Abstract
Proteomics study of pancreatic cancer using bodily fluids emphasizes biomarker discovery and clinical application, presenting unique prospect and challenges. Depending on the physiological nature of the bodily fluid and its proximity to pancreatic cancer, the proteomes of bodily fluids, such as pancreatic juice, pancreatic cyst fluid, blood, bile and urine, can be substantially different in terms of protein constitution and the dynamic range of protein concentration. Thus, a comprehensive discovery and specific detection of cancer-associated proteins within these varied fluids is a complex task, requiring rigorous experiment design and a concerted approach. While major challenges still remain, fluid proteomics studies in pancreatic cancer to date have provided a wealth of information in revealing proteome alterations associated with pancreatic cancer in various bodily fluids.
Keywords: proteomics, mass spectrometry, pancreatic cancer, pancreatic ductal adenocarcinoma, plasma, serum, pancreatic juice, pancreatic cyst fluid, bile, urine
Introduction
Pancreatic cancer is a highly lethal malignancy that is difficult to detect at early stages when curable treatments are most effective. It is the twelfth most common cancer and the seventh leading cause of cancer death in the world [1]. In the US, pancreatic cancer is the fourth leading cause of cancer death with 6% 5-year survival rate, and its incidence is on the rise [2]. Pancreatic ductal adenocarcinoma (PDAC) - an aggressive and devastating disease characterized by its poor prognosis and resistance to chemotherapy, accounts for 80–90% of pancreatic malignancies [2] (the term ‘pancreatic cancer’ refers to PDAC in this review). The high mortality of pancreatic cancer is predominantly due to the advanced stage of the disease at the time of initial diagnosis and a lack of effective treatments. Despite the advances in tomography techniques, such as computed tomography (CT) and endoscopic ultrasonography (EUS), it has been a challenge for diagnosing pancreatic cancer at early preinvasive stages, such as high-grade pancreatic intraepithelial neoplasia (PanIN 3) and invasive intraductal papillary mucinous neoplasms (IPMNs). Common genetic changes associated with pancreatic cancer, such as mutations in KRAS and TP53, are not reliable clinical parameters for predicting invasive pancreatic cancer. While mutations in KRAS present in >90% of PDAC, it alone is not sufficient for malignant transformation [3–5]. TP53 mutations also occur at high frequency in PDAC, but arise relatively late in the neoplastic progress towards the invasive pancreatic cancer [6–8]. CA19-9 is the current clinical blood-based biomarkers for pancreatic cancer testing – mainly used for monitoring the progress of pancreatic cancer treatment, but does not provide sufficient accuracy for diagnosis of pancreatic cancer. Biomarkers that can facilitate current pancreatic cancer diagnosis and early detection are desperately needed and would benefit the outcome of this deadly disease [9–11].
Pancreatic cancer-associated molecular events and signaling can induce pathway-driven changes in protein synthesis, post-translational modifications (PTMs), structure, degradation, subcellular localization, and impact protein interactions with proteins and other biomolecules, profoundly influencing cellular functions and biological processes at different levels. Such disease-associated protein alterations, discrete from genomic changes, are structurally or quantitatively reflected at the proteome level and may be detectable in relevant bodily fluids. In such context, proteomics, especially quantitative proteomics, has been increasingly applied in pancreatic cancer studies with research interests ranging from investigating disease mechanism to biomarker discovery [12–20]. These bodily fluids may include blood (plasma/serum), pancreatic juice, pancreatic cyst fluids, urine and bile. Depending on the nature of the bodily fluid, its proximity to the pancreatic neoplastic lesions, and its physiological functions, the proteome of different bodily fluids can vary significantly. Certain protein species that are characteristic to a specific aspect of the disease pathogenesis may be inherently enriched in one bodily fluid but less so in others, posing a variety of technical and analytical challenges to the discovery and detection of disease-associated protein alterations.
Mass spectrometry methods in fluid proteomics
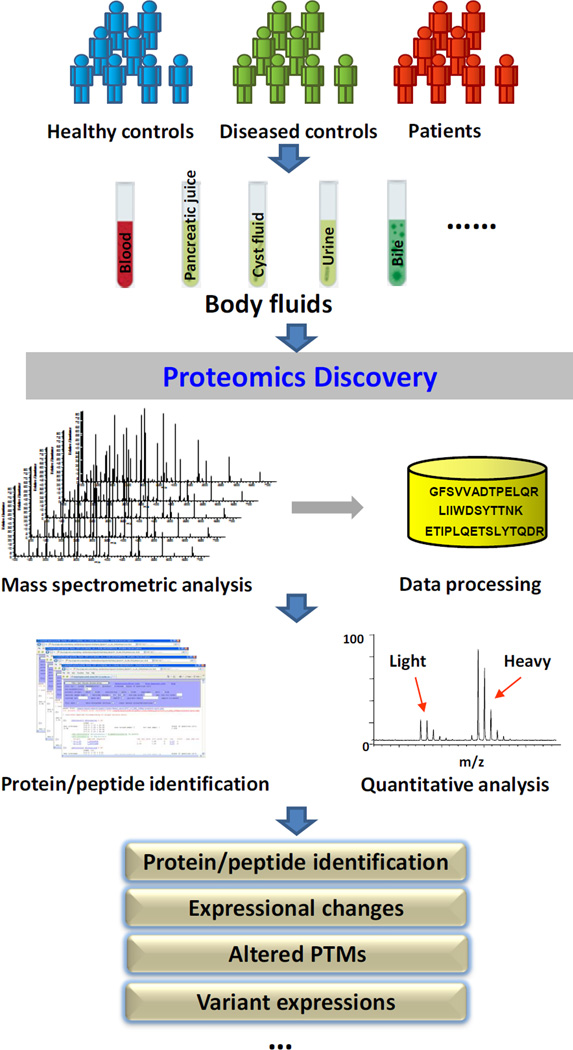
Proteomics has been widely applied in clinical studies, and in-depth overview of proteomics technology can be found in excellent reviews in the literature [21–28]. Unlike genomic analysis, in which the copy number of genes can be amplified using PCR to enhance analytical sensitivity, in proteomics, no such tool is feasible. Analysis of low abundant species in a complex sample requires high sensitivity and specificity, as well as sufficient dynamic range, posing unique analytical challenges for bodily fluid analysis. Mass spectrometry-based fluid proteomics can broadly be categorized into two platforms based on their applications: 1) data-intensive discovery-based proteomics which allows non-biased profiling of a proteome at a global scale; and 2) targeted proteomics which affords highly specific quantitative detection of selected candidates within a biological sample. As illustrated in Figure 1, proteomics discovery of bodily fluids for the study of pancreatic cancer involves a comparison of the proteomes of bodily fluid specimens from pancreatic cancer patients with diseased and non-diseased controls; and such comparison is based on the registry of protein and peptide species reliably identified in the specimens. A non-biased global profiling of diseased versus control samples can potentially reveal aberrant proteins associated with the disease, including proteins with abnormal expression levels, altered PTMs and sequence variants due to polymorphisms, alternative splicing, gene mutation or other DNA sequence alterations. Such a complex task is supported by not only an integrated analytical platform, but also a collection of sophisticated bioinformatics tools [25] for data transformation, database search, statistical validation and quantitative computation. While various experimental designs may be adapted to accommodate different sample types and study purposes, a discovery-based shotgun proteomics pipeline typically consists of the following modules: sample preparation, protein/peptide fractionation, mass spectrometric analysis, and peptide/protein identification and quantification. In the quantitative analysis of clinical specimens, chemical derivatization has been the most relevant and widely used method to label proteins or peptides from different sample origins with unique stable isotope tags to facilitate mass spectrometric analysis. The labeling methods may include isotopic techniques such as isotope-coded affinity tags (ICAT) [29], isotope-coded protein label (ICPL) [30] and dimethyl labeling [31], as well as isobaric techniques such as isobaric tags for relative and absolute quantitation (iTRAQ) [32] and tandem mass tags (TMT) [33]. O18 enzymatic labeling [34] can also be used in bodily fluid proteomics for quantitative analysis. In addition, label-free approaches have been increasingly applied in clinical proteomics studies. Separation techniques, such as electrophoreses, reverse-phase liquid chromatography (LC), ion-exchange and size-exclusion chromatography, as well as immuno-enrichment or -depletion are commonly applied to fractionate or enrich proteins or peptides prior to mass spectrometric analysis to reduce sample complexity, accommodate mass spectrometry capacity and enhance overall analytical sensitivity and depth. Recent technical advances in mass spectrometry, which is the most sophisticated part and critical aspect of a proteomics analysis, have significantly improved the instrumentation with better design, higher acquisition frequency and mass accuracy, positively impacting fluid proteomics analysis. For instance, the number of proteins reliably identified in human plasma has increased from hundreds a decade ago to thousands to date, with many low abundant tissue proteins now being uncovered, substantially increasing the depth and breadth of plasma proteomics. In addition, other recent innovations including ion mobility technique, which affords an additional dimension for further resolving a complex biological sample within a mass spectrometer [35], and soft ionization methods, such as electron-transfer dissociation (ETD) and electron-capture dissociation (ECD) [36] have enhanced proteomics analysis in many ways, including protein PTM characterization.
Figure 1.
Proteomics discovery of bodily fluids relevant to pancreatic cancer.
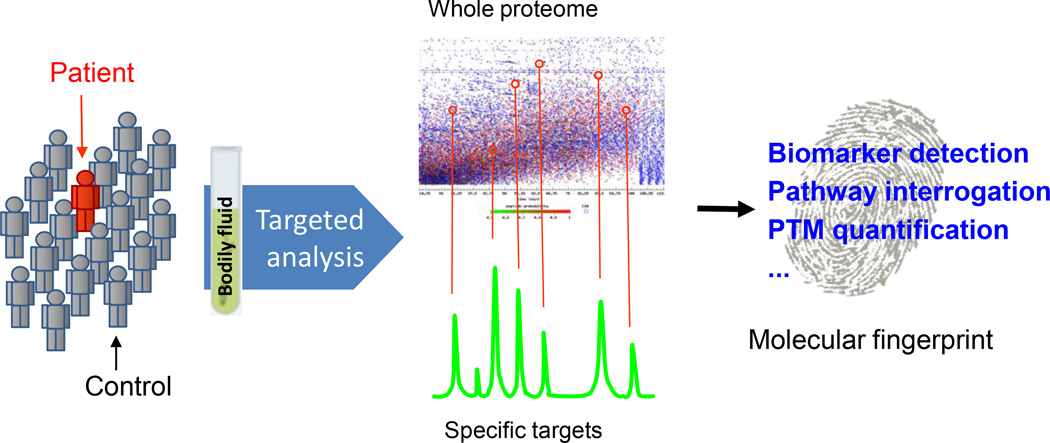
In contrast to non-biased protein profiling typically used for discovery studies, targeted analysis is a candidate-based proteomics technique, which can specifically detect selected peptides representing targeted proteins within a complex background, based on the mass and/or fragmentation signatures of a peptide [24,37–42]. This technique has rapidly emerged as an important tool for protein/peptide quantification, providing an alternative protein detection mechanism from antibody-based measurements, such as the enzyme-linked immunosorbent assay (ELISA). Figure 2 demonstrates the strategy of targeted proteomics, in which mass spectrometry assays can be developed and deployed to quantitatively interrogate an assortment of targeted proteins to measure their concentrations, to monitor their PTM levels and to detect their aberrant sequences in bodily fluids. While different instruments have been used for targeted peptide/protein analysis, currently, the triple quadrupole-based selected (or multiple) reaction monitoring (SRM or MRM) technique has been the most widely used approach [38,43–45]. Recently, the concept of data-independent-acquisition (DIA) has emerged and being increasingly applied for targeted proteomics analysis, representing a paradigm shift from conventional data-dependent-acquisition (DDA) approach [46,47]. The DIA methodologies have been implemented on multiple mass spectrometric platforms, including triple TOF-based SWATH (Sequential Window Acquisition of all Theoretical fragment-ion spectra) [48], Q-TOF-based MSE [49], and Orbitrap-based multiplexing strategy (MSX) [50]. For bodily fluid analysis, targeted proteomics renders a universal approach to develop highly specific, multiplexed mass spectrometry assays to assess a wide variety of protein/peptide candidates and their PTMs in a biological specimen, offering great potential for clinical application.
Figure 2.
Targeted proteomics for quantitative detection of selected candidates in bodily fluids.
Plasma/serum
Global protein profiling of blood plasma or serum from pancreatic cancer patients in comparison to diseased and non-diseased controls allows for identification of cancer-associated proteins or peptides. The major challenge in plasma/serum proteomics arises from its enormous complexity in protein constitution with the addition of PTMs and protein variants, as well as the vast dynamic range in protein concentration (>10-orders of magnitude) [51,52]. Many tissue proteins leaked or discharged from neoplastic lesions may be of low abundance in the circulatory system, challenging the scope and depth of current proteomics technology. Various proteomics work-flows have been applied to discover serological protein biomarkers associated with pancreatic cancer in plasma or serum.
Two-dimensional gel electrophoresis (2-DE) coupled with mass spectrometry has been used in several studies [53–60], to identify proteins with elevated concentration in the plasma/serum of pancreatic cancer patients, including haptoglobin [55], alpha-1-antitrypsin [55,58,59], leucine-rich alpha-2-glycoprotein [55,56], mannose-binding lectin 2 [57], myosin light chain kinase 2 [57], fibrinogen gamma [54], cyclin I [58], Rab GDP dissociation inhibitor beta [58], apolipoprotein E [60,61], alpha −1-antichymotrypsin [60], inter- alpha -trypsin inhibitor [60], apolipoprotein A1 [61], and transthyretin [61]. Many of the proteins identified by 2-DE approach are present at medium to high concentration in plasma/serum, reflecting the challenges in using 2-DE approach for plasma/serum profiling. Among these proteins, one study suggested that the changes in plasma concentration of apolipoprotein A1and apolipoprotein E might be associated with the confounding effects related to jaundice due to bile duct-obstruction [61].
More recently, shotgun proteomics has been applied to profile the plasma proteome of patients with pancreatic cancer compared to plasma samples from non-diseased and diseased controls. While some studies focused on developing quantitative techniques, such as iTRAQ and TMT, for plasma/serum profiling using pancreatic cancer as a disease model [62–64], other studies were aimed at translational study and sought to discover biomarkers associated with pancreatic cancer [65–67]. Two studies have quantitatively profiled the plasma proteomes of mouse models of human PDAC from different perspectives. In one study, the plasma samples from an engineered mouse model of human pancreatic cancer at early and advanced stage of disease were profiled in comparison to matched non-cancerous controls [65]. Based on the mouse proteomics results, the study then evaluated a group of the up-regulated protein candidates in the serum samples of newly diagnosed human PDAC patients; and demonstrated that measurements of ALCAM protein (ALCAM), intercellular adhesion molecule 1 (ICAM1), neutrophil gelatinase-associated lipocalin (LCN2), tissue inhibitor of metalloproteinase 1 (TIMP1), lithostathine 1 (REG1A), regenerating islet-derived protein 3 (REG3), and insulin-like growth factor-binding protein 4 (IGFBP4) as a panel had similar performance as CA19-9 when comparing PDAC to non-diseased controls, and outperformed CA19-9 when comparing PDAC to pancreatitis. The second mouse model study sought to identify alternative splice variant (ASV) proteins in the plasma proteome using a mouse model of PDAC [66]. The study identified 420 splice variant proteins in mouse plasma, including peptides from novel variants of muscle pyruvate kinase (Pkm), malate dehydrogenase 1 (Mdh1), glyceraldehyde-3-phosphate dehydrogenase (Gapdh), proteoglycan 4 (Prg4), minichromosome maintenance complex component 9 (Mcm9), high mobility group box 2 (Hmgb2), and hepatocyte growth factor activator (Hgfac).
Human plasma samples from patients with pancreatic cancer were also analyzed in comparison to plasma samples from non-diseased and chronic pancreatitis controls using quantitative proteomics. In one study, more than 1300 proteins were identified in human plasma with protein concentrations spanning across 8-orders of magnitude [67]. Many of these proteins were also found in pancreatic tissue (76%), reflecting the feasibility of detecting cancer-associated tissue leakage proteins in the blood [67]. Among the differentially expressed proteins identified in the plasma of PDAC patients in comparison with the non-disease and chronic pancreatitis controls, nine protein candidates, including TIMP1, ICAM1, C-C motif chemokine 5 (CCL5), zinc-alpha 2-glycoprotein 1 (AZGP1), lactotransferrin (LTF), apolipoprotein A-II (APOA2), thrombospondin-1 (THBS1), lipopolysaccharide-binding protein (LBP) and platelet basic protein (PPBP), were evaluated with an independent cohort of PDAC patients as well as diseased and non-diseased controls. The study demonstrated that a combination of TIMP1 and ICAM1 achieved a better accuracy than CA19-9 in distinguishing patients with PDAC from the non-disease and chronic pancreatitis controls; AZGP1 might be also valuable as a biomarker candidate for chronic pancreatitis if further validated. In a different study, high-definition mass spectrometry (HDMSE) was applied to identify serum alterations associated with patients with resectable pancreatic cancer compared to non-diseased controls and patients with benign pancreatic diseases using label-free quantification [68]. Among the 715 proteins identified, 40 showed a significant overexpression in the pancreatic cancer group based on hierarchical clustering analysis, including several p53 associated proteins, such as bromodomain adjacent to zinc finger domain protein 2A (BAZ2A), cell division protein kinase 13 (CDK13), death-associated protein kinase 1 (DAPK1), bullous pemphigoid antigen 1 (DST), exosome complex exonuclease RRP40 (EXOSC3), inhibin beta E chain (INHBE), histone acetyltransferase KAT2B (KAT2B), kinesin-like protein KIF20B (KIF20B), structural maintenance of chromosomes protein 1B (SMC1B), and astrin (SPAG5). In a different label-free proteomics study, CXC chemokine ligand 7 (CXCL7) were identified with a decreased level in the plasma of pancreatic cancer patients [69]. Many of the proteins elevated in the plasma from pancreatic cancer patients, such as TIMP1 [70–74], DAPK1 [75–79] and ICAM1 [80–84] have been previously associated with malignancy, and play multifaceted roles implicating the complex molecular events in cancer progression and metastasis. In addition to the global profiling studies, a combination of immuno-precipitation and mass spectrometry has been used to investigate the blood protein carriers of the CA19-9 antigen and identified several proteins associated with CA19-9, including apolipoprotein B-100 (APOB), kininogen (KNG1), armadillo repeat gene deleted in Velo-Cardio-Facial syndrome (ARVCF), and apolipoprotein E (APOE) [85]. In a different study, using immunoblot assays followed by mass spectrometric analysis, the reactivity of autoantibodies against DEAD-box protein 48 (DDX48) was detected in the sera of 33% pancreatic cancer patients [86].
Pancreatic juice
Pancreatic juice is secreted directly into the pancreatic ducts, making it a rich resource for proteins that are associated with PDAC. Although the content of pancreatic juice has been extensively studied decades ago and several major pancreatic enzymes have been identified [87,88], it was not until recently that a more comprehensive characterization of the pancreatic juice proteome was available owing to the development of proteomics techniques. The proteome of pancreatic juice has been characterized in patients with pancreatic adenocarcinoma [89–94], pre-cancer [95] and benign pancreatic diseases [90,96], as well as in individuals with no apparent pancreatic pathology [97]. These studies have identified hundreds of proteins in human pancreatic juice, including pancreatic enzymes and other pancreas associated proteins. We have shown that nearly 60% of the proteins identified in pancreatic juice were also present in plasma proteome (unpublished data). In comparison of pancreatic juices from patients with PDAC with non-cancerous controls, one study [89] identified a group of differential proteins in cancer samples, including kallikrein 1 (KLK1), IGFBP2, lithostathine 1 (REG1A, REG1B), pancreatic secretory granule membrane major glycoprotein (GP2), tumor-associated trypsin inhibitor (SPINK1), pancreatitis-associated protein 1 (PAP1), pancreatic ribonuclease (RNASE1), and T-cell receptor beta chain (TCRB). In addition, a differential gel electrophoresis (DIGE) based study identified the over-expression of matrix metalloproteinase-9 (MMP9), oncogene DJ1 (PARK7) and alpha-1B–glycoprotein (A1BG) in pancreatic juices from pancreatic cancer patients [94]. An iTRAQ-based quantitative proteomics study investigated the proteome of pancreatic juice samples collected from patients with primary precancerous PanIN 3 (high-grade dysplasia) lesions compared to non-diseased controls, and found anterior gradient-2 (AGR2) overexpressed in the juice of the PanIN 3 cases [95]. This finding corresponds to a different study that demonstrated the overexpression of AGR2 in the tissues of both PanIN lesions and pancreatic cancer [98]. AGR2 was further evaluated in the pancreatic juice of an independent cohort consisting of patients with PDAC, pre-malignant lesions (including PanIN 3, PanIN 2, IPMNs) and benign pancreatic diseases (including chronic pancreatitis) [95]. The elevated levels of AGR2 in the pancreatic juice were significantly associated with patients with pre-malignant conditions (PanIN lesions and IPMNs) and patients with PDAC comparing to benign controls, suggesting its potential value as a pancreatic juice biomarker for early detection of pancreatic cancer. Moreover, despite its low abundance in blood, a more recent investigation observed an increased AGR2 concentration in the plasma from PDAC patients compared to non-diseased controls [93].
Pancreatic cyst fluid
Pancreatic cystic lesions are increasingly detected with the widespread use of advanced imaging techniques. However, clinical management of pancreatic cystic lesions, such as IPMN, remains imprecise due to difficulties in accurately distinguishing lesions with high-grade dysplasia or invasive carcinoma from those that are lower grade and more benign. Currently available tests, including cross-sectional imaging, cytology and carcinoembryonic antigen (CEA), are inadequate to accurately diagnose pancreatic cyst lesions with malignant potential [99]. The development of proteomics techniques has stimulated considerable interest to comprehensively profile the proteome of pancreatic cyst fluids and identify potential biomarkers that can be used to guide early detection and surgical intervention. One study applied 2-DE and LC MS/MS to investigate the peptidome and proteome of cyst fluids collected from 20 patients using endoscopy ultrasound-fine needle aspiration [100]. The study identified more than 350 free peptides and 462 proteins in the cyst fluids. Approximately 137 common plasma proteins were identified in 13 out of the 20 pancreatic cyst fluids. The study suggested that the detection of protein family members of amylase, mucins, CEACAMs, and S100 proteins might facilitate the diagnosis of pancreatic cyst with malignant potential. In a different study, cyst fluids from eight patients with symptomatic pancreatic cystic lesions were collected by direct puncture during open surgery [101]. The cyst fluids were immunodepleted, fractionated with SDS-PAGE gels and analyzed by LC MS/MS. The study identified 220 to 727 proteins in the cyst fluids analyzed, and indicated that olfactomedin-4 (OLFM4) was differentially expressed in cyst fluids from potentially malignant lesions of mucinous cystic neoplasms (MCN) and IPMN, and mucin-18 (MUC18) was differentially expressed in neuroendocrine tumors (NET) cyst fluids.
Urine
Urine is an easily accessible bodily fluid that has been used for clinical testing. Although urine is derived from the kidneys, it can be viewed as an ultrafiltrate of plasma and also may contain valuable biomarkers that could assist with pancreatic cancer diagnosis. Using 2D DIGE and MALDI TOF mass spectrometer, proteomics analysis of urine samples collected from patients with PDAC, chronic pancreatitis and non-diseased controls have identified several differential proteins associated with PDAC from 127 statistically validated protein spots [102]. These proteins include CD59 glycoprotein (CD59), annexin A2 (ANXA2), 21 kDa gelsolin (GSN) fragment, protein S100-A9 (S100A9) and tumor necrosis factor alpha-induced protein 3 (TNFAIP3). Some of these proteins, such as ANXA2 and GSN, have been found overexpressed in pancreatic cancer tissues [17]. In a different study, immunoaffinity chromatography and mass spectrometry were applied to detect different variants of SPINK1 – a protein that has long been associated with chronic pancreatitis, in the urine samples collected from pancreatitis patients and non-diseased controls [103]. Although the pathophysiologic implications of SPINK1 variants in pancreatic cancer remains to be determined, the study demonstrated the precise detection of distinct SPINK1 genetic variants at the protein level in urine using a proteomics approach.
Bile
Bile is produced by the liver and discharged by biliary ducts into the duodenum to aid with the digestion of lipids. Biliary stenosis can be caused by pancreatic diseases, such as pancreatic adenocarcinoma (malignant) or chronic pancreatitis (benign). Proteomics investigation was carried out to profile the proteome of bile specimens from patients with pancreatic adenocarcinoma, cholangiocarcinoma, chronic pancreatitis and gallstones [104]. The study identified 127 proteins in bile using SDS-PAGE and LC MS/MS, and showed the overexpression of CEACAM6 and MUC1 in pancreatic cancer and cholangiocarcinoma samples based on the Western Blot analysis. This study confirmed the feasibility of detecting pancreatic cancer-associated proteins in bile fluid.
Analysis of protein glycosylation
Protein glycosylation plays pivotal roles in regulating a variety of protein functions involved in malignancy. Aberrant protein glycosylation has long been recognized as a molecular feature in epithelial cancer, including PDAC [105–108]. Identification of abnormal protein glycosylation associated with pancreatic cancer in bodily fluids may present meaningful targets for cancer detection. Using double lectin affinity chromatography and mass spectrometry, one study profiled N-linked glycans in pancreatic cancer serum compared with normal controls, identifying 44 oligosaccharides that were distinct in the pancreatic cancer serum, and observed increased protein fucosylation and sialylation associate with pancreatic cancer [109]. In a different investigation, a specific anti-sialyl Lewis X (SLex) antibody and mass spectrometry analysis were used to study the glycosylation of major serum acute-phase proteins (APP), including alpha-1-acid glycoprotein (AGP1), haptoglobin (HP), fetuin (AHSG), alpha-1-antitrypsin (SERPINA1) and transferrin (TF) [110]. The study observed an increase in SLex level and N-glycan branching on several APP in the sera from patients with advanced pancreatic cancer and chronic pancreatitis, and an increase in core fucosylation on AGP1 and HP in advanced pancreatic cancer patients. The sialylated N-glycopeptide levels in sera from pancreatic cancer patient were also evaluated compared to non-diseased controls and acute pancreatitis patients using Sambucus nigra lectin affinity chromatography [111]. The study identified 13 glycoforms, mainly from high-abundant serum proteins, with significant changes associated with pancreatic cancer group. The glycoproteome has also been investigated in cyst fluids collected from patients with MCN and IPMN, leading to the identification of 80 N-linked glycans, and several hyper-fucosylated glycoproteins, including triacylglycerol lipase and pancreatic α-amylase [112]. Notably, in addition to the alterations of glycan composition and structure, cancer-associated aberrant glycosylation can also involve changes in glycosylation site occupancy, which has been observed in pancreatic tumor tissues using quantitative glycoproteomics [113].
Targeted proteomics detection
Targeted proteomics was applied to assess the plasma concentration of five pancreatic cancer associated proteins, including LUM, GSN, TIMP1, transglutaminase 2 (TGM2) and SFN, using a plasma cohort consisting of patients with early stage PDAC (stage 1 and 2), chronic pancreatitis and non-diseased controls [114]. Four proteins except TGM2, which has a plasma concentration below the detection limit, were quantitatively measured by SRM. The receiver operating characteristic (ROC) analysis indicated that TIMP1, LUM and GSN had an area-under-curve (AUC) value greater than 0.75 in distinguishing pancreatic cancer plasma from the diseased and non-diseased controls. Targeted proteomics was also applied to analyze proline-hydroxylated α-fibrinogen in plasma using the SRM technique [115]. By measuring the concentration of proline-hydroxylated and unmodified α-fibrinogen in the plasma samples from pancreatic cancer patients and non-diseased controls, the study indicated that the percent hydroxylation of α-fibrinogen and concentration of hydroxylated α-fibrinogen were both significantly greater in the plasma of pancreatic cancer patients, including some of those with negative CA19-9 results.
Challenges and emerging technology
To date, the proteomics studies in blood and other pancreas-related bodily fluids have revealed a wealth of information regarding proteome alterations associated with pancreatic cancer. Some of these studies pioneered the methodology and discovery in fluid proteomics, providing a better understanding of the proteome of blood and other bodily fluids, as well as the intrinsic alterations relevant to pancreatic tumorigenesis. However, major challenges still remain in fluid proteomics. While a few protein targets have been the focus of further investigation for their potential clinical utility, much work remains on the majority of the protein candidates identified by the proteomics studies. Identification of an optimal surrogate marker that can provide satisfactory accuracy for general population screening of pancreatic cancer is not a trivial task, in part, due to the fact that pancreatic cancer is a low prevalent disease (4.2 per 100,000) [1] with almost no symptoms at its early stages. In addition, other pancreas diseases, such as chronic pancreatitis, diabetes and jaundice can confound the performance of a protein biomarker in pancreatic cancer detection. It has been demonstrated that the accuracy of a protein biomarker that had high specificity and sensitivity in distinguishing pancreatic cancer from normal control can be affected when chronic pancreatitis patients were included in the testing cohort [114,116]. This is because the two diseases share many common clinical and molecular features [90,117–120]. In fact, nearly 50% of differential proteins identified in pancreatic cancer tissue were also identified with concurrent expression in severe chronic pancreatitis in a tissue proteomics study [121]. Due to the complex mechanisms involved in pancreatic tumorigenesis, many oncoproteins participate in multiple molecular events implicating in pancreatic cancer, such as cancer initiation, metastasis, inflammation, fibrosis and immunoresponse, making it particularly challenging for developing a single protein biomarker for pancreatic cancer detection. It is generally agreed that a composite biomarker with an integration of a panel of complementary proteins may enhance the detection accuracy and provide better robustness. Pancreatic cancer biomarker candidates that can distinguish cancer from normal controls with high accuracy, but are associated with some confounding effects may still be valuable for biomarker panel construction, or alternatively, can be used to build a two-stage test that requires reflexive testing for those patients who test positive on the first stage of the test. In addition, targeting high-risk population, such as patients with chronic pancreatitis, genetic susceptibility and/or diabetes, may provide better efficiency and economic effect for early detection of pancreatic cancer [119,122].
Factors that can potentially influence the outcome of a fluid proteomics study also include biological heterogeneity, the nature of a bodily fluid, sampling and storage protocols, as well as differences inherent to the analytical methodologies. For example, the proteome of pancreatic juice can be influenced by the level of obstruction of the main pancreatic ducts due to cancer, inflammation or other diseases [123], or contamination due to the presence of bile or blood in the sample. The outcome of a fluid proteomics study can also be biased by the limited number of cases and controls included in the study. Biobanking, which includes specimen collection, transportation, processing and storage, is therefore an integrated component of clinical proteomics research. The availability and quality of specimens can greatly impact the progress in disease and biomarker study. Strategies, guidelines and standard operation protocols have been provided to facilitate biorepository effort to enhance clinical proteomics research [124–128]. For fluid proteomics studies that involve specimens from multiple institutions, a standardization process can be applied to account for the potential bias due to the various sample collection site [114,129,130]. While the discrepancies observed in some studies may be due, in part, to the factors aforementioned and the current limitations of proteomics technology, it also reflects the heterogeneous and dynamic nature of protein profiles present in these bodily fluids. The advances in mass spectrometry instrumentation, methodologies and bioinformatic tools are expected to improve the depth, breadth as well as reproducibility of fluid proteomics for future studies.
Acknowledgment
The author thanks Dr. Lisa Ann Lai for proofreading the manuscript. This work was supported in part with federal funds from the National Institutes of Health under grants K25CA137222, R21CA149772, R21CA161575 and R01CA180949, and funds from the Canary Foundation and Swim Across America.
Reference
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int.J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J.Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Berger DH, Chang H, Wood M, Huang L, Heath CW, Lehman T, Ruggeri BA. Mutational activation of K-ras in nonneoplastic exocrine pancreatic lesions in relation to cigarette smoking status. Cancer. 1999;85:326–332. doi: 10.1002/(sici)1097-0142(19990115)85:2<326::aid-cncr9>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 4.Hruban RH, Wilentz RE, Kern SE. Genetic progression in the pancreatic ducts. Am.J Pathol. 2000;156:1821–1825. doi: 10.1016/S0002-9440(10)65054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luttges J, Schlehe B, Menke MA, Vogel I, Henne-Bruns D, Kloppel G. The K-ras mutation pattern in pancreatic ductal adenocarcinoma usually is identical to that in associated normal, hyperplastic, and metaplastic ductal epithelium. Cancer. 1999;85:1703–1710. [PubMed] [Google Scholar]
- 6.Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 7.Redston MS, Caldas C, Seymour AB, Hruban RH, da Costa L, Yeo CJ, Kern SE. p53 mutations in pancreatic carcinoma and evidence of common involvement of homocopolymer tracts in DNA microdeletions. Cancer Res. 1994;54:3025–3033. [PubMed] [Google Scholar]
- 8.Ruggeri B, Zhang SY, Caamano J, DiRado M, Flynn SD, Klein-Szanto AJ. Human pancreatic carcinomas and cell lines reveal frequent and multiple alterations in the p53 and Rb-1 tumor-suppressor genes. Oncogene. 1992;7:1503–1511. [PubMed] [Google Scholar]
- 9.Ballehaninna UK, Chamberlain RS. Biomarkers for pancreatic cancer: promising new markers and options beyond CA 19-9. Tumour.Biol. 2013;34:3279–3292. doi: 10.1007/s13277-013-1033-3. [DOI] [PubMed] [Google Scholar]
- 10.Chakraborty S, Baine MJ, Sasson AR, Batra SK. Current status of molecular markers for early detection of sporadic pancreatic cancer. Biochim.Biophys.Acta. 2011;1815:44–64. doi: 10.1016/j.bbcan.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goggins M. Molecular markers of early pancreatic cancer. J.Clin.Oncol. 2005;23:4524–4531. doi: 10.1200/JCO.2005.19.711. [DOI] [PubMed] [Google Scholar]
- 12.Cecconi D, Palmieri M, Donadelli M. Proteomics in pancreatic cancer research. Proteomics. 2011;11:816–828. doi: 10.1002/pmic.201000401. [DOI] [PubMed] [Google Scholar]
- 13.Chen R, Pan S, Aebersold R, Brentnall T. Proteomics studies of pancreatic cancer. Proteomics Clin.Appl. 2007;1:1582–1591. doi: 10.1002/prca.200700414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen R, Pan S, Brentnall TA, Aebersold R. Proteomic profiling of pancreatic cancer for biomarker discovery. Mol.Cell Proteomics. 2005;4:523–533. doi: 10.1074/mcp.R500004-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Grantzdorffer I, Carl-McGrath S, Ebert MP, Rocken C. Proteomics of pancreatic cancer. Pancreas. 2008;36:329–336. doi: 10.1097/MPA.0b013e31815cc452. [DOI] [PubMed] [Google Scholar]
- 16.Omenn GS, Yocum AK, Menon R. Alternative splice variants, a new class of protein cancer biomarker candidates: findings in pancreatic cancer and breast cancer with systems biology implications. Dis.Markers. 2010;28:241–251. doi: 10.3233/DMA-2010-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan S, Brentnall TA, Kelly K, Chen R. Tissue proteomics in pancreatic cancer study: discovery, emerging technologies, and challenges. Proteomics. 2013;13:710–721. doi: 10.1002/pmic.201200319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun C, Rosendahl AH, Ansari D, Andersson R. Proteome-based biomarkers in pancreatic cancer. World J Gastroenterol. 2011;17:4845–4852. doi: 10.3748/wjg.v17.i44.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tonack S, Aspinall-O'Dea M, Neoptolemos JP, Costello E. Pancreatic cancer: proteomic approaches to a challenging disease. Pancreatology. 2009;9:567–576. doi: 10.1159/000212083. [DOI] [PubMed] [Google Scholar]
- 20.Vimalachandran D, Costello E. Proteomic technologies and their application to pancreatic cancer. Expert.Rev.Proteomics. 2004;1:493–501. doi: 10.1586/14789450.1.4.493. [DOI] [PubMed] [Google Scholar]
- 21.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 22.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 23.Gstaiger M, Aebersold R. Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat.Rev.Genet. 2009;10:617–627. doi: 10.1038/nrg2633. [DOI] [PubMed] [Google Scholar]
- 24.Huttenhain R, Malmstrom J, Picotti P, Aebersold R. Perspectives of targeted mass spectrometry for protein biomarker verification. Curr.Opin.Chem.Biol. 2009;13:518–525. doi: 10.1016/j.cbpa.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mueller LN, Brusniak MY, Mani DR, Aebersold R. An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J Proteome Res. 2008;7:51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- 26.Pan S, Chen R, Aebersold R, Brentnall TA. Mass spectrometry based glycoproteomics--from a proteomics perspective. Mol.Cell Proteomics. 2011;10:R110. doi: 10.1074/mcp.R110.003251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu.Rev.Biomed.Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Fonslow BR, Shan B, Baek MC, Yates JR., III Protein analysis by shotgun/bottom-up proteomics. Chem.Rev. 2013;113:2343–2394. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat.Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt A, Kellermann J, Lottspeich F. A novel strategy for quantitative proteomics using isotope-coded protein labels. Proteomics. 2005;5:4–15. doi: 10.1002/pmic.200400873. [DOI] [PubMed] [Google Scholar]
- 31.Hsu JL, Huang SY, Chow NH, Chen SH. Stable-isotope dimethyl labeling for quantitative proteomics. Anal.Chem. 2003;75:6843–6852. doi: 10.1021/ac0348625. [DOI] [PubMed] [Google Scholar]
- 32.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol.Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal.Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 34.Mirgorodskaya OA, Kozmin YP, Titov MI, Korner R, Sonksen CP, Roepstorff P. Quantitation of peptides and proteins by matrix-assisted laser desorption/ionization mass spectrometry using (18)O-labeled internal standards. Rapid Commun.Mass Spectrom. 2000;14:1226–1232. doi: 10.1002/1097-0231(20000730)14:14<1226::AID-RCM14>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 35.Baker ES, Burnum-Johnson KE, Jacobs JM, Diamond DL, Brown RN, Ibrahim YM, Orton DJ, Piehowski PD, Purdy DE, Moore RJ, Danielson WF, III, Monroe ME, Crowell KL, Slysz GW, Gritsenko MA, Sandoval JD, Lamarche BL, Matzke MM, Webb-Robertson BJ, Simmons BC, McMahon BJ, Bhattacharya R, Perkins JD, Carithers RL, Jr, Strom S, Self SG, Katze MG, Anderson GA, Smith RD. Advancing the high throughput identification of liver fibrosis protein signatures using multiplexed ion mobility spectrometry. Mol.Cell Proteomics. 2014;13:1119–1127. doi: 10.1074/mcp.M113.034595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JE, Shabanowitz J, Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochim.Biophys.Acta. 2006;1764:1811–1822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallien S, Duriez E, Domon B. Selected reaction monitoring applied to proteomics. J.Mass Spectrom. 2011;46:298–312. doi: 10.1002/jms.1895. [DOI] [PubMed] [Google Scholar]
- 38.Pan S, Aebersold R, Chen R, Rush J, Goodlett DR, McIntosh MW, Zhang J, Brentnall TA. Mass spectrometry based targeted protein quantification: methods and applications. J.Proteome.Res. 2009;8:787–797. doi: 10.1021/pr800538n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schiess R, Wollscheid B, Aebersold R. Targeted proteomic strategy for clinical biomarker discovery. Mol.Oncol. 2009;3:33–44. doi: 10.1016/j.molonc.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boja ES, Rodriguez H. Mass spectrometry-based targeted quantitative proteomics: achieving sensitive and reproducible detection of proteins. Proteomics. 2012;12:1093–1110. doi: 10.1002/pmic.201100387. [DOI] [PubMed] [Google Scholar]
- 41.Elschenbroich S, Kislinger T. Targeted proteomics by selected reaction monitoring mass spectrometry: applications to systems biology and biomarker discovery. Mol.Biosyst. 2011;7:292–303. doi: 10.1039/c0mb00159g. [DOI] [PubMed] [Google Scholar]
- 42.Meng Z, Veenstra TD. Targeted mass spectrometry approaches for protein biomarker verification. J Proteomics. 2011;74:2650–2659. doi: 10.1016/j.jprot.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 43.Gallien S, Duriez E, Domon B. Selected reaction monitoring applied to proteomics. J Mass Spectrom. 2011;46:298–312. doi: 10.1002/jms.1895. [DOI] [PubMed] [Google Scholar]
- 44.Maiolica A, Junger MA, Ezkurdia I, Aebersold R. Targeted proteome investigation via selected reaction monitoring mass spectrometry. J.Proteomics. 2012;75:3495–3513. doi: 10.1016/j.jprot.2012.04.048. [DOI] [PubMed] [Google Scholar]
- 45.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat.Methods. 2012;9:555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 46.Chapman JD, Goodlett DR, Masselon CD. Multiplexed and data-independent tandem mass spectrometry for global proteome profiling. Mass Spectrom.Rev. 2013 doi: 10.1002/mas.21400. [DOI] [PubMed] [Google Scholar]
- 47.Sajic T, Liu Y, Aebersold R. Using data-independent, high resolution mass spectrometry in protein biomarker research: Perspectives and clinical applications. Proteomics Clin.Appl. 2014 doi: 10.1002/prca.201400117. [DOI] [PubMed] [Google Scholar]
- 48.Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol.Cell Proteomics. 2012;11:O111. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li GZ, McKenna T, Nold MJ, Richardson K, Young P, Geromanos S. Quantitative proteomic analysis by accurate mass retention time pairs. Anal.Chem. 2005;77:2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 50.Egertson JD, Kuehn A, Merrihew GE, Bateman NW, MacLean BX, Ting YS, Canterbury JD, Marsh DM, Kellmann M, Zabrouskov V, Wu CC, Maccoss MJ. Multiplexed MS/MS for improved data-independent acquisition. Nat.Methods. 2013;10:744–746. doi: 10.1038/nmeth.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol.Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 52.Omenn GS. THE HUPO Human Plasma Proteome Project. Proteomics Clin.Appl. 2007;1:769–779. doi: 10.1002/prca.200700369. [DOI] [PubMed] [Google Scholar]
- 53.Abulaizi M, Tomonaga T, Satoh M, Sogawa K, Matsushita K, Kodera Y, Obul J, Takano S, Yoshitomi H, Miyazaki M, Nomura F. The application of a three-step proteome analysis for identification of new biomarkers of pancreatic cancer. Int.J.Proteomics. 2011;2011:628787. doi: 10.1155/2011/628787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bloomston M, Zhou JX, Rosemurgy AS, Frankel W, Muro-Cacho CA, Yeatman TJ. Fibrinogen gamma overexpression in pancreatic cancer identified by large-scale proteomic analysis of serum samples. Cancer Res. 2006;66:2592–2599. doi: 10.1158/0008-5472.CAN-05-3659. [DOI] [PubMed] [Google Scholar]
- 55.Deng R, Lu Z, Chen Y, Zhou L, Lu X. Plasma proteomic analysis of pancreatic cancer by 2-dimensional gel electrophoresis. Pancreas. 2007;34:310–317. doi: 10.1097/MPA.0b013e31802f2483. [DOI] [PubMed] [Google Scholar]
- 56.Kakisaka T, Kondo T, Okano T, Fujii K, Honda K, Endo M, Tsuchida A, Aoki T, Itoi T, Moriyasu F, Yamada T, Kato H, Nishimura T, Todo S, Hirohashi S. Plasma proteomics of pancreatic cancer patients by multi-dimensional liquid chromatography and two-dimensional difference gel electrophoresis (2D–DIGE): up-regulation of leucine-rich alpha-2-glycoprotein in pancreatic cancer. J.Chromatogr.B Analyt.Technol.Biomed.Life Sci. 2007;852:257–267. doi: 10.1016/j.jchromb.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rong Y, Jin D, Hou C, Hu J, Wu W, Ni X, Wang D, Lou W. Proteomics analysis of serum protein profiling in pancreatic cancer patients by DIGE: up-regulation of mannose-binding lectin 2 and myosin light chain kinase 2. BMC.Gastroenterol. 2010;10:68. doi: 10.1186/1471-230X-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun ZL, Zhu Y, Wang FQ, Chen R, Peng T, Fan ZN, Xu ZK, Miao Y. Serum proteomic-based analysis of pancreatic carcinoma for the identification of potential cancer biomarkers. Biochim.Biophys.Acta. 2007;1774:764–771. doi: 10.1016/j.bbapap.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 59.Wang Y, Kuramitsu Y, Yoshino S, Takashima M, Zhang X, Ueno T, Suzuki N, Oka M, Nakamura K. Screening for serological biomarkers of pancreatic cancer by two-dimensional electrophoresis and liquid chromatography-tandem mass spectrometry. Oncol.Rep. 2011;26:287–292. doi: 10.3892/or.2011.1278. [DOI] [PubMed] [Google Scholar]
- 60.Yu KH, Rustgi AK, Blair IA. Characterization of proteins in human pancreatic cancer serum using differential gel electrophoresis and tandem mass spectrometry. J.Proteome.Res. 2005;4:1742–1751. doi: 10.1021/pr050174l. [DOI] [PubMed] [Google Scholar]
- 61.Yan L, Tonack S, Smith R, Dodd S, Jenkins RE, Kitteringham N, Greenhalf W, Ghaneh P, Neoptolemos JP, Costello E. Confounding effect of obstructive jaundice in the interpretation of proteomic plasma profiling data for pancreatic cancer. J.Proteome Res. 2009;8:142–148. doi: 10.1021/pr800451h. [DOI] [PubMed] [Google Scholar]
- 62.Sinclair J, Timms JF. Quantitative profiling of serum samples using TMT protein labelling, fractionation and LC-MS/MS. Methods. 2011;54:361–369. doi: 10.1016/j.ymeth.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 63.Tonack S, Aspinall-O'Dea M, Jenkins RE, Elliot V, Murray S, Lane CS, Kitteringham NR, Neoptolemos JP, Costello E. A technically detailed and pragmatic protocol for quantitative serum proteomics using iTRAQ. J.Proteomics. 2009;73:352–356. doi: 10.1016/j.jprot.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 64.Zhou C, Simpson KL, Lancashire LJ, Walker MJ, Dawson MJ, Unwin RD, Rembielak A, Price P, West C, Dive C, Whetton AD. Statistical considerations of optimal study design for human plasma proteomics and biomarker discovery. J.Proteome Res. 2012;11:2103–2113. doi: 10.1021/pr200636x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Faca VM, Song KS, Wang H, Zhang Q, Krasnoselsky AL, Newcomb LF, Plentz RR, Gurumurthy S, Redston MS, Pitteri SJ, Pereira-Faca SR, Ireton RC, Katayama H, Glukhova V, Phanstiel D, Brenner DE, Anderson MA, Misek D, Scholler N, Urban ND, Barnett MJ, Edelstein C, Goodman GE, Thornquist MD, McIntosh MW, DePinho RA, Bardeesy N, Hanash SM. A mouse to human search for plasma proteome changes associated with pancreatic tumor development. PLoS.Med. 2008;5:e123. doi: 10.1371/journal.pmed.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Menon R, Zhang Q, Zhang Y, Fermin D, Bardeesy N, DePinho RA, Lu C, Hanash SM, Omenn GS, States DJ. Identification of novel alternative splice isoforms of circulating proteins in a mouse model of human pancreatic cancer. Cancer Res. 2009;69:300–309. doi: 10.1158/0008-5472.CAN-08-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pan S, Chen R, Crispin DA, May D, Stevens T, McIntosh MW, Bronner MP, Ziogas A, Anton-Culver H, Brentnall TA. Protein alterations associated with pancreatic cancer and chronic pancreatitis found in human plasma using global quantitative proteomics profiling. J.Proteome Res. 2011;10:2359–2376. doi: 10.1021/pr101148r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ansari D, Andersson R, Bauden MP, Andersson B, Connolly JB, Welinder C, Sasor A, Marko-Varga G. Protein deep sequencing applied to biobank samples from patients with pancreatic cancer. J Cancer Res Clin.Oncol. 2014 doi: 10.1007/s00432-014-1817-x. [DOI] [PubMed] [Google Scholar]
- 69.Matsubara J, Honda K, Ono M, Tanaka Y, Kobayashi M, Jung G, Yanagisawa K, Sakuma T, Nakamori S, Sata N, Nagai H, Ioka T, Okusaka T, Kosuge T, Tsuchida A, Shimahara M, Yasunami Y, Chiba T, Hirohashi S, Yamada T. Reduced plasma level of CXC chemokine ligand 7 in patients with pancreatic cancer. Cancer Epidemiol.Biomarkers Prev. 2011;20:160–171. doi: 10.1158/1055-9965.EPI-10-0397. [DOI] [PubMed] [Google Scholar]
- 70.Chirco R, Liu XW, Jung KK, Kim HR. Novel functions of TIMPs in cell signaling. Cancer Metastasis Rev. 2006;25:99–113. doi: 10.1007/s10555-006-7893-x. [DOI] [PubMed] [Google Scholar]
- 71.Grunnet M, Mau-Sorensen M, Brunner N. Tissue inhibitor of metalloproteinase 1 (TIMP-1) as a biomarker in gastric cancer: a review. Scand.J Gastroenterol. 2013;48:899–905. doi: 10.3109/00365521.2013.812235. [DOI] [PubMed] [Google Scholar]
- 72.Nielsen HJ, Brunner N, Frederiksen C, Lomholt AF, King D, Jorgensen LN, Olsen J, Rahr HB, Thygesen K, Hoyer U, Laurberg S, Christensen IJ. Plasma tissue inhibitor of metalloproteinases-1 (TIMP-1): a novel biological marker in the detection of primary colorectal cancer. Protocol outlines of the Danish-Australian endoscopy study group on colorectal cancer detection. Scand.J Gastroenterol. 2008;43:242–248. doi: 10.1080/00365520701523439. [DOI] [PubMed] [Google Scholar]
- 73.Ries C. Cytokine functions of TIMP-1. Cell Mol.Life Sci. 2014;71:659–672. doi: 10.1007/s00018-013-1457-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wurtz SO, Schrohl AS, Mouridsen H, Brunner N. TIMP-1 as a tumor marker in breast cancer--an update. Acta Oncol. 2008;47:580–590. doi: 10.1080/02841860802022976. [DOI] [PubMed] [Google Scholar]
- 75.Bialik S, Kimchi A. DAP-kinase as a target for drug design in cancer and diseases associated with accelerated cell death. Semin.Cancer Biol. 2004;14:283–294. doi: 10.1016/j.semcancer.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 76.Chen HY, Lee YR, Chen RH. The functions and regulations of DAPK in cancer metastasis. Apoptosis. 2014;19:364–370. doi: 10.1007/s10495-013-0923-6. [DOI] [PubMed] [Google Scholar]
- 77.Gozuacik D, Kimchi A. DAPk protein family and cancer. Autophagy. 2006;2:74–79. doi: 10.4161/auto.2.2.2459. [DOI] [PubMed] [Google Scholar]
- 78.Michie AM, McCaig AM, Nakagawa R, Vukovic M. Death-associated protein kinase (DAPK) and signal transduction: regulation in cancer. FEBS J. 2010;277:74–80. doi: 10.1111/j.1742-4658.2009.07414.x. [DOI] [PubMed] [Google Scholar]
- 79.Ng MH. Death associated protein kinase: from regulation of apoptosis to tumor suppressive functions and B cell malignancies. Apoptosis. 2002;7:261–270. doi: 10.1023/a:1015364104672. [DOI] [PubMed] [Google Scholar]
- 80.Johnson JP. The role of ICAM-1 in tumor development. Chem.Immunol. 1991;50:143–163. [PubMed] [Google Scholar]
- 81.Kotteas EA, Boulas P, Gkiozos I, Tsagkouli S, Tsoukalas G, Syrigos KN. The intercellular cell adhesion molecule-1 (icam-1) in lung cancer: implications for disease progression and prognosis. Anticancer Res. 2014;34:4665–4672. [PubMed] [Google Scholar]
- 82.Rosette C, Roth RB, Oeth P, Braun A, Kammerer S, Ekblom J, Denissenko MF. Role of ICAM1 in invasion of human breast cancer cells. Carcinogenesis. 2005;26:943–950. doi: 10.1093/carcin/bgi070. [DOI] [PubMed] [Google Scholar]
- 83.Shi X, Jiang J, Ye X, Liu Y, Wu Q, Wang L. Prognostic prediction and diagnostic role of intercellular adhesion molecule-1 (ICAM1) expression in clear cell renal cell carcinoma. J Mol.Histol. 2014;45:427–434. doi: 10.1007/s10735-014-9568-1. [DOI] [PubMed] [Google Scholar]
- 84.Zhu XW, Gong JP. Expression and role of icam-1 in the occurrence and development of hepatocellular carcinoma. Asian Pac.J Cancer Prev. 2013;14:1579–1583. doi: 10.7314/apjcp.2013.14.3.1579. [DOI] [PubMed] [Google Scholar]
- 85.Yue T, Partyka K, Maupin KA, Hurley M, Andrews P, Kaul K, Moser AJ, Zeh H, Brand RE, Haab BB. Identification of blood-protein carriers of the CA 19-9 antigen and characterization of prevalence in pancreatic diseases. Proteomics. 2011;11:3665–3674. doi: 10.1002/pmic.201000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xia Q, Kong XT, Zhang GA, Hou XJ, Qiang H, Zhong RQ. Proteomics-based identification of DEAD-box protein 48 as a novel autoantigen, a prospective serum marker for pancreatic cancer. Biochem.Biophys.Res Commun. 2005;330:526–532. doi: 10.1016/j.bbrc.2005.02.181. [DOI] [PubMed] [Google Scholar]
- 87.Goke B, Keim V, Dagorn JC, Arnold R, Adler G. Resolution of human exocrine pancreatic juice proteins by reversed-phase high performance liquid chromatography (HPLC) Pancreas. 1990;5:261–266. doi: 10.1097/00006676-199005000-00004. [DOI] [PubMed] [Google Scholar]
- 88.Beaudoin AR, St Jean P, Grondin G. Pancreatic juice composition: new views about the cellular mechanisms that control the concentration of digestive and nondigestive proteins. Dig.Dis. 1989;7:210–220. doi: 10.1159/000171221. [DOI] [PubMed] [Google Scholar]
- 89.Chen R, Pan S, Yi EC, Donohoe S, Bronner MP, Potter JD, Goodlett DR, Aebersold R, Brentnall TA. Quantitative proteomic profiling of pancreatic cancer juice. Proteomics. 2006;6:3871–3879. doi: 10.1002/pmic.200500702. [DOI] [PubMed] [Google Scholar]
- 90.Chen R, Pan S, Cooke K, Moyes KW, Bronner MP, Goodlett DR, Aebersold R, Brentnall TA. Comparison of pancreas juice proteins from cancer versus pancreatitis using quantitative proteomic analysis. Pancreas. 2007;34:70–79. doi: 10.1097/01.mpa.0000240615.20474.fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gronborg M, Bunkenborg J, Kristiansen TZ, Jensen ON, Yeo CJ, Hruban RH, Maitra A, Goggins MG, Pandey A. Comprehensive Proteomic Analysis of Human Pancreatic Juice. J.Proteome Res. 2004;3:1042–1055. doi: 10.1021/pr0499085. [DOI] [PubMed] [Google Scholar]
- 92.Lv S, Gao J, Zhu F, Li Z, Gong Y, Xu G, Ma L. Transthyretin, identified by proteomics, is overabundant in pancreatic juice from pancreatic carcinoma and originates from pancreatic islets. Diagn.Cytopathol. 2011;39:875–881. doi: 10.1002/dc.21484. [DOI] [PubMed] [Google Scholar]
- 93.Makawita S, Smith C, Batruch I, Zheng Y, Ruckert F, Grutzmann R, Pilarsky C, Gallinger S, Diamandis EP. Integrated proteomic profiling of cell line conditioned media and pancreatic juice for the identification of pancreatic cancer biomarkers. Mol.Cell Proteomics. 2011;10:M111. doi: 10.1074/mcp.M111.008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tian M, Cui YZ, Song GH, Zong MJ, Zhou XY, Chen Y, Han JX. Proteomic analysis identifies MMP-9, DJ-1 and A1BG as overexpressed proteins in pancreatic juice from pancreatic ductal adenocarcinoma patients. BMC.Cancer. 2008;8:241. doi: 10.1186/1471-2407-8-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen R, Pan S, Duan X, Nelson BH, Sahota RA, de Rham S, Kozarek RA, McIntosh M, Brentnall TA. Elevated level of anterior gradient-2 in pancreatic juice from patients with pre-malignant pancreatic neoplasia. Mol.Cancer. 2010;9:149. doi: 10.1186/1476-4598-9-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Paulo JA, Kadiyala V, Banks PA, Steen H, Conwell DL. Mass Spectrometry-Based (GeLC-MS/MS) Comparative Proteomic Analysis of Endoscopically (ePFT) Collected Pancreatic and Gastroduodenal Fluids. Clin.Transl.Gastroenterol. 2012;3:e14. doi: 10.1038/ctg.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Doyle CJ, Yancey K, Pitt HA, Wang M, Bemis K, Yip-Schneider MT, Sherman ST, Lillemoe KD, Goggins MD, Schmidt CM. The proteome of normal pancreatic juice. Pancreas. 2012;41:186–194. doi: 10.1097/MPA.0b013e31822862f6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ramachandran V, Arumugam T, Wang H, Logsdon CD. Anterior gradient 2 is expressed and secreted during the development of pancreatic cancer and promotes cancer cell survival. Cancer Res. 2008;68:7811–7818. doi: 10.1158/0008-5472.CAN-08-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kwon RS, Simeone DM. The use of protein-based biomarkers for the diagnosis of cystic tumors of the pancreas. Int.J.Proteomics. 2011;2011:413646. doi: 10.1155/2011/413646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ke E, Patel BB, Liu T, Li XM, Haluszka O, Hoffman JP, Ehya H, Young NA, Watson JC, Weinberg DS, Nguyen MT, Cohen SJ, Meropol NJ, Litwin S, Tokar JL, Yeung AT. Proteomic analyses of pancreatic cyst fluids. Pancreas. 2009;38:e33–e42. doi: 10.1097/MPA.0b013e318193a08f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cuoghi A, Farina A, Z’graggen K, Dumonceau JM, Tomasi A, Hochstrasser DF, Genevay M, Lescuyer P, Frossard JL. Role of proteomics to differentiate between benign and potentially malignant pancreatic cysts. J.Proteome Res. 2011;10:2664–2670. doi: 10.1021/pr2000557. [DOI] [PubMed] [Google Scholar]
- 102.Weeks ME, Hariharan D, Petronijevic L, Radon TP, Whiteman HJ, Kocher HM, Timms JF, Lemoine NR, Crnogorac-Jurcevic T. Analysis of the urine proteome in patients with pancreatic ductal adenocarcinoma. Proteomics Clin.Appl. 2008;2:1047–1057. doi: 10.1002/prca.200780164. [DOI] [PubMed] [Google Scholar]
- 103.Valmu L, Paju A, Lempinen M, Kemppainen E, Stenman UH. Application of proteomic technology in identifying pancreatic secretory trypsin inhibitor variants in urine of patients with pancreatitis. Clin.Chem. 2006;52:73–81. doi: 10.1373/clinchem.2005.056861. [DOI] [PubMed] [Google Scholar]
- 104.Farina A, Dumonceau JM, Frossard JL, Hadengue A, Hochstrasser DF, Lescuyer P. Proteomic analysis of human bile from malignant biliary stenosis induced by pancreatic cancer. J.Proteome Res. 2009;8:159–169. doi: 10.1021/pr8004925. [DOI] [PubMed] [Google Scholar]
- 105.Brooks SA, Carter TM, Royle L, Harvey DJ, Fry SA, Kinch C, Dwek RA, Rudd PM. Altered glycosylation of proteins in cancer: what is the potential for new anti-tumour strategies. Anticancer Agents Med.Chem. 2008;8:2–21. doi: 10.2174/187152008783330860. [DOI] [PubMed] [Google Scholar]
- 106.Dennis JW, Granovsky M, Warren CE. Glycoprotein glycosylation and cancer progression. Biochim.Biophys.Acta. 1999;1473:21–34. doi: 10.1016/s0304-4165(99)00167-1. [DOI] [PubMed] [Google Scholar]
- 107.Kobata A, Amano J. Altered glycosylation of proteins produced by malignant cells, and application for the diagnosis and immunotherapy of tumours. Immunol.Cell Biol. 2005;83:429–439. doi: 10.1111/j.1440-1711.2005.01351.x. [DOI] [PubMed] [Google Scholar]
- 108.Maupin KA, Sinha A, Eugster E, Miller J, Ross J, Paulino V, Keshamouni VG, Tran N, Berens M, Webb C, Haab BB. Glycogene expression alterations associated with pancreatic cancer epithelial-mesenchymal transition in complementary model systems. PLoS.One. 2010;5:e13002. doi: 10.1371/journal.pone.0013002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao J, Qiu W, Simeone DM, Lubman DM. N-linked glycosylation profiling of pancreatic cancer serum using capillary liquid phase separation coupled with mass spectrometric analysis. J.Proteome.Res. 2007;6:1126–1138. doi: 10.1021/pr0604458. [DOI] [PubMed] [Google Scholar]
- 110.Sarrats A, Saldova R, Pla E, Fort E, Harvey DJ, Struwe WB, de Llorens R, Rudd PM, Peracaula R. Glycosylation of liver acute-phase proteins in pancreatic cancer and chronic pancreatitis. Proteomics Clin.Appl. 2010;4:432–448. doi: 10.1002/prca.200900150. [DOI] [PubMed] [Google Scholar]
- 111.Kontro H, Joenvaara S, Haglund C, Renkonen R. Comparison of sialylated N-glycopeptide levels in serum of pancreatic cancer patients, acute pancreatitis patients, and healthy controls. Proteomics. 2014;14:1713–1723. doi: 10.1002/pmic.201300270. [DOI] [PubMed] [Google Scholar]
- 112.Mann BF, Goetz JA, House MG, Schmidt CM, Novotny MV. Glycomic and proteomic profiling of pancreatic cyst fluids identifies hyperfucosylated lactosamines on the N-linked glycans of overexpressed glycoproteins. Mol.Cell Proteomics. 2012;11:M111. doi: 10.1074/mcp.M111.015792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pan S, Chen R, Tamura Y, Crispin DA, Lai LA, May DH, McIntosh MW, Goodlett DR, Brentnall TA. Quantitative glycoproteomics analysis reveals changes in N-glycosylation level associated with pancreatic ductal adenocarcinoma. J Proteome Res. 2014;13:1293–1306. doi: 10.1021/pr4010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pan S, Chen R, Brand RE, Hawley S, Tamura Y, Gafken PR, Milless BP, Goodlett DR, Rush J, Brentnall TA. Multiplex targeted proteomic assay for biomarker detection in plasma: a pancreatic cancer biomarker case study. J.Proteome Res. 2012;11:1937–1948. doi: 10.1021/pr201117w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yoneyama T, Ohtsuki S, Ono M, Ohmine K, Uchida Y, Yamada T, Tachikawa M, Terasaki T. Quantitative targeted absolute proteomics-based large-scale quantification of proline-hydroxylated alpha-fibrinogen in plasma for pancreatic cancer diagnosis. J.Proteome Res. 2013;12:753–762. doi: 10.1021/pr3008144. [DOI] [PubMed] [Google Scholar]
- 116.Chen R, Crispin DA, Pan S, Hawley S, McIntosh MW, May D, Anton-Culver H, Ziogas A, Bronner MP, Brentnall TA. Pilot study of blood biomarker candidates for detection of pancreatic cancer. Pancreas. 2010;39:981–988. doi: 10.1097/MPA.0b013e3181dac920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen R, Brentnall TA, Pan S, Cooke K, Moyes KW, Lane Z, Crispin DA, Goodlett DR, Aebersold R, Bronner MP. Quantitative proteomics analysis reveals that proteins differentially expressed in chronic pancreatitis are also frequently involved in pancreatic cancer. Mol.Cell Proteomics. 2007;6:1331–1342. doi: 10.1074/mcp.M700072-MCP200. [DOI] [PubMed] [Google Scholar]
- 118.Lowenfels AB, Maisonneuve P, Cavallini G, Ammann RW, Lankisch PG, Andersen JR, Dimagno EP, Andren-Sandberg A, Domellof L. Pancreatitis and the risk of pancreatic cancer. International Pancreatitis Study Group. N.Engl.J.Med. 1993;328:1433–1437. doi: 10.1056/NEJM199305203282001. [DOI] [PubMed] [Google Scholar]
- 119.Malka D, Hammel P, Maire F, Rufat P, Madeira I, Pessione F, Levy P, Ruszniewski P. Risk of pancreatic adenocarcinoma in chronic pancreatitis. Gut. 2002;51:849–852. doi: 10.1136/gut.51.6.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rosty C, Geradts J, Sato N, Wilentz RE, Roberts H, Sohn T, Cameron JL, Yeo CJ, Hruban RH, Goggins M. p16 Inactivation in pancreatic intraepithelial neoplasias (PanINs) arising in patients with chronic pancreatitis. Am.J.Surg.Pathol. 2003;27:1495–1501. doi: 10.1097/00000478-200312000-00001. [DOI] [PubMed] [Google Scholar]
- 121.Pan S, Chen R, Stevens T, Bronner MP, May D, Tamura Y, McIntosh MW, Brentnall TA. Proteomics portrait of archival lesions of chronic pancreatitis. PLoS.One. 2011;6:e27574. doi: 10.1371/journal.pone.0027574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pannala R, Basu A, Petersen GM, Chari ST. New-onset diabetes: a potential clue to the early diagnosis of pancreatic cancer. Lancet Oncol. 2009;10:88–95. doi: 10.1016/S1470-2045(08)70337-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhou L, Lu Z, Yang A, Deng R, Mai C, Sang X, Faber KN, Lu X. Comparative proteomic analysis of human pancreatic juice: methodological study. Proteomics. 2007;7:1345–1355. doi: 10.1002/pmic.200600086. [DOI] [PubMed] [Google Scholar]
- 124.LaBaer J. Improving international research with clinical specimens: 5 achievable objectives. J Proteome Res. 2012;11:5592–5601. doi: 10.1021/pr300796m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Moore HM, Kelly AB, Jewell SD, McShane LM, Clark DP, Greenspan R, Hayes DF, Hainaut P, Kim P, Mansfield E, Potapova O, Riegman P, Rubinstein Y, Seijo E, Somiari S, Watson P, Weier HU, Zhu C, Vaught J. Biospecimen reporting for improved study quality (BRISQ) J Proteome Res. 2011;10:3429–3438. doi: 10.1021/pr200021n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Moore HM. The NCI Biospecimen Research Network. Biotech.Histochem. 2012;87:18–23. doi: 10.3109/10520295.2011.591833. [DOI] [PubMed] [Google Scholar]
- 127.Schrohl AS, Wurtz S, Kohn E, Banks RE, Nielsen HJ, Sweep FC, Brunner N. Banking of biological fluids for studies of disease-associated protein biomarkers. Mol.Cell Proteomics. 2008;7:2061–2066. doi: 10.1074/mcp.R800010-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tuck MK, Chan DW, Chia D, Godwin AK, Grizzle WE, Krueger KE, Rom W, Sanda M, Sorbara L, Stass S, Wang W, Brenner DE. Standard operating procedures for serum and plasma collection: early detection research network consensus statement standard operating procedure integration working group. J Proteome Res. 2009;8:113–117. doi: 10.1021/pr800545q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McIntosh MW, Drescher C, Karlan B, Scholler N, Urban N, Hellstrom KE, Hellstrom I. Combining CA 125 and SMR serum markers for diagnosis and early detection of ovarian carcinoma. Gynecol.Oncol. 2004;95:9–15. doi: 10.1016/j.ygyno.2004.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pepe MS, Longton G. Standardizing diagnostic markers to evaluate and compare their performance. Epidemiology. 2005;16:598–603. doi: 10.1097/01.ede.0000173041.03470.8b. [DOI] [PubMed] [Google Scholar]