Abstract
Background
Little is known about genetic factors associated with nasopharyngeal carcinoma (NPC). To gain insight into NPC etiology, we performed whole exome sequencing on germline and tumor DNA from three closely related family members with NPC.
Methods
The family was ascertained through the Pediatric Familial Cancer Clinic at The University of Chicago. The diagnosis of NPC was confirmed pathologically for each individual. For each sample sequenced, 97.3% of the exome was covered at 5×, with an average depth of 44×. Candidate germline and somatic variants associated with NPC were identified and prioritized using a custom pipeline.
Results
We discovered 72 rare deleterious germline variants in 56 genes shared by all three individuals. Of these, only three are in previously identified NPC-associated genes, all of which are located within MLL3, a gene known to be somatically altered in NPC. One variant introduces an early stop codon in MLL3, which predicts complete loss-of-function. Tumor DNA analysis revealed somatic mutations and EBV integration events; none, however, were shared among all three individuals.
Conclusions
These data suggest that inherited mutations in MLL3 may have predisposed these three individuals from a single family to develop NPC, and may cooperate with individually-acquired somatic mutations or EBV integration events in NPC etiology.
Impact
Our finding is the first instance of a plausible candidate high penetrance inherited mutation predisposing to NPC.
Keywords: Nasopharyngeal carcinoma, cancer genetics, cancer risk, whole exome sequencing, bioinformatics
INTRODUCTION
Nasopharyngeal carcinoma (NPC) is a rare malignancy arising from epithelial cells of the head and neck. Although worldwide incidence of NPC is under 1:100,000, rates vary by geography and ethnicity. In Southern Italy, Greece, Turkey, Northern Africa, and among Alaskan Eskimos, rates range from 15-20:100,000. Incidence peaks in Southeastern China and Southeast Asia at 25:100,000, whereas NPC is rare in the United States and Western Europe, with an incidence of only 0.5-2:100,000 (1, 2). NPC etiology is multi-factorial, and includes exposure to nitrosamines found in, for example, tobacco, salted fish, and cosmetics and pesticide manufacturing; exposure to formaldehyde; infection with Epstein-Barr Virus (EBV); and genetic susceptibilities (3-5).
Although little is known about the genetic contribution to NPC risk, that there is a genetic component to susceptibility was demonstrated in two separate studies, one performed in South Asian individuals and the other in European individuals from Greenland and Denmark. In both, it was found that individuals with a first-degree relative with NPC were at an 8.0-fold greater relative risk for developing NPC as compared to the general population (6). To identify genetic factors underlying NPC susceptibility, only a small number of studies have thus far been performed. In one candidate gene study, the association between NPC and polymorphic variation in base excision repair genes, the pathway required for repair of nitrosamine-induced DNA damage, was investigated. Variants in XRCC1 and hOGG1 were found to be associated with NPC; these findings, however, await replication (7). In another study, a genome-wide linkage scan of familial NPC in 54 affected individuals from 20 families led to the discovery of a susceptibility locus at chromosome 4p15.1-q12 (8). More recently, four Genome Wide Association Studies (GWAS) of NPC have been performed and have led to the identification of 20 variants associated with NPC (9-12). The full list of associated variants is in Supplementary Table 1.
Because the genetic architecture of familial disease is vastly simplified relative to that of sporadic disease, studying the genetics of NPC in families with multiple affected individuals is an attractive strategy for discovering high penetrance susceptibility variants. Towards this end, we performed WES on germline DNA from three related individuals of Italian descent, two full siblings and a half nephew, all of whom developed NPC (Figure 1). Additionally, we performed WES on their tumor DNA to determine whether their shared predispositions are associated with the acquisition of shared somatic mutations. Finally, we scanned their germline and tumor exomes for sites of EBV integration to investigate the possibility that patterns of EBV insertion were common among all three individuals. This is the first study of the genetic etiology of NPC undertaken in individuals of European ancestry.
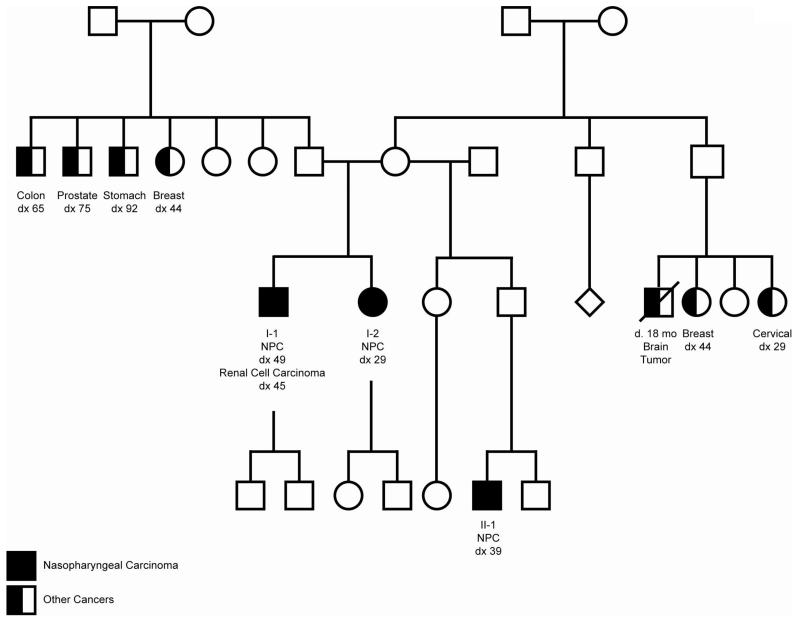
Figure 1. Nasopharyngeal Carcinoma family pedigree.
Shown is a four-generation pedigree of the family. Germline and tumor DNA of individuals with NPC (Individuals I-1, I-2, and II-1) were analyzed by WES.
MATERIALS AND METHODS
Study subjects
The family investigated was ascertained by the Pediatric Familial Cancer Clinic at The University of Chicago. All study subjects provided written informed consent to participate in a study of NPC genetics that was approved by the local institutional review board. The pedigree is presented in Figure 1. To protect the anonymity of the study subjects, the family pedigree was altered in ways that did not affect the genetic analysis.
Exome capture and sequencing
Germline DNA for WES was obtained from whole blood. Tumor DNA was isolated from FFPE scrolls after evaluation by a pathologist (>80% tumor). At least 1 ug of DNA was used for whole exome capture using SureSelect Human All Exon V4 50 Mb kit (Agilent Technologies, Santa Clara, USA). Sequence reads were generated on an Illumina HiSeq2000 instrument (Illumina, San Diego, USA). An average of 63 million 2×100bp paired-end (PE) reads were generated for each sample.
Variant calling and quality control
The quality of raw reads was assessed by FastQC (13), followed by adapter clipping and 3′ overlap mate merging. Processed reads were aligned to the human reference genome assembly (hg19) using three short-read aligners: BWA (14), Bowtie2 (15), and Novoalign (16). Exon coverage was calculated using BEDTools (17). Read duplicates were removed using the Picardtools MarkDuplicates program (18). The alignment was post-processed by GATK v1.6 (19) for InDel realignment and base quality score calibration. For each alignment, GATK UnifiedGenotyper (19, 20), FreeBayes (21), Atlas2 (22), and SAMtools mpileup/bcftools (23) were used to detect variants. Variant calls passing the internal quality filters of each caller were then filtered to remove potential false positives based upon: 1) variant quality score <50; 2) read coverage ≤5; or 3) location within a single nucleotide variant (SNV) cluster in which >3 SNVs were called within a 10bp window. After combining results from the three aligners and four callers, variants called by at least two callers using the aligned sequence from at least two aligners were carried forward for annotation using ANNOVAR (24). Population minor allele frequencies (MAF) were derived from The 1000 Genomes Project database (25) (phase 1, release v3, 20101123) and the Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP, version ESP6500-V2-SSA137, Seattle, WA; 06/2012 accessed) (26).
Each variant was annotated for pathogenicity using: SIFT (27); Polyphen-2 (28); MutationTaster (29); MutationAssessor (30); FATHMM (31); LR and LRT (32); and Radial SVM (24, 33). They were assessed for multispecies conservation using GERP++ (34), and PhyloP (35).
Germline mutations were defined as variants with a population allele frequency of <0.01 or which were unobserved in either The 1000 Genomes Project or The ESP databases.
Prioritization of candidate rare germline variants associated with NPC
To investigate rare variation, we required that variants passing our QC pipeline: 1) have a population MAF ≤0.01 in the European subset from either The ESP or The 1000 Genomes Project; 2) be either non-synonymous, a splice site modifier, an insertion or deletion creating a frameshift, or create a stop codon; 3) and be deleterious as predicted by one of the pathogenicity prediction algorithms. For each variant identified, we confirmed its presence in the tumor sample of each individual.
We compiled a list of NPC-associated genes from a thorough literature review, a previously published catalog of genes associated with NPC (36), and 4 NPC GWAS listed in the NHGRI resource (9, 11, 12, 37). Variants were prioritized as likely to be NPC-associated if they were found in genes either associated with NPC risk or somatically mutated in NPC (Supplementary Table 1).
Identification of somatic mutations
To identify somatic variants, we analyzed matched normal/tumor pairs for all three individuals using MuTect (38), Strelka (39), Virmid (40) and VarScan2 (41). All four programs detect somatic SNVs. Strelka and VarScan2 also detect somatic InDels. Variants passing the internal quality control of each caller were retrieved and filtered for high-confidence calls based upon: 1) variant quality score ≥20; 2) sequencing read depth ≥8 and; 3) allele fraction in the tumor sample of >0.20 and allele fraction in the germline sample of <0.05. We then combined somatic variants identified by any of the four calling algorithms for downstream analysis. Somatic mutations in each tumor were manually inspected using Integrative Genome Viewer (42) to confirm that the variant allele was not present in the matched normal sample.
Identification of EBV insertion sites
Using Novoalign, sequencing reads were mapped to the human reference genome assembly hg19, and to the type I, type I-HKNPC1 and type II EBV reference genomes (GenBank Accession IDs: NC_007605.1, JQ009376.1 and NC_009334.1). Read duplicates were removed, and alignments with mapping quality scores <20 were excluded. EBV insertion sites were identified using chimeric read pairs, which have one mate mapped to the human genome and the other mapped to at least one of the three EBV reference genomes. The insertion sites were approximated as the 3′ end of the chimeric mate mapped to the human genome, and annotated for nearby genes.
RESULTS
Familial WES pipeline
We performed WES on germline and tumor DNA from three members of a single family of Italian ancestry, all of whom were affected with NPC. For all individuals, the tumors were EBV-positive, as confirmed by EBER in situ hybridization. Two individuals are full siblings and the third is their half nephew (Figure 1). Individual I-1 was diagnosed with renal clear cell carcinoma at age 45 and with NPC at age 49. He lived in Argentina, worked as a beautician for 30 years, and smoked regularly (half a pack for over 30 years). Individual I-2 is the sister of Individual I-1 and was diagnosed with NPC at age 29. She is 12 years younger than Individual I-1, worked in a nail salon for 7 years prior to her diagnosis, and did not smoke or chew tobacco. Individual II-1 is the maternal half nephew of Individuals I-1 and I-2, and was diagnosed with NPC at age 39. Over a period of 8 years, he worked in construction and then went on to be a line cook at a restaurant. He did not smoke or chew tobacco. He died from metastatic NPC at 41 years of age. Of note, two siblings who were first cousins of the NPC-affected sibling pair and in the same blood lineage as Individual II-1 also developed cancer, one a brain tumor at 18 months, and the other early-onset breast cancer (at age 44). No samples from these individuals were available for WES.
Following WES and quality control, for each sample sequenced, 97.3% of the exome was covered at 5× and 86.4% of the exome was covered at 20×, with an average coverage depth of 44× or greater across the exome (Supplementary Table 2). Variants of low quality score, coverage depth less than 5, and those that were not called by at least two genotype callers within the aligned sequence generated by at least two aligners were removed. After filtering out non-exonic variants and those leading to synonymous amino acid changes, an average of 8,767 variants in each germline sample and 8,832 variants in each tumor sample were identified. The mutational spectrum of the variants for each sample (germline and tumor) for each individual is summarized in Supplementary Table 3.
Identification of candidate familial NPC-predisposing germline mutations
To discover candidate NPC-predisposing mutations in this family, we first identified rare exonic germline variants (MAF ≤0.01) shared by all three individuals. We found an average of 780 rare or novel variants in each individual, of which 190 were shared by all three individuals. Variants were categorized as: non-frameshift insertion/deletions (n = 34), non-synonymous single nucleotide variants (SNVs, n = 113), frameshift insertion/deletions (n = 3), stop gain (n = 1), and unannotated (n = 39). Of these, we predicted 72 variants in 56 genes to be deleterious (Supplementary Table 4).
To prioritize among these 72 candidates, we filtered them against a list of 76 previously identified NPC-associated genes (Supplementary Table 1) (9, 11, 12, 36, 37). This reduced the list to only three variants in a single gene, MLL3 (rs150073007; rs4024453; and rs10454320) (Table 1). All three deleterious variants are unique to this family and not found in any of the 9007 individuals sequenced as part of The 1000 Genomes or Exome Sequencing Projects. rs4024453 (c.2315c>t), results in a serine to leucine change that occurs in a serine rich domain, and therefore may affect protein-protein interactions. rs10454320 (c.946a>t), is a threonine to serine change at amino acid 316, upstream of any known functional domain. Most compellingly, rs150073007 results in the introduction of a premature stop codon at position 816, N-terminal to most functional domains within the gene product.
TABLE 1. Familial deleterious germline mutations in MLL3 identified by WES.
All three variants were observed in all three NPC-affected family members.
| Gene | Chr | Position | Reference Allele |
Variant Allele |
Exonic Function |
rsID | Nucleotide Change |
Protein Change |
|---|---|---|---|---|---|---|---|---|
| MLL3 | 7 | 151945071 | - | T | stopgain SNV |
rs150073007 | 2447dupA | Y816* |
| MLL3 | 7 | 151945204 | G | A | ns SNV | rs4024453 | C2315T | S772L |
| MLL3 | 7 | 151970856 | T | A | ns SNV | rs10454320 | A946T | T316S |
Chr: Chromosome; SNV: single nucleotide variant; ns: nonsynonymous
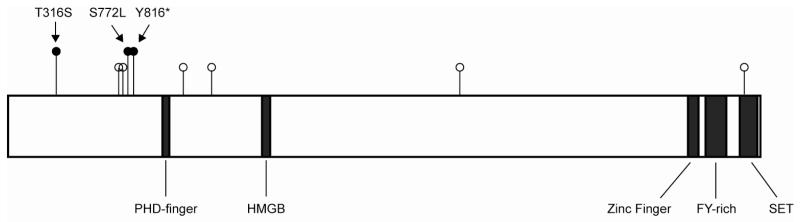
The location of these three variants, in particular the early stop-gain mutation, suggests that they completely abrogate MLL3 protein function, leading to the hypothesis that one or more of these mutations predisposed these three family members to develop NPC. Supporting this contention is the observation that somatic mutations in MLL3 have been previously reported in 4% of NPC cases, including a recurrent mutation introducing a premature stop codon at amino acid 728, near the site of the germline premature stop codon observed in this family (Figure 2) (36).
Figure 2. MLL3 mutations in NPC.
Diagram of MLL3 with functional domains. Black lollipops indicate germline mutations identified in this study. White lollipops indicate previously identified somatic mutations.
Somatic mutation analysis of familial NPC
We then investigated somatic mutations in the exome of the tumor DNA from each individual to determine whether a common “second hit” had occurred in all three individuals in MLL3 or any of the 56 genes containing a candidate deleterious germline mutation.
Individual I-1 had 90 somatic mutations of which 36 were predicted to be deleterious. Individual I-2 had 68 somatic mutations of which 24 were predicted to be deleterious. Individual II-1 had 110 somatic mutations of which 47 were predicted to be deleterious. No individual had acquired a somatic mutation in MLL3. Among the three individuals, six of the overall set of 56 genes with deleterious germline mutations had also acquired somatic mutations. Specifically, Individual I-1 acquired somatic mutations in MUC2 and MUC6; Individual I-2 acquired somatic mutations in MUC6; and Individual II-1 acquired somatic mutations in in HRNR, KCNJ12, PABPC1, and PCMTD1. Notably, mutations in these genes have not previously been implicated in NPC (36). Additionally, mutations in MUC genes are frequently observed as false-positives in next-generation sequencing studies and must be interpreted with caution (43). There were, however, two de novo somatic mutations in genes previously implicated in NPC. Individual I-1 acquired a somatic mutation in NRAS (Q61R) (44-48), a well-characterized mutation observed in numerous cancers, and Individual II-1 acquired a somatic mutation in PIK3CG (X87Y), a mutation not previously reported.
Thus, we did not find overlap in the spectrum of somatic mutations among the three individuals. Results are summarized in Supplementary Table 5.
EBV integration analysis
There is a strong association between NPC and EBV integration (3). To determine whether there are shared patterns of EBV integration among the three NPC-affected family members, we mapped EBV integration sites in the germline and tumor DNA of each individual. We found that the germline exome from the two siblings, Individuals I-1 and I-2, did not contain any EBV DNA. Individual I-1 had only a single somatic insertion event in his tumor, while Individual I-2 had nine somatic insertion events. In contrast, Individual II-1 had one EBV insertion event in his germline exome and 42 somatic insertions in his tumor. EBV integration events were not found in any of the 56 genes with deleterious germline mutations or in any gene previously associated with NPC in any individual. Results are summarized in Supplementary Table 6.
DISCUSSION
In this study, we employed a family-based WES strategy to discover germline variants predisposing to NPC. We hypothesized that the three affected individuals in this family were predisposed to NPC through the shared inheritance of a single or small number of highly penetrant mutations. We found 72 rare deleterious germline variants in 56 genes shared by all three family members, three of which are in known NPC-associated genes. All three are located within a single gene, MLL3, which is recurrently mutated somatically in NPC. While all germline MLL3 mutations are predicted to attenuate MLL3 function, one mutation, rs150073007, is a particularly compelling candidate as causative because it results in the introduction of a stop codon near the N-terminus of the protein. The observation that none of these three variants is reported in large population databases such as The 1000 Genomes Project and The ESP leads us to speculate that they originated in this family. Based on our analysis, we propose MLL3 as the candidate NPC-predisposition gene in this family.
MLL3 is a histone lysine methyltransferase that functions in transcriptional co-activation of nuclear receptor targets. It is mutated not only in NPC, but in a variety of other cancers as well (49-53). As a component of the ASCOM complex, MLL3 is a co-activator of p53, and deletion of its catalytic domain results in the development of kidney and ureter epithelial tumors in mice (54). Additionally, MLL3 functions to regulate enhancer activity. Since enhancers play an important role in the tissue-specific expression of genes, mutations in MLL3 may affect tumorigenesis in a tissue-dependent manner (53). Functional studies will be necessary to determine the consequences of the mutations identified here on MLL3 activity.
We did not find any combination of germline variants, somatic mutations and/or EBV integration events common among all three individuals that altered the function of any gene other than MLL3. The fact that we did not observe any recurrent acquired genetic changes in the tumor DNA of the three individuals suggests that either: 1) shared acquired mutations may have occurred outside of the exome; 2) the mutations we did identify are unique to each individual but converge on and deregulate common pathways; or 3) other factors such as differences in exposures, variation in regulatory molecules such as miRNAs or lncRNAs, or epigenetic changes may also have contributed to the excess of NPC in this family. The complexity of the mutational landscape and lack of concordant acquired changes among all three individuals underscores the difficulties inherent in the genomic analysis of even highly penetrant families.
While NPC etiology depends on multiple factors such as environmental exposures, geography, diet, and EBV, the co-occurrence of the disease in three closely related individuals from a single family is strongly suggestive of a common genetic etiology. Recently, a germline mutation in MLL3 that introduced a premature stop codon at amino acid 827 was reported in a Chinese family with colorectal cancer and acute myeloid leukemia (55). This mutation is located very close to the premature stop codon at amino acid 816 identified in all three NPC-affected family members described in this study. Importantly, in addition to the three NPC-affected individuals we sequenced, there are two other closely related cancer-affected individuals following the same blood lineage in this family; one is an individual with early onset breast cancer (diagnosed at age 44), and the other is a baby with a brain tumor who died at age 18 months. Unfortunately, hospital records and samples from these two individuals are not available. Taken together, the finding of familial mutations predicted to abolish MLL3 function in two unrelated families with multiple cancer-affected members leads us to the intriguing hypothesis that inactivating mutations of MLL3 may be associated with a highly penetrant and previously unsuspected cancer-predisposition syndrome. In other studies, the familial aggregation of other cancers with NPC remains controversial (6, 56). It will be of interest to determine whether inactivating mutations in MLL3 are found in other families in which NPC is one of several cancer types observed.
In summary, we have identified the first instance of a plausible high penetrance inherited mutation predisposing to NPC. This study indicates that by performing WES on just a few affected individuals from a single well-chosen family, it is possible to generate a small list of highly likely disease-causing germline mutations that are amenable to future functional investigation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank E Bartom for development of the WES analysis pipelines; M Jarsulic for technical assistance with job execution on high-performance computing clusters.
Financial support: This work was supported by grants from the National Institutes of Health (HD0433871, CA129045 and CA40046 to K Onel); the American Cancer Society – Illinois Division (K Onel); the Cancer Research Foundation (K Onel); and The University of Chicago GREAT KIDS (Genomics for Risk Evaluation and Anticancer Therapy in Kids) Program (K Onel, AD Skol). The Center for Research Informatics is funded by the Biological Science Division and The Institute for Translational Medicine/CTSA (NIH UL1 RR024999) at The University of Chicago.
Footnotes
All authors declare no potential conflicts of interest
REFERENCES
- 1.Chang ET, Adami HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2006;15:1765–77. doi: 10.1158/1055-9965.EPI-06-0353. [DOI] [PubMed] [Google Scholar]
- 2.Eduardo B, Raquel C, Rui M. Nasopharyngeal carcinoma in a south European population: epidemiological data and clinical aspects in Portugal. European archives of oto-rhino-laryngology : official journal of the European Federation of Oto-Rhino-Laryngological Societies. 2010;267:1607–12. doi: 10.1007/s00405-010-1258-3. [DOI] [PubMed] [Google Scholar]
- 3.Chu EA, Wu JM, Tunkel DE, Ishman SL. Nasopharyngeal carcinoma: the role of the Epstein-Barr virus. Medscape journal of medicine. 2008;10:165. [PMC free article] [PubMed] [Google Scholar]
- 4.Vaughan TL, Stewart PA, Teschke K, Lynch CF, Swanson GM, Lyon JL, et al. Occupational exposure to formaldehyde and wood dust and nasopharyngeal carcinoma. Occupational and environmental medicine. 2000;57:376–84. doi: 10.1136/oem.57.6.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ward MH, Pan WH, Cheng YJ, Li FH, Brinton LA, Chen CJ, et al. Dietary exposure to nitrite and nitrosamines and risk of nasopharyngeal carcinoma in Taiwan. International journal of cancer Journal international du cancer. 2000;86:603–9. doi: 10.1002/(sici)1097-0215(20000601)86:5<603::aid-ijc1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 6.Friborg J, Wohlfahrt J, Koch A, Storm H, Olsen OR, Melbye M. Cancer susceptibility in nasopharyngeal carcinoma families--a population-based cohort study. Cancer research. 2005;65:8567–72. doi: 10.1158/0008-5472.CAN-04-4208. [DOI] [PubMed] [Google Scholar]
- 7.Cho EY, Hildesheim A, Chen CJ, Hsu MM, Chen IH, Mittl BF, et al. Nasopharyngeal carcinoma and genetic polymorphisms of DNA repair enzymes XRCC1 and hOGG1. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2003;12:1100–4. [PubMed] [Google Scholar]
- 8.Feng BJ, Huang W, Shugart YY, Lee MK, Zhang F, Xia JC, et al. Genome-wide scan for familial nasopharyngeal carcinoma reveals evidence of linkage to chromosome 4. Nature genetics. 2002;31:395–9. doi: 10.1038/ng932. [DOI] [PubMed] [Google Scholar]
- 9.Bei JX, Li Y, Jia WH, Feng BJ, Zhou G, Chen LZ, et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nature genetics. 2010;42:599–603. doi: 10.1038/ng.601. [DOI] [PubMed] [Google Scholar]
- 10.Ng CC, Yew PY, Puah SM, Krishnan G, Yap LF, Teo SH, et al. A genome-wide association study identifies ITGA9 conferring risk of nasopharyngeal carcinoma. Journal of human genetics. 2009;54:392–7. doi: 10.1038/jhg.2009.49. [DOI] [PubMed] [Google Scholar]
- 11.Tang M, Lautenberger JA, Gao X, Sezgin E, Hendrickson SL, Troyer JL, et al. The principal genetic determinants for nasopharyngeal carcinoma in China involve the HLA class I antigen recognition groove. PLoS genetics. 2012;8:e1003103. doi: 10.1371/journal.pgen.1003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tse KP, Su WH, Chang KP, Tsang NM, Yu CJ, Tang P, et al. Genome-wide association study reveals multiple nasopharyngeal carcinoma-associated loci within the HLA region at chromosome 6p21.3. American journal of human genetics. 2009;85:194–203. doi: 10.1016/j.ajhg.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andrews S. FastQC: A quality control application for high throughput sequence data. 2012 cited; Available from: http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc.
- 14.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cabana MD, Kunselman SJ, Nyenhuis SM, Wechsler ME. Researching asthma across the ages: insights from the National Heart, Lung, and Blood Institute’s Asthma Network. The Journal of allergy and clinical immunology. 2014;133:27–33. doi: 10.1016/j.jaci.2013.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Picard Tools. cited; Available from: http://broadinstitute.github.io/picard/
- 19.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. 2012 [Google Scholar]
- 22.Challis D, Yu J, Evani US, Jackson AR, Paithankar S, Coarfa C, et al. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012;13:8. doi: 10.1186/1471-2105-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.NHLBI Exome Sequencing Project Exome Variant Server. cited; Available from: http://evs.gs.washington.edu/EVS/
- 27.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–6. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 30.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Human mutation. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–61. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Consortium UIG. Barrett JC, Lee JC, Lees CW, Prescott NJ, Anderson CA, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–4. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–13. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–21. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin DC, Meng X, Hazawa M, Nagata Y, Varela AM, Xu L, et al. The genomic landscape of nasopharyngeal carcinoma. Nature genetics. 2014;46:866–71. doi: 10.1038/ng.3006. [DOI] [PubMed] [Google Scholar]
- 37.Bhat M, Nguyen GC, Pare P, Lahaie R, Deslandres C, Bernard EJ, et al. Phenotypic and genotypic characteristics of inflammatory bowel disease in French Canadians: comparison with a large North American repository. Am J Gastroenterol. 2009;104:2233–40. doi: 10.1038/ajg.2009.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature biotechnology. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–7. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 40.Kim S, Jeong K, Bhutani K, Lee J, Patel A, Scott E, et al. Virmid: accurate detection of somatic mutations with sample impurity inference. Genome biology. 2013;14:R90. doi: 10.1186/gb-2013-14-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–76. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nature biotechnology. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fukushima T, Suzuki S, Mashiko M, Ohtake T, Endo Y, Takebayashi Y, et al. BRAF mutations in papillary carcinomas of the thyroid. Oncogene. 2003;22:6455–7. doi: 10.1038/sj.onc.1206739. [DOI] [PubMed] [Google Scholar]
- 45.Fukushima T, Takenoshita S. Roles of RAS and BRAF mutations in thyroid carcinogenesis. Fukushima journal of medical science. 2005;51:67–75. doi: 10.5387/fms.51.67. [DOI] [PubMed] [Google Scholar]
- 46.Omholt K, Karsberg S, Platz A, Kanter L, Ringborg U, Hansson J. Screening of N-ras codon 61 mutations in paired primary and metastatic cutaneous melanomas: mutations occur early and persist throughout tumor progression. Clinical cancer research : an official journal of the American Association for Cancer Research. 2002;8:3468–74. [PubMed] [Google Scholar]
- 47.Tone AA, McConechy MK, Yang W, Ding J, Yip S, Kong E, et al. Intratumoral heterogeneity in a minority of ovarian low-grade serous carcinomas. BMC cancer. 2014;14:982. doi: 10.1186/1471-2407-14-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu S, Kuo H, Li WQ, Canales AL, Han J, Qureshi AA. Association between BRAFV600E and NRASQ61R mutations and clinicopathologic characteristics, risk factors and clinical outcome of primary invasive cutaneous melanoma. Cancer causes & control : CCC. 2014;25:1379–86. doi: 10.1007/s10552-014-0443-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nature genetics. 2011;43:875–8. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–9. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song Y, Li L, Ou Y, Gao Z, Li E, Li X, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91–5. doi: 10.1038/nature13176. [DOI] [PubMed] [Google Scholar]
- 53.Herz HM, Hu D, Shilatifard A. Enhancer malfunction in cancer. Molecular cell. 2014;53:859–66. doi: 10.1016/j.molcel.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee S, Kim DH, Goo YH, Lee YC, Lee SK, Lee JW. Crucial roles for interactions between MLL3/4 and INI1 in nuclear receptor transactivation. Molecular endocrinology. 2009;23:610–9. doi: 10.1210/me.2008-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li WD, Li QR, Xu SN, Wei FJ, Ye ZJ, Cheng JK, et al. Exome sequencing identifies an MLL3 gene germ line mutation in a pedigree of colorectal cancer and acute myeloid leukemia. Blood. 2013;121:1478–9. doi: 10.1182/blood-2012-12-470559. [DOI] [PubMed] [Google Scholar]
- 56.Yu KJ, Hsu WL, Chiang CJ, Cheng YJ, Pfeiffer RM, Diehl SR, et al. Cancer patterns in nasopharyngeal carcinoma multiplex families in Taiwan. International journal of cancer Journal international du cancer. 2009;124:1622–5. doi: 10.1002/ijc.24051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.