prognostic significance in univariate analysis (Supplementary Table S8).
Potential clinical applications of LMS subtyping
To explore the potential for clinical implications in the recognition of LMS subtypes we compared the genes that are specifically overexpressed in each LMS subtype with genes from the TARGET V2 database known to be activated by mutations or amplifications (35). This database contains genes for which targeted therapy is either currently available or under development. A large number of these genes were found to be expressed at different levels between the 3 molecular subtypes (Table 2). This suggests that LMS subtypes may respond differentially to those targeted therapies, an observation that may play a role in determining the success rate of novel therapies in clinical trials. For several of these targets there may be an incomplete correlation between mRNA levels for the mutated or amplified target gene and response to drug, but this analysis nevertheless forms a first step towards personalization of LMS patient care.
Table 2.
Targets unique to LMS subtypes.
| Gene | Therapeutic Agents |
|---|---|
| Genes overexpressed in subtype I | |
| ARAF | Sorafenib, Vemurafenib, Dabrafenib, RAF inhibitors |
| CCNE1 | CDK2 inhibitor |
| KDR | KDR inhibitors |
| NOTCH1 | Notch Inhibitors |
| FGFR2 | FGFR Inhibitors |
| FGFR1 | FGFR Inhibitors |
| Genes overexpressed in subtype II | |
| MCL1 | Tubulins |
| CDK4 | CDK4/6 inhibitors |
| CTNNB1 | WNT inhibitors |
| AURKA | AURKA Inhibitors |
| RHEB | MTOR inhibitors |
| CCND2 | CDK4/6 inhibitors |
| CCND1 | Hormone therapy, CDK4/6 inhibitors |
| MTOR | Everolimus, Temsirolimus, MTOR inhibitors |
| MAPK3 | Erlotinib, Gefitinib, EGFR Inhibitors |
| MAPK1 | Erlotinib, Gefitinib, EGFR Inhibitors |
| CCND3 | CDK4/6 inhibitors |
| NOTCH2 | Notch Inhibitors |
| MAP2K2 | MEK inhibitors |
| Genes overexpressed in subtype III | |
| MDM4 | MDM4 inhibitors |
| ERBB3 | Pertuzumab |
| EPHA3 | Dasatinib, Ephrin inhibitors |
| ESR1 | Hormonal therapy |
| Genes overexpressed in subtype II and III | |
| EGFR | Erlotinib, Gefitinib, EGFR Inhibitors |
In addition to the genes identified from the TARGET V2, we studied the expression level of ROR2, a receptor tyrosine kinase that plays a role in tumor progression and that has a low level of expression in normal human tissues. This molecule has been proposed as a novel therapeutic target in LMS (34) and other tumors (36–38) and was found to be more frequently expressed in subtype II LMS than in the other subtypes (FDR < 0.05). Immunostaining for the ROR2 protein on the TMA containing cases analyzed by 3SEQ similarly showed a correlation with subtype II LMS (p = 0.0347, Chi-square test).
Discussion
Leiomyosarcoma is a malignant soft tissue tumor with complex genetic abnormalities. In clinical practice the tumors are treated differently depending on whether they originate from the uterus or from extrauterine sites. However, this distinction is not based on a molecular rationale and appears based, at least in part, on clinical sub-specialization. Recently Italiano et al. demonstrated a molecular difference between LMS occurring in the retroperitoneum and extremities (39), but this paper does not include uterine lesions. Leiomyosarcomas are tumors that exhibit varying degrees of smooth muscle differentiation and often appear to be derived from smooth muscle cells in the myometrium. They can also arise from the walls of blood vessels or connective tissues throughout the body. Conventional chemotherapy and radiation therapy only have limited effects and surgical resection remains the best option for treatment. Characterization of molecular subtypes in malignancies can lead to better individualization of treatment. In a prior study on 51 cases of LMS for which frozen tissue was available, we used 44k spotted cDNA arrays for gene expression profiling and obtained data that indicated that at least 3 molecular subtypes exist in LMS (13). This study relied on the availability of fresh frozen tissues and as a result only small numbers of LMS cases could be studied. The 3SEQ approach can use FFPE materials and renders a digital rather than an analogue readout for gene expression levels with a higher signal to noise ratio and allows for accurate measurement of genes expressed at a wide range of levels, including those with low levels of expression. 3SEQ analysis on 99 samples of LMS and data on a third cohort of 82 LMS cases from the TCGA study were obtained. Despite the differences in methodology and sample selection between these three cohorts, remarkable similarities were apparent and indicated the existence of three molecular subtypes in LMS.
Subtype I LMS expresses most genes associated with muscle function by Gene Ontology annotation. The similarity of subtype I LMS to smooth muscle differentiation was also supported by the fact that subtype I cases clustered with samples of leiomyoma and myometrium. Finally, we observed high expression levels for LMOD1 in subtype I LMS, a smooth muscle cell-restricted gene that is preferentially expressed in differentiated smooth muscle cells (40). The expression of smooth muscle markers in subtype I suggest a tumor subtype that is closer to the smooth muscle cells than subtype II and III. However this similarity is not captured by the objective measurement of histologic grade, as no correlation between grade and molecular subtype was found. Others have noticed the existence of a subset of LMS that express muscle function related genes at higher levels than other LMS cases. Francis et al. (41) identified a 200-gene signature for LMS by expression profiling different subtypes of 177 soft tissue sarcomas, this signature was characterized by overexpression of muscle specific genes and was shown to be highly expressed in 12 of 40 LMS cases by supervised clustering. Villacis et al. (42) performed gene expression profiling and supervised clustering with 587 genes expressed differentially between UPS and LMS, and found that a subset of LMS that highly expressed muscle function associated genes clustered separately from UPS and other LMS cases.
In contrast to subtype I LMS, subtype II LMS showed no significant smooth muscle differentiation by GO analysis and therefore represents a less differentiated form of LMS. Undifferentiated pleomorphic sarcoma (UPS) is a type of sarcoma that lacks any indication of differentiation by histologic examination and by immunohistochemical analysis. Hierarchical clustering of the core LMS cases with 6 cases of UPS showed co-clustering of UPS with subtype II LMS. Others have studied the relation between LMS and UPS by molecular studies but did not take into account the existence of LMS subtypes (43). Villacis et al. performed gene expression profiling of 22 LMS and 22 UPS, and did not identify a clear distinction of expression profiles of UPS and LMS by either unsupervised clustering or even supervised clustering using a gene list derived from SAM analysis. However, when performing supervised clustering with 587 genes expressed differentially between UPS and LMS, a subset of LMS that consisted predominantly of retroperitoneal LMS clustered separately from UPS and the remaining LMS, in part based on higher expression of muscle function associated genes in the retroperitoneal LMS (42). These findings partially overlap with our results; in our study 6 retroperitoneal LMS were analyzed, 4 of which grouped in subtype I with 2 cases grouping in subtype II, the subtype most associated with UPS.
Subtype III LMS is the only subtype that shows a preference for a specific anatomic site and was more likely to be from uterus (p = 0.005, Fishers exact test). Only one of 13 subtype III LMS did not originate from the uterus and presented in the scrotum. However, uterine LMS as a group was evenly distributed over the 3 subtypes.
Neither uterine nor extrauterine LMS have clinically accepted prognostic molecular markers, and histological grading to predict clinical outcome is controversial in uterine LMS. Several molecular prognosticators (28, 39, 41, 44, 45) have previously been reported in LMS. By genomic and expression profiling of 183 sarcomas, Chibon et al. established a prognostic signature called Complexity Index in Sarcoma (CINSARC), that includes 67 genes related to mitosis and chromosome management. This signature was shown to be associated with metastatic outcome in sarcomas including leiomyosarcoma, and even in lymphomas and breast carcinoma. The CINSARC signature is a powerful predictor of tumor metastasis and could improve the patient selection for chemotherapy (28). To investigate the significance of LMS subtypes on clinical outcome, we stained a TMA containing cores from 127 LMS cases with clinical outcome with the IHC markers for subtype I and II LMS. Subtype I LMS was associated with good outcome in extrauterine LMS, while subtype II was associated with poor outcome in univariate analysis. In this context it is interesting to note that the association between muscle gene expression and better clinical outcome is a finding that was also reported in various ways by other immunohistochemical studies (18, 45). While subtyping of LMS by immunohistochemistry, can help predict clinical outcome, in multivariate analysis the subtype I and II biomarkers were outperformed by the previously described markers ROR2 and the “CSF1 protein response signature” (27, 34). The association between immunohistochemically defined subtype II LMS with poor outcome was consistent with previously published gene expression profiling data. The CINSARC signature was significantly associated with subtype II LMS but not with the other subtypes (p = 0.0279, by Fisher’s exact test).
At this time, no targeted therapies exist for LMS. The recognition of molecular subtypes in malignancy has led to the identification of targets that are unique to a subset of the cases in a variety of tumors that include breast cancer, colon cancer, bladder cancer and glioblastoma and others (3–12). In previous studies we have identified ROR2, a receptor tyrosine kinase (34) and the presence of intratumoral macrophages (27) as prognostic markers in both uterine and extrauterine LMS. Based on our finding Subtype II LMS, which has high levels of ROR2 expression, would benefit most from drugs targeting this molecule either as a single drug or in combination with CD47 based therapy (34, 46, 47).
Here we report that the molecular subtypes in LMS show differential expression of a large number of genes for which therapeutics either currently exist or are under development. These subtype specific targets may not show a significant response when LMS cases are considered as a single homogenous entity in a clinical trial. Recognition of molecular subtypes in LMS may highlight preferential response in that subtype for which the target gene is expressed at higher levels.
In summary, we used gene expression profiling and immunohistochemical assays to demonstrate the existence of 3 distinct molecular subtypes in LMS in two independent sets of cases and showed that they are associated with distinct clinical outcomes. These findings indicate distinct biological subclasses in LMS that may respond differently to novel therapeutic approaches.
Supplementary Material
Statement of translational relevance.
Leiomyosarcoma is a malignant neoplasm with varying degrees of smooth muscle differentiation and complex genetic abnormalities. It can occur in a wide range of sites but is usually managed as a single disease in conventional therapy and in clinical trials. The recognition of molecular subtypes can lead to the identification of novel therapies that may affect one of these subtypes preferentially. We confirmed the results of an earlier study and determined the existence of 3 molecular subtypes in LMS using two independent cohorts. We identified two new FFPE tissue-compatible diagnostic immunohistochemical markers for two of these subtypes: LMOD1 for subtype I and ARL4C for subtype II, and showed that these molecular subtypes are associated with distinct clinical outcomes. Our study offers an opportunity for a LMS subtype-specific targeted treatment.
Acknowledgments
Financial support
MvdR was supported by the National Institutes of Health (CA 112270) and IE was supported by the Fund for Health Research (FIS PI12/01645).
We thank the Stanford Center for Genomics and Personalized Medicine with help on Illumina sequencing, and thank Alayne Brunner, Patricia Marian Robinson, Joanna Przybyl and other labmates for sample collection, helpful discussions and experimental design.
Grant Support
This study was supported by the National Institutes of Health (Grant No CA 112270) and the Fund for Health Research (FIS PI12/01645).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
XG, MVD designed the research and wrote the paper. XG, VYJ, AM, CHL, IE, MRN, EF, TH, SA, KG, AHB, RBW, CF analyzed and interpreted the data. XG, SXZ and SV performed the experiments. MVD supervised the research. All authors read and approved the final manuscript.
References
- 1.Gustafson P, Willen H, Baldetorp B, Ferno M, Akerman M, Rydholm A. Soft tissue leiomyosarcoma. A population-based epidemiologic and prognostic study of 48 patients, including cellular DNA content. Cancer. 1992;70:114–9. doi: 10.1002/1097-0142(19920701)70:1<114::aid-cncr2820700119>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 2.Toro JR, Travis LB, Wu HJ, Zhu K, Fletcher CD, Devesa SS. Incidence patterns of soft tissue sarcomas, regardless of primary site, in the surveillance, epidemiology and end results program, 1978–2001: An analysis of 26,758 cases. Int J Cancer. 2006;119:2922–30. doi: 10.1002/ijc.22239. [DOI] [PubMed] [Google Scholar]
- 3.Witt H, Mack SC, Ryzhova M, Bender S, Sill M, Isserlin R, et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell. 2011;20:143–57. doi: 10.1016/j.ccr.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marisa L, de Reynies A, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A. 2004;101:811–6. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shai R, Shi T, Kremen TJ, Horvath S, Liau LM, Cloughesy TF, et al. Gene expression profiling identifies molecular subtypes of gliomas. Oncogene. 2003;22:4918–23. doi: 10.1038/sj.onc.1206753. [DOI] [PubMed] [Google Scholar]
- 7.Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, et al. Identification of distinct Basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25:152–65. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damrauer JS, Hoadley KA, Chism DD, Fan C, Tiganelli CJ, Wobker SE, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci U S A. 2014;111:3110–5. doi: 10.1073/pnas.1318376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beck AH, Lee CH, Witten DM, Gleason BC, Edris B, Espinosa I, et al. Discovery of molecular subtypes in leiomyosarcoma through integrative molecular profiling. Oncogene. 2010;29:845–54. doi: 10.1038/onc.2009.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beck AH, Weng Z, Witten DM, Zhu S, Foley JW, Lacroute P, et al. 3′-end sequencing for expression quantification (3SEQ) from archival tumor samples. PLoS One. 2010;5:e8768. doi: 10.1371/journal.pone.0008768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunner AL, Beck AH, Edris B, Sweeney RT, Zhu SX, Li R, et al. Transcriptional profiling of lncRNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012;13:R75. doi: 10.1186/gb-2012-13-8-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee CH, Ou WB, Marino-Enriquez A, Zhu M, Mayeda M, Wang Y, et al. 14-3-3 fusion oncogenes in high-grade endometrial stromal sarcoma. Proc Natl Acad Sci U S A. 2012;109:929–34. doi: 10.1073/pnas.1115528109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunner AL, Li J, Guo X, Sweeney RT, Varma S, Zhu SX, et al. A shared transcriptional program in early breast neoplasias despite genetic and clinical distinctions. Genome Biol. 2014;15:R71. doi: 10.1186/gb-2014-15-5-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demicco EG, Boland GM, Brewer Savannah KJ, Lusby K, Young ED, Ingram D, et al. Progressive Loss Of Myogenic Differentiation In Leiomyosarcoma Has Prognostic Value. Histopathology. 2015;66:627–38. doi: 10.1111/his.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo X, Zhu SX, Brunner AL, van de Rijn M, West RB. Next generation sequencing-based expression profiling identifies signatures from benign stromal proliferations that define stromal components of breast cancer. Breast Cancer Res. 2013;15:R117. doi: 10.1186/bcr3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee CH, Ali RH, Rouzbahman M, Marino-Enriquez A, Zhu M, Guo X, et al. Cyclin D1 as a diagnostic immunomarker for endometrial stromal sarcoma with YWHAE-FAM22 rearrangement. Am J Surg Pathol. 2012;36:1562–70. doi: 10.1097/PAS.0b013e31825fa931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–3. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peter RJ. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. Journal of Computational and Applied Mathematics. 1987;20:5365. [Google Scholar]
- 23.Hoshida Y, Brunet JP, Tamayo P, Golub TR, Mesirov JP. Subclass mapping: identifying common subtypes in independent disease data sets. PLoS One. 2007;2:e1195. doi: 10.1371/journal.pone.0001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Tibshirani R. Finding consistent patterns: A nonparametric approach for identifying differential expression in RNA-Seq data. Stat Methods Med Res. 2013;22:519–36. doi: 10.1177/0962280211428386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Espinosa I, Beck AH, Lee CH, Zhu S, Montgomery KD, Marinelli RJ, et al. Coordinate expression of colony-stimulating factor-1 and colony-stimulating factor-1-related proteins is associated with poor prognosis in gynecological and nongynecological leiomyosarcoma. Am J Pathol. 2009;174:2347–56. doi: 10.2353/ajpath.2009.081037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chibon F, Lagarde P, Salas S, Perot G, Brouste V, Tirode F, et al. Validated prediction of clinical outcome in sarcomas and multiple types of cancer on the basis of a gene expression signature related to genome complexity. Nat Med. 2010;16:781–7. doi: 10.1038/nm.2174. [DOI] [PubMed] [Google Scholar]
- 29.Chen JL, Espinosa I, Lin AY, Liao OY, van de Rijn M, West RB. Stromal responses among common carcinomas correlated with clinicopathologic features. Clin Cancer Res. 2013;19:5127–35. doi: 10.1158/1078-0432.CCR-12-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beck AH, Espinosa I, Edris B, Li R, Montgomery K, Zhu S, et al. The macrophage colony-stimulating factor 1 response signature in breast carcinoma. Clin Cancer Res. 2009;15:778–87. doi: 10.1158/1078-0432.CCR-08-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–4. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 32.Liu CL, Prapong W, Natkunam Y, Alizadeh A, Montgomery K, Gilks CB, et al. Software tools for high-throughput analysis and archiving of immunohistochemistry staining data obtained with tissue microarrays. Am J Pathol. 2002;161:1557–65. doi: 10.1016/S0002-9440(10)64434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mills AM, Beck AH, Montgomery KD, Zhu SX, Espinosa I, Lee CH, et al. Expression of subtype-specific group 1 leiomyosarcoma markers in a wide variety of sarcomas by gene expression analysis and immunohistochemistry. Am J Surg Pathol. 2011;35:583–9. doi: 10.1097/PAS.0b013e318211abd6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edris B, Espinosa I, Muhlenberg T, Mikels A, Lee CH, Steigen SE, et al. ROR2 is a novel prognostic biomarker and a potential therapeutic target in leiomyosarcoma and gastrointestinal stromal tumour. J Pathol. 2012;227:223–33. doi: 10.1002/path.3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Allen EM, Wagle N, Stojanov P, Perrin DL, Cibulskis K, Marlow S, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20:682–8. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rebagay G, Yan S, Liu C, Cheung NK. ROR1 and ROR2 in Human Malignancies: Potentials for Targeted Therapy. Front Oncol. 2012;2:34. doi: 10.3389/fonc.2012.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morioka K, Tanikawa C, Ochi K, Daigo Y, Katagiri T, Kawano H, et al. Orphan receptor tyrosine kinase ROR2 as a potential therapeutic target for osteosarcoma. Cancer Sci. 2009;100:1227–33. doi: 10.1111/j.1349-7006.2009.01165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright TM, Brannon AR, Gordan JD, Mikels AJ, Mitchell C, Chen S, et al. Ror2, a developmentally regulated kinase, promotes tumor growth potential in renal cell carcinoma. Oncogene. 2009;28:2513–23. doi: 10.1038/onc.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Italiano A, Lagarde P, Brulard C, Terrier P, Lae M, Marques B, et al. Genetic profiling identifies two classes of soft-tissue leiomyosarcomas with distinct clinical characteristics. Clin Cancer Res. 2013;19:1190–6. doi: 10.1158/1078-0432.CCR-12-2970. [DOI] [PubMed] [Google Scholar]
- 40.Nanda V, Miano JM. Leiomodin 1, a new serum response factor-dependent target gene expressed preferentially in differentiated smooth muscle cells. J Biol Chem. 2012;287:2459–67. doi: 10.1074/jbc.M111.302224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Francis P, Namlos HM, Muller C, Eden P, Fernebro J, Berner JM, et al. Diagnostic and prognostic gene expression signatures in 177 soft tissue sarcomas: hypoxia-induced transcription profile signifies metastatic potential. BMC Genomics. 2007;8:73. doi: 10.1186/1471-2164-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Villacis RA, Silveira SM, Barros-Filho MC, Marchi FA, Domingues MA, Scapulatempo-Neto C, et al. Gene expression profiling in leiomyosarcomas and undifferentiated pleomorphic sarcomas: SRC as a new diagnostic marker. PLoS One. 2014;9:e102281. doi: 10.1371/journal.pone.0102281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carneiro A, Francis P, Bendahl PO, Fernebro J, Akerman M, Fletcher C, et al. Indistinguishable genomic profiles and shared prognostic markers in undifferentiated pleomorphic sarcoma and leiomyosarcoma: different sides of a single coin? Lab Invest. 2009;89:668–75. doi: 10.1038/labinvest.2009.18. [DOI] [PubMed] [Google Scholar]
- 44.Lee YF, John M, Falconer A, Edwards S, Clark J, Flohr P, et al. A gene expression signature associated with metastatic outcome in human leiomyosarcomas. Cancer Res. 2004;64:7201–4. doi: 10.1158/0008-5472.CAN-04-1673. [DOI] [PubMed] [Google Scholar]
- 45.Perot G, Mendiboure J, Brouste V, Velasco V, Terrier P, Bonvalot S, et al. Smooth muscle differentiation identifies two classes of poorly differentiated pleomorphic sarcomas with distinct outcome. Mod Pathol. 2014;27:840–50. doi: 10.1038/modpathol.2013.205. [DOI] [PubMed] [Google Scholar]
- 46.Edris B, Weiskopf K, Volkmer AK, Volkmer JP, Willingham SB, Contreras-Trujillo H, et al. Antibody therapy targeting the CD47 protein is effective in a model of aggressive metastatic leiomyosarcoma. Proc Natl Acad Sci U S A. 2012;109:6656–61. doi: 10.1073/pnas.1121629109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willingham SB, Volkmer JP, Gentles AJ, Sahoo D, Dalerba P, Mitra SS, et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci U S A. 2012;109:6662–7. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.