Abstract
Adipose tissue engineered models are needed to enhance our understanding of disease mechanisms and for soft tissue regenerative strategies. Perfusion systems generate more physiologically relevant and sustainable adipose tissue models, however adipocytes have unique properties that make culturing them in a perfusion environment challenging. In this paper we describe the methods involved in the development of two perfusion culture systems (2D and 3D) to test their applicability for long term in vitro adipogenic cultures. It was hypothesized that a silk protein biomaterial scaffold would provide a 3D framework, in combination with perfusion flow, to generate a more physiologically relevant sustainable adipose tissue engineered model than 2D cell culture. Consistent with other studies evaluating 2D and 3D culture systems for adipogenesis we found that both systems successfully model adipogensis, however 3D culture systems were more robust, providing the mechanical structure required to contain the large, fragile adipocytes that were lost in 2D perfused culture systems. 3D perfusion also stimulated greater lipogenesis and lipolysis and resulted in decreased secretion of LDH compared to 2D perfusion. Regardless of culture configuration (2D or 3D) greater glycerol was secreted with the increased nutritional supply provided by perfusion of fresh media. These results are promising for adipose tissue engineering applications including long term cultures for studying disease mechanisms and regenerative approaches, where both acute (days to weeks) and chronic (weeks to months) cultivation are critical for useful insight.
Keywords: adipose tissue engineering, three dimensional culture, silk scaffold, long term culture, perfusion
1. Introduction
Adipose tissue engineering is a multifaceted research initiative that will address current clinical needs related to filling various adipose tissue defects and for study disease pathologies in vitro. Defects in adipose tissue are caused by congenital abnormalities, traumatic injuries and tumor resections in various areas of the body and require better tissue engineering strategies for repair [1, 2]. Physiologically relevant and sustainable adipose models will also enhance our understanding of cell biology and provide novel experimental systems in vitro to study metabolism and metabolic disease mechanisms, such as obesity and type II diabetes [3].
In general, cells require and rely on fluid flow for mechanotransduction and nutrient supply in vivo. Similarly, perfusion systems provide control of the physiological environment in vitro (i.e. mechanical signals, pH, temperature, nutrient supply, oxygen tension, waste removal) and play a significant role in useful tissue engineering outcomes by improving the quality of engineered tissues, correcting for insufficient nutrient and metabolite transport present in static culture, improving mass transport, and generating more homogenous cell distributions [4–9]. The key cells in adipose tissue (adipocytes), however have unique properties that make culturing more challenging in a perfusion environment. Adipocytes are buoyant and their lipid laden morphology makes them fragile [10, 11]. Additionally, differentiating adipocyte cell lines requires lengthy culture times (at least 3 weeks) which needs to occur within the cultivation system [12, 13] or differentiated cells need to be perfused into the device [14]. Finally, adipocytes secrete hydrophobic metabolites that can accumulate in microscale devices blocking fluid flow [11].
To create more physiologically relevant tissues we sought to create a perfusion system for enhancing adipogenic potential in long term culture. Two dimensional (2D) culture systems have utility when compared to 3D culture systems, including direct visualization of cells and the lack of a complex three dimensional (3D) matrix. However, cells respond differently in 2D versus 3D environments [15]. In particular, 3D culture systems improve adipogenic differentiation [16, 17], including increased secretion [16] and production [18] of relevant proteins and gene expression patterns which are more similar to in vivo patterns [19]. Therefore, we describe the development of two perfusion culture systems (2D and 3D) to evaluate their applicability for long term in vitro adipogenic culture. Long term culture is particularly relevant for adipose tissue systems to create fully differentiated tissues [20] and will be required to generate an adequate supply of tissue mass required to implant in vivo. Additionally, diseases that affect adipose dysfunction, such as obesity and type II diabetes, are chronic conditions that develop in vivo over weeks to months [21], suggesting long term cultures will be required to create accurate in vitro models.
It was hypothesized that silk protein scaffolds would provide a 3D framework that in combination with perfusion flow would establish more physiologically relevant sustainable adipose tissue engineered systems. Silk is a naturally occurring and clinically acceptable biocompatible polymeric biomaterial that has tunable mechanical strength, low inflammatory and immunogenic responses, an absence of cell-specific signaling domains, and can be tailored to degrade slowly to support the needs of long term cultures [22–24]. Additionally, silk as a biomaterial scaffold has demonstrated compatibility with adipose tissue engineering applications [25–30].
2. Material and Methods
2.1 Materials
Bombyx mori silkworm cocoons were obtained from Tajima Shoji Co (Yokohama, Japan). Unless otherwise noted all cell culture supplies were purchased from Invitrogen (Carlsbad, CA). Additionally, phalloidin, 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI), Picogreen kits and collagenase type I were purchased from Invitrogen. Human recombinant insulin, 3-isobutyl-1-methylxanthine (IBMX), dexamethasone, indomethacin, laminin and bovine serum albumin (BSA) were purchased from Sigma–Aldrich (St. Louis, MO). AdipoRed was purchased from Lonza (Walkersville, MD). Triglyceride and Glycerol quantification kits were purchased from BioAssay Systems (Hayward, CA). A Lactate Dehydrogenase (LDH) Assay Kit was puchased from Abcam (Cambridge, MA). Polyethylene molds (Catalog number: 03-338-1E), 5 mm biopsy punches (Catalog number: NC9151828), needle-free valves (Catalog number: NC0332521), 4 way stopcocks (Catalog number: NC9052592), hydrophobic filters (SLFGL25BS), hydrophilic filters (SLMPL25SS), Polydimethylsiloxane (PDMS, Catalog number: NC9644388), tridecafluoro-1,1,2,2-tetrahydrooctyl trichlorosilane and 60 mL syringes were purchased from Fisher Scientific (Waltham, MA). Tubing (Catalog number: 5054K304) and male and female adaptors were purchased from McMaster Carr (Robbinsville, NJ). Uncoated glass bottom 96 well plates were purchased from MatTek Corp (Ashland, MA). 3D Kube perfusion chambers were obtained from Kiyatec (Greenville, SC).
2.2 Isolation and expansion of preadipocytes
Subcutaneous adipose tissue was obtained from elective abdominoplasty procedures (Tufts University IRB Protocol #0906007, first patient – 30 year old female with a BMI of 28, second patient – 50 year old female with a BMI of 27, both non diabetic) and was processed the same day of surgery as described previously [31] with some modifications. The adipose tissue was separated from the skin and the fascia of Scarpa by blunt dissection. The remaining adipose tissue was liquefied in a blender by successive short pulses to break up the tissue. An equal volume of pre-warmed phosphate buffered saline (PBS) was added to the blended adipose tissue and the resulting solution was allowed to phase separate for 3–5 min at room temperature. The lipid laden tissue floated above the aqueous phase and the infranatant (lower liquid phase) was aspirated. This wash step was repeated until the infranatant was clear of blood (~ 2 times). An equal volume of warm collagenase (PBS+1%BSA+0.1% collagenase type I) was then added, and the tubes were incubated for one hour (37°C, 5% CO2). Following the incubation step, the solution was centrifuged at 300 × g for 5 min at room temperature. The stromal vascular fraction pelleted at the bottom of the tube and the supernatant containing oil, primary adipocytes (yellow layer of floating cells) and the collagenase solution was aspirated. The pellet was resuspended in warm PBS with 1% BSA and was centrifuged again (300 × g for 5 min at room temperature). The pellet was resuspended in maintenance media (DMEM/F12, 10% fetal bovine serum, 1× Antibiotic-Antimycotic) and centrifuged for the final time (300 × g for 5 min at room temperature) and the cells were plated in flasks so that 0.16 mL of the original liquefied tissue was plated per cm2. Fourty-eight hours after plating, the media was aspirated and the flasks were washed with pre-warmed PBS before adding maintenance media. The media was changed every 2–3 days until cells were 80–90% confluent. Cells were then released from the plastic with trypsin, counted, and used for 2D and 3D experiments at passage 2. As the cells were obtained from patients on different days, all of the experiments were independently repeated at different times.
2.3 2D device fabrication and cell seeding
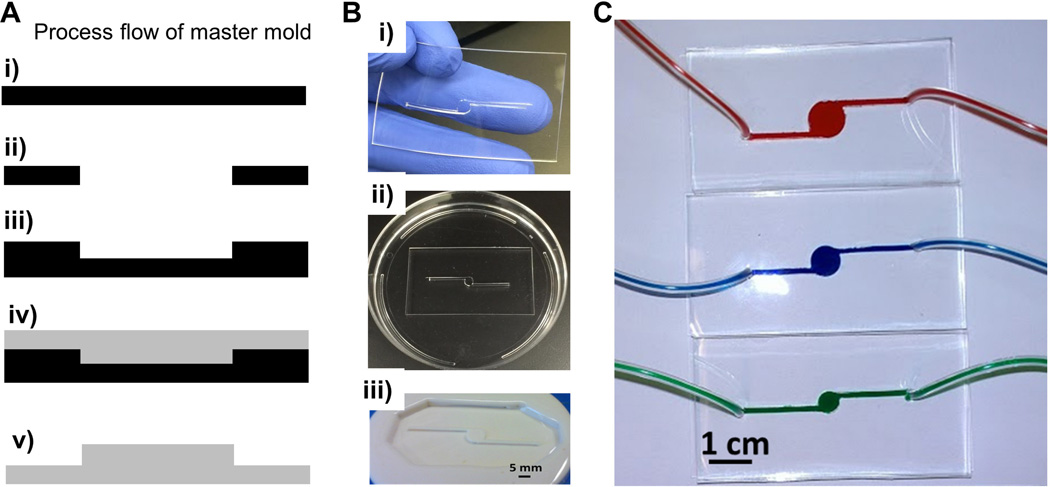
An epoxy mold was fabricated as shown in Figure 1. First, a thin film (1 mm) of polydimethylsiloxane (PDMS or Sylgard® 184) was formed by mixing the silicon elastomeric base with the curing agent at a 10:1 ratio. The solution was cured for 6 hours at 60°C. The desired feature of the device, a circular chamber with one inlet and outlet, was cut from the PDMS film using a 5 mm biopsy punch and a sharp blade. In this study, the large chamber size (5 mm) was chosen based on the concept that adipocytes secrete hydrophobic metabolites which may be difficult to transport through microscale PDMS-based devices [11]. Additionally, 5 mm was chosen to match the diameter of the glass bottom 96 well plates used as a static control. After treating both surfaces with an oxygen plasma gun (Rotaloc: Littleton, Colorado) for 30 seconds the PDMS film was bonded to another layer of PDMS. The bond was cured for 2 hours at 60°C. The surface of the PDMS mold was modified using a silanizing agent (tridecafluoro-1,1,2,2-tetrahydrooctyl trichlorosilane) to facilitate the de-molding process for the next step. Next, the design was transferred to an epoxy to create a master mold for the device. This process was essential for creating multiple devices with a consistent design. To create an epoxy mold, a two component fast curing epoxy - Smooth-Cast® 321 - was mixed at a 1:1 ratio and blended thoroughly to avoid bubbles in the plastic disposable container. The mixture was then poured over the PDMS mold and cured for 40 minutes at room temperature. The epoxy mold was then used to create PDMS devices by casting PDMS over the mold and allowing the solution to cure for 6 hr at 60°C. The PDMS was then peeled off from the epoxy mold and inlet/ outlet holes were punched out of the PDMS. The PDMS mold was bonded to a glass substrate by treating both surfaces (PDMS and Glass) with the oxygen plasma gun for 30 seconds. Devices were annealed for 2 hours at 60°C and autoclaved. Prior to cell seeding the devices were coated with laminin (2 µg/cm2) for 2 hours. The devices were rinsed with PBS and seeded with cells. An approximate density of 9,000 cells/cm2 [32] was seeded in the 2D devices and static culture (96 well glass bottom plates coated with laminin). Cells were incubated in the device for 2 hours before perfusion was initiated. Both static and perfused 2D cultures were grown in maintenance media to confluence and then differentiated for the remainder of the culture in differentiation media which consisted of DMEM/F12 supplemented with 10% fetal bovine serum, 1× antibioticantimycotic, 1 µM insulin, 0.5 mM IBMX, 1 µM dexamethasone and 0.05 mM indomethacin. Static culture media was changed twice a week (200 µL of media was used in each well of a 96 well plate).
Figure 1. 2D device fabrication for perfusion culture.
(A) Schematic illustration of the master mold fabrication process: (i) side view of the PDMS thin film, (ii) the desired feature was cut out of the PDMS, (iii) the PDMS with cutout was transferred to another PDMS surface, (iv) an epoxy was cured onto the PDMS mold and (v) the epoxy was de-molded from the PDMS serving as a master mold for the PDMS device. (B) Representative images of the (i) cut out feature in the PDMS film which was bonded to (ii) another layer of PDMS in a dish and used to create (iii) an epoxy mold with the desired feature. (C) The PDMS fluidic devices bonded to the glass substrate perfused with colored solutions through different diameter chambers demonstrating a leak proof device.
2.4 Silk scaffold fabrication
Silk solution was extracted from B. mori silkworm cocoons as described previously [33]. The aqueous silk solution was lyophilized and re-solubilized in a 17% w/v hexafluoro-2-propanol (HFIP) solution. To create a porous sponge structure, sieved salt crystals were used as a porogen (500–600 µm). Two mL of the silk/HFIP solution was poured over 6.8 grams of salt in a 20 mm diameter polyethylene vial. The hinged caps on the vials were closed and left in a fume hood for 2 days to ensure the HFIP/silk/salt was evenly distributed. To allow the HFIP to evaporate, the vials were opened for 1 day. After the HFIP had evaporated the vials were immersed in methanol overnight to induce a β sheet structure in the silk. The vials were then removed from the methanol and left in the hood to allow the methanol to evaporate for 1 day. Next the vials were placed in water to leach out the salt particles. Once the scaffolds were devoid of salt, they were removed from the containers, cut to size (5 mm height × 5 mm diameter) and autoclaved. Scaffolds were soaked overnight in maintenance media prior to cell seeding.
2.5 Seeding cells in the scaffolds and 3D perfusion devices
Media was aspirated out of the scaffolds immediately prior to cell seeding. The silk scaffolds are sponges that will absorb the media containing cells if they are dry. However, the scaffolds become brittle when they dry out, so this step has to be performed quickly. Twenty µL of maintenance media containing 200,000 cells was added to the top and bottom of each scaffold, for a total of 400,000 cells per scaffold. The cell seeded scaffolds were placed into 24 well plates for 2 hours in an incubator (37°C, 5% CO2) without media to allow the cells to attach to the scaffold. After the cells had time to attach, maintenance media was added. Following three days of growth in the maintenance media (statically) the scaffolds were either moved to the 3D Kube perfusion system or remained in static culture. Both static and perfused cultures were differentiated with the same differentiation media as 2D cultures for the remainder of the culture (3 or 6 weeks). Static culture media was changed twice a week (1mL of media was used in each well of a 24 well plate).
2.6 Perfusion of 2D and 3D devices
Perfusion systems were set up with a syringe pump (Pump Systems Inc.: Catalog number NE 1800, Farmingdale, NY) providing the differentiation media in line with a four way valve consisting of a hydrophilic filter, two hydrophobic filters for gas exchange, and a needle free valve. Tubes connected the gas exchanger to the inlet of either the 2D PDMS device or the 3D Kube device obtained commercially. From the outlet another tube was connected to a waste container where supernatant samples were collected to determine secretion of LDH and glycerol throughout culture. Since adipocytes are not directly exposed to shear forces in vivo, a low flow rate of 1.6 µL/min was used. In the literature flow rates for adipogenesis can vary from 0.3 µL/min [12] to 2–4 µL/min [14, 34, 35] in 2D perfusion devices. When chambers are inset and cells are not directly exposed to the flow, higher rates can be used ranging from 8 µL/min [36] to 80 µL/min [11]. Static and perfused systems were cultured for 3 or 6 weeks and then either processed for immunostaining, or lysed to determine DNA and triglyceride content.
2.7 Immunostaining
Cellular morphology was visualized in samples that were fixed with 10% neutral buffered formalin for 30 minutes. Samples were washed twice with phosphate buffered saline (PBS) containing 0.2% Triton-X100 and stained with AdipoRed (1:35), DAPI (1:1000), and Alexa flour 488 phalloidin (1:40) for 1 hour and rinsed with phosphate buffered saline twice. Constructs were imaged with an inverted Leica DMIRE2 confocal microscope PMTs (photomultiplier tubes) collected fluorescence signal from DAPI (405 nm/420–440 nm), Phalloidin (488 nm/500–520 nm), AdipoRed (488/564–616 nm), and naturally-autofluorescent scaffolds (all PMTs), which were given pseudocolors as described: DAPI/Blue, Phalloidin/Green, AdipoRed/Red, and Scaffold/appearing as violet. Z-stack images were acquired and processed using LeicaLAS software to create single average projection images.
2.8 DNA and Triglyceride Content
2D and 3D scaffold cultures were lysed in a Tris-EDTA buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.5) and finely minced using micro-dissection scissors. Samples were stored at −20°C until the assays were performed. Frozen samples were thawed and immediately assayed according to the manufacturer's protocol for DNA and triglyceride content with the Picogreen assay and with the EnzyChrom Triglyceride Assay Kit, respectively.
2.9 Quantification of secretion
Supernatant samples were collected and stored at −20°C until the assays were performed. Frozen samples were thawed and immediately assayed according to the manufacturer's protocol with the EnzyChrom Adipolysis Assay Kit for glycerol secretion and the LDH Assay Kit for LDH secretion.
2.10 Statistics
All experiments were repeated with 2 different patient samples (at least N=5 devices and static cultures for each). Statistics were performed with GraphPad Prism software (GraphPad, CA, USA). A two way Analysis of Variance (ANOVA) was performed for DNA content, triglyceride content and glycerol secretion, where the factors were culture condition (static vs perfusion culture in 2D and 3D) and time in culture (3 weeks vs 6 weeks). When there was a significant effect of either factor, a Tukey post-hoc test was performed between the different groups within the factors, respectively. When there was a significant interaction between the two factors in the two way ANOVA all of the groups were compared individually with a Tukey posthoc test. Significant differences were always defined as p<0.05.
3. Results and Discussion
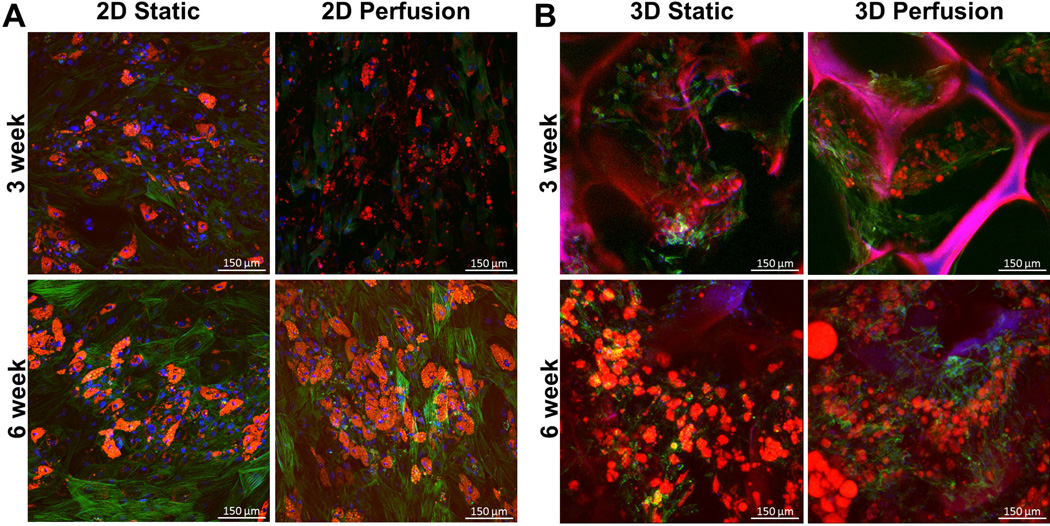
Maintenance of mature adipocytes ex vivo is difficult due to their fragile and buoyant nature. Therefore preadipocytes are often chosen for tissue engineering approaches to enhance their longevity in culture [2]. Preadipocytes undergo dramatic changes in morphology and accumulate lipids under adipogenic differentiation conditions. Staining with AdipoRed, a lipophilic dye was used to test the adipogenic potential of the 2D and 3D perfusion devices. Figure 2 demonstrates that adipogenesis was supported in both platforms (2D and 3D) at 3 and 6 weeks. Consistent with other findings [20] long term 3D perfusion supported larger, fused lipids more similar to the unilocular morphology seen in situ.
Figure 2. Adipogenesis is supported in 2D and 3D perfusion cultures at 3 and 6 weeks.
(A) 2D and (B) 3D preadipocytes were cultured in static or perfused environments and were stained with a lipophilic dye (AdipoRed, red = lipids), nuclear dye (DAPI, blue = nuclei), and cytoskeletal dye (phalloidin, green = actin). Silk fluorescence was visible in the blue and red wavelengths. All groups resulted in lipid laden cells. Scale bars are 150 µm.
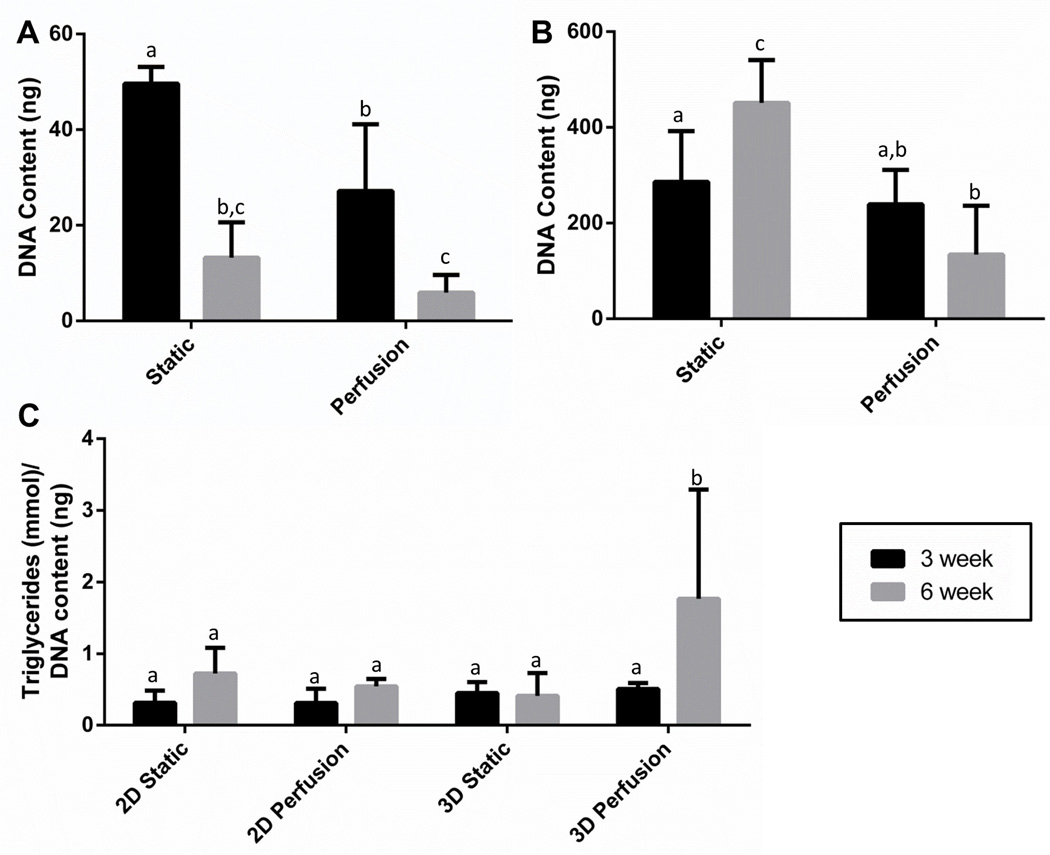
Next, we wanted to test if cell number was affected by extended culture or perfusion. Since 2D and 3D culture systems were seeded with different numbers of cells initially, changes in DNA content were not compared directly. As shown in Figure 3A, 2D cultures had a significant decrease in DNA content between static and perfused cultures. This is likely related to some of the cells detaching due to the shearing effects of fluid flow. This has been avoided in other culture platforms by creating inset chambers so the cells are not directly exposed to the shear stress [11, 36]. The current design could be easily amended to avoid shear stress by adding another PDMS chamber below the flow to create an inset design. However, in 2D cultures lipid laden cells also become buoyant, and detach from the surface and are often lost in media changes [10], which is consistent with the significant decrease observed in DNA content from 3 weeks to 6 weeks in 2D static and perfused culture systems. This drop in DNA content suggests that long term culture of adipogenic cells in 2D cultures will remain challenging with or without perfusion.
Figure 3. 3D cultures maintained DNA content and had greater triglyceride content after 6 weeks in perfusion culture, while 2D DNA content decreased in response to perfusion flow and over time.
(A) 2D cultures had decreased DNA content at longer time points (p<0.001) and due to perfusion (p<0.001) with no significant effect of the two factors interacting (p=0.052). (B) 3D cultures had a significant effect of the two factors interacting (p<0.001) and demonstrated an increase in DNA content in the static group and maintenance of DNA content in the perfusion group. (C) Triglyceride content also had a significant effect of the two factors interacting (p=0.007) and was highest in the 3D perfused group at 6 weeks. Groups sharing a common letter were not significantly different. Results represent combined data from repeated experiments with cells obtained from different patients. Error bars represent standard deviation.
In 3D matrices large lipid laden adipocytes are embedded in the matrix and are not lost in media changes [10]. In static cultures there was an increase in DNA content from 3 weeks to 6 weeks, while perfusion maintained DNA content over time (Figure 3B). This suggests that cells were not lost as was the case in the 2D conditions. In addition, reaching a steady state level in DNA content in the perfusion system is essential for studying disease mechanisms where cellular fluctuations will affect readouts. As the goal of this study was to compare culture conditions, DNA content was not measured at the time of seeding. Therefore, it’s unknown whether the cells proliferated initially or simply maintained cell numbers throughout culture in the 3D perfusion group.
Subsequently, we wanted to test whether the culture conditions could support the metabolic activity of adipocytes over long term culture. A hallmark of adipocyte lipid metabolism includes the storage of triglycerides (lipogenesis) as well as the breakdown of triglycerides into glycerol and free fatty acids (lipolysis) to meet energy needs [37]. In this study, 3D perfused scaffolds at 6 weeks had the highest triglyceride content compared to 3D static cultures, and 2D static and perfused cultures (Figure 3C). Additionally, perfused 3D cultures had an increase in triglyceride content from 3 weeks to 6 weeks that was not accompanied by an increase in cell number, suggesting additional lipogenesis was occurring with time in the 3D perfused culture system. The 3D static group on the other hand had an increase in DNA content without an increase in the ratio of triglyceride/DNA content indicating there were either more non-differentiated cells in the 3D static cultures or there were greater numbers of smaller adipocytes compared to the perfusion group.
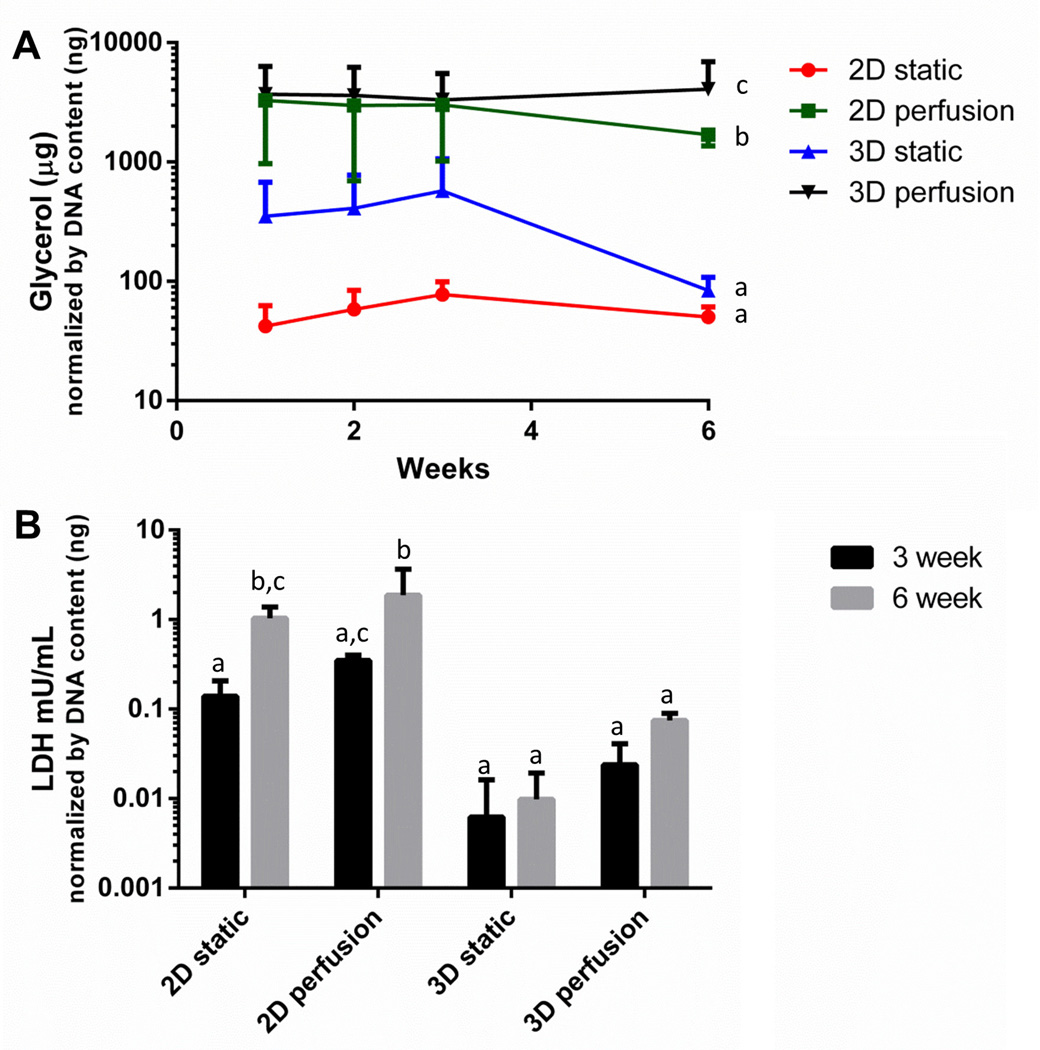
Supporting the concept that there were more differentiated cells in 3D perfused cultures, glycerol secretion was greatest in the 3D perfused group (Figure 4A). Additionally, there was an observed increase in glycerol secretion in 3D over 2D culture configurations, consistent with prior studies (without perfusion) that showed enhanced glycerol secretion in 3D relative to 2D [16]. Interestingly, regardless of culture configuration (2D or 3D) greater glycerol was secreted with the increased nutritional supply provided by perfusion of fresh media.
Figure 4. Glycerol secretion over 6 weeks of culture was the highest in the 3D perfusion culture condition, with minimal LDH release.
(A) Glycerol secretion was significantly affected by culture condition (p<0.001) but was not affected by time in culture (p=0.750) or significantly affected by the two factors interacting (p=0.711). (B) LDH is a stable cytoplasmic enzyme that is only released when the plasma membrane is damaged and was significantly affected by perfusion (p<0.001) and time in culture (p<0.001) with a significant effect of the two factors interacting (p<0.001). In general 3D cultures had lower LDH release. Groups sharing a common letter were not significantly different. Results represent combined data from repeated experiments with cells obtained from different patients. Error bars represent standard deviation.
Finally, LDH secretion was used as a marker of cell death in response to perfusion and length of culture (Figure 4B). LDH is a stable cytoplasmic enzyme that is only released when the plasma membrane is damaged [38]. Generally, 3D cultures secreted less LDH than 2D cultures. In addition, 2D cultures secreted higher amounts of LDH secretion with time in culture, reinforcing the concept that long term culture of adipogenic cells in 2D cultures with or without perfusion is not ideal.
While the long term culture of 2D differentiated preadipocytes in this study resulted in decreases in DNA content, lower levels of stored triglycerides and released glycerol, and higher LDH secretion; there are applications where longer term cultures may not be necessary. For instance, adipocytes have been included in microfluidic devices that isolate cell types to different compartments and fluidically link them with the ultimate goal of performing pharmacokinetic studies [13, 14]. 2D devices have also included adipocytes towards “human on a chip” applications with lung, liver and other tissue systems [14, 34] including kidney [14]. As shown in Figure 1 this process can be used to fabricate perfused devices of variable sizes, demonstrating utility for other applications at different scales.
4. Conclusions
We have established 2D and 3D perfusion culture techniques that support adipogenesis of preadipocytes. Consistent with other studies evaluating 2D and 3D culture systems for adipogenesis [10, 15–19] we found that both systems successfully model adipogensis, however 3D culture systems were more robust. 3D perfused cultures provided the mechanical structure required to contain large, fragile adipocytes that were lost in the 2D perfused culture system. 3D perfusion stimulated greater lipogenesis and lipolysis and resulted in decreased secretion of LDH compared to 2D perfusion. Regardless of culture configuration (2D or 3D) greater glycerol was secreted with the increased nutritional supply provided by perfusing fresh media constantly. These results are promising for adipose tissue engineering applications including long term cultures for studying disease mechanisms and regenerative approaches.
Highlights.
2D and 3D perfusion culture techniques supported adipogenesis of preadipocytes.
3D perfusion contained adipocytes that were lost in the 2D perfused cultures.
3D perfusion stimulated greater lipogenesis and lipolysis compared to 2D perfusion.
3D perfusion stimulated decreased amounts of LDH compared to 2D perfusion.
Greater glycerol was secreted with perfusion versus static cultures.
Acknowledgements
The authors wish to thank Dr. Sonal Pandya for providing the surgical specimens. The authors would also like to thank Dr. Irene Georgakoudi for the use of her confocal microscope. Finally, the authors would like to thank Francis Borowsky and Elisa Clark for technical assistance and Dr. Teresa DesRochers and Kiyatec for providing the 3D Kube culture systems. This work was funded by the NIH Tissue Engineering Resource Center (P41 EB002520) and AFIRM (W81XWH-08-2-0032).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
No conflicts of interest.
References
- 1.Patrick CW., Jr The Anatomical record. 2001;263:361–366. doi: 10.1002/ar.1113. [DOI] [PubMed] [Google Scholar]
- 2.Gomillion CT, Burg KJ. Biomaterials. 2006;27:6052–6063. doi: 10.1016/j.biomaterials.2006.07.033. [DOI] [PubMed] [Google Scholar]
- 3.Minteer DM, Gerlach JC, Marra KG. Journal of diabetes science and technology. 2014;8:1227–1232. doi: 10.1177/1932296814548215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaspar DA, Gomide V, Monteiro FJ. Biomatter. 2012;2:167–175. doi: 10.4161/biom.22170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mabvuure N, Hindocha S, Khan WS. Current stem cell research & therapy. 2012;7:287–292. doi: 10.2174/157488812800793018. [DOI] [PubMed] [Google Scholar]
- 6.Salter E, Goh B, Hung B, Hutton D, Ghone N, Grayson WL. Tissue engineering. Part B, Reviews. 2012;18:62–75. doi: 10.1089/ten.TEB.2011.0209. [DOI] [PubMed] [Google Scholar]
- 7.Oragui E, Nannaparaju M, Khan WS. The open orthopaedics journal. 2011;5(Suppl 2):267–270. doi: 10.2174/1874325001105010267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin I, Wendt D, Heberer M. Trends in biotechnology. 2004;22:80–86. doi: 10.1016/j.tibtech.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Sistino JJ. The Journal of extra-corporeal technology. 2003;35:200–202. [PubMed] [Google Scholar]
- 10.Daya S, Loughlin AJ, Macqueen HA. Differentiation; research in biological diversity. 2007;75:360–370. doi: 10.1111/j.1432-0436.2006.00146.x. [DOI] [PubMed] [Google Scholar]
- 11.Clark AM, Sousa KM, Jennings C, MacDougald OA, Kennedy RT. Analytical chemistry. 2009;81:2350–2356. doi: 10.1021/ac8026965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ni XF, Crozatier C, Sensebe L, Langonne A, Wang L, Fan Y, He PG, Chen Y. Microelectron Eng. 2008;85:1330–1333. [Google Scholar]
- 13.Viravaidya K, Shuler ML. Biotechnology progress. 2004;20:590–597. doi: 10.1021/bp034238d. [DOI] [PubMed] [Google Scholar]
- 14.Zhang C, Zhao Z, Abdul Rahim NA, van Noort D, Yu H. Lab on a chip. 2009;9:3185–3192. doi: 10.1039/b915147h. [DOI] [PubMed] [Google Scholar]
- 15.Cukierman E, Pankov R, Yamada KM. Current opinion in cell biology. 2002;14:633–639. doi: 10.1016/s0955-0674(02)00364-2. [DOI] [PubMed] [Google Scholar]
- 16.Stacey DH, Hanson SE, Lahvis G, Gutowski KA, Masters KS. Tissue engineering. Part A. 2009;15:3389–3399. doi: 10.1089/ten.TEA.2008.0293. [DOI] [PubMed] [Google Scholar]
- 17.Girandon L, Kregar-Velikonja N, Bozikov K, Barlic A. Folia biologica. 2011;57:47–56. doi: 10.14712/fb2011057020047. [DOI] [PubMed] [Google Scholar]
- 18.Grayson WL, Ma T, Bunnell B. Biotechnology progress. 2004;20:905–912. doi: 10.1021/bp034296z. [DOI] [PubMed] [Google Scholar]
- 19.Chun TH, Hotary KB, Sabeh F, Saltiel AR, Allen ED, Weiss SJ. Cell. 2006;125:577–591. doi: 10.1016/j.cell.2006.02.050. [DOI] [PubMed] [Google Scholar]
- 20.Gerlach JC, Lin YC, Brayfield CA, Minteer DM, Li H, Rubin JP, Marra KG. Tissue engineering. Part C, Methods. 2012;18:54–61. doi: 10.1089/ten.tec.2011.0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawasaki N, Asada R, Saito A, Kanemoto S, Imaizumi K. Scientific reports. 2012;2:799. doi: 10.1038/srep00799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Kim HJ, Vunjak-Novakovic G, Kaplan DL. Biomaterials. 2006;27:6064–6082. doi: 10.1016/j.biomaterials.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 23.Altman GH, Horan RL, Lu HH, Moreau J, Martin I, Richmond JC, Kaplan DL. Biomaterials. 2002;23:4131–4141. doi: 10.1016/s0142-9612(02)00156-4. [DOI] [PubMed] [Google Scholar]
- 24.Meinel L, Hofmann S, Karageorgiou V, Kirker-Head C, McCool J, Gronowicz G, Zichner L, Langer R, Vunjak-Novakovic G, Kaplan DL. Biomaterials. 2005;26:147–155. doi: 10.1016/j.biomaterials.2004.02.047. [DOI] [PubMed] [Google Scholar]
- 25.Choi JH, Bellas E, Gimble JM, Vunjak-Novakovic G, Kaplan DL. Tissue engineering. Part A. 2011;17:1437–1444. doi: 10.1089/ten.tea.2010.0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi JH, Gimble JM, Vunjak-Novakovic G, Kaplan DL. Tissue engineering. Part C, Methods. 2010;16:1157–1165. doi: 10.1089/ten.tec.2009.0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang JH, Gimble JM, Kaplan DL. Tissue engineering. Part A. 2009;15:2227–2236. doi: 10.1089/ten.tea.2008.0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward A, Quinn KP, Bellas E, Georgakoudi I, Kaplan DL. PloS one. 2013;8:e55696. doi: 10.1371/journal.pone.0055696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellas E, Marra K, Kaplan DLP. Tissue engineering. Part C, Methods. 2013 doi: 10.1089/ten.tec.2012.0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellas E, Panilaitis BJ, Glettig DL, Kirker-Head CA, Yoo JJ, Marra KG, Rubin JP, Kaplan DL. Biomaterials. 2013;34:2960–2968. doi: 10.1016/j.biomaterials.2013.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dubois SG, Floyd EZ, Zvonic S, Kilroy G, Wu X, Carling S, Halvorsen YD, Ravussin E, Gimble JM. Methods Mol Biol. 2008;449:69–79. doi: 10.1007/978-1-60327-169-1_5. [DOI] [PubMed] [Google Scholar]
- 32.Wagner W, Wein F, Seckinger A, Frankhauser M, Wirkner U, Krause U, Blake J, Schwager C, Eckstein V, Ansorge W, Ho AD. Experimental hematology. 2005;33:1402–1416. doi: 10.1016/j.exphem.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Rockwood DN, Preda RC, Yucel T, Wang X, Lovett ML, Kaplan DL. Nature protocols. 2011;6:1612–1631. doi: 10.1038/nprot.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viravaidya K, Shuler ML. Biotechnology progress. 2004;20:590–597. doi: 10.1021/bp034238d. [DOI] [PubMed] [Google Scholar]
- 35.Lai N, Sims JK, Jeon NL, Lee K. Tissue Eng Part C-Me. 2012;18:958–967. doi: 10.1089/ten.tec.2012.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark AM, Sousa KM, Chisolm CN, MacDougald OA, Kennedy RT. Analytical and bioanalytical chemistry. 2010;397:2939–2947. doi: 10.1007/s00216-010-3897-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen MD. The Journal of clinical endocrinology and metabolism. 2008;93:S57–S63. doi: 10.1210/jc.2008-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goergen JL, Marc A, Engasser JM. Cytotechnology. 1993;11:189–195. doi: 10.1007/BF00749869. [DOI] [PubMed] [Google Scholar]