SUMMARY
Experimental structure determination remains very difficult for G protein-coupled receptors (GPCRs). We propose a new hybrid protocol to construct GPCR structure models that integrates experimental mutagenesis data with ab initio transmembrane (TM) helix assembly simulations. The method was tested on 24 known GPCRs where the ab initio TM-helix assembly procedure constructed the correct fold for 20 cases. When combined with weak-homology and sparse mutagenesis restraints, the method generated correct folds for all the tested cases with an average C-alpha RMSD 2.4 Å in the TM-regions. The new hybrid protocol was applied to model all 1026 GPCRs in the human genome, where 923 have a high confidence score that are expected to have correct folds; these contain many pharmaceutically important families with no previously solved structures, including Trace amine, Prostanoids, Releasing hormones, Melanocortins, Vasopressin and Neuropeptide Y receptors. The results demonstrate new progress on genome-wide structure modeling of transmembrane proteins.
Keywords: G protein-coupled receptors, protein structure prediction, ab initio folding, human genome, mutagenesis experimental data
INTRODUCTION
G protein–coupled receptors (GPCRs) are integral membrane proteins which transmit chemical signals into a wide array of different cell types. Many diseases, including those associated with differentiation, proliferation, angiogenesis, cancer, development and cell survival, involve malfunctions of the receptors, which make GPCRs one of the most widely-used drug targets - accounting for over 40% of all FDA approved pharmaceuticals (Eglen et al., 2007). While knowledge of GPCR structures provides important information for function elucidation and drug design, experimental determination of 3D structures of GPCR proteins has proved to be extremely difficult. Significant efforts have been made on the technical improvement of GPCR expression and crystallization, which resulted in successful solution of 15 human GPCRs in the last eight years since 2007 (Jaakola et al., 2008; Rasmussen et al., 2007). Although remarkable, these only account for a small portion of all GPCRs in the human genome, which is estimated to be approximately one thousand (Takeda et al., 2002). The lack of atomic-level protein structure information for GPCRs has considerably hindered function annotation and structure-based drug discovery.
Significant efforts have also been made recently in the computational structure modeling of GPCR proteins with progress witnessed on both new method development and modeling accuracy (Fanelli and De Benedetti, 2011). For instance, Barth et al. developed a structure modeling method to assemble helix–helix packing of membrane proteins with limited constraints. In 4 out of 12 proteins, the method produced models of RMSD <4 Å to the X-ray structure (Barth et al., 2009). Chen et al. presented an interesting attempt to assemble protein transmembrane (TM) helices using distance restraints from sparse NMR paramagnetic relaxation enhancement data. Constrained with a simple geometry pattern, TM helix bundles up to 7 helices can be correctly constructed using 1 to 3 restraints (Chen et al., 2011). Yang et al. combined multiple machine learning classifiers for generating inter-TM helix contact predictions which have an average accuracy of 62% in the top L/5 predictions. When incorporated in fragment assembly simulations, the predicted inter-helix contact restraints increased the TM-score of the final GPCR models by 37% (Yang et al., 2013). The contact-assisted structure assembly approach has also been exploited by several recent modeling studies for GPCR and other TM proteins (Hopf et al., 2012; Nugent and Jones, 2012).
Despite these advances, the majority of computational approaches to GPCR modeling rely on the detection of homologous templates (Fanelli and De Benedetti, 2011; Zhang et al., 2006). It is well-known that pair-wise sequence identity between GPCR families is low, and close homologous templates are not available for most of the unknown GPCR families (Archer et al., 2003). Despite the limited availability of global X-ray structures, numerous mutagenesis experiments have been performed on GPCRs to identify the critical residues and motifs, which contain spatial information for improving the modeling accuracy of GPCR structures. For example, the coupled activation and deactivation of residues in mutagenesis experiments usually indicate that the residues are spatially adjacent because they are binding to common ligands (Shi and Javitch, 2002). Furthermore, the orientation of mutated functional residues is usually towards the inside core of the seven-helix bundle due to the conservation of inter-helix contacts (Schushan et al., 2010). Thus, specific contacts and distance maps and residue orientations can be derived from the mutagenesis experimental data and converted into 3D restraints to guide the GPCR structure modeling simulations. This is particularly helpful for the modeling of structurally variable regions that cannot be directly transferred by homology inference.
In this work, we aim to develop a new hybrid structure assembly algorithm, GPCR-I-TASSER, by extending the iterative threading assembly method (I-TASSER). The major advantages of GPCR-I-TASSER over existing homology-based methods are:
A new GPCR-specific database, GPCR-RD (Zhang and Zhang, 2010a) containing experimental contact and helix orientation data from the literature and database mining, is exploited to improve the structural assembly accuracy.
When homology templates are unavailable, a new ab initio folding method is introduced for assembling the TM-helix bundle topology from scratch.
A set of new GPCR- and transmembrane-specific energy terms is developed and incorporated into the I-TASSER force field to improve the structure assembly and refinement of both ab initio and threading template models. The major focus of this work is to construct reliable models for the GPCRs that lack close homologous templates.
To examine the efficiency, we first test GPCR-I-TASSER on all known GPCRs in the PDB and report the blind test results from the community-wide GPCRDock experiments. It was found that the new pipeline can significantly improve the modeling accuracy of template structure identified from threading. For GPCRs without homologous templates, the ab initio folding process can construct an approximately correct fold for all receptors with assistance from sparse mutagenesis data. The algorithm was finally applied to the modeling of all putative GPCRs in the human genome. The comparison with new mutagenesis data and confidence scoring system showed that nearly 90% of targets are expected to have correct folds, including many GPCRs from the families that have no previously solved experimental structures.
RESULTS
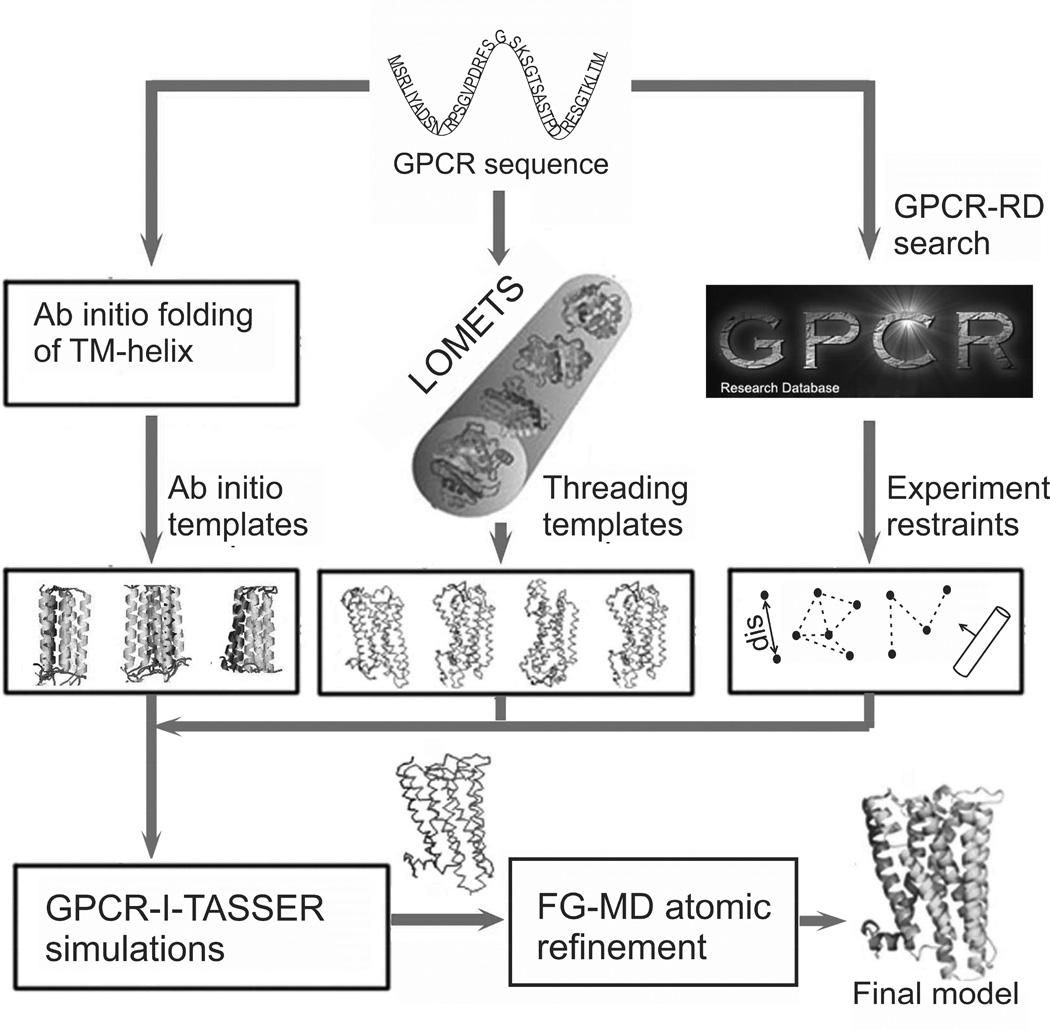
GPCR-I-TASSER, as depicted in Figure 1, has three steps consisting of template identification (or ab initio TM-helix construction) and experimental restraint collection, Monte Carlo fragment assembly simulation, and atomic-level structural refinement (see EXPERIMENTAL PROCEDURES (EP) and SUPPLEMENTAL EXPERIMENTAL PROCEDURES (SEP), for details).
Figure 1.
Flowchart of the GPCR-I-TASSER protocol for GPCR structure modeling.
Benchmark Test on 24 Solved GPCRs
To benchmark GPCR-I-TASSER, we collected a set of test structures containing all 24 GPCRs solved so far in the PDB. Since there are multiple entries solved for single GPCRs, we used CD-HIT (Fu et al., 2012) to remove the redundancy of these entries, which retains the entries having the longest structural coverage for each GPCR family. Table S1 lists the name and organism of the test GPCRs. Since many GPCRs were solved with fused external domains for facilitating crystal nucleation and structure determination, these domains have been excluded in our structure modeling. Table S2 lists the GPCR domains after manual trimming and the TM-helix annotations taken either from the original literature source or from manual inspection of the PDB structure. An updated list of all GPCRs solved in the PDB can be found at http://zhanglab.ccmb.med.umich.edu/GPCR-EXP/.
Distant homology modeling
We first tested GPCR-I-TASSER by excluding all homology templates which have a sequence identity to target >30% or are detectable by PSI-BLAST with an E-value <0.05. Despite the relatively stringent filters, many GPCR targets still have some analogous templates which can be detected by LOMETS (Wu and Zhang, 2007). The threading search generated templates with an average RMSD=5.74 (or 3.7) Å to the entire chain (or the TM-helix domains) of the native. The average TM-score of the templates is 0.675 (or 0.755). Here and afterwards, the RMSD is calculated on C-alpha atoms only. TM-score is a sequence length-independent metric for measuring structure similarity with a range [0, 1]. A TM-score >0.5 generally corresponds to similar structures in the same SCOP/CATH fold family (Xu and Zhang, 2010). Such a high TM-score of the template detection by LOMETS probably reflects the focus of the experimental efforts that have been made on a set of similar GPCRs; therefore templates can be inferred easily for the benchmark targets from other solved homologous GPCR structures. In fact, we have conducted a simple exercise by counting the homologous templates defined by the LOMETS alignments. The average number of homologous templates with a LOMETS Z-score above the confidence Z-score cutoff is 3.9 in this benchmark set, which is 2.4 times higher than the average for all other human GPCRs (1.6).
Despite the good quality of the threading alignments, GPCR-I-TASSER repacked the structure of the TM helices and drew the threading templates considerably closer to the native. Compared to the experimental structure, the first GPCR-I-TASSER models have the average RMSD reduced from 5.74 Å to 4.22 Å by 1.52 Å in the same threading alignment regions. The TM-score increased from 0.675 to 0.806 by 19.4%. A detailed list of the threading templates and GPCR-I-TASSER models is given in Table S3, where values in the parenthesis are RMSD and TM-score data in the TM-regions, and values after ‘/’ are RMSD of the GPCR-I-TASSER models in the threading aligned regions. A summary of the results is presented in Table 1.
Table 1.
Summary of the structure modeling on 24 benchmark GPCRs. Values in parenthesis are for the transmembrane region (see also Tables S1–S4 in Supplemental Information).
| Template filter | Methods | <RMSD> (Å) | <TM-score> |
|---|---|---|---|
| Excluding all homologous templates | Threadinga | 5.74 (3.70) | 0.675 (0.755) |
| MODELLERb | 8.07 (3.85) | 0.694 (0.764) | |
| GPCR-I-TASSER (ali)c | 4.22 (2.32) | ||
| GPCR-I-TASSERd | 5.09 (2.40) | 0.806 (0.868) | |
| Excluding all homologous & membrane protein templates | Threadinga | 12.46 (10.25) | 0.096 (0.102) |
| MODELLERb | 21.74 (11.42) | 0.142 (0.149) | |
| Ab Initio Folding (1)e | 11.39 (8.96) | 0.389 (0.389) | |
| Ab Initio Folding (B)f | 10.81 (8.31) | 0.412 (0.419) | |
| GPCR-I-TASSER (1)e | 8.57 (6.37) | 0.517 (0.517) | |
| GPCR-I-TASSER (B)f | 8.35 (6.25) | 0.524 (0.526) |
Best template by LOMETS;
MODELLER model based on the best template;
RMSD of the first model in the threading aligned region;
RMSD and TM-score of the first model in entire chain;
First model;
The best in top five models.
In Table S3, we also present the results by the widely-used comparative modeling tool, MODELLER (Sali and Blundell, 1993), based on the best LOMETS templates. Since MODELLER is designed to construct models by optimally satisfying spatial restraints from templates, there is not much improvement of the final models over templates. Compared to LOMETS templates, the average RMSD of the MODELLER models increases from 5.74 to 8.07 Å and TM-score increases from 0.675 to 0.694 in the TM region; these moderate RMSD/TM-score increases are probably mainly due to the length increase in the MODELLER modeling.
Goddard and colleagues developed a program MembStruk for GPCR structure prediction (Vaidehi et al., 2002). At the time of the MembStruk modeling, there was only one GPCR with experimental structure available (i.e., bovine rhodopsin). MembStruk built a model with a RMSD =3.1 Å in the TM-helix region and a RMSD =8.3 Å in full-length regions of bovine rhodopsin. As the models generated by MembStruk are not available publicly, we compare GPCR-I-TASSER with MembStruk on this GPCR only. As shown in Table S3, the RMSD of the GPCR-I-TASSER model for bovine rhodopsin (2hpyB) is 1.35/5.25 Å in the TM-helix/all regions, which is 1.75/3.05 Å lower than the MembStruk model. However, we note that this comparison might not be entirely fair because there are now more GPCR structures that can serve as templates. We have re-run GPCR-I-TASSER by excluding all GPCR templates (but keeping other membrane structures) in the template library, which resulted in the first predicted model of bovine rhodopsin with 1.82/6.31 Å in the TM-helix/all regions; these RMSD values are slightly higher than the data in Table S3 but still considerably lower than that of the MembStruck results.
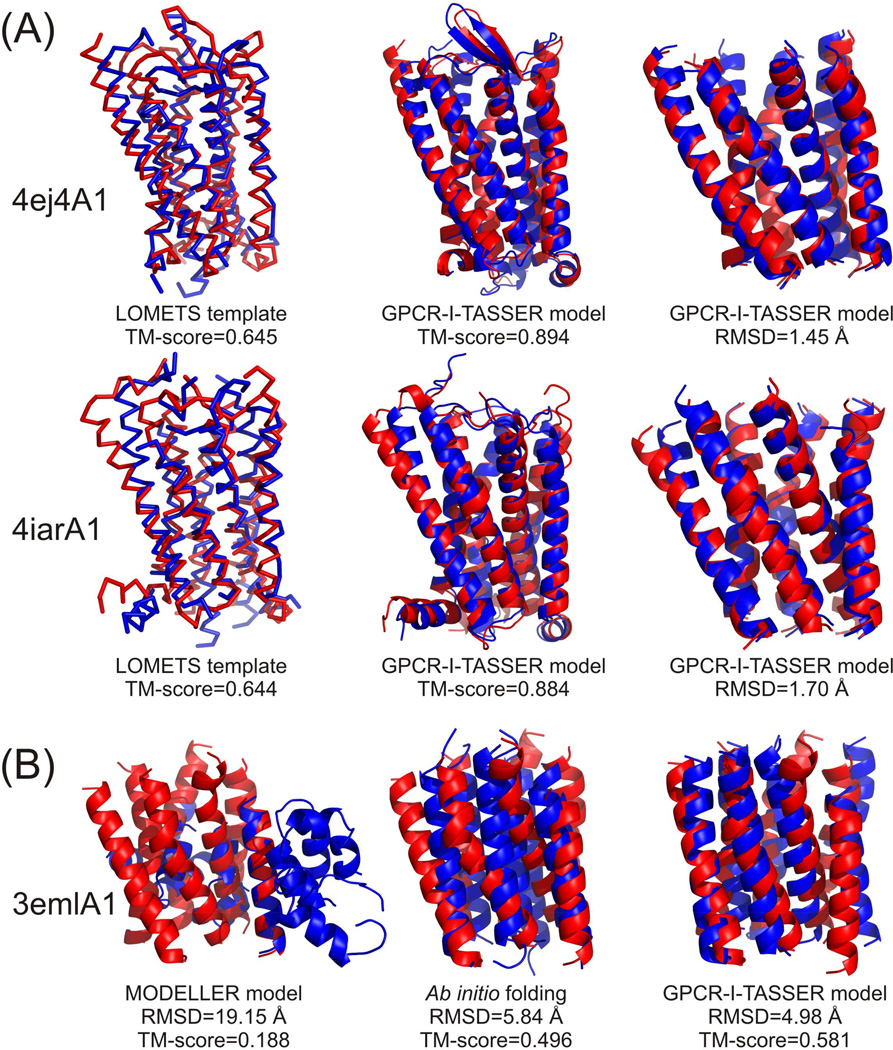
In Figure 2A, we present two examples from human opioid receptor (PDB ID:4ej4A1) (Granier et al., 2012) and human serotonin receptor (PDB ID: 4iarA1) (Wang et al., 2013), which represent two targets with the most significant structure refinements, where the threading templates have a TM-score=0.644 and 0.645 but GPCR-I-TASSER refined the models to TM-score=0.894 and 0.884, respectively. The major improvement occurs at the TM-helix regions, where the RMSD was reduced from 4.66 and 4.67 Å to 1.44 and 1.7 Å, respectively. This improvement is mainly attributed to the new GPCR-specific helical packing potential and the atomic level FG-MD refinements.
Figure 2.
Illustrative examples of GPCR structural modeling with blue and red representing model and experimental structure respectively. (A) Template based modeling on human opioid (PDB ID:4ej4A1) (Granier et al., 2012) and serotonin (PDB ID: 4iarA1) (Wang et al., 2013) receptors with close homologous templates detectable by PSI-BLAST or sequence identity >30% excluded. Left to right columns are the LOMETS templates and GPCR-I-TASSER models overlaid on the native in the whole chain and TM-helix regions respectively. (B) Ab initio modeling on human Adenosine A2a receptor (PDB ID: 3emlA1) (Jaakola et al., 2008) with all homologous and membrane templates excluded. Left to right columns are the models built by MODELLER, ab initio assembly and GPCR-I-TASSER models overlaid with TM regions of the native structure, respectively.
Compared to the TM-helix regions, the modeling of loop structure is more challenging since these regions are less conserved and the threading programs often have alignment gaps. In the 24 proteins, there are on average 7.9% of residues without threading alignments, which are mainly located on the loops/tails. The GPCR-I-TASSER pipeline constructs models for these regions by a lattice-based, ab initio structure assembly procedure extended from the I-TASSER protocol, which resulted in models with an average RMSD=5.37 Å for the 6 intra- and extra-cellular loops. For the functionally important second extracellular loop (EL2) that is often involved in ligand recognition and receptor activation, the average RMSD is 3.85 Å with an average length of 20.4 amino acids in this test.
It should be mentioned that the quality of template-based structure modeling is sensitive to the level of homologous template filtering. For instance, if we only filtered out the templates of sequence identity >30% (i.e., dropping off the PSI-BLAST E-value filter) as done in many previous benchmark experiments of structure prediction (Simons et al., 1999; Zhang and Skolnick, 2004a), the TM-score and RMSD of the threading templates will increase to 0.756 and 4.65 Å, respectively, while the quality of the GPCR-I-TASSER models will be improved accordingly with an average TM-score=0.912 and RMSD=3.21 Å (or 1.57 Å in the TM-helix and 3.35 Å in the loop regions).
Ab initio GPCR folding
Most GPCRs in the human genome are not closely homologous to the solved GPCRs in the PDB. To examine the ability of GPCR-I-TASSER in ab initio structure assembly, we exploited a second level of template filtering, i.e., to regenerate the models by excluding all GPCR and membrane proteins from our template library.
Since all correct templates have been excluded, it is expected that the templates detected by threading will now have a completely different topology from the native structures. The average TM-score of the templates with the highest Z-score is 0.096, which is well below the average of random structure pairs (0.17) (Xu and Zhang, 2010; Zhang and Skolnick, 2004b). When we applied MODELLER (Sali and Blundell, 1993) to these templates for full-length model construction using the default setting, a similar set of random models were obtained with an average TM-score=0.142 and RMSD =21.74 Å (Table 1). This is expected again because MODELLER was designed to construct structure models by satisfying spatial restraints from templates, an approach best suitable to the targets with close homologous templates.
To build a de novo TM-helix bundle topology, GPCR-I-TASSER first performs a rapid ab initio Monte Carlo assembly simulation with the conformational search guided mainly by the generic atomic contact and membrane transfer potentials (Eqs. S2 and S3 in SEP). The structural decoys were clustered by SPICKER (Zhang and Skolnick, 2004c), which resulted in the first ab initio models with an average TM-score=0.389 and RMSD=11.39 Å (Table 1). In 9 cases, the models have a TM-score >0.4, which indicates an approximately correct topology of the TM-helix assembly (Xu and Zhang, 2010). If we consider the best in the top five models, this number increases to 17 (see Table S4).
Starting from the ab initio TM-helix models and the low-resolution threading template alignments, GPCR-I-TASSER Monte Carlo simulations were conducted to reassemble the TM helices which have the relative orientations restricted by the loop structures. Meanwhile, 294 spatial restraints were extracted from the GPCR-RD database for the 24 test GPCRs. On average, 7 residue-residue contact restraints and 5 helix orientation restraints per target were used to constrain the simulations. This procedure generated full-length models with an average TM-score=0.517, which is 32% higher than that of the models created by ab initio folding. All the targets have a TM-score >0.4, and 20 out of 24 targets have a TM-score >0.5 (Table S4).
To test the effect of the mutagenesis restraints, we also ran a version of GPCR-I-TASSER without restraints from GPCR-RD. The average TM-score of the final model decreased by 3.9%. The TM-score reduction in this set of models was found considerably larger than that of the template-based models from the last section (1.4%); this is understandable because the mutagenesis restraints are implemented using a relatively large distance cutoff (i.e. dij <10 Å in Eq. S9) or with helix orientation adjustment (Eq. S10), which should have a stronger effect on refining models with low resolution.
To illustrate the procedure of ab initio folding, in Figure 2B we show the structural superposition of the predicted models for the Adenosine A2a receptor over the experimental structure (PDB ID: 3emlA1) (Jaakola et al., 2008) from the three modeling steps. The LOMETS programs hit incorrect templates which resulted in the MODELLER model with a different topology (TM-score=0.188). The ab initio folding procedure rearranged artificial helices and constructed a TM-helix bundle with approximately correct topology (TM-score=0.496). Finally, the GPCR-I-TASSER refinement simulations improved the structural model to a TM-score =0.581.
The data for the 24 benchmark proteins, including template alignments, ab initio folding and GPCR-I-TASSER models, are downloadable at http://zhanglab.ccmb.med.umich.edu/GPCR-I-TASSER/benchmark.
Blind Test in the GPCRDock Experiment
As a blind test of GPCR-I-TASSER, we participated (as “UMich/0460”) in the community-wide GPCR Structure-based Homology Modeling and Docking Assessment 2010 (or GPCRDock2010), organized by Kufareva et al (Kufareva et al., 2011). In the experiment, the organizers requested structure predictions for three GPCR-ligand complexes which were solved by Stevens and coworkers (Chien et al., 2010; Wu et al., 2010): the human CXCR4 chemokine receptor bound either to the small molecule antagonist IT1t or to the peptide antagonist CVX15, and the human dopamine D3 receptor with eticlopride. The predictions were blind, as the target structures were not released until the predictions were completed.
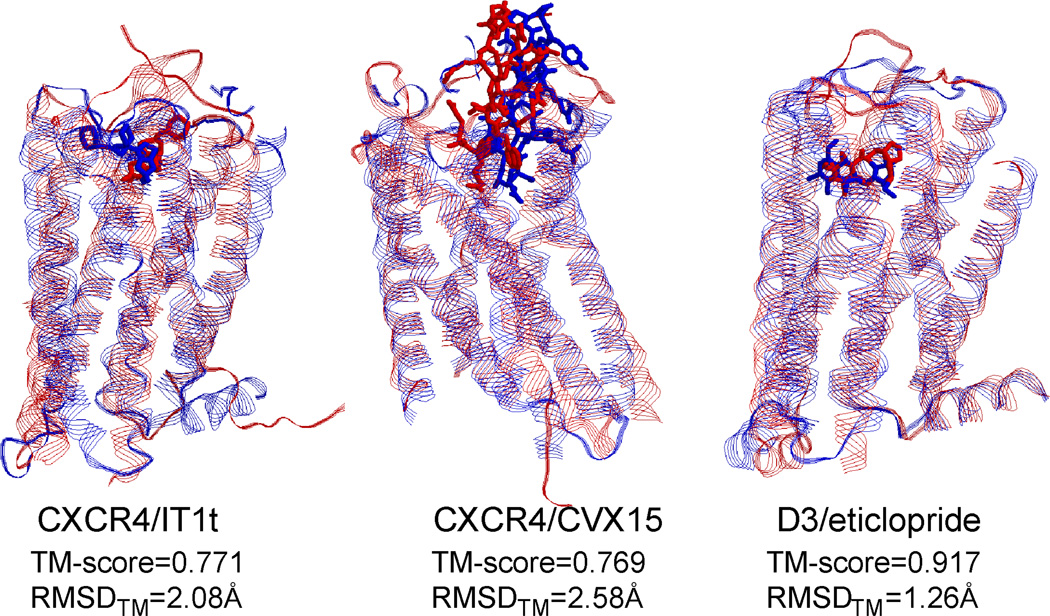
In Figure 3, we show the GPCR models built by the GPCR-I-TASSER pipeline in GPCRDock2010, where the GPCR-RD restraint data were not exploited. First, LOMETS threading identified B1AR and B2AR as the templates for the CXCR4 and D3 receptors, respectively, which have a TM-score of 0.695 and 0.627, respectively. The RMSDs of the templates in the threading-aligned region of the TM helices are 3.06 Å and 1.61 Å, respectively. After GPCR-I-TASSER reassembly, the final models have a TM-score =0.771, 0.768, 0.917 for CXCR4/IT1t, CXCR4/CVX15, and D3/eticlopride, respectively, which are 11%, 11% and 46% higher than the initial templates. In the same threading-aligned TM-region, RMSDs of the final models are 2.08 Å, 2.58 Å, and 1.26 Å, respectively, which are 0.98 Å, 0.48 Å, and 0.35 Å lower than the initial templates. These results confirm that GPCR-I-TASSER has the ability to draw threading templates considerably closer to the native structure.
Figure 3.
Ligand-receptor docking models generated by GPCR-I-TASSER and BSP-SLIM in GPCR-Dock 2010 (Kufareva et al., 2011). Blue and red represent predicted models and experimental structures, respectively. Left to right columns are models for CXCR4 chemokine receptor with IT1t (PDB ID: 3oe6) (Wu et al., 2010), CXCR receptor with CVX15 (PDB ID: 3oe0) (Wu et al., 2010), and dopamine D3 with eticlopride (PDB ID: 3pbl) (Chien et al., 2010), respectively. TM-score listed is for the whole-chain model and RMSDTM listed is the deviation of the model in the TM region relative to the native.
The ligand-bound GPCR models were generated by BSP-SLIM (Lee and Zhang, 2012), which first identified the ligand-binding pocket positions on the receptor protein by structurally aligning the receptor models to known complex structures in the PDB using TM-align (Zhang and Skolnick, 2005). The ligand-docking models were then generated from a conformational search with ligands constrained in the predicted binding pocket. The ligand-GPCR binding energy in BSP-SLIM consists of hydrogen-bonding, statistical contact potential, solvation and van der Waals interactions. The RMSDs of the final ligand models by BSP-SLIM are 9.61, 7.35, and 3.51 Å, for CXCR4/IT1t, CXCR4/CVX15, and D3/eticlopride, respectively (Figure 3).
Table S5 lists the top ten groups in GPCRDock2010 based on the cumulative Z-scores of the receptor and ligand models for all three targets. Among the 35 participant groups, the UMich-Zhang/0460 groups using GPCR-I-TASSER had the highest Z-score in the receptor models and the second highest in the ligand-docking positions, which resulted in the highest total Z-score of receptor and ligand models, according to the analysis by Kufareva et al. (Kufareva et al., 2011). The most noticeable success is on the distant homologous target CXCR4/CVX15, whereby the assessors commented in the assessment article that “Modeling the CXCR4/CVX15 peptide complex represented the biggest challenge of GPCR Dock 2010. The top model of this complex (#5 by UMich-Zhang, Figure 8C) has the Z-score of 2.4, thus far exceeding other models in accuracy” (Kufareva et al., 2011). For the two other less challenging targets (CXCR4/IT1t and D3/eticlopride), however, although the TM-backbone RMSD of the receptor models is ranked at the top for both targets, the accuracy of the functionally important EL2 and the ligand docking score are considerably worse than the top performing groups (http://ablab.ucsd.edu/GPCRDock2010/), highlighting the need to improve EL2 modeling and BSP-SLIM docking.
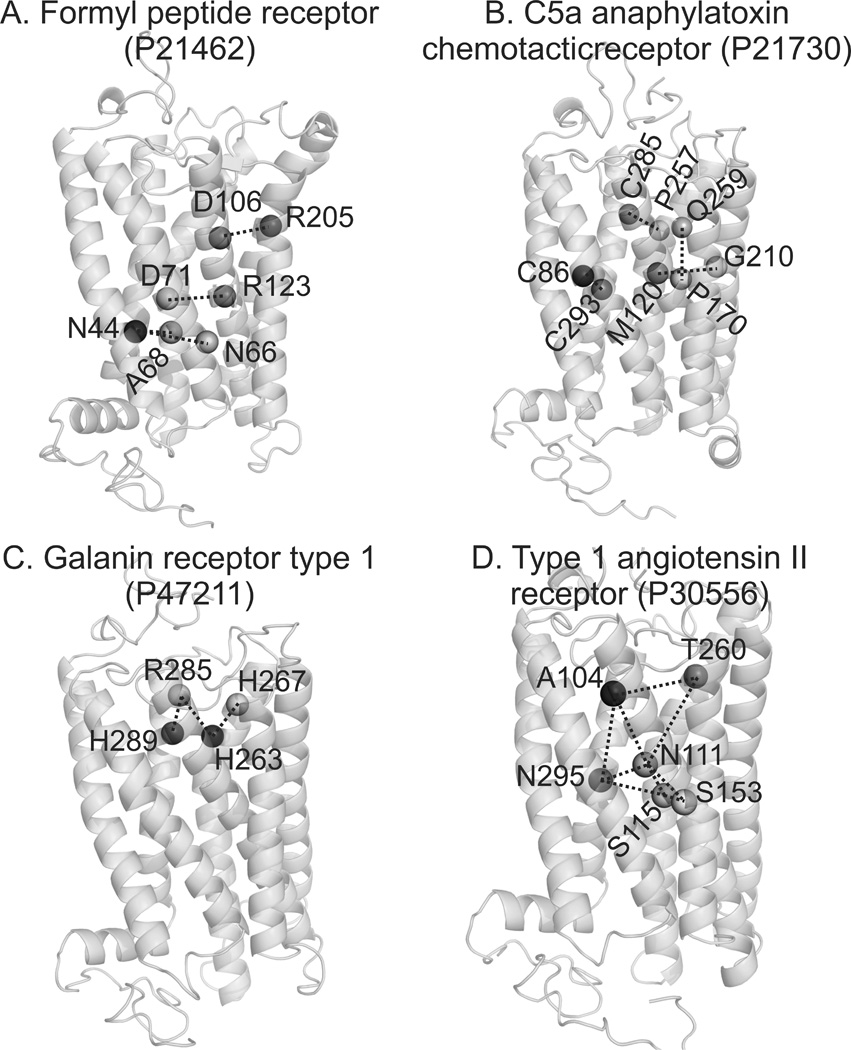
Figure 8.
Examples of the first GPCR-I-TASSER models in comparison with experimental mutagenesis data. (A) Formyl peptide receptor; (B) C5a anaphylatoxin chemotactic receptor; (C) Type 1 galanin receptor; (D) Type-1 aniotensin II receptor. Dashed lines connect the residue pairs supposed to be in contact in the mutagenesis data which are all within 10 Å in the GPCR-I-TASSER models.
We note that the GPCRDock experiment aims to benchmark the modeling of GPCR-ligand complexes with an emphasis on the ligand-docking technique. The receptor structure submitted by the other groups may not reflect the best receptor models due to the consideration of ligand-docking interactions. Nevertheless, the data provides a partial but independent assessment of GPCR-I-TASSER on the GPCR structure modeling in comparison with other state-of-the-art approaches.
Structure Modeling of 1026 GPCRs in the Human Genome
GPCR-I-TASSER modeling
A total of 1063 distinct GPCR sequences in the human genome were collected by scanning the databases GPCRDB (http://www.gpcr.org/7tm/data/) and UNIPROT (http://www.uniprot.org/docs/7tmrlist). Since errors often exist in automated data collection, we used a semi-manual procedure to examine these GPCR sequences: First, we generated TM-helix prediction by three TM prediction programs from HMMTOP (Tusnady and Simon, 1998), MEMSAT (Jones et al., 1994) and TMHMM (Krogh et al., 2001). If the number of TM helices predicted by any of the programs is <7 or the number of overlapped residues between the TM regions by the three programs is <5, we manually examined these sequences (about 400) by checking the UniProt annotation on the TM helices. In case there is no UniProt annotation, we used the GPCR-I-TASSER structure models to extract the TM helices. With this manual verification, we identified 37 non-GPCR sequences where most of them are extracellular domains attached to the receptor but mis-classified as GPCRs. Finally, 1026 validated GPCR sequences were retained for GPCR-I-TASSER modeling.
The GPCR sequences were first threaded through the PDB library using LOMETS (Wu and Zhang, 2007). In 862 cases, at least one of the programs used by LOMETS identified template structures with a significant Z-score above the corresponding program’s confidence cutoff. For the rest of proteins, we constructed the initial TM-helix bundle conformations using the ab initio folding procedure.
In the next step, we collected the sparse experimental data from GPCR-RD (Zhang and Zhang, 2010a), a manually curated database containing multiple GPCR data on site-directed mutagenesis, electron microscopy, neutron diffraction, FTIR, and disulfide bridge. The experimentally identified disulfide bridges and functionally important residues (binding to a particular ligand) indicate that these residues should be close to each other to perform their functions. So we applied contact restraints to these residue pairs as described in Eq. S9. Besides, the majority of the functionally related point mutations should face to the inside core of the TM-helix bundle (Schushan et al., 2010), which are used to guide the packing of helix orientations as described in Eq. S10. These resulted in 3425 contacts and 1401 orientation restraints for the 1026 human GPCRs. These restraints, together with the threading alignments and ab initio TM-helix models, were used to guide the GPCR-I-TASSER assembly simulations. The atomic details were finally refined by the fragment-guided molecular dynamics simulation program, FG-MD (Zhang et al., 2011).
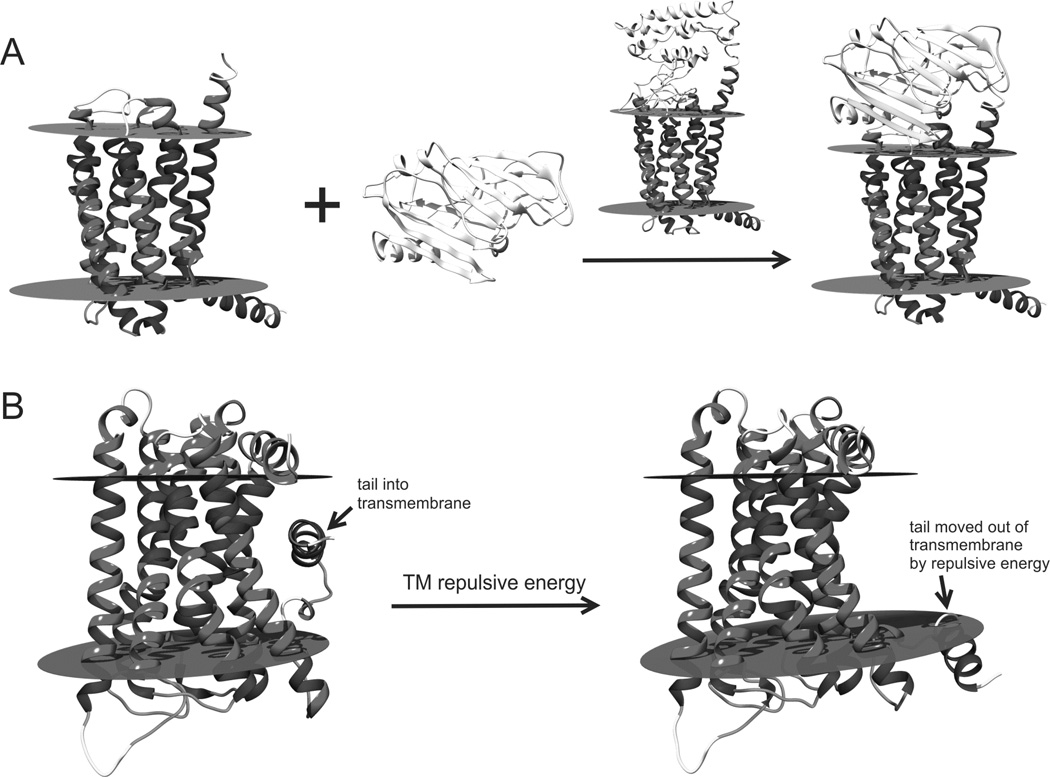
For the sequences containing extra domains, which are detected by TheaDom (Xue et al., 2013), models are created for each domain individually using GPCR-I-TASSER (for transmembrane domain) or I-TASSER (for globular domain). The full-length GPCR models are then constructed by assembling the domain structures as described in SEP. This domain parsing and assembly procedure can improve the confidence score and modeling accuracy of the individual domains as demonstrated in previous benchmark tests (Zhang, 2014). A multiple domain example from Q6ZMI9, which contains a transmembrane and a globular domain, is presented in Figure 4A, where the domain parsing and assembly procedure increased the C-score (defined below) from −1.79 of the full-chain GPCR-I-TASSER model to 1.11 for the globular domain and 1.32 for the GPCR domain, respectively.
Figure 4.
Illustration examples of the multi-domain assembly and TM-structure packing. (A) Multi-domain modeling for UniProt ID: Q6ZMI9. The N-terminal domain was excised from the UniProt sequence following the ThreaDom linker prediction. This globular domain and the core TM-domain were modeled separately by I-TASSER and GPCR-I-TASSER; the final model was assembled from the structure of two domains with the domain orientation decided by the full-chain GPCR-I-TASSER model. A FG-MD refinement was finally conducted on the full-chain models to eliminate possible steric clashes. (B) GPCR-I-TASSER models for UniProt ID: Q8NGQ3. The left panel shows the model generated when the membrane repulsive energy was turned off, where the N-terminal tail entered into the transmembrane region. The right panel shows that the tails were moved out of the membrane when the membrane repulsive energy in Eq. S4 was included.
All the models for the 1026 human GPCRs by GPCR-I-TASSER, together with the template alignment, local- and global-confidence scoring annotations, and the secondary structure and solvation predictions, are deposited in the GPCR-HGmod database (http://zhanglab.ccmb.med.umich.edu/GPCR-HGmod/). Due to the sensitivity of the model quality to the templates in the PDB, the model prediction for all human GPCRs will be updated every 12 months (the old models will be archived in the on-line database for tracking progress).
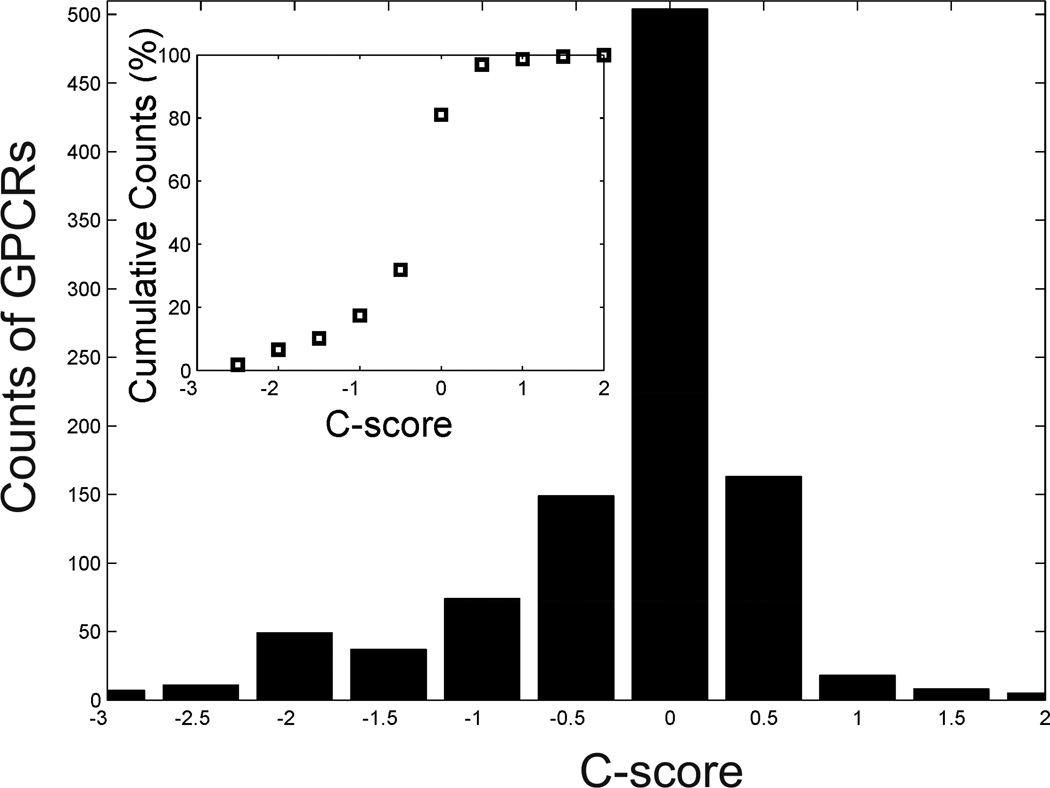
Global confidence score analyses
In Figure 5, we present a histogram distribution of the confidence scores (C-score) of the GPCR-I-TASSER models. Here, C-score is defined as the product of the normalized Z-score from LOMETS threading and the cluster density from SPICKER, i.e.
| (1) |
where M/Mtot is the normalized multiplicity of the structure decoys in the cluster, 〈RMSD〉 is the average RMSD of the decoys to the cluster centroid, Zi is the highest Z-score of the template detected by the ith threading program in LOMETS, and is the corresponding Z-score cutoff for distinguishing between good and bad template alignments (see SEP). The C-score has a strong correlation coefficient (0.91) to the actual TM-score of the predicted models based on large-scale benchmark tests (Zhang, 2008).
Figure 5.
C-score distribution of GPCR-I-TASSER models for 1026 GPCRs in the human genome. Inset: Percentage of the cumulative counts along with the C-score distribution.
From the histogram of TM-score data obtained from the benchmark study, we roughly estimated the number of GPCRs expected to have a TM-score >0.5 which indicates a similar fold to the target, i.e., where Mbin=15 is the number of bins split in the C-score space, Nm is the number of GPCR-I-TASSER models in mth C-score bin, and rm is the folding rate for the GPCR-I-TASSER/I-TASSER models in the mth bin based on large-scale benchmark tests on 1107 known proteins, including the 24 GPCR proteins from the PDB library. We found that there are 923 cases out of the 1026 GPCRs which should have the highest ranked model with a correct topology (TM-score >0.5). This number is similar to the direct counting of GPCRs with a C-score >−1.5, which is a cutoff that approximately corresponds to the correct models in the benchmark data (Zhang, 2008). In addition, all the models predicted by GPCR-I-TASSER have the typical 7-TM helix bundle topology due to the ab intio folding algorithm and the GPCR-RD experimental restraints, although a number of GPCRs (~200) did not have any TM templates detected by the threading search. Here, we note that the C-score histogram in Figure 5 was calculated based on the whole-chain GPCR sequences, which may contain multiple domains. If we only count for the TM domains, the number of folded cases should be slightly higher since the domain parsing and assembly procedure can increase the C-score and modeling accuracy of individual domains as illustrated in Figure 4.
The 923 high C-score GPCRs cover 53 out of the 54 families in the human genome - the only missed family is “Family 3 metabotropic glutamate and calcium receptors” where none of the four members (Q8NFJ5, Q9NQ84, Q9NZD1, Q9NZH0) have a confident prediction from GPCR-I-TASSER. Since the experimentally solved GPCRs cover only 16 families (Table S1), such a high family coverage partly demonstrates the ability of GPCR-I-TASSER to model distant-homology proteins across different families.
In Table S6, we list the top 20 families which have the highest number of GPCRs with a C-score >−1.5. As expected, for the families which have some members with experimentally solved structures, all the GPCRs have high C-score models generated due to the easily-detected homologous templates. While most of the high C-score GPCRs are from the Odorant/olfactory and gustatory family, GPCR-I-TASSER also generated models of high C-scores for many families which have no experimentally solved members but are pharmaceutically important drug targets, including Trace amine-associated (brain monoamine regulation (Panas et al., 2012)), Prostanoids (initiating cancer and inflammation pathways (Breyer et al., 2001)), Releasing hormones (progression of cancers (Harrison et al., 2004)), Melanocortins (familial glucocorticoid deficiency type 1 (Vassart and Costagliola, 2011)), Vasopressin (nephrogenic diabetes insipidus (Vassart and Costagliola, 2011)), and Neuropeptide Y (anxiety and pain (Brothers and Wahlestedt, 2010)) receptors.
Residue-level local quality and B-factor estimation
While C-score is designed to assess the confidence of the global topology, the accuracy of local structures also needs to be assessed because it is important for function annotation and virtual screening. We developed a procedure, called ResQ, to estimate the residue-level quality of the GPCR models based on large-scale support vector regression training of decoy 3D models. The training features of ResQ include (1) structure variation of GPCR-I-TASSER assembly simulation; (2) consistency between model and sequence-based feature prediction; (3) threading alignment coverage; (4) B-factor of threading templates; (5) sequence profile (see SEP). A benchmark test on 635 non-redundant proteins showed that the residue-level accuracy can be estimated with an average error ~2.15 Å and the estimated B-factor has a Pearson’s correlation coefficient 0.58 with the X-ray crystallography data (Yang et al., 2015).
The local structure quality estimates on the GPCR-I-TASSER models showed that 89% out of the 365,343 residues in the 1026 GPCRs are correctly modeled if we consider a distance tolerance <2 Å. The majority of the incorrectly predicted residues are located in the loop or tail regions which have an average local error 3.62 Å higher than the residues in the conserved TM helices. Interestingly, the EL2 loops have an average error 2.56 Å lower than other loop and tail residues, which is probably due to the detection of better structure profiles for these loops. While these local structure analysis data highlighted uncertainties in the unaligned regions, the functionally important EL2 loops were modeled with higher certainty than the other unaligned non-TM-helix regions.
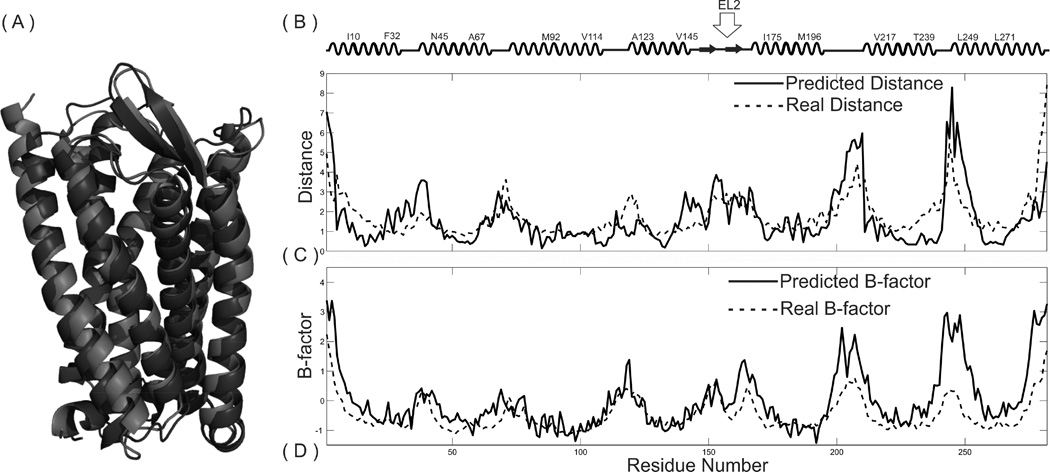
In Figure 6, we present an example of the estimated local structure accuracy and B-factor profiles, in control with the X-ray crystallography data from the histamine H1 receptor (PDB ID: 3rzeA1) (Shimamura et al., 2011). The actual distance errors are mainly located in the unaligned loop and tail regions, which are highly consistent with the ResQ estimation. These profiles are provided for each of the GPCR-I-TASSER models in the GPCR-HGmod database.
Figure 6.
An illustration of ResQ based local structure quality estimation on the human histamine H1 receptor (PDB ID: 3rzeA1) (Shimamura et al., 2011). (A) Overlay of the GPCR-I-TASSER model (blue) and the X-ray structure; (B) secondary structure assignment by STRIDE; (C) predicted and actual distance deviations of the model from the X-ray structure; (D) predicted and actual B-factor profiles of the target.
Sequence and structure networks of human GPCRs
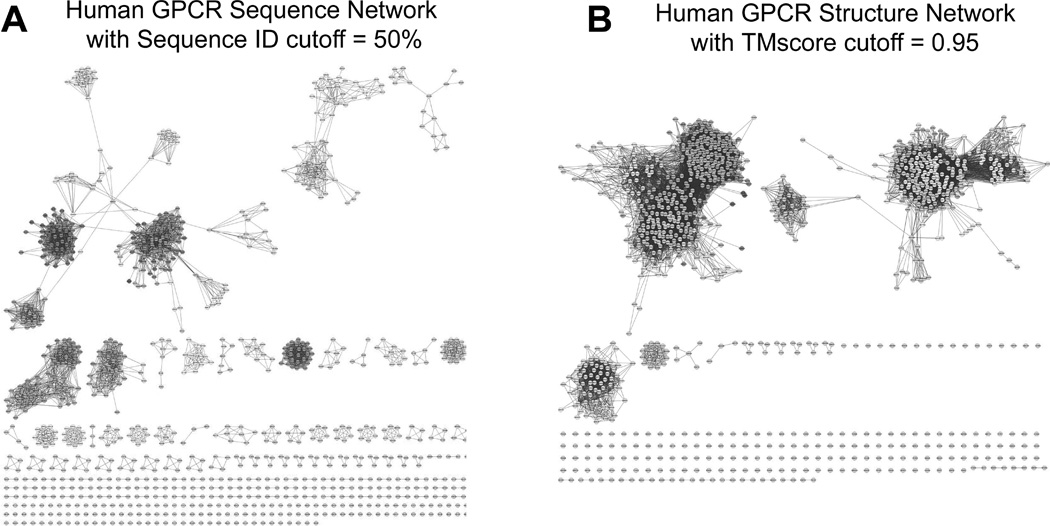
Given the sequence and structural models generated, we present in Figure 7 a 2-dimensional view of the sequence and structure distributions of all 1026 GPCRs in human prepared using Cytoscape 2.8 (Cline et al., 2007). The sequence similarity matrix is measured by pair-wise sequence identity calculated by NW-align (http://zhanglab.ccmb.med.umich.edu/NW-align/), where a cutoff of 50% is used to ensure that the connected nodes have conserved functionality. There are 151 GPCR clusters or orphans with an average number of neighbors =8.9, and the average number of neighbors of non-orphan clusters is 10.9.
Figure 7.
GPCR sequence network (A) and structure network (B) in human genome. The nodes with more connections are shown in red and those with fewer connections in green. The plot is generated using Cytoscape 2.8 (Cline et al., 2007).
In the structure space, the distance matrix is measured by the pair-wise TM-score of the GPCR-I-TASSER models, where a cutoff of TM-score >0.95 is used for node connections to distinguish subtle structural similarity. The total number of the clusters or orphans is 171 in structure space, similar to the number in sequence space. However, the average number of neighbors for the non-orphan clusters is 41.2, which is much higher than that in the sequence space, despite the stringent TM-score cutoff. This data suggests that human GPCRs are much more converged in structure space than in sequence space. We have re-examined the data using more permissive sequence identity cutoffs in the 30–50% range or TM-score cutoffs in 0.6–0.95 but the clustering data did not qualitatively change.
The high degree of conservation in structure space is partly due to the fact that GPCR structures are largely constrained by the 7-TM helix bundle topology, despite considerable variations existing in the relative location and orientation of helices and arrangement of loops. There are however a few big families, such as olfactory receptors, which have a highly similar structure but with very diverse pair-wise sequence identity. In fact, the biggest cluster in structure space includes 711 members which all belong to the Class A Rhodopsin-like receptors and have a sequence identity as low as 17%. Thus, high-resolution structure modeling should serve as a useful complement to sequence-based analysis for GPCR function annotation.
Cross-validation of GPCR-I-TASSER Models with Experimental Mutagenesis Data
Although the number of experimental 3D structures for GPCRs is low, numerous experiments have been performed on GPCRs to identify the critical residues and motifs from site-directed mutagenesis, solid-state NMR, and neutron diffraction data. Many of these data have been collected in the GPCR-RD database (Zhang and Zhang, 2010a) and converted into the 3D spatial restraints to guide the GPCR-I-TASSER structure modeling. To validate the GPCR-I-TASSER structure models, we compared the predictions with recently collected mutagenesis data that had not yet been incorporated into the GPCR-RD at the time of modeling.
To test the high-confidence models, we collected 58 GPCR-I-TASSER models which have a C-score >1.0 and at least one contact residue pair from the new mutagenesis experiments. Excluding the N- and C-terminal tails, we found that all the first models by GPCR-I-TASSER have their residue contacts consistent with the mutagenesis data, i.e., with C-alpha distance <10 Å for contact restraints or Eorientation <0 from Eq. S10 for the orientation restraints. In Figure 8, we present a set of randomly selected examples from the high-confident GPCR-I-TASSER models where the key functional residues are highlighted.
Figure 8A shows the GPCR-I-TASSER model for the formyl peptide receptors, which respond to chemokines and chemoattractants found on the surface of phagocytes. There are three residues pairs (D106-R205, A68-N44 and N44-N66) and two functionally related residues (D71 and R123) which are supposed to be in contact with each other based on the mutation and ligand binding analysis experiments (Lala et al., 1993; Mills et al., 2000; Prossnitz et al., 1999). These residue pairs are all in contact in our formyl peptide receptor model with distances <10 Å (Figure 8A).
Figure 8B shows a second example from the C5a anaphylatoxin chemotactic receptors that mediate cell activation and receptor desensitization. One disulfide bond (C293-C86), two residue pairs (P257-C285 and G210-M120), and two functionally important residues (P170 and Q259) should be in contact according to the experimental data (Baranski et al., 1999; Giannini et al., 1995; Kolakowski et al., 1995; Raffetseder et al., 1996), which is also consistent with the GPCR-I-TASSER models.
Figures 8C and 8D are two other examples from the galanin receptor and the Type-1 angiotensin II receptor, respectively. In Figure 8C, one contact pair (H263-R285) and four functional residues (H263, H267, H285 and H289) from the mutagenesis experiments (Berthold et al., 1997; Kask et al., 1996) are all consistent with the GPCR-I-TASSER model. In Figure 8D, six function-related residues (N111, A104, S115, W153, T260 and N295) form a well-shaped binding pocket in the GPCR-I-TASSER model, which were identified in the mutagenesis experiments as critical binding residues with the non-peptide ligands (Perlman et al., 1997; Perlman et al., 1995; Schambye et al., 1994).x
CONCLUSIONS
Progress in experimental GPCR structure determination has been slow due to difficulties in acquiring high-resolution experimental data. Computational approaches can also produce high-resolution models, but so far they have been limited to cases where a homologous template is available. To address these limitations, we have developed a new hybrid method, GPCR-I-TASSER, that can exploit distant-homology templates and spatial restraints from low-resolution but more easy-to-acquire experimental data to assist high-resolution GPCR structure modeling.
In addition to the generic knowledge-based force field, a set of new GPCR- and TM-protein-specific energy terms, including membrane repulsion, hydrophobic moment, and enhanced aromatic and cation-π interactions, were introduced to guide the GPCR-I-TASSER structure assembly simulations. Our unpublished results showed that the inclusion of these TM-specific potentials resulted in a TM-score increase of the GPCR structure models by 3.5% on the test proteins with a P-value <10−5. For the targets that do not have close homologies, a new ab initio folding procedure was developed to construct the TM-helix bundles from scratch, which are further refined by the fragment assembly simulations. This hybrid pipeline enables the structure construction of different families of GPCRs, which is essential for genome-wide GPCR modeling and GPCR-ligand screening. Although progress was made to advance computational methods for GPCR modeling, accuracy can still be limited, especially in the de novo cases and in the loop and tail regions. We provide local confidence scores to help identify these uncertain regions.
The GPCR-I-TASSER method was tested on two benchmarks. First, it was tested on 24 GPCR proteins which have an experimentally-solved structure. After excluding all homologous proteins with a sequence identity >30% and templates detectable by PSI-BLAST, the threading programs successfully identified templates of correct topology with an average TM-score=0.675 and RMSD=5.74 Å. After the GPCR-I-TASSER structural reassembly refinement, the TM-score of final models increased to 0.806 by 19.4% and RMSD reduced to 4.22 Å by 1.52 Å in the same threading aligned region (or 2.40 Å in the TM helix region). Even with the most stringent template filtering, i.e., excluding all GPCR and TM proteins from the template library, the ab initio folding procedure constructed correct folds for 20 cases with a TM-score >0.5 (or 22 cases in the TM regions). This data demonstrates a significant advantage over the traditional homology-based approaches such as MODELLER (Sali and Blundell, 1993), where none of the models can have a TM-score >0.25 without using the GPCR templates in our tests.
Second, we tested GPCR-I-TASSER in the community-wide blind GPCRDock experiment. The final models of the CXCR4 and D3 receptors have a TM-score 11% and 46% higher than the threading templates. The RMSD of the TM regions was 2.08, 2.58 and 1.26 Å, which are 0.98, 0.48, and 0.35 Å lower than the corresponding initial templates, respectively. These predictions have a higher average significance score (Z-score) than the other 34 predictor groups.
Finally, we applied the GPCR-I-TASSER pipeline to the modeling of all 1026 putative GPCR proteins collected from the UNIPROT and GPCRDB databases. There are 923 cases which are expected to have a correct global fold with a predicted TM-score >0.5, based on the correlation between C-score and TM-score. The targets with high-confidence models include many unsolved but pharmaceutically important GPCR families including Trace amine, Prostanoids, Releasing hormones, Melanocortins, Vasopressin and Neuropeptide Y receptors. The sequence and structure-based clustering studies showed that the structures of GPCRs are much more conserved than the sequences during evolution. As part of cross-validations, we compared the GPCR-I-TASSER models with experimental mutagenesis data, which was not used in our structure modeling. Consistency with the experimental data was demonstrated in all GPCR-I-TASSER models which have a confidence score above 1.0. These results demonstrated new progress on genome-wide structure modeling of G protein-coupled receptors.
EXPERIMENTAL PROCEDURES
GPCR-I-TASSER is designed to construct 3D models of G protein-coupled receptors and consists of three steps of TM-helix assembly, full-length structure reassembly simulations, and model selection and atomic-level structure refinement (Figure 1). The processes are outlined below, with detailed procedures described in Supplemental Experimental Procedures (SEP).
Generation of transmembrane helix framework
The query GPCR sequence is threaded through the PDB by LOMETS (Wu and Zhang, 2007), a meta-threading approach containing nine cutting-edge threading programs, to identify appropriate structure templates. The regions of extra/intra-cellular loops and TM-helices are predicted separately and introduced as additional alignment constraints to enhance the accuracy of the threading alignments for GPCRs (see Eq. S1 in SEP).
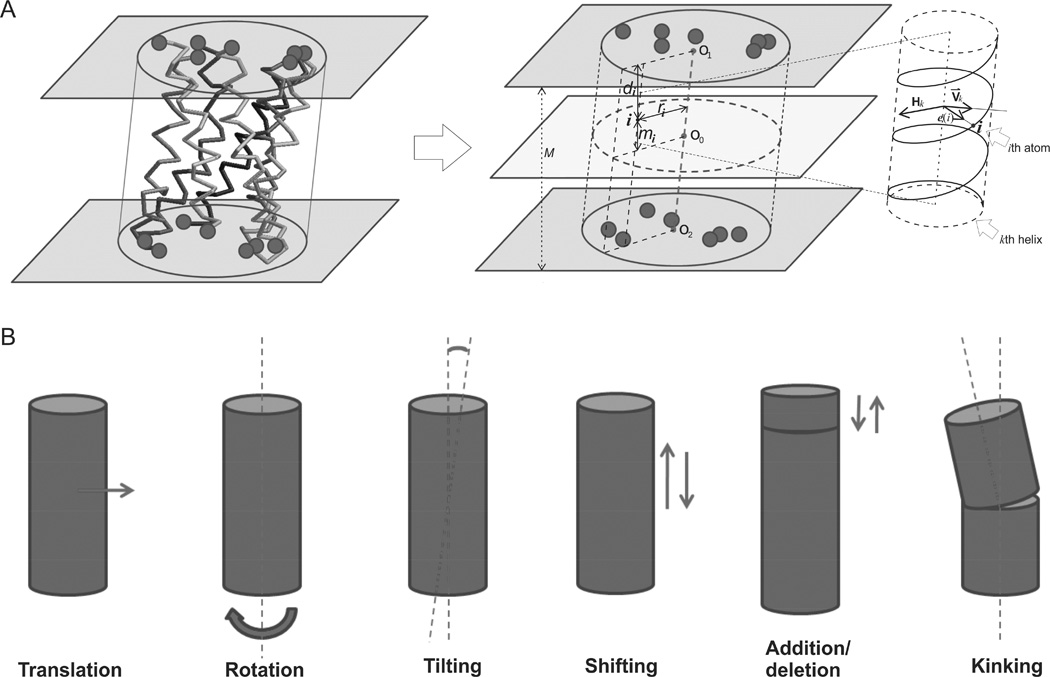
In case that no significant template is identified, a new ab initio folding approach is developed to construct the TM framework by Replica-exchange Monte Carlo (REMC) simulation, starting from seven ideal helices located sequentially along a perimeter of 8 Å. The MC movements involve translation, rotation and tilting of the helices, and sequence shifts along the helix, addition/deletion of residues, and helix kinking (Figure 9). The simulations are guided by a simple force field consisting of a knowledge-based, distance-specific contact potential, RW (Zhang and Zhang, 2010b) and the free energy change of GPCR and water/lipid interactions (Lomize et al., 2006) (see Eqs. S2–3 in SEP).
Figure 9.
Illustrations for ab initio transmembrane helix folding. (A) Initial conformation and variable definitions. (B) Monte Carlo movements in the ab initio TM-helix folding.
Template-based fragment assembly simulations
Full-length GPCR models were constructed by reassembling the continuous fragments (mainly TM-helices) excised from LOMETS threading alignments or ab initio TM helix models, following the I-TASSER protocol (Roy et al., 2010; Yang et al., 2015). The force field of the GPCR-I-TASSER simulation consists of three components. The first component is a generic knowledge-based potential extended from I-TASSER which includes statistical Cα and side-chain contact potentials, backbone-orientation specific hydrogen-bond, solvation from neural network prediction, and predicted secondary structure propensities; the second is spatial restraints derived from LOMETS templates and/or ab initio TM-helix models, which consists of Cα distance maps and Cα and side-chain contacts; and the third component consists of six GPCR- and/or transmembrane-specific energy terms as described in Eqs. S4–10 in SEP. Two types of spatial restraints are derived from the site-directed mutagenesis and affinity labeling experiments collected from the GPCR-RD database (Zhang and Zhang, 2010a). These include contact restraints accounting for the experimentally identified disulfide bridges and the functionally important residues (Eq. S9), and an orientation restraint of TM helix to count for the functionally related point mutations (Eq. S10). A general membrane repulsive potential is introduced in Eq. S4 to enhance the GPCR-specific topology, i.e., all non-TM-helix residues should be excluded from the TM regions (see Figure 4B).
Model selection and fragment-guided structure refinement
Structure decoys generated in GPCR-I-TASSER are submitted to SPICKER (Zhang and Skolnick, 2004c) for structure clustering. The decoys with the highest number of structural neighbors are selected, with full-atomic models refined by the fragment-guided molecule dynamic (FG-MD) simulations (Zhang et al., 2011). Furthermore, the SPICKER centroid model is used as a probe to identify analog fragments from the PDB by TM-align (Zhang and Skolnick, 2005), which provides additional spatial restraints to improve the energy landscape funnel in atomic-level structure refinements in FG-MD.
Multiple-domain assembly
For the GPCRs of multi-domains, we first use ThreaDom (Xue et al., 2013) to identify the domain boundary and then use GPCR-I-TASSER and I-TASSER to fold the receptor and globular domains separately. The full-length models are finally built by docking the domain models using the whole-chain model as a reference template.
Supplementary Material
Highlights.
New approach to ab initio GPCR structure assembly
Use of mutagenesis data to assist 3D structure construction
High-resolution structure models for 923 human GPCRs
Provide reliably model for GPCR families that have no experimental structure
ACKNOWLEDGEMENTS
We are grateful to Dr. Jeffrey Brender for critical reading of the manuscript. The project is supported in part by the NIGMS (GM083107 and GM084222)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTROBUTION YZ conceived the project; JZ, JY and RJ conduct the calculation and data analysis; JZ, JY and YZ wrote the paper.
REFERENCES
- Archer E, Maigret B, Escrieut C, Pradayrol L, Fourmy D. Rhodopsin crystal: new template yielding realistic models of G-protein-coupled receptors? Trends Pharmacol Sci. 2003;24:36–40. doi: 10.1016/s0165-6147(02)00009-3. [DOI] [PubMed] [Google Scholar]
- Baranski TJ, Herzmark P, Lichtarge O, Gerber BO, Trueheart J, Meng EC, Iiri T, Sheikh SP, Bourne HR. C5a receptor activation. Genetic identification of critical residues in four transmembrane helices. J Biol Chem. 1999;274:15757–15765. doi: 10.1074/jbc.274.22.15757. [DOI] [PubMed] [Google Scholar]
- Barth P, Wallner B, Baker D. Prediction of membrane protein structures with complex topologies using limited constraints. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1409–1414. doi: 10.1073/pnas.0808323106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold M, Kahl U, Jureus A, Kask K, Nordvall G, Langel U, Bartfai T. Mutagenesis and ligand modification studies on galanin binding to its GTP-binding-protein-coupled receptor GalR1. Eur J Biochem. 1997;249:601–606. doi: 10.1111/j.1432-1033.1997.00601.x. [DOI] [PubMed] [Google Scholar]
- Breyer RM, Bagdassarian CK, Myers SA, Breyer MD. Prostanoid receptors: subtypes and signaling. Annu Rev Pharmacol Toxicol. 2001;41:661–690. doi: 10.1146/annurev.pharmtox.41.1.661. [DOI] [PubMed] [Google Scholar]
- Brothers SP, Wahlestedt C. Therapeutic potential of neuropeptide Y (NPY) receptor ligands. EMBO molecular medicine. 2010;2:429–439. doi: 10.1002/emmm.201000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Ji F, Olman V, Mobley CK, Liu Y, Zhou Y, Bushweller JH, Prestegard JH, Xu Y. Optimal mutation sites for PRE data collection and membrane protein structure prediction. Structure. 2011;19:484–495. doi: 10.1016/j.str.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien EY, Liu W, Zhao Q, Katritch V, Han GW, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eglen RM, Bosse R, Reisine T. Emerging concepts of guanine nucleotide-binding protein-coupled receptor (GPCR) function and implications for high throughput screening. Assay Drug Dev Technol. 2007;5:425–451. doi: 10.1089/adt.2007.062. [DOI] [PubMed] [Google Scholar]
- Fanelli F, De Benedetti PG. Update 1 of: computational modeling approaches to structure-function analysis of G protein-coupled receptors. Chem Rev. 2011;111:PR438–PR535. doi: 10.1021/cr100437t. [DOI] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannini E, Brouchon L, Boulay F. Identification of the major phosphorylation sites in human C5a anaphylatoxin receptor in vivo. J Biol Chem. 1995;270:19166–19172. doi: 10.1074/jbc.270.32.19166. [DOI] [PubMed] [Google Scholar]
- Granier S, Manglik A, Kruse AC, Kobilka TS, Thian FS, Weis WI, Kobilka BK. Structure of the delta-opioid receptor bound to naltrindole. Nature. 2012;485:400–404. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison GS, Wierman ME, Nett TM, Glode LM. Gonadotropin-releasing hormone and its receptor in normal and malignant cells. Endocr Relat Cancer. 2004;11:725–748. doi: 10.1677/erc.1.00777. [DOI] [PubMed] [Google Scholar]
- Hopf TA, Colwell LJ, Sheridan R, Rost B, Sander C, Marks DS. Three-dimensional structures of membrane proteins from genomic sequencing. Cell. 2012;149:1607–1621. doi: 10.1016/j.cell.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry-Us. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- Kask K, Berthold M, Kahl U, Nordvall G, Bartfai T. Delineation of the peptide binding site of the human galanin receptor. Embo J. 1996;15:236–244. [PMC free article] [PubMed] [Google Scholar]
- Kolakowski LF, Jr, Lu B, Gerard C, Gerard NP. Probing the "message:address" sites for chemoattractant binding to the C5a receptor. Mutagenesis of hydrophilic and proline residues within the transmembrane segments. J Biol Chem. 1995;270:18077–18082. doi: 10.1074/jbc.270.30.18077. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of molecular biology. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kufareva I, Rueda M, Katritch V, Stevens RC, Abagyan R. Status of GPCR modeling and docking as reflected by community-wide GPCR Dock 2010 assessment. Structure. 2011;19:1108–1126. doi: 10.1016/j.str.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lala A, Sharma A, Sojar HT, Radel SJ, Genco RJ, De Nardin E. Recombinant expression and partial characterization of the human formyl peptide receptor. Biochim Biophys Acta. 1993;1178:302–306. doi: 10.1016/0167-4889(93)90208-7. [DOI] [PubMed] [Google Scholar]
- Lee HS, Zhang Y. BSP-SLIM: a blind low-resolution ligand-protein docking approach using predicted protein structures. Proteins. 2012;80:93–110. doi: 10.1002/prot.23165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomize AL, Pogozheva ID, Lomize MA, Mosberg HI. Positioning of proteins in membranes: a computational approach. Protein science : a publication of the Protein Society. 2006;15:1318–1333. doi: 10.1110/ps.062126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JS, Miettinen HM, Cummings D, Jesaitis AJ. Characterization of the binding site on the formyl peptide receptor using three receptor mutants and analogs of Met-Leu-Phe and Met-Met-Trp-Leu-Leu. J Biol Chem. 2000;275:39012–39017. doi: 10.1074/jbc.M003081200. [DOI] [PubMed] [Google Scholar]
- Nugent T, Jones DT. Accurate de novo structure prediction of large transmembrane protein domains using fragment-assembly and correlated mutation analysis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E1540–E1547. doi: 10.1073/pnas.1120036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas MW, Xie Z, Panas HN, Hoener MC, Vallender EJ, Miller GM. Trace amine associated receptor 1 signaling in activated lymphocytes. J Neuroimmune Pharmacol. 2012;7:866–876. doi: 10.1007/s11481-011-9321-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S, Costa-Neto CM, Miyakawa AA, Schambye HT, Hjorth SA, Paiva AC, Rivero RA, Greenlee WJ, Schwartz TW. Dual agonistic and antagonistic property of nonpeptide angiotensin AT1 ligands: susceptibility to receptor mutations. Mol Pharmacol. 1997;51:301–311. doi: 10.1124/mol.51.2.301. [DOI] [PubMed] [Google Scholar]
- Perlman S, Schambye HT, Rivero RA, Greenlee WJ, Hjorth SA, Schwartz TW. Non-peptide angiotensin agonist. Functional and molecular interaction with the AT1 receptor. J Biol Chem. 1995;270:1493–1496. doi: 10.1074/jbc.270.4.1493. [DOI] [PubMed] [Google Scholar]
- Prossnitz ER, Gilbert TL, Chiang S, Campbell JJ, Qin S, Newman W, Sklar LA, Ye RD. Multiple activation steps of the N-formyl peptide receptor. Biochemistry-Us. 1999;38:2240–2247. doi: 10.1021/bi982274t. [DOI] [PubMed] [Google Scholar]
- Raffetseder U, Roper D, Mery L, Gietz C, Klos A, Grotzinger J, Wollmer A, Boulay F, Kohl J, Bautsch W. Site-directed mutagenesis of conserved charged residues in the helical region of the human C5a receptor. Arg2O6 determines high-affinity binding sites of C5a receptor. Eur J Biochem. 1996;235:82–90. doi: 10.1111/j.1432-1033.1996.00082.x. [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Schambye HT, von Wijk B, Hjorth SA, Wienen W, Entzeroth M, Bergsma DJ, Schwartz TW. Mutations in transmembrane segment VII of the AT1 receptor differentiate between closely related insurmountable and competitive angiotensin antagonists. Br J Pharmacol. 1994;113:331–333. doi: 10.1111/j.1476-5381.1994.tb16899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schushan M, Barkan Y, Haliloglu T, Ben-Tal N. C(alpha)-trace model of the transmembrane domain of human copper transporter 1, motion and functional implications. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10908–10913. doi: 10.1073/pnas.0914717107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Javitch JA. The binding site of aminergic G protein-coupled receptors: the transmembrane segments and second extracellular loop. Annu Rev Pharmacol Toxicol. 2002;42:437–467. doi: 10.1146/annurev.pharmtox.42.091101.144224. [DOI] [PubMed] [Google Scholar]
- Shimamura T, Shiroishi M, Weyand S, Tsujimoto H, Winter G, Katritch V, Abagyan R, Cherezov V, Liu W, Han GW, et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons KT, Bonneau R, Ruczinski I, Baker D. Ab initio protein structure prediction of CASP III targets using ROSETTA. Proteins. 1999;(Suppl 3):171–176. doi: 10.1002/(sici)1097-0134(1999)37:3+<171::aid-prot21>3.3.co;2-q. [DOI] [PubMed] [Google Scholar]
- Takeda S, Kadowaki S, Haga T, Takaesu H, Mitaku S. Identification of G protein-coupled receptor genes from the human genome sequence. Febs Lett. 2002;520:97–101. doi: 10.1016/s0014-5793(02)02775-8. [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. Journal of molecular biology. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- Vaidehi N, Floriano WB, Trabanino R, Hall SE, Freddolino P, Choi EJ, Zamanakos G, Goddard WA., 3rd Prediction of structure and function of G protein-coupled receptors. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12622–12627. doi: 10.1073/pnas.122357199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassart G, Costagliola S. G protein-coupled receptors: mutations and endocrine diseases. Nature reviews. Endocrinology. 2011;7:362–372. doi: 10.1038/nrendo.2011.20. [DOI] [PubMed] [Google Scholar]
- Wang C, Jiang Y, Ma J, Wu H, Wacker D, Katritch V, Han GW, Liu W, Huang XP, Vardy E, et al. Structural basis for molecular recognition at serotonin receptors. Science. 2013;340:610–614. doi: 10.1126/science.1232807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Zhang Y. LOMETS: A local meta-threading-server for protein structure prediction. Nucl. Acids. Res. 2007;35:3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhang Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics. 2010;26:889–895. doi: 10.1093/bioinformatics/btq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Xu D, Wang Y, Zhang Y. ThreaDom: extracting protein domain boundary information from multiple threading alignments. Bioinformatics. 2013;29:i247–i256. doi: 10.1093/bioinformatics/btt209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Jang R, Zhang Y, Shen HB. High-accuracy prediction of transmembrane inter-helix contacts and application to GPCR 3D structure modeling. Bioinformatics. 2013;29:2579–2587. doi: 10.1093/bioinformatics/btt440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nature Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liang Y, Zhang Y. Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure. 2011;19:1784–1795. doi: 10.1016/j.str.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang Y. GPCRRD: G protein-coupled receptor spatial restraint database for 3D structure modeling and function annotation. Bioinformatics. 2010a;26:3004–3005. doi: 10.1093/bioinformatics/btq563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang Y. A Novel Side-Chain Orientation Dependent Potential Derived from Random-Walk Reference State for Protein Fold Selection and Structure Prediction. Plos One. 2010b;5 doi: 10.1371/journal.pone.0015386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Interplay of I-TASSER and QUARK for template-based and ab initio protein structure prediction in CASP10. Proteins. 2014;82(Suppl 2):175–187. doi: 10.1002/prot.24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Devries ME, Skolnick J. Structure modeling of all identified G protein-coupled receptors in the human genome. PLoS computational biology. 2006;2:e13. doi: 10.1371/journal.pcbi.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on a genomic scale. Proc. Natl. Acad. Sci. USA. 2004a;101:7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004b;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. SPICKER: A clustering approach to identify near-native protein folds. J Comput Chem. 2004c;25:865–871. doi: 10.1002/jcc.20011. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic. Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.