SUMMARY
CBR hydroxamidines are small-molecule inhibitors of bacterial RNA polymerase (RNAP) discovered through high-throughput-screening of synthetic-compound libraries. CBR pyrazoles are structurally related RNAP inhibitors discovered through “scaffold hopping” from CBR hydroxamidines. CBR hydroxamidines and pyrazoles selectively inhibit Gram-negative bacterial RNAP and exhibit selective antibacterial activity against Gram-negative bacteria. Here, we report crystal structures of the prototype CBR hydroxamidine, CBR703, and two CBR pyrazoles in complex with E. coli RNAP holoenzyme. In addition, we define the full resistance determinant for CBR703, show that the binding site and resistance determinant for CBR703 do not overlap the binding sites and resistance determinants of other characterized RNAP inhibitors, show that CBR703 exhibits no or minimal cross-resistance with other characterized RNAP inhibitors, and show that co-administration of CBR703 with other RNAP inhibitors results in additive antibacterial activities. The results set the stage for structure-based optimization of CBR inhibitors as antibacterial drugs.
INTRODUCTION
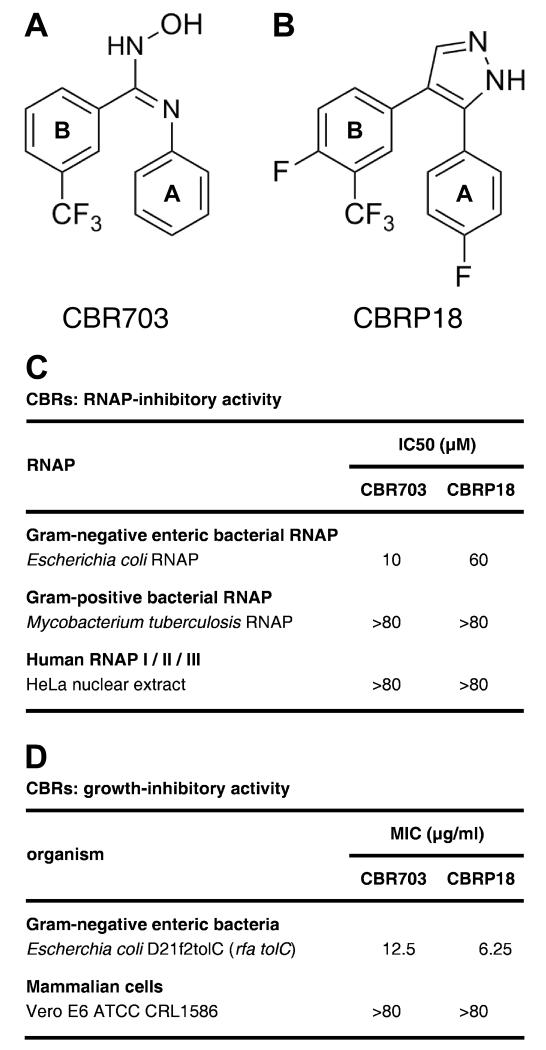
CBR703 is the prototype of the “CBR hydroxamidine” class of small-molecule inhibitors of bacterial RNA polymerase (RNAP; Figure 1A; Li et al., 2001a; Artsimovitch et al., 2003). CBR703 was discovered by the Cumbre, Inc. division of Tularik, Inc. by high-throughput screening of synthetic-compound libraries for novel small-molecule inhibitors of Escherichia coli RNAP (Artsimovitch et al., 2003). CBR703 is a relatively small (MW = 280 Da) and relatively simple compound comprising two aromatic rings, one with a 3-trifluomethyl substituent, and an amidoxime linker (Figure 1A). The compound inhibits Gram-negative enteric bacterial RNAP (e.g., E. coli RNAP) but not Gram-positive bacterial RNAP (e.g., Mycobacterium tuberculosis RNAP) or human RNAP I, II, and III (Figure 1C), and exhibits antibacterial activity against efflux-deficient strains of Gram-negative enteric bacteria, but does not exhibit cytotoxic activity against mammalian cells in culture (Figure 1D).
Figure 1. CBR inhibitors.
(A) Structure of the CBR hydroxamidine inhibitor CBR703 (compound of Example 1 of Li et al., 2001a).
(B) Structure of the CBR pyrazole inhibitor CBRP18 (compound of Example 18 of Li et al., 2001b).
(C) RNAP-inhibitory activities. IC50: concentration resulting in 50% inhibition.
(D) Growth-inhibitory activities. MIC: minimum inhibitory concentration. Antibacterial activities against Gram-negative enteric bacteria are limited to efflux-deficient strains (e.g., E. coli D21f2tolC). MICs against wild-type strains (e.g., E. coli type strain ATCC 25922) are >50 μg/ml.
The “CBR pyrazole” class of small-molecule inhibitors of bacterial RNAP are closely structurally related to CBR hydroxamidines but contain a cyclic conformational constraint (replacement of the amidoxime linker by a pyrazole linker, which prevents cis-trans isomerization; Figure 1B; Li et al., 2001b; Artsimovitch et al., 2003). CBR pyrazoles were identified by “scaffold hopping” from the CBR hydroxamidine scaffold. CBR pyrazoles, like CBR hydroxamidines, exhibit Gram-negative-enteric-selective RNAP-inhibitory activity and Gram-negative-enteric-selective antibacterial activity (Figures 1C-D).
CBR hydroxamidines and pyrazoles have been shown to inhibit both transcription initiation by RNAP and transcription elongation by RNAP (Artsimovitch et al., 2003; Malinen et al. 2014). Reaction-step-specific assays suggest that CBR hydroxamidines and pyrazoles inhibit the translocation step and/or bond-formation step of the nucleotide-addition cycle--comprising RNAP translocation, NTP binding, bond formation, and pyrophosphate release--in transcription initiation and transcription elongation (Artsimovitch et al., 2003; Malinen et al. 2014). These properties of CBR hydroxamidines and pyrazoles differ from the properties of the best-known small-molecule inhibitor of bacterial RNAP, rifampin (Rif), which inhibits solely transcription initiation, and which does so by sterically preventing the extension of short RNA products (Campbell et al., 2001; Feklistov et al., 2008; Ho et al., 2009).
CBR hydroxamidines and pyrazoles have been shown to inhibit RNAP derivatives containing amino acid substitutions in the Rif binding site that confer resistance to Rif, suggesting that CBR hydroxamidines and pyrazoles inhibit RNAP through a binding site different from the Rif binding site (Artsimovitch et al., 2003). Isolation and sequencing of CBR-hydroxamidine-resistant and CBR-pyrazole-resistant mutants indicates that CBR hydroxamidines and pyrazoles function through a determinant on RNAP--the “CBR target”--that does not overlap the Rif binding site and is distant from the RNAP active center (Artsimovitch et al., 2003). The CBR target is located at the N-terminus of the RNAP “bridge helix,” a long α-helix that spans nearly the full width of RNAP (Artsimovitch et al., 2003). The C-terminal part of the bridge-helix forms one wall of the RNAP active center and is thought to undergo conformational cycling--bending and unbending--in each nucleotide-addition cycle in transcription (Weinzierl, 2010; Hein and Landick, 2010). Accordingly, it is thought that CBR hydroxamidines and pyrazoles inhibit RNAP by binding to the CBR target and allosterically affecting conformational cycling of the bridge-helix and/or associated structural elements (Artsimovitch et al., 2003; Malinen et al. 2014). A structural model of RNAP bound to a CBR inhibitor has been proposed based on in silico docking (Malinen et al. 2014).
There is an extremely urgent need for new antibacterial drugs effective against Gram-negative bacteria. However, the potential development of CBR hydroxamidines and pyrazoles as Gram-negative antibacterial drugs has been thwarted by the low potencies of known CBR hydroxamidines and pyrazoles against wild-type, efflux-proficient Gram-negative bacterial strains (MICs > 50 μg/ml; legend to Figure 1; Zhu et al., 2014), and by the absence of structural information on RNAP-CBR interactions that could be used to inform and direct medicinal chemistry efforts to improve potency (Li et al., 2001a,b; Zhu et al., 2014).
In this work, we report crystal structures of bacterial RNAP bound to CBR inhibitors, and we show--by analyzing bromine anomalous diffraction from a bromine-containing CBR inhibitor--that RNAP binds CBR inhibitors in an orientation opposite to that predicted by Malinen et al. 2014 based on in silico docking. In addition, we define the full resistance determinant for a CBR inhibitor, show that the binding site and resistance determinant for the CBR inhibitor do not overlap the binding site and resistance determinant of previously characterized RNAP inhibitors, show that the CBR inhibitor does not exhibit cross-resistance with previously characterized RNAP inhibitors, and show that co-administration of the CBR inhibitor and other RNAP inhibitors results in additive antibacterial activities.
RESULTS
Structural basis of transcription inhibition by CBR inhibitors: crystal structures of E. coli RNAP holoenzyme in complex with CBR inhibitors
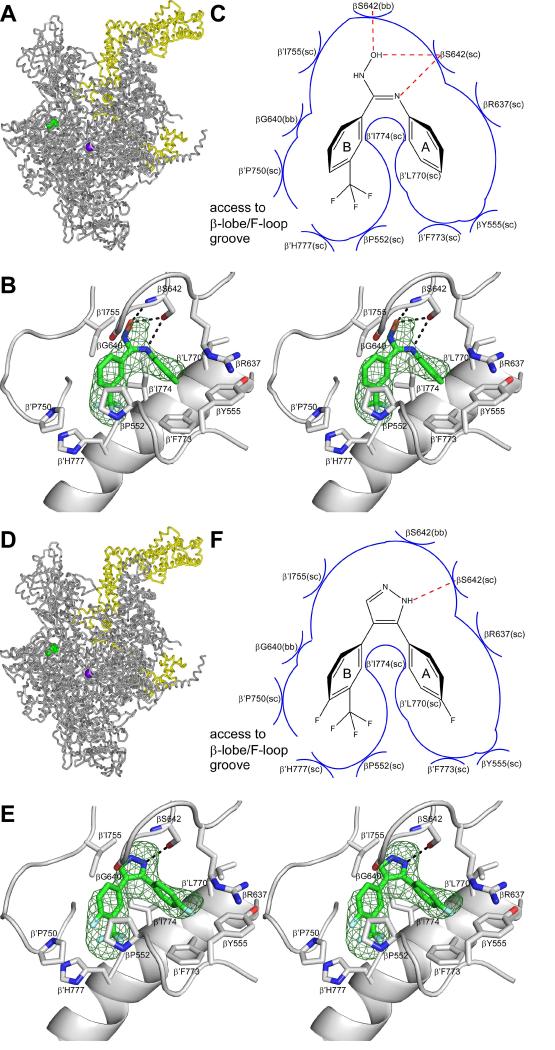
To define the structural basis of transcription inhibition by CBR inhibitors, we determined crystal structures of E. coli RNAP holoenzyme in complex with CBR inhibitors. We soaked the CBR hydroxamidine CBR703 and the CBR pyrazole CBRP18 into pre-formed crystals of E. coli RNAP holoenzyme, collected X-ray diffraction data, solved structures by molecular replacement, and refined structures (Table 1). The resulting crystal structures of E. coli RNAP holoenzyme in complex with CBR703 at 4.2 Å resolution and E. coli RNAP holoenzyme in complex with CBRP18 at 4.0 Å resolution are shown in Figure 2A-C and Figure 2D-F, respectively.
Table 1. Crystallographic data and refinement statistics.
| dataset | Eco RNAP-CBR703 | Eco RNAP-CBRP18 | Eco RNAP-CBRH16-Br |
| beamline | BNL-X29A | APS-19-ID | APS-19-ID |
| space group | P212121 | P212121 | P212121 |
| resolution range | 50.00-4.20 Å (4.27-4.20 Å) | 50.00-4.00 Å (4.07-4.00 Å) | 50.00-4.10 Å (4.17-4.10 Å) |
| cell parameters (Å, °) |
a=185.8, b=205.8, c=307.5 α=90.0, β=90.0, γ=90.0 |
a=185.6, b=204.1, c=308.0 α=90.0, β=90.0, γ=90.0 |
a=186.3, b=205.8, c=307.6 α=90.0, β=90.0, γ=90.0 |
| completeness | 1.000 (1.000) | 1.000 (1.000) | 1.000 (1.000) |
| multiplicity | 8.7 (7.8) | 14.4 (13.8) | 13.2 (13.2) |
| mean I/σ | 15.6 (2.1) | 14.6 (2.9) | 10.1 (3.8) |
| Rmerge | 0.126 (0.980) | 0.176 (0.998) | 0.234 (0.761) |
| Rwork | 0.270 | 0.252 | 0.241 |
| Rfree | 0.296 | 0.286 | 0.271 |
| bond-length rmsd | 0.004 Å | 0.005 Å | 0.005 Å |
| bond-angle rmsd | 0.913° | 0.983° | 0.970° |
| PDB code | 4ZH2 | 4ZH4 | 4ZH3 |
Figure 2. Crystal structures of E. coli RNAP holoenzyme in complex with CBR inhibitors.
Upper panels (A-C), data for the CBR hydroxamidine CBR703.
Lower panels (D-F), data for the CBR pyrazole CBRP18.
(A),(D) Structure of E. coli RNAP holoenzyme in complex with the CBR inhibitor. Gray ribbon: RNAP core. Yellow ribbon: σ70. Violet sphere: active-center catalytic Mg2+. Green: CBR inhibitor.
(B),(E) Contacts between RNAP and the CBR inhibitor(stereodiagram). Green mesh: NCS-averaged mFo-DFc electron density omit map for the CBR inhibitor (contoured at 2.5σ). Green, red, blue, and cyan: carbon, oxygen, nitrogen, and fluorine atoms of the CBR inhibitor. Gray ribbons: RNAP. Gray helical ribbon: RNAP bridge helix (N-terminus at right).
(C),(F) Schematic summary of inferred contacts between RNAP and the CBR inhibitor. Red dashed lines: H-bonds. Blue arcs: van der Waals interactions.
The structures establish that the CBR hydroxamidine CBR703 and the CBR pyrazole CBRP18 bind to the genetically defined CBR target located at the N-terminal end of the RNAP bridge-helix (Figure 2A-B, D-E) and bind in the identical binding location and in the identical binding pose (Figure 2B,E). For each CBR inhibitor, clear electron density features are observed for each of the two aromatic rings and for the linker (Figure 2 B,E).
For the CBR hydroxamidine CBR703, the shape of the electron density feature indicates that the double bond of the linker adopts the cis configuration (Figure 2B), as predicted based on the similarity of RNAP-inhibitory potencies of CBR hydroxamidines to those of CBR pyrazoles, in which double bond is constrained to a cis configuration by a cyclic conformational constraint (Li et al., 2001a,b).
For each compound, one of the electron density features corresponding to the two aromatic rings is substantially larger than the other (Figure 2B,E). For each compound, we assign the larger density feature to ring B--which contains an electron-dense trifluoromethyl substituent. This assignment defines the orientation of each compound relative to the binding site (orientation as shown in Figure 2B-C,E-F). The orientation defined in this manner is opposite to the orientation that was predicted by Malinen et al,.. 2014 based on in silico docking.
Structural basis of transcription inhibition by CBR inhibitors: crystal structure of E. coli RNAP holoenzyme in complex with a bromine-containing CBR inhibitor
In view of the relatively low resolution of the structures (4.2 Å resolution and 4.0 Å), in view of the relatively high degree of symmetry of the observed electron density features (Figure 2B,E), in view of relatively high degree of symmetry of the compounds (Figure 1A,B), and in view of the conflict between the binding orientation inferred from our electron density maps and the binding orientation predicted by Malinen et al., 2014 from in silico docking, we deemed it imperative to obtain an independent experimental confirmation of the binding orientation.
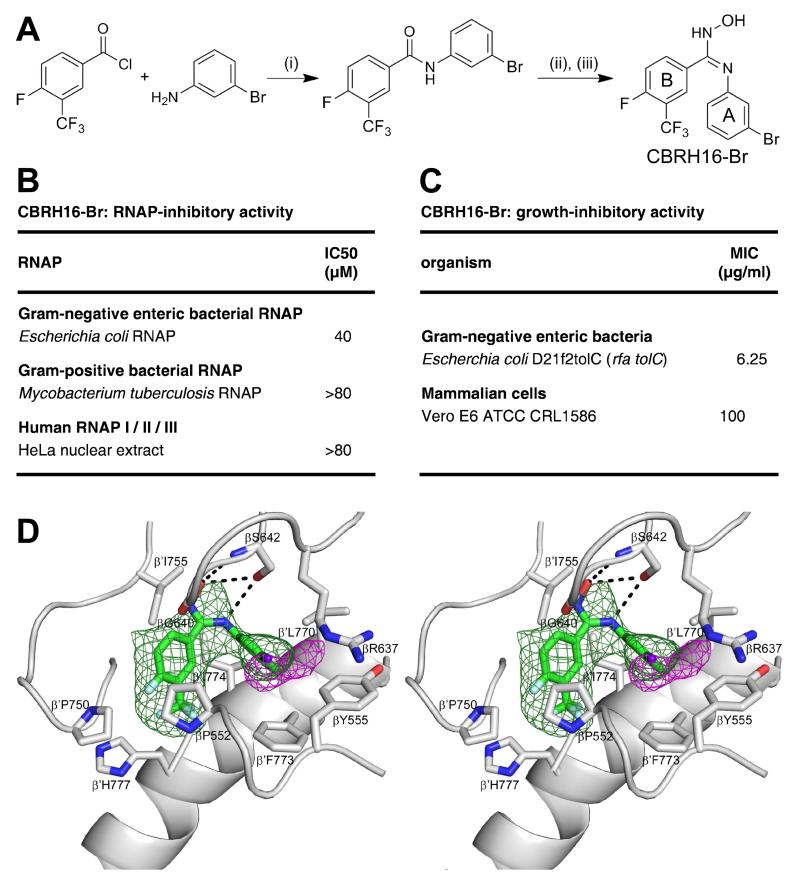
To confirm the binding position and binding orientation of CBR inhibitors, we synthesized a bromine-containing CBR derivative and collected X-ray diffraction data, including bromine anomalous diffraction, for crystals of E. coli RNAP holoenzyme in complex with the bromine-containing CBR derivative (Figure 3 Table 1). We synthesized CBR703 derivative containing bromine at position 3 of ring A and fluorine at position 4 of ring B (CBRH16-Br; designed based on the structure activity relationships in Li et al. 2001a indicating that bromine at position 3 of ring A is tolerated without major loss of RNAP-inhibitory activity and that fluorine at position 4 of ring B enhances RNAP-inhibitory activity; Figure 3A). CBRH16-Br exhibited essentially full RNAP-inhibitory activity and essentially full antibacterial activity (Figure 3B). The RNAP-CBRH16-Br complex exhibited electron density for CBRH16-Br matching that of the RNAP-CBR703 complex (green mesh in Figures 2B and 3D), and exhibited a single peak of bromine anomalous difference density adjacent to the electron density for CBRH16-Br in the position expected for a bromine atom at position 3 of ring A (Figure 3D). The results unequivocally confirm the binding position and binding orientation of CBR inhibitors is as shown in Figure 2 and show that the orientation predicted by Malinen et al., 2014 from in silico docking is incorrect.
Figure 3. Crystal structure of E. coli RNAP holoenzyme in complex with a bromine-containing CBR inhibitor.
(A) Synthesis and structure of CBRH16-Br (derivative of compound of Example 16 of Li et al. 2001a having bromine at position 3 of ring A). (i), (CH3CH2)3N/CH2Cl2; 0°C; 1 h. (ii), PCl5/C2H4Cl2; 70°C; 5 h. (iii), (CH3CH2)3N/NH2OH-HCl/CH3CN; 0-24°C.
(B) RNAP-inhibitory activities of CBRH16-Br.
(C) Growth-inhibitory activities of CBRH16-Br.
(D) Electron density, bromine anomalous difference density, and atomic model for CBRH16-Br. Green mesh: NCS-averaged mFo-DFc omit map for CBRH16-Br (contoured at 2.5σ). Pink mesh: bromine anomalous difference density for CBRH16-Br (contoured at 4.5σ). Other colors and labels as in Figure 2B,E.
Structural basis of transcription inhibition by CBR inhibitors: RNAP-CBR interactions
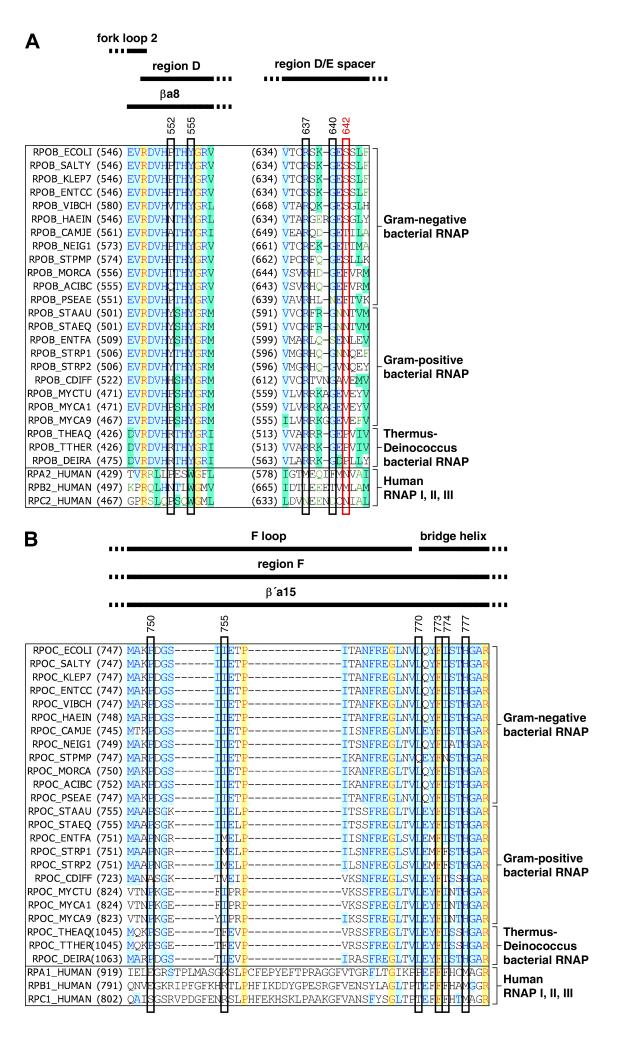
The CBR binding site on RNAP is located at an interface between the RNAP β subunit and the RNAP β′ subunit (Figure 2B-C,E-F). The CBR binding site comprises residues of the RNAP β subunit “region D” and the “D-E spacer” and residues of the RNAP β′ subunit “F-loop” and bridge helix (Figure 4; conserved-region nomenclature as in Sweester et al., 1987; Weinzierl, 2010; and Hein and Landick, 2010).
Figure 4. Interactions between RNAP and CBR inhibitors: sequence alignments.
Locations of residues that contact CBR inhibitors in the sequences of RNAP β subunit (A) and RNAP β′ subunit (B). Sequence alignments for the β and β′ subunits of bacterial RNAP (top 24 sequences in each panel) and corresponding subunits of human RNAP I, RNAP II, and RNAP III (bottom three sequences in each panel), showing locations of residues that contact CBR inhibitors in crystal structures of RNAP-CBR703 and CBRP18 (red rectangle for residue that makes H-bonded interactions; rectangles for residues that make van der Waal interactions; identities from Figure 2B-C,E-F), locations of RNAP structural elements (top row of black bars; boundaries from Weinzierl, 2010 and Hein and Landick, 2010), and RNAP conserved regions (next two rows of black bars; boundaries from Sweetser et al., 1987 and Lane and Darst, 2010). Species are as follows: E. coli (ECOLI), Salmonella typhimurium (SALTY), Klebsiella pneumoniae (KLEP7), Enterococcus cloacae (ENTCC), Vibrio cholerae (VIBCH), Haemophilus influenzae (HAEIN), Campylobacter jejuni (CAMJE), Neisseria gonorrhoeae (NEIG1), Stenotrophomonas maltophilia (STPMP), Moraxella catarrhalis (MORCA), Acinetobacter baumannii (ACIBC), Pseudomonas aeruginosa (PSEAE), Staphylococcus aureus (STAAU), Staphylococcus epidermidis (STAEQ), Enterococcus faecalis (ENTFA), Streptococcus pyogenes (STRP1), Streptococcus pneumoniae (STRP2), Clostridium difficile (CDIFF), Mycobacterium tuberculosis (MYCTU), Mycobacterium avium (MYCA1), Mycobacterium abscessus (MYCA9), Thermus thermophilus (THETH), Thermus aquaticus (THEAQ), Deinococcus radiodurans (DEIRA), and Homo sapiens (HUMAN).
The binding site contains two pocket-shaped hydrophobic subsites, which interact with the two aromatic rings of the CBR inhibitor (Figure 2B-C,E-F). The CBR inhibitor inserts into the two subsites in a manner reminiscent of that in which an electric plug (two-prong; US) plugs into a socket (Figure 2B-C,E-F).
The first subsite, “subsite A,” is centered on the first turn of the RNAP bridge helix, extending from β′L770 to β′I774. The second subsite, “subsite B,” is centered on the second turn of the RNAP bridge helix, extending from β′I774 to β′H777. Residue β′I774 is shared by the two subsites, and forms a wall that separates the two subsites. This residue wedges between aromatic rings A and B of the CBR inhibitor, interacting with both rings and with the connector between the rings.
In subsite A, the sidechains of β′L770 and β′I774 stack on opposite faces of ring A of the CBR inhibitor, the sidechain of β′F773 makes an aromatic-aromatic face-edge interaction with ring A, the sidechain of βY555 makes an aromatic-aromatic edge-edge interaction with ring A, and the aliphatic portion of the sidechain of βR637 makes van der Waals interaction with ring A (Figure 2B-C,E-F).
In subsite B, the sidechains of βP552 and β′P750 sandwich aromatic ring B of the CBR inhibitor, the backbone of βG640 and the sidechain of β′I774 make van der Waals interactions with ring B, and the sidechain of β′H777 makes van der Waals interactions with the trifluoromethyl substituent on ring B (Figure 2B-C,E-F). The portion of subsite B closest to atoms 4 and 5 of aromatic ring B of the CBR inhibitor contains an opening that affords access to bulk solvent, located in a deep groove formed between the RNAP “β lobe” and the RNAP F-loop (“β-lobe/F-loop groove”; Figure 2C,F). This opening, affords access to bulk solvent and accounts for structure-activity relationships in Li et al. 2001a,b indicating that long sidechains can be incorporated at atoms 4 and 5 of ring B without loss of RNAP-inhibitory activity (Li et al. 2001a,b).
Additional interactions between RNAP and the CBR inhibitor--including the only H-bonded interactions between RNAP and the CBR inhibitors--involve the connector between rings A and B of the CBR inhibitor (Figure 2B-C,E-F). In the case of the CBR hydroxamidine CBR703, the sidechain hydroxyl of βS642 is inferred to make H-bonds to the oxygen atom and one nitrogen atom of the amidoxime connector, the backbone amide of βS642 is inferred to make an H-bond to the oxygen atom of the amidoxime connector, and the sidechain of β′I755 makes van der Waals interactions with the amidoxime connector (Figure 2B-C). In the case of the CBR pyrazole CBRP18, the sidechain hydroxyl of βS642 is inferred to make an H-bond to one nitrogen atom of the pyrazole, and the sidechain of β′I755 makes van der Waals interactions with the pyrazole (Figure 2E-F).
Residues of the CBR binding site from RNAP β-subunit region D and the RNAP β′-subunit bridge helix are conserved in RNAP from bacteria and RNAP I, II and III from humans (Figure 4). In contrast, residues of the CBR binding site from the β′-subunit F-loop are conserved in RNAP from bacteria but are not conserved in RNAP I, II and III from humans, and residues of the CBR binding site from the β-subunit D-E spacer--especially β residue 642, the residue that forms H-bonds with CBR inhibitors in the structures in Figures 2-3--exhibit distinct patterns of sequence conservation in RNAP from Gram-negative enteric bacteria (e.g., E. coli, Salmonella typhimurium, Klebsiella pneumoniae, and Enterobacter cloacae), RNAP from other bacteria, and RNAP I, II, and III from humans (Figure 4). Taken together, these patterns of residue conservation are consistent with, and account for, the observed selectivity of CBR inhibitors for RNAP from Gram-negative enteric bacteria (Figure 1B).
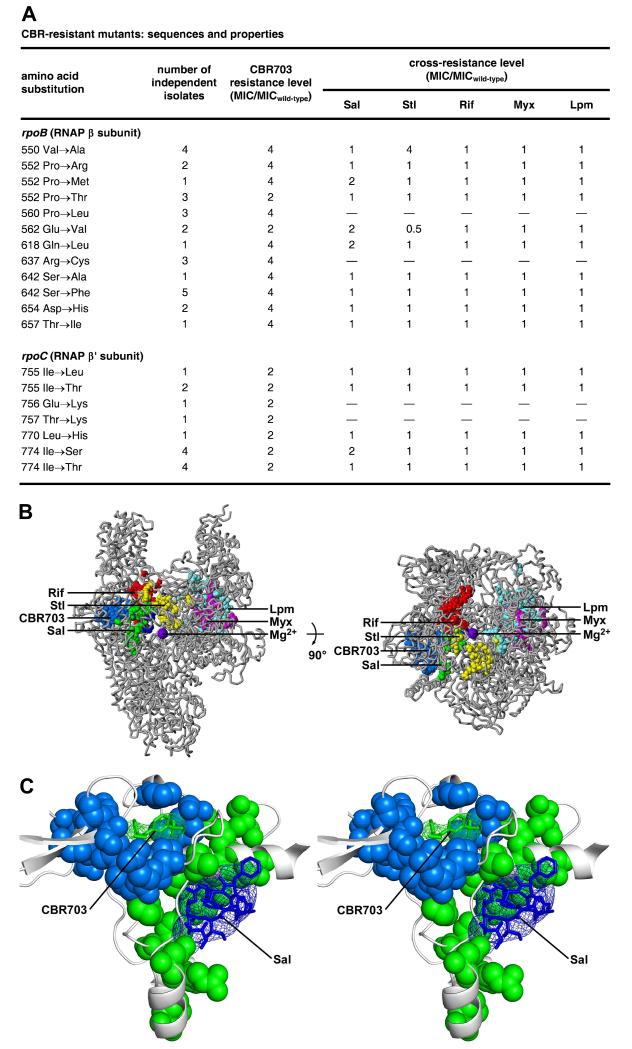
Resistance determinant of CBR703
The CBR target originally was identified by isolating and sequencing a small collection of CBR703-resistant mutants of an efflux-deficient strain of E. coli (Artsimovitch et al., 2003). The collection comprised substitutions at three sites: βP560, βR637 and βS642 (Artsimovitch et al., 2003).
Here, in order to provide a comprehensive description of the resistance determinant for a CBR inhibitor, we isolated and sequenced a large collection of CBR703-resistant mutants of an efflux-deficient, outer-membrane-disrupted strain of E. coli (Figure 5). We performed high-level saturation mutagenesis targeting all codons for all residues of the RNAP β and β′ subunits located within 30 Å of residues of the RNAP bridge helix and/or of the catalytic Mg2+ ion of the RNAP active center, and we isolated and sequenced mutants resistant to CBR703. We identified nineteen different single-amino-acid substitutions that conferred resistance to CBR703: twelve different substitutions affecting nine sites in RNAP β subunit, and different seven substitutions affecting five sites in RNAP β′ subunit (Figure 5A, column 1, and Figure S1). The indentified resistance sites included the three resistance sites of Artsimovitch et al., 2003 (βP560, βR637, and βS642) and also included six new resistance sites in RNAP β subunit (βV550, βP552, βQ618, βD654, and βT657) and five new resistance sites in RNAP β′ subunit (β′I755, β′Q756, β′T757, β′L770, and β′I774; Figure 5A, column 1 and Figure S1). The statistics of the analysis, specifically the isolation of multiple independent isolates of most resistance substitutions (Figure 5A, column 2), indicate that the analysis is likely to have identified the large majority of, if not all, potential resistance sites.
Figure 5. Relationship between the binding sites and resistance determinant of CBR703 and the binding sites and resistance determinants of other RNAP inhibitors.
(A) Sequences, isolation statistics, CBR703-resistance levels, and Sal-, Stl-, Rif-, Myx- and Lpm-cross-resistance levels of CBR703-resistant mutants. MICwild-type,CBR703 = 6.25 μg/ml; MICwild-type,Sal = 0.049 μg/ml; MICwild-type,Stl = 1.56 μg/ml.; MICwild-type,Rif = 0.20 μg/ml; MICwild-type,Myx = 0.20 μg/ml; MICwild-type,Lpm = 1.56 μg/ml.
(B) The CBR703 resistance determinant does not overlap the resistance determinants of Sal, Rif, Stl, Myx, and Lpm. Structure of bacterial RNAP (two orthogonal views; gray ribbons for RNAP; violet sphere for active-center catalytic Mg2+; PDB: 2CW0), showing sites of substitutions that confer resistance to CBR703 (blue; Figure 5A; Artsimovitch et al., 2003), Sal (green; Degen et al, 2014), Stl (yellow; Lisitsyn et al., 1985; Heisler et al., 1993; Severinov et al., 1993, 1995; Tuske et al., 2005), Rif (red; Ovchinnikov et al., 1981, 1983; Lisitsyn et al., 1984; Jin and Gross, 1988; Severinov et al., 1993,1994; Campbell et al., 2001; Garibyan et al., 2003), Myx (magenta; Mukhopadhyay et al., 2008; Srivastava et al., 2011), and Lpm (cyan; Ebright, 2005; Srivastava et al., 2011).
(C) The CBR703 binding site and resistance determinant do not overlap the binding site and resistance determinant of Sal. Stereodiagram. Green mesh, green sticks, gray ribbons, and gray helical ribbon: mFo-DFc omit map for CBR703, atomic model for CBR703, RNAP, RNAP bridge helix (N-terminus at left) from the crystal structure of RNAP-CBR703 (Fig. 2A-C). Blue mesh and blue sticks: mFo-DFc omit map for Sal and atomic model for Sal from the crystal structure of RNAP-Sal (PDB: 4MEX). Green surfaces: residues at which substitutions confer CBR703 resistance. Blue surfaces: residues at which substitutions confer Sal-resistance. See also Table S1 and Figure S1.
Most resistance substitutions in RNAP β subunit conferred 4-fold resistance to CBR703 (Figure 5A, column 3). Most resistance substitutions in RNAP β′ subunit conferred 2-fold resistance to CBR703 (Figure 5A, column 3).
When mapped on to the three-dimensional structure of bacterial RNAP, the identified resistance sites form a determinant of ~30 Å × ~20 Å × ~10 Å centered on the CBR target of Artsimovitch et al. 2003 and centered on the crystallographically defined binding site for CBR inhibitors of this work (Figure 5B). The identified resistance sites include six RNAP residues that make direct contacts with CBR703 in the crystal structure of the RNAP-CBR703 complex (βP552, βR637, βS642, β′I755, β′L770 and β′I774; Figures 2, 4, 5A,C) and include five other RNAP residues that contact RNAP residues that, in turn, contact CBR inhibitors (βV550, βP560, βQ618, βD654, and β′T757; Figure 5A,C).
Relationship between the binding site and resistance determinant of CBR703 and the binding sites and resistance determinants of other RNAP inhibitors
The binding site and resistance determinant for CBR inhibitors are located adjacent to, but do not overlap, the binding site and resistance determinant for the previously characterized RNAP inhibitor salinamide A (Sal; Figure 5B,C). CBR inhibitors and Sal both interact with the RNAP bridge helix, but they interact with different segments of the RNAP bridge helix--bridge-helix turns 1 and 2 for CBR inhibitors vs. bridge-helix turns 3 and 4 for Sal--and they interact with different, nearly opposite, faces of the RNAP bridge helix (Figure 5B,C). We previously have designated the Sal binding site as the RNAP “bridge-helix cap” inhibitor binding site (Degen et al., 2014). Here, we designate the CBR inhibitor binding site as the RNAP “bridge helix N-terminus” inhibitor binding site.
The binding site and resistance determinant for CBR inhibitors is distant from, and does not overlap, the binding sites and resistance determinants for other characterized RNAP inhibitors, including streptolydigin (Stl; which interacts with the central and C terminal segments of the bridge helix), GE23077 (GE; which interacts with RNAP active center i and i+1 nucleotide-addition sites), Rif (which interacts with a site along the path of RNA from the RNAP active center), myxopyronin (Myx; which interacts with RNAP switch region, SW1/SW2 subregion), and lipiarmycin (Lpm; which interacts with RNAP switch region, SW2/S3 subregion; Figure 5B).
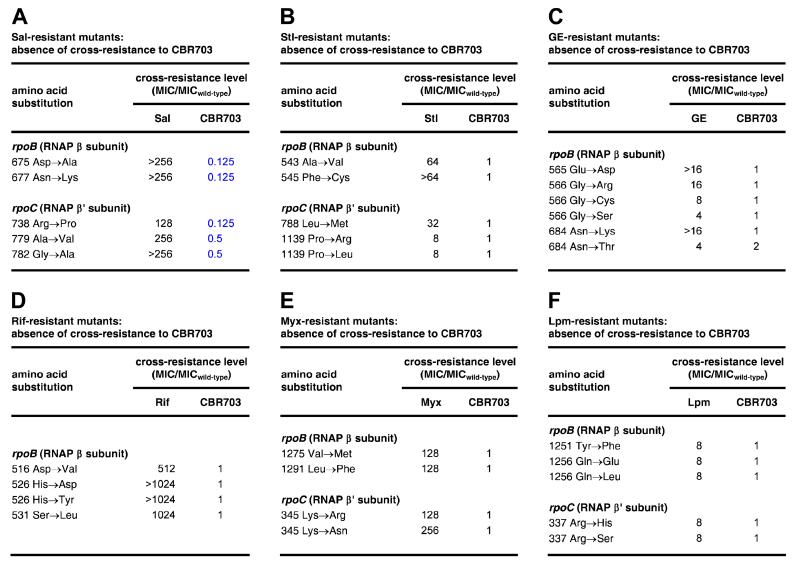
The absence of overlap between the binding site and resistance determinant of CBR inhibitors and those of other previously characterized RNAP inhibitors raises the possibility that CBR inhibitors will not exhibit cross-resistance with previously characterized RNAP inhibitors. We have tested--and validated--this possibility by performing antibacterial susceptibility assays assessing cross-resistance of CBR703-resistant mutants to Sal, Stl, Rif, Myx, and Lpm, and assessing cross-resistance of Sal-resistant, Stl-resistant, GE-resistant, Rif-resistant, Myx-resistant, and Lpm-resistant mutants to CBR703 (Figures, 5A, 6). The results in columns 4-8 of Figure 5A show that CBR703-resistant mutants exhibit no or minimal cross-resistance to Sal, Stl, Rif, Myx, and Lpm. The results in Figure 6 show--reciprocally--that Sal-resistant, Stl-resistant, GE-resistant, Rif-resistant, Myx-resistant, and Lpm-resistant mutants exhibit no significant cross-resistance to CBR703; and further show that many Sal-resistant mutants exhibit hyper-susceptibility, by factors of 2-8, to CBR703.
Figure 6. Absence of cross-resistance between CBR703 and other RNAP inhibitors.
(A) Absence of cross-resistance, and presence of hyper-susceptibility, between Sal-resistant mutants (Degen et al., 2014) and CBR703. Blue: hyper-susceptibility (MIC ratio ≤1). MICwild-type,CBR703 = 6.25 μg/ml; MICwild-type,Sal = 0.049 μg/ml.
(B) Absence of cross-resistance between Stl-resistant mutants (Tuske et al., 2005) and CBR703. MICwild-type,Stl = 1.56 μg/ml.
(C) Absence of cross-resistance between GE-resistant mutants (Zhang et al., 2014) and CBR703. MICwild-ype,GE = 500 μg/ml.
(D) Absence of cross-resistance between Rif-resistant mutants (Jin and Gross, 1988; Garibyan et al., 2003; Zhang et al., 2014) and CBR703. MICwild-type,Rif = 0.20 μg/ml.
(E) Absence of cross-resistance between Myx-resistant mutants (Mukhopadhyay et al., 2008; Degen et al., 2014) and CBR703. MICwild-type,Myx = 0.20 μg/ml.
(F) Absence of cross-resistance between Lpm -resistant mutants (Ebright, 2005; Degen et al., 2014) and CBR703. MICwild-type,Lpm = 1.56 μg/ml.
The absence of overlap between the binding site of CBR inhibitors and those of previously characterized RNAP inhibitors further raises the possibility that CBR inhibitors and previously characterized RNAP inhibitors may be able to bind simultaneously to RNAP and thus that co-administration of a CBR inhibitor with another RNAP inhibitor may result in additive or super-additive activity. We have tested--and obtained evidence for--this possibility by performing “checkerboard” assays (Horrevorts et al., 1987; White et al., 1996; Meletiadis et al., 2010) assessing antibacterial activities against E. coli D21f2tolC of combinations of CBR703 and Sal, CBR703 and Stl, CBR703 and Rif, and CBR703 Myx (Table 2). The results show that co-administration of CBR703 with Sal, Stl, Rif, or Myx yields minimum fractional inhibitory concentration indices (FICImin) of 0.6-0.99 and maximum fractional inhibitory concentration indices (FICImax) of 1.1-1.5 (Figure 4D)--all of which are within the ranges of >0.5-1.0 for FICImin and 1.0-4.0 for FICImax deemed indicative of additive interactions (Table 2; White et al., 1996; Meletiadis et al., 2010).
Table 2.
Additive interaction of CBR703 with other bacterial RNAP inhibitors
| antibacterial agents | minimum FIC index (FICImin) | maximum FIC index (FICImax) | interaction type |
|---|---|---|---|
| CBR703 + Sal | 0.99 | 1.5 | additive |
| CBR703 + Stl | 0.60 | 1.1 | additive |
| CBR703 + Rif | 0.97 | 1.4 | additive |
| CBR703 + Myx | 0.98 | 1.4 | additive |
The observation that co-administration of CBR703 and other RNAP inhibitors results in additive antibacterial activity (Table 2) has two important implications for the potential development of CBR inhibitors as antibacterial therapeutic agents. First, combining a CBR inhibitor with another RNAP inhibitor should increase therapeutic efficacy. Second, combining a CBR inhibitor with another RNAP inhibitor should suppress the emergence of spontaneous resistance (since resistance to a combination of two compounds with different binding sites requires two mutational hits and therefore is much rarer than resistance to one compound with one binding site).
Relationship between the binding site and resistance determinant of CBR703 and the RNAP active center
The CBR binding site and resistance determinant are distant from the RNAP active center (30 Å from the RNAP active center catalytic Mg2+ ion; blue surface and violet sphere in Figures 2A,D and 5B) and also are distant from the RNAP channels that bind DNA and RNA. The position of the CBR binding site and resistance determinant are consistent with the proposal of Artsimovitch et al., 2003 and Malinen et al. 2014 that CBR inhibitors inhibit RNAP through an allosteric, rather than a direct steric mechanism;
CBR inhibitors make extensive direct interactions with the N-terminus of the RNAP bridge helix; aromatic rings A and B of the CBR inhibitor are inserted into the grooves formed by the first and second turns of the bridge helix in a manner that resembles insertion of a plug in a socket (Figure 2). The position of the CBR binding site and resistance determinant, and the binding mode of the CBR inhibitor are consistent with the specific proposal of Artsimovitch et al. 2003 and Malinen et al. 2014 that CBR inhibitors inhibit RNAP allosterically by interfering with conformational cycling of the RNAP bridge helix and/or associated RNAP structural elements.
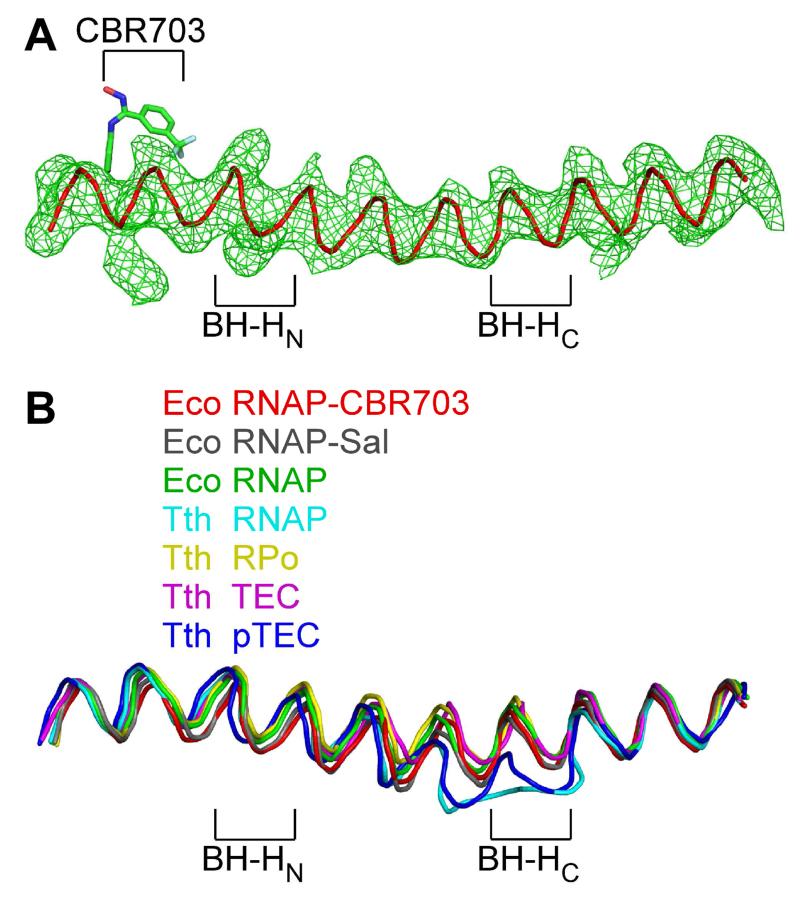
With respect to the hypothesis that CBR inhibitors interfere with conformational cycling of the RNAP bridge helix, we note that, in our crystal structures of E. coli RNAP holoenzyme bound to CBR inhibitors, the bridge helix exhibits the same straight, unbent conformation as in E. coli RNAP holoenzyme in the absence of CBR inhibitors (Figure 7), indicating that CBR inhibitors bind to and stabilize a straight, unbent conformation of the bridge helix. We further note that, within the limits of resolution, all three classes of bacterial RNAP inhibitors that have bridge-helix binding sites--CBR inhibitors, Sal, and Stl--bind to and stabilize the same straight, unbent conformation of the bridge helix (Figure 7B; Degen et al., 2014; Tuske et al. 2005).
Figure 7. Relationship between the binding site and resistance determinant of CBR703 and the RNAP active center: CBR inhibitors interact with a straight, unbent state of the RNAP bridge helix.
(A) Electron density and model for bridge helix in crystal structure of RNAP-CBR703. Green mesh: mFo-DFc omit map for bridge helix (contoured at 2.5σ). Red ribbon: bridge-helix backbone. Green, red, blue, and cyan: CBR703 carbon, oxygen, nitrogen, and fluorine atoms. BH-HN: bridge-helix N-terminal hinge. BH-HC: bridge-helix C-terminal hinge.
(B) Superimposition of bridge helices of E. coli RNAP-CBR703 (red; unbent BH-HN and BH-HC), RNAP-Sal (black; PDB: 4MEX), E. coli RNAP (green; PDB: 4MEY), T. thermophilus RNAP (cyan; PDB: 1IW7), T. thermophilus RPo (yellow; PDB: 4G7H), T. thermophilus transcription elongation complex (pink; PDB: 2O5J), and paused T. thermophilus transcription elongation complex (violet; PDB: 4GZY).
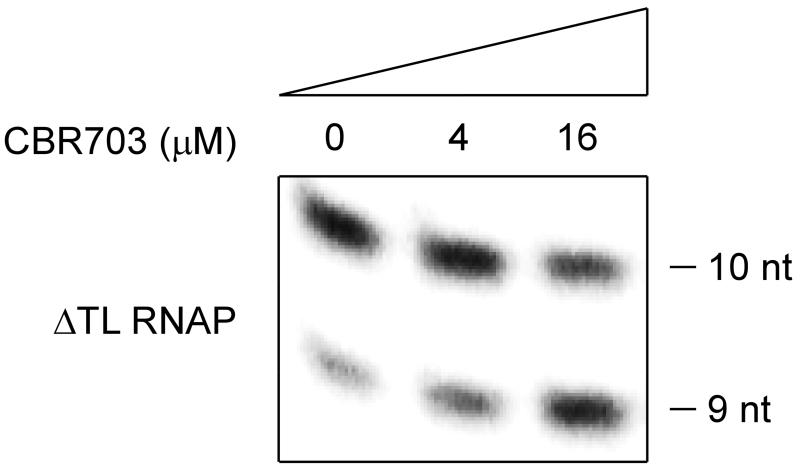
With respect to the hypothesis that CBR inhibitors interfere with conformational cycling of an RNA structural element associated with the bridge helix--e.g., the “trigger loop,” F-loop, or β lobe (Weinzierl., 2010; Hein and Landick, 2010)--we note that CBR inhibitors are able to inhibit transcription by an RNAP derivative that lacks the RNAP trigger loop, indicating that transcription inhibition by CBR inhibitors does not require the RNAP trigger loop, and suggesting that transcription inhibition by CBR inhibitors involves mechanisms that are, at least in part, independent of trigger-loop conformation (Figure 8). Of three classes of bacterial RNAP inhibitors that have bridge-helix binding sites--CBR inhibitors, Sal, and Stl--CBR inhibitors and Sal are able to inhibit transcription by an RNAP derivative that lacks the trigger loop (Figure 8; Degen et al., 2014), but Stl is not (Temiakov et al. 2005).
Figure 8. Relationship between the binding site and resistance determinant of CBR703 and the RNAP active center: transcription inhibition by CBR703 does not require the RNAP trigger loop.
Effects of CBR703 on nucleotide addition by an E. coli RNAP derivative lacking the RNAP trigger loop (ΔTL RNAP; Temiakov et al., 2005).
The proximity of the binding sites for CBR inhibitors and Sal (Figure 5B-C), together with the similar bridge-helix conformations in crystal structures of RNAP bound to CBR inhibitors and Sal (Figure 7) and similar trigger-loop-independence of transcription inhibition by CBR inhibitors and Sal (Figure 8), suggest that the mechanisms of inhibition by CBR inhibitors and Sal may be similar.
DISCUSSION
The results presented here define the structural basis of transcription inhibition by CBR inhibitors, define comprehensively, or nearly comprehensively, the resistance determinant for CBR inhibitors, and define the relationship between the binding sites and resistance determinants for CBR inhibitors and those for previously characterized RNAP inhibitors.
The structural information in this work provides a foundation for structure-based design of novel CBR inhibitors with increased RNAP-inhibitory and antibacterial potency. The structures suggest strategies to modify CBR inhibitors in order to: (1) increase potency specifically against Gram-negative enteric bacterial RNAP and other bacterial RNAP having Ser or Thr at β position 642 (e.g., RNAP from E. coli, K. pneumoniae, E. cloacae, V. cholerae, Haemophilus influenzae, C. jejuni, and Neisseria gonorrhoeae; Figure 4); (2) increase potency specifically against bacterial RNAP having an apolar amino acid at RNAP β position 642 (e.g., RNAP form Moraxella catarrhalis, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterococcus faecalis, Mycobacterium tuberculosis, Mycobacterium avium, and Mycobacterium abscessus; Figure 4), and (3) to increase potency against any bacterial RNAP.
The structures suggest that potency against Gram-negative enteric bacterial RNAP and other bacterial RNAP having Ser or Thr at β position 642 could be increased by replacing the pyrazole moiety of a CBR pyrazole with a heterocycle able to form an additional H-bond with the sidechain hydroxyl of Ser or Thr at RNAP β residue 642 (Figure 2E-F).
The structures suggest that potency against bacterial RNAP species having an apolar amino acid at β position 642 could be increased by replacing the pyrazole moiety of a CBR pyrazole with a heterocycle that positions apolar atoms, rather than polar atoms, adjacent to the sidechain of β residue 642.
The structures suggest that potency against RNAP from most or all bacterial species could be increased by replacing the pyrazole moiety of a CBR pyrazole with a heterocycle able to donate an H-bond to the backbone carbonyl group of β residue 642 (Figs. 2B-C,E-F); by incorporating a positively charged substituent at position 6 of ring B of a CBR inhibitor in order to enable a salt bridge with βE641; appending a sidechain with metal-coordinating functionality at position 4 or position 5 of ring B to enable metal-ion-mediated interactions with a pair of adjacent histidine sidechains in the β-lobe/F-loop groove (βH447 of β conserved region C and β551 of β conserved region D); appending a sidechain with positively-charged functionality at position 4 or position 5 of ring B to enable interactions with negatively charged sidechains in the β-lobe/F-loop groove (βD444 and βD446 of region C and βE546 of the region C/D spacer); or appending a sidechain with aromatic functionality to the connector between rings A and B to enable interactions with a hydrophobic pocket formed by the bridge helix and F-loop (β′I755, β′T757, β′I759, β′L770, and β′I774.
We note that we were able to isolate CBR703-resistant mutants in E. coli strain D21f2tolC when we used merodiploid RNAP subunit genes (i.e., mutant RNAP subunit gene on a plasmid and corresponding wild-type RNAP subunit gene on the chromosome; Figure 4), but we were unable to isolate CBR703-resistant mutants in E. coli strain D21f2tolC when we used haploid RNAP subunit genes (i.e., mutant RNAP subunit gene on the chromosome and no corresponding wild-type RNAP subunit gene; Table S1). The observed spontaneous resistance rate for CBR703 in E. coli strain D21f2tolC in haploid was <1×10−12, which was at least 2 to 4 orders of magnitude less than the spontaneous resistance rates for other characterized RNAP inhibitors (Table S1). We attribute the low observed spontaneous resistance rate to high fitness costs, and large growth defects, for CBR703-resistant mutants in E. coli strain D21f2tolC in haploid.
The ability to perform structure-based design of improved CBR inhibitors--made possible by the structural studies in this work--the potential ability to increase antibacterial activity by co-administering a CBR inhibitor with another RNAP inhibitor, the potential ability to suppress the emergence of spontaneous resistance by co-administering a CBR inhibitor with another RNAP inhibitor, and the apparent low spontaneous resistance frequencies and high fitness costs of CBR-resistant mutants, make the CBR hydroxamidine and CBR pyrazole chemical scaffolds attractive chemical scaffolds for antibacterial drug discovery.
EXPERIMENTAL PROCEDURES
E. coli RNAP
E. coli RNAP σ70 holoenzyme, RNAP core enzyme, and ΔTL RNAP core enzyme were prepared as in Degen et al., 2014.
RNAP-inhibitory activity
Fluorescence-detected RNAP-inhibition assays were performed using the pro-fluorescent substrate γ-[2′-(2-benzothiazoyl)-6′-hydroxybenzothiazole]-ATP (BBT-ATP; Niyomrattanakit et al., 2011). Reaction mixtures contained (20 μl): 0-100 μM test compound, bacterial RNAP holoenzyme [75 nM E. coli RNAP holoenzyme, or 75 nM Mycobacterium tuberculosis RNAP core enzyme and 300 nM M. tuberculosis σA (prepared as in Srivastava et al., 2011)], 20 nM DNA fragment containing positions −42 to +426 of the lacUV5(ICAP) promoter (Naryshkin et al., 2001), 25 μM BBT-ATP (Jena Bioscience), 100 μM GTP, 100 μM UTP, and 100 μM CTP, in TB (50 mM Tris-HCl, pH 8.0, 100 mM KCl, 10 mM MgCl2, 1 mM DTT, 10 μg/ml bovine serum albumin, 5% methanol, and 5.5% glycerol). Reaction components other than DNA and NTPs were pre-incubated 10 min at 37°C. Reactions were carried out by addition of DNA and incubation 15 min at 37°C, followed by addition of NTPs and incubation 60 min at 37°C. Reactions were terminated and profluorescent BBT-diphosphate produced during reactions was hydrolyzed to fluorescent BBT by addition of 1 μl 0.5 M AMPSO (Sigma-Aldrich) containing 0.5 U calf-intestinal alkaline phosphatase (New England BioLabs) and incubation 20 min at 37°C. Fluorescence emission intensities were measured using a GENios Pro microplate reader (Tecan; excitation wavelength = 415 nm; emission wavelength = 535 nm). Half-maximal inhibitory concentrations (IC50s) were calculated by non-linear regression in SigmaPlot (SPSS).
RNAP -inhibitory activities against human RNAP I, II, and III were determined as in Degen et al., 2014.
Nucleotide addition by ΔTL RNAP was assayed as in Degen et al., 2014, but using a reaction time of 2 min at 37°C.
Growth-inhibitory activity
Minimum inhibitory concentrations (MICs) were determined as in Degen et al., 2014, but using an initial cell density of ~1 × 105 cells/ml.
CBR703-resistant mutants
CBR703-resistant mutants were isolated and sequenced essentially as described for Stl-resistant mutants in Tuske et al., 2005. Saturation mutagenesis of plasmid pRL706 carrying E. coli rpoB (Severinov et al. 1997) and plasmid pRL663 carrying E. coli rpoC (Wang et al. 1995) was performed by use of PCR amplification with “doped” oligodeoxyribonucleotide primers spanning codons within 30 Å of residues of the RNAP bridge helix or the RNAP active center catalytic Mg2+ ion. Mutagenesis reactions were performed using the QuikChange Site-Directed Mutagenesis Kit (Agilent/Stratagene) with a “doped” oligodeoxyribonucleotide primer, a complementary oligodeoxyribonucleotide primer, and pRL706 or pRL663 as template (primers at 75 nM; all other components at concentrations as specified by the manufacturer). Mutagenized plasmid DNA was introduced by transformation into E. coli XL1-Blue (Agilent/Stratagene). Transformants (~104 cells) were applied to LB-agar plates (Sambrook and Russell, 2001) containing 200 μg/ml ampicillin, plates were incubated 16 h at 37°C, and plasmid DNA was prepared from the pooled resulting colonies. The resulting passaged mutagenized plasmid DNA was introduced by transformation into E. coli D21f2tolC (Fralick and Burns-Keliher, 1994). Transformants (~104 cells) were applied to LB-agar plates containing 7 μg/ml CBR703, 200 μg/ml ampicillin, and 1 mM IPTG; and plates were incubated 16 h at 37°C. CBR-resistant mutants were identified by the ability to form colonies on this medium and were confirmed by re-streaking on the same medium. For each confirmed mutant, the nucleotide sequence of the mutagenized rpoB or rpoC segment was determined by Sanger sequencing.
Structure determination: crystallization, crystal soaking, and cryo-cooling
Crystallization and crystal handling were performed essentially as in Degen et al. 2014. Crystallization drops contained 1 μl E. coli RNAP holoenzyme in 10 mM Tris-HCl, pH 7.9, 100 mM NaCl, and 1% glycerol, and 1 μl reservoir buffer (RB; 100 mM HEPES, pH 7.0, 200 mM CaCl2, and 18% PEG400), and were equilibrated against 500 μl RB in a vapor-diffusion hanging-drop tray at 22°C. Crystals formed and grew to a final size of 0.2 mm × 0.2 mm × 0.2 mm within one week. CBR703, CBRP18 and CBRH16-Br were soaked into crystals by adding 0.2 μl 20 mM of CBR703, CBRP18, orCBRH16-Br in (2R, 3R)-(−)- 2,3-butanediol to the crystallization drop and incubating 30 min at 22°C. Crystals were transferred to RB containing 15% (v/v) (2R, 3R)-(−)-2,3-butanediol and then flash-cooled with liquid nitrogen.
Structure determination: data collection and reduction
Diffraction data were collected from cryo-cooled crystals at Brookhaven National Laboratory (BNL) beamline X29A and Argonne National Laboratory (ANL) beamline 19ID. Data were processed using HKL2000 (Otwinowski et al., 1997).
Structure determination: structure solution and refinement
The structures of E. coli RNAP-CBR703, RNAP-CBRP18, and RNAP-CBRH16-Br were solved by molecular replacement with AutoMR in Phenix (McCoy et al., 2007; Adams et al., 2010), using a crystal structure of E. coli RNAP holoenzyme as the search model (PDB: 4LK1; Bae et al, 2013). Early-stage refinement included rigid-body refinement of the RNAP molecule, followed by rigid-body refinement of each subunit of RNAP molecule. Cycles of iterative model building with Coot (Emsley et al., 2010) and refinement with Phenix (Adams et al., 2010) were performed. Atomic models of CBR703, CBRP18, and CBRH16-Br were built into mFo-DFc difference maps, and subsequent cycles of refinement and model building were performed. The final E. coli RNAP-CBR703, RNAP-CBRP18 and RNAP-CBRH16-Br models were deposited in the PDB with accession codes PDB: 4ZH2, 4ZH4, and 4ZH3, respectively (Table 1).
Supplementary Material
HIGHLIGHTS.
Crystal structures of CBR inhibitors bound to E. coli RNA polymerase are presented
The resistance spectrum of CBR inhibitors is defined
CBR inhibitors show no cross-resistance with other RNA polymerase inhibitors
CBR inhibitors function additively with other RNA polymerase inhibitors
ACKNOWLEDGEMENTS
This work was supported by NIH grant GM041376. We thank W. Fenical for Sal and the Brookhaven National Synchrotron Light Source, and the Argonne Photon Source for beamline access for X-ray data collection.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
YF determined structures. DD, XW, and MG isolated and characterized resistant mutants. DD, XW, MG, SL, YZ, MT, and NC determined RNAP-inhibitory and growth-inhibitory properties. DD, TM, and YE synthesized compounds. RHE conceived the research, directed the research, and wrote the manuscript.
REFERENCES
- Adams P, Afonine P, Bunkóczi G, Chen V, Davis I, Echols N, Head J, Hung L, Kapral G, Grosse-Kunstleve R, McCoy A, Moriarty N, Oeffner R, Read R, Richardson D, Richardson J, Terwilliger T, Zwart P. PHENIX: a comprehensive Python-based system for macromolecular structure. Acta Cryst. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Chu C, Lynch A, Landick R. A new class of bacterial RNA polymerase inhibitor affects nucleotide addition. Science. 2003;302:650–654. doi: 10.1126/science.1087526. [DOI] [PubMed] [Google Scholar]
- Bae B, Davis E, Brown D, Campbell E, Wigneshweraraj S, Darst SA. Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma70 domain 1.1. Proc. Natl. Acad. Sci. USA. 2013;110:19772–19777. doi: 10.1073/pnas.1314576110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell E, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst S. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- Degen D, Feng Y, Zhang Y, Ebright KY, Ebright YW, Gigliotti M, Vahedian-Movahed H, Mandal S, Talaue M, Connell N, Arnold E, Fenical W, Ebright RH. Transcription inhibition by the depsipeptide antibiotic salinamide A. eLife. 2014;3:e02451. doi: 10.7554/eLife.02451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright R. RNA exit channel--target and method for inhibition of bacterial RNA polymerase. 2005 WO/2005/001034.
- Ebright R, Ebright Y. Antibacterial agents: high-potency myxopyronin derivatives. 2013 WO/2012/037508.
- Emsley P, Lohkamp B, Scott W, Cowtan K. Features and development of Coot. Acta Cryst. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feklistov A, Mekler V, Jiang Q, Westblade L, Irschik H, Jansen R, Mustaev A, Darst S, Ebright R. Rifamycins do not function by allosteric modulation of binding of Mg2+ to the RNA polymerase active center. Proc. Natl. Acad. Sci. USA. 2008;105:14820–14825. doi: 10.1073/pnas.0802822105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fralick J, Burns-Keliher L. Additive effect of tolC and rfa mutations on the hydrophobic barrier of the outer membrane of E. coli K-12. J. Bacteriol. 1994;176:6404–6406. doi: 10.1128/jb.176.20.6404-6406.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garibyan L, Huang T, Kim M, Wolff E, Nguyen A, Nguyen T, Diep A, Hu K, Iverson A, Yang H, Miller JH. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair (Amst.) 2003;2:593–608. doi: 10.1016/s1568-7864(03)00024-7. [DOI] [PubMed] [Google Scholar]
- Hein P, Landick R. The bridge helix coordinates movements of modules in RNA polymerase. BMC Biol. 2010;8:141. doi: 10.1186/1741-7007-8-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler LM, Suzuki H, Landick R, Gross C. Four contiguous amino acids define the target for streptolydigin resistance in the β subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 1993;268:25369–25375. [PubMed] [Google Scholar]
- Horrevorts A, Michel M, Kerrebijn K. Antibiotic interaction: interpretation of fractional inhibitory and fractional bactericidal concentration indices. Eur. J. Clin. Microbiol. 1987;6:502–503. doi: 10.1007/BF02013128. [DOI] [PubMed] [Google Scholar]
- Jin DJ, Gross C. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J. Mol. Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- Lane W, Darst S. Molecular evolution of multisubunit RNA polymerases: sequence analysis. J. Mol. Biol. 2010;395:671–685. doi: 10.1016/j.jmb.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Chen X, Fan P, Mihalic J, Cutler S. Antibacterial agents. 2001 WO/2001/051456.
- Li L, Chen X, Cutler S, Mann J. Pyrazole antimicrobial agents. 2001 WO/2001/082930.
- Lisitsyn N, Sverdlov E, Moiseeva E, Nikiforov V. Localization of mutation leading to resistance of E. coli RNA polymerase to the antibiotic streptolydigin in the gene rpoB coding for the β subunit of the enzyme. Bioorg. Khim. 1985;11:132–134. [PubMed] [Google Scholar]
- Ma W, Sandri G, Sarkar S. Analysis of the Luria-Delbrück distribution using discrete convolution powers. J. Appl. Probab. 1992;29:255–267. [Google Scholar]
- Malinen A, Nandymazumdar M, Turtola M, Malmi H, Grocholski T, Artsimovitch I, Belogurov G. CBR antimicrobials alter coupling between the bridge helix and the beta subunit in RNA polymerase. Nature Commun. 2014;5:3408. doi: 10.1038/ncomms4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A, Grosse-Kunstleve R, Adams P, Winn M, Storoni L, Read R. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletiadis J, Pournaras S, Roilides E, Walsh TJ. Defining fractional inhibitory concentration index cutoffs for additive interactions based on self-drug additive combinations, Monte Carlo simulation analysis, and in vitro-in vivo correlation data for antifungal drug combinations against Aspergillus fumigatus. Antimicrob. Agents Chemother. 2010;54:602–609. doi: 10.1128/AAC.00999-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Trischman J, Dieter Seng D, Kho D, Jensen P, Fenical W. Salinamides: antiinflammatory depsipeptides from a marine streptomycete. J. Org. Chem. 1999;64:1145–1150. [Google Scholar]
- Mukhopadhyay J, Das K, Ismail S, Koppstein D, Jang M, Hudson B, Sarafianos S, Tuske S, Patel J, Jansen R, Irschik H, Arnold E, Ebright R. The RNA polymerase “switch region” is a target for inhibitors. Cell. 2008;135:295–307. doi: 10.1016/j.cell.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naryshkin N, Kim Y, Dong Q, Ebright R. Site-specific protein-DNA photocrosslinking: analysis of bacterial transcription initiation complexes. Meths. Mol. Biol. 2001;148:337–361. doi: 10.1385/1-59259-208-2:337. [DOI] [PubMed] [Google Scholar]
- Niyomrattanakit P, Abas SN, Lim CC, Beer D, Shi P-Y, Chen Y-L. A fluorescence-based alkaline phosphatase-coupled polymerase assay for identification of inhibitors of dengue virus RNA-dependent RNA polymerase. J. Biomol. Screen. 2011;16:201–210. doi: 10.1177/1087057110389323. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meths. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov Y, Monastyrskaya G, Gubanov V, Lipkin V, Sverdlov E, Kiver I, Bass I, Mindlin S, Danilevskaya O, Khesin R. Primary structure of Escherichia coli RNA polymerase nucleotide substitution in the β subunit gene of the rifampicin resistant rpoB255 mutant. Mol. Gen. Genet. 1981;184:536–538. doi: 10.1007/BF00352535. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov Y, Monastyrskaya G, Guriev S, Kalinina N, Sverdlov E, Gragerov A, Bass I, Kiver I, Moiseyeva E, Igumnov V, Mindlin S, Nikiforov V, Khesin R. RNA polymerase rifampicin resistance mutations in Escherichia coli: sequence changes and dominance. Mol. Gen. Genet. 1983;190:344–348. doi: 10.1007/BF00330662. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Ma W, Sandri G. On fluctuation analysis: a new, simple and efficient method for computing the expected number of mutants. Genetica. 1992;85:173–179. doi: 10.1007/BF00120324. [DOI] [PubMed] [Google Scholar]
- Severinov K, Soushko M, Goldfarb A, Nikiforov V. Rifampicin region revisited: new rifampicin-resistant and streptolydigin-resistant mutants of the β subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 1993;268:14820–14825. [PubMed] [Google Scholar]
- Severinov K, Soushko M, Goldfarb A, Nikiforov V. RifR mutations in the beginning of the Escherichia coli rpoB gene. Mol. Gen. Genet. 1994;244:120–126. doi: 10.1007/BF00283512. [DOI] [PubMed] [Google Scholar]
- Severinov K, Markov D, Severinova E, Nikiforov V, Landick R, Darst S, Goldfarb A. Streptolydigin-resistant mutants in an evolutionarily conserved region of the β′ subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 1995;270:23926–23929. doi: 10.1074/jbc.270.41.23926. [DOI] [PubMed] [Google Scholar]
- Severinov K, Mooney R, Darst S, Landick R. Tethering of the large subunits of Escherichia coli RNA polymerase. J. Biol. Chem. 1997;272:24137–24140. doi: 10.1074/jbc.272.39.24137. [DOI] [PubMed] [Google Scholar]
- Srivastava A, Talaue M, Liu S, Degen D, Ebright RY, Sineva E, Chakraborty A, Druzhinin S, Chatterjee S, Mukhopadhyay J, Ebright YW, Zozula A, Shen J, Sengupta S, Niedfeldt R, Xin C, Kaneko T, Irschik H, Jansen R, Donadio S, Connell N, Ebright RH. New target for inhibition of bacterial RNA polymerase: “switch region”. Curr. Opin. Microbiol. 2011;14:532–543. doi: 10.1016/j.mib.2011.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetser D, Nonet M, Young R. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc. Natl. Acad. Sci. USA. 1987;84:1192–6. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temiakov D, Zenkin N, Vassylyeva M, Perederina A, Tahirov T, Kaihatsu K, Savkina M, Zorov S, Nikiforov V, Igarashi N, Matsugaki N, Wakatsuki S, Severinov K, Vassylyev D. Structural basis of transcription inhibition by antibiotic streptolydigin. Mol. Cell. 2005;19:655–666. doi: 10.1016/j.molcel.2005.07.020. [DOI] [PubMed] [Google Scholar]
- Tuske S, Sarafianos S, Wang X, Hudson B, Sineva E, Mukhopadhyay J, Birktoft J, Leroy O, Ismail S, Clark A, Dharia C, Napoli A, Laptenko O, Lee J, Borukhov S, Ebright R, Arnold E. Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation. Cell. 2005;122:541–552. doi: 10.1016/j.cell.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Meier T, Chan C, Feng G, Lee D, Landick R. Discontinuous movements of DNA and RNA in RNA polymerase accompany formation of a paused transcription complex. Cell. 1995;81:341–350. doi: 10.1016/0092-8674(95)90387-9. [DOI] [PubMed] [Google Scholar]
- Weinzierl R. The nucleotide addition cycle of RNA polymerase is controlled by two molecular hinges in the Bridge Helix domain. BMC Biol. 2010;8:134. doi: 10.1186/1741-7007-8-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R, Manduru M, Bosso J. Comparison of three different in vitro methods of detecting synergy: time-kill, checkerboard, and E test. Antimicrob. Agents Chemother. 1996;40:1914–1918. doi: 10.1128/aac.40.8.1914. D., B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Degen D, Ho M, Sineva E, Ebright K, Ebright Y, Mekler V, Vahedian-Movahed H, Feng Y, Yin R, Tuske S, Irschik H, Jansen R, Maffioli S, Donadio S, Arnold E, Ebright RH. GE23077 binds to the RNA polymerase ‘i’ and ‘i+1’ sites and prevents the binding of initiating nucleotides. eLife. 2014;3:e02450. doi: 10.7554/eLife.02450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Haupenthal J, Groh M, Fountain M, Hartmann R. New insights into the bacterial RNA polymerase inhibitor CBR703 as a starting point for optimization as an anti-infective agent. Antimicrob. Agents Chemother. 2014;58:4242–4245. doi: 10.1128/AAC.02600-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.