Abstract
Lung carcinogenesis is a multistep process in which normal lung epithelial cells are converted to cancer cells through the sequential acquisition of multiple genetic or epigenetic events. Despite the utility of current genetically engineered mouse (GEM) models of lung cancer, most do not allow temporal dissociation of the cardinal events involved in lung tumor initiation and cancer progression. Here we describe a novel two-switch GEM model for BRAFV600E-induced lung carcinogenesis allowing temporal dissociation of these processes. In mice carrying a Flp recombinase-activated allele of Braf (BrafFA) in conjunction with Cre-regulated alleles of Trp53, Cdkn2a or c-MYC, we demonstrate that secondary genetic events can promote bypass of the senescence-like proliferative arrest displayed by BRAFV600E-induced lung adenomas leading to malignant progression. Moreover, restoring or activating TP53 in cultured BRAFV600E/TP53Null or BRAFV600E/INK4A-ARFNull lung cancer cells triggered a G1 cell cycle arrest regardless of p19ARF status. Perhaps surprisingly, neither senescence nor apoptosis was observed upon TP53 restoration. Our results establish a central function for the TP53 pathway in restricting lung cancer development, highlighting the mechanisms that limit malignant progression of BRAFV600E-initiated tumors.
INTRODUCTION
Lung cancer will kill ~160,000 people in the United States in 2015, which is more than the death rates of the next four most common cancers combined (1). Aberrant proliferation of non-small cell lung cancers (NSCLC) is frequently associated with mutational activation of receptor tyrosine kinase (RTK) signaling including genes encoding transmembrane receptor tyrosine kinases (ALK, EGFR, MET, ROS1) or intracellular signaling proteins such as KRAS or its effectors A- B- or C-RAF or PIK3CA (encoding PI3’-kinase-α) (2,3). BRAF is estimated to be mutated in ~9% of NSCLCs, 25% of which express the BRAFT1799A oncogene encoding the BRAFV600E oncoprotein kinase (4–6). However, like many cancers, mutational activation of proto-oncogenes such as ERBB1, KRAS or BRAF is generally accompanied by silencing of tumor suppressor genes (TSGs) such as TP53, CDKN2A or PTEN that cooperatively serve to promote the step-wise malignant transformation of normal lung epithelial cells to malignant lung cancer cells (5,6).
To model BRAFV600E-induced lung tumorigenesis, we previously generated BrafCA mice engineered to carry a Cre-activated Braf allele that allows conversion of normal BRAF to BRAFV600E following exposure of cells to viruses encoding Cre recombinase (7,8). Expression of BRAFV600E in the distal lung epithelium results in development of benign lung tumors that fail to progress to lung cancer due to the onset of a senescence-like proliferative arrest (8, 9). Importantly, when TSGs or proto-oncogenes (Trp53, Cdkn2a, Ctnnb1, Pik3ca) are mutated simultaneously in conjunction with BRAFV600E expression, progression to NSCLC is readily observed (8, 9).
Despite the utility of BrafCA mice to model BRAFV600E-induced tumorigenesis (8–10), we are constrained by our inability to temporally dissociate genetic events that contribute to cancer progression. Somatic recombination of conditional alleles by Cre recombinase is such that oncogene activation and TSG silencing occur simultaneously – a situation that rarely occurs in humans (11). We therefore wanted to generate a new GEM model of lung cancer in which expression of BRAFV600E could be temporally dissociated from cooperating genetic events that contribute to malignant progression. To do so, we generated mice carrying a Flp-activated allele of Braf (BrafFA) that allows conversion of normal BRAF to BRAFV600E in response to Flp recombinase. This allele allows investigator control of the order and timing of BRAFV600E expression in combination with mutational alteration of cooperating genes.
Here, we observed that temporal dissociation of BRAFV600E expression from the silencing of TP53 or INK4A-ARF expression resulted in lung adenocarcinomas. Furthermore, TP53 silencing was able to bypass the senescence-like proliferative arrest observed in late-stage BRAFV600E-initiated benign lung tumors. Since it is possible to lose TP53 function prior to acquiring oncogenic mutations such as that observed in Li-Fraumeni patients (12,13), we next modeled this phenomenon by inducing BRAFV600E expression after TP53 silencing. This order of events appeared to modestly enhance the aggressiveness of the disease. To explore the consequences of TP53 silencing in lung cancer cells, we generated BRAFV600E/TP53Null lung cancer cell lines in which we could restore TP53 activity. Restoration of TP53 activity did not result in senescence or apoptosis, but in a reversible G1 cell cycle arrest that was independent of p19ARF expression. These results highlight the growing sophistication of GEM models of human cancer and demonstrate the importance of TP53 signaling in restricting malignant progression of BRAFV600E-induced benign lung tumors.
MATERIALS AND METHODS
Strains of mice and Adenoviral Infections
The following strains of mice have been previously described: Braftm1Mmcm aka BrafCA (8)), Trp53tm1Brn (Trp53flox, (14)), Cdkn2atm4Cjs (Cdkn2aflox (15)), Trp53tm3Tyj (Trp53lox-R270H, (16)), Gt(ROSA)26Sortm1Sor (R26R::galflox, (17)), Gt(ROSA)26Sortm(myc*T58A)Rcse (R26MYC(T58A), (18)), Sftpctm1(cre/ERT2,rtTA)Hap aka SPCCreER, (19)). Mice were bred and maintained in a pathogen-free facility and experiments were performed according to the protocols approved by the Institutional Animal Care and Use Committee at the University of California San Francisco. Mice were infected with adenoviruses encoding -galactosidase (Ad-gal), Cre recombinase (Ad-Cre) or a codon optimized, enhanced activity Flp recombinase (Ad-FlpO/E). Adenoviruses were purchased from Viraquest. All experiments contained at least 15 mice per group and repeated once.
Derivation of lung cancer cell lines, culture and analysis
BRAFV600E lung cancer derived cell lines were generated by mincing tumor tissue with a sterile razor blade and placing them into media and pelleted by centrifugation. The tissue pellet was re-suspended in 5–10ml of 0.25%(w/v) Trypsin-EDTA and digested at 37C for 20 minutes with intermittent mixing. The digested cell suspension was filtered sequentially through a 100, 70 and finally a 40µm filter. The final cell pellet was re-suspended in 10ml of DF12/Glutamax (Invitrogen) media supplemented with 10%(v/v) FCS and 1%(v/v) Pen-Strep on collagen-coated plates. Genetic alterations were confirmed either by PCR or by immunoblot.
Proliferation was assayed by plating cells in triplicate and cultured in various combinations of vehicle, 4-hydroxytamoxifen (4-HT), nutlin-3a (Cayman Chemical, 10–20µM) and PD0325901 (Hansun Trading Co., 1µM) for 48–72hrs and counted using a Countess cell counter (Invitrogen). A two-tailed Student’s t-test was used to determine significance. Crystal Violet assays were also used to assess proliferation. Cells were washed with cold PBS twice and fixed with cold methanol for 10 minutes on ice. Following the removal of methanol, cells were stained with 0.05%(w/v) Crystal Violet in methanol for 10 minutes at room temperature (RT) with unbound Crystal Violet removed by extensive washing with ddH2O.
For cell cycle analysis, cells were seeded at the appropriate densities and treated with various agents for 48 hours. Cells were incubated with 10µM BrdU for 4 hours prior to harvest and analyzed as previously described (20). All cell culture experiments were repeated at least three times.
Immunoblotting
Immunoblotting was conducted as previously described (9, 20, 21). 50µg (tissue culture cells) to 150µg (mouse tissue) aliquots of protein were analyzed by immunoblotting with the following antisera from Cell Signaling Technologies unless otherwise indicated: BRAF (Epitomics); BIM (Abcam); Cleaved Caspase-3; MDM2 (2A10, Abcam); Puma, pS216-MDM2; p19ARF (DC3, Abcam); p21CIP1 (ThermoPierce); pT202/Y204-ERK1/2, total ERK1/2, pS473-AKT, total AKT, and p53 (FL-393, Santa Cruz). Antigen-antibody complexes were detected using a LiCOR Odyssey Fluorescent System with fluorescent secondary antisera (LiCOR) and analyzed using ImageStudio software.
Immunohistochemistry
Lung tissues sections were prepared as described previously (9, 20). In addition, endogenous peroxidase activity was quenched with 3%(v/v) H202 in methanol for 20 min and then blocked in either 5% goat serum or serum-free block (DAKO) for 30 min at RT. Antibodies were applied overnight at 4C: CC10, SP-C and TTF1 (Santa Cruz); BrdU (Serotec); p19Arf and Ki67 (Abcam); pT202/Y204-ERK1/2, p53 (Novocastra), p21 (Abcam) and p27KIP1 (CST). The appropriate secondary was applied followed either by an anti-peroxidase or ABC amplification for 3–60 minutes at room temperature. Antigen-antibody complexes were detected using a DAB chromagen system (Sigma). PBS with 0.1%(v/v) Tween 20 was used for all washes.
Quantification of Tumor Burden
Tumor burden was quantified as described previously (9, 20). Tumor burden was calculated by measuring the area of all tumors and averaged. At least 3 or more animals were used for each genotype.
RESULTS
Generation and characterization of BrafFA mice
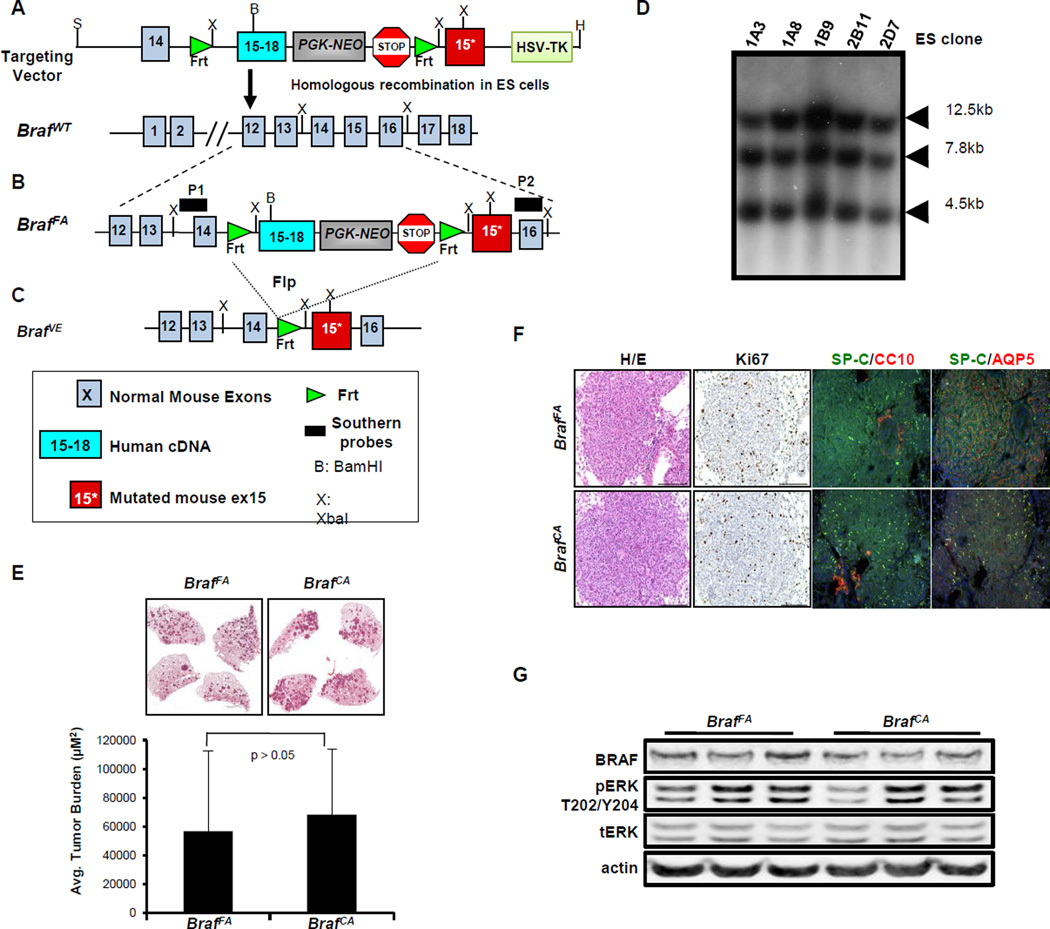
To generate BrafFA mice, a targeting vector was made by exchanging the two loxP sites in the original BrafCA targeting vector with Frt sites using standard cloning techniques (Fig. 1A) (8). By homologous recombination, we generated BrafFA/+ ES cells and confirmed correct targeting of Braf by Southern blot analysis of ES cell genomic DNA as described previously (Fig. 1B–D) (8). One of these ES clones was injected into mouse blastocysts, which in turn gave rise to a chimeric mouse that transmitted the BrafFA allele through the germ-line. The resulting progeny were used for further experimental studies.
Figure 1. Generation of BrafFA mice.
A. Schematic representation of the targeting construct used to generate the BrafFA mouse through homologous recombination into the endogenous Braf genomic locus of 129SvEv derived ES cells.
B. Schematic of the structure of the BrafFA allele that expresses normal BRAF and NEO prior to Flp-mediated recombination. P1 and P2 indicate the location of Southern blotting probes used to confirm appropriate targeting of the Braf gene.
C. Schematic of the structure of the BrafFA allele following Flp-mediated recombination that now expresses mutationally activated BRAFV600E.
D. Southern blot analysis to identify ES clones with the BrafFA targeting construct.
E. H&E stained sections of BrafFA (left) and BrafCA (right) 10 weeks p.i. with Ad-Flp or Ad-Cre respectively with average tumor burden indicated in the bar graph.
F. Lung sections of tumor-bearing BrafFA or BrafCA mice stained with H&E, Ki67, anti-SPC/CC10 or AQP5 using either IHC and IF methods as indicated.
G. Immunoblot analysis of lung tumor lysates from BrafFA or BrafCA mice 10 weeks p.i.
To benchmark the utility of BrafFA mice, we conducted a head-to-head comparison of lung tumorigenesis induced by Ad-Cre or Ad-Flp into BrafCA and BrafFA mice respectively. To facilitate these studies, we generated a new adenoviral vector expressing a codon optimized, activity enhanced form of Flp recombinase (Ad-FlpO/E) for expression in mammalian cells (Not shown) (21). BRAFV600E expression was initiated in the distal lung epithelium of either BrafFA or BrafCA mice by intranasal instillation of 107 plaque-forming units (pfu) of Ad-Flp or Ad-Cre (7,8). Mice were euthanized at 10 weeks post-initiation (p.i.) and analyzed for tumor burden, grade and expression of lung epithelial markers (Fig. 1E–F).
Initiated BrafFA and BrafCA mice developed a similar spectrum of atypical adenomatous hyperplasias (AAHs) and benign adenomas as assessed by the grading criteria established by others and adapted by us (20,22). Average tumor burden of BrafFA and BrafCA mice was not significantly different (56906µM2 and 68387µM2 respectively, Fig. 1E). Furthermore, BrafFA and BrafCA adenomas displayed a similar proliferative index (Ki67), co-expressed Surfactant Protein-C (SP-C) and Aquaporin V (AQP5) and were negative for Clara Cell antigen (CCA) expression (Fig. 1F). Immunoblot analysis of tumor lysates from BrafFA or BrafCA mice revealed equivalent activation of the MEK1/2→ERK1/2 MAP kinase pathway (Fig. 1G). Similar to previous observations, very few BRAFV600E-induced lung tumors arising in BrafFA mice displayed spontaneous progression to adenocarcinoma even at late time points (6 months p.i.). As anticipated, there were no significant differences in BRAFV600E-induced lung tumorigenesis arising in BrafFA versus BrafCA mice.
BRAFV600E-initiated lung tumors cells can be infected with Ad-Cre
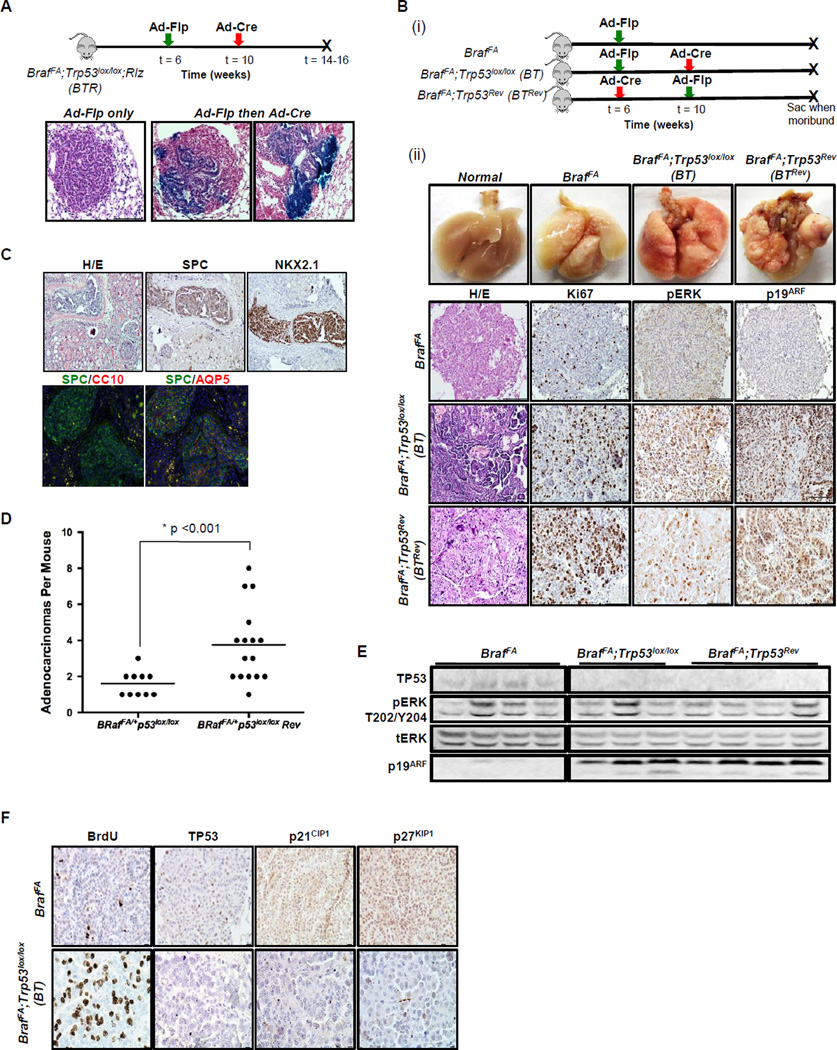
In order to temporally dissociate Ad-Flp induced BRAFV600E expression from additional genetic events, it was necessary to activate Cre recombinase in pre-existing Ad-Flp induced BRAFV600E expressing lung tumors in vivo. To test the feasibility and the efficiency of Ad-Cre infection, we generated BrafFA;Trp53lox/lox mice carrying a Rosa26Cre-Bgeo (Rlz) reporter (BrafFA;Trp53lox/lox;Rlz, BTR), which expresses β-galactosidase following Cre-mediated recombination (14,17). BRAFV600E-induced lung tumorigenesis was initiated in BTR mice using Ad-Flp (107 pfu). Four weeks p.i., mouse lungs were instilled with 107 pfu of Ad-Cre to elicit simultaneous Cre-mediated silencing of TP53 and expression of β-galactosidase (-gal) from the Rlz reporter. Mice were euthanized for analysis 4–6 weeks after Ad-Cre infection with lung tissues examined for -gal activity as a measure of successful Cre-mediated recombination of the Rlz reporter (Fig. 2A).
Figure 2. Temporal dissociation of BRAFV600E expression from additional genetic events.
A. Lung infection schematic of BrafFA;Trp53lox/lox; Rlz (BTR) mice to test the efficiency of Ad-Cre mediated recombination in established BRAFV600E-driven tumors initiated with Ad-Flp. Below are H&Es stained with β-galactosidase.
B. (i) Schematic of infection (ii) Comparison of temporal separation of TP53 silencing and BRAFV600E activation to BRAFV600E alone using two infection strategies. 1st row, photographs of the macro lung. 2nd–4th rows, paraffin lung sections of BrafFA, BT and BrafFA; Trp53Rev (BTRev) stained with H&E, Ki67, pERK, and p19ARF to compare proliferation and MAPK signaling.
C. Paraffin sections of a kidney metastasis detected in a BTRev mouse stained with H&E, SPC, NKX2.1, CC10 and AQP5 by IHC and IF.
D. Dot plot showing the average number of lung adenocarcinomas in BT or BTRev mice in which BRAFV600E was activated either prior to or after silencing of TP53.
E. Immunoblot analysis of BRAFV600E or BRAFV600E/TP53Null lung tumor lysates as indicated.
F. Sections of lung tumors from BrafFA or BT mice were stained to detect BrdU incorporation or for expression of TP53, p21CIP1 or p27KIP1 as indicated.
Control lung tumor-bearing BTR mice displayed no evidence of -gal activity in the airways, lung epithelium or in BRAFV600E-induced lung tumors indicating a low rate of spontaneous recombination of the Rlz reporter. By contrast, -gal activity was observed in tumor bearing BTR mice infected with Ad-Cre specifically in the airways (data not shown), lung epithelium and in a variable, but generally small percentage of cells in BRAFV600E adenomas (Fig. 2A). These results demonstrate the feasibility of infecting BRAFV600E-induced tumor cells for conditional manipulation of Cre-regulated alleles, such as Trp53lox in a manner temporally dissociated from BRAFV600E expression.
Silencing of TP53 expression prior to BRAFV600E expression only modestly enhances BRAFV600E-driven lung tumorigenesis
To assess the effects of temporal dissociation of BRAFV600E activation from TP53 silencing, six-week old BrafFA;Trp53lox/lox (BT) or BrafFA mice were first infected with Ad-Flp (4×106 or 107 pfu). Four weeks later, these mice were re-infected with Ad-Cre (107 pfu) and the lungs were harvested for analysis when the mice were at end-stage. As a control for Flp- or Cre-mediated recombination, BT mice were infected with an adenovirus that expressed -gal (Ad-gal) in lieu of either Ad-Flp or Ad-Cre. At necropsy, the lungs of BT mice had noticeably larger lung tumors compared to BrafFA controls (Fig. 2B), while BT mice infected with Ad-Flp and Ad-gal had tumors similar to those induced in BrafFA mice. Histopathological analysis confirmed that these large tumors were indeed adenocarcinomas that were highly proliferative (Ki67 expression or BrdU incorporation, not shown) relative to BrafFA benign adenomas. Immunohistochemical analysis of phospho-ERK1/2 (pERK) was performed to assess activation of BRAFV600E→MEK→ERK signaling. The level of pERK was generally low in BRAFV600E adenomas arising in either BT or BrafFA mice. By contrast, the majority of BRAFV600E-induced adenocarcinomas arising in BT mice had readily detectable pERK staining. Often these lung cancers displayed variable pERK staining with higher levels of activation detected around apparent invasive edges. Consistent with the results of others, we detected p19ARF expression in malignant regions corresponding to high pERK staining (Fig. 2B) (22,23). Similar results were obtained in BrafFA; Trp53LSL-R270H/+ mice heterozygous for a conditional dominant-negative allele of Trp53 that encodes TP53R270H in response to Cre recombinase (Supp. Fig 1) (16).
Humans with Li-Fraumeni syndrome are often heterozygous for a mutated germ-line allele of TP53 and are at high risk for solid tumors, including lung cancer (12,13). Consequently, we decided to model this by silencing TP53 expression in the mouse lung prior to the activation of BRAFV600E. BT mice were first infected with Ad-Cre (107 pfu) at six weeks of age to generate a field of lung epithelial cells lacking TP53 expression. Importantly, TP53 silencing in the lung is insufficient to induce any overt lung pathology. Four weeks after Ad-Cre infection, the mice were re-infected with Ad-Flp (4×106 or 107 pfu) and euthanized for analysis at end-stage (BrafFA;Trp53Rev:BTRev mice). A number of the BTRev mice developed multiple large adenocarcinomas in contrast to BT mice, with an average 3.75 versus 1.6 adenocarcinomas/mouse (Fig. 2D, p<0.001, 2-tailed t-test). Although BTRev mice had an increased multiplicity of large adenocarcinomas, the overall latency of tumor formation was unchanged compared to BT mice in which BRAFV600E expression was induced prior to TP53 silencing (data not shown). Moreover, similar to lung cancers arising in BT mice, these lesions displayed elevated levels of pERK, p19ARF and were highly proliferative (Fig. 2B). Interestingly, we observed a single kidney metastasis in a BTRev mouse. The lung cancer origin of this macroscopic lesion was confirmed by its expression of lung epithelium markers: NKX2.1, AQP5 and SP-C (Fig. 2C). These markers were not detected in the surrounding kidney tissue.
Next we generated tumor lysates from BTRev, BT and BrafFA mice to assess pERK1/2 and expression of other proteins by immunoblotting. There were variable levels of pERK across all genotypes, possibly due to contribution of non-tumor tissue. Consistent with the IHC analysis (Fig. 2B), we observed expression of p19ARF and the absence of TP53 expression in BTRev and BT mice, but not from BRAFV600E-driven lung tumors from BrafFA mice (Fig. 2E).
TP53 silencing bypasses the senescence-like proliferative arrest in BRAFV600E-induced lung adenomas
The ability to temporally dissociate BRAFV600E expression from TSG silencing led us to ask whether TP53 silencing could bypass the senescence-like proliferative arrest displayed by late-stage BRAFV600E-induced benign lung tumors (8). To that end, BRAFV600E expression was initiated in six-week old BT mice with Ad-Flp and then subsequently infected with Ad-Cre at 4, 6 or 10 weeks. We also included a cohort of BTRev mice for this experiment in which we silenced TP53 first followed by BRAFV600E activation using the same defined time-points.
Approximately 10 weeks p.i., BRAFV600E-induced adenomas display evidence of a senescence-like proliferative arrest as evidenced by decreased Ki67 expression (Fig 2B) or BrdU incorporation (Fig. 2E). These adenomas express inhibitors of the cell division cycle: TP53, p21CIP1 and p27KIP1, with no evidence of cell death (data not shown). Incidence of adenocarcinomas was highest when TP53 was silenced four weeks following initiation of BRAFV600E expression (68%, Supp. Table 1). As the time between recombination events increased, the incidence of adenocarcinomas decreased, dropping to 14.8% at 10+ weeks post initiation of BRAFV600E expression. This decrease, however, may simply reflect possibly reduced efficiency of infection of established BRAFV600E-induced lung tumors by Ad-Cre. A similar decrease in adenocarcinoma frequency was observed with increased time between TP53 silencing and BRAFV600E expression in the cohort of BTRev mice. Although low in frequency, these data strongly suggest that delayed TP53 silencing can bypass the senescence-like proliferative arrest observed in BRAFV600E-induced lung tumors to promote malignant progression.
BRAFV600E cooperates with INK4A/ARF silencing to elicit lung adenocarcinomas
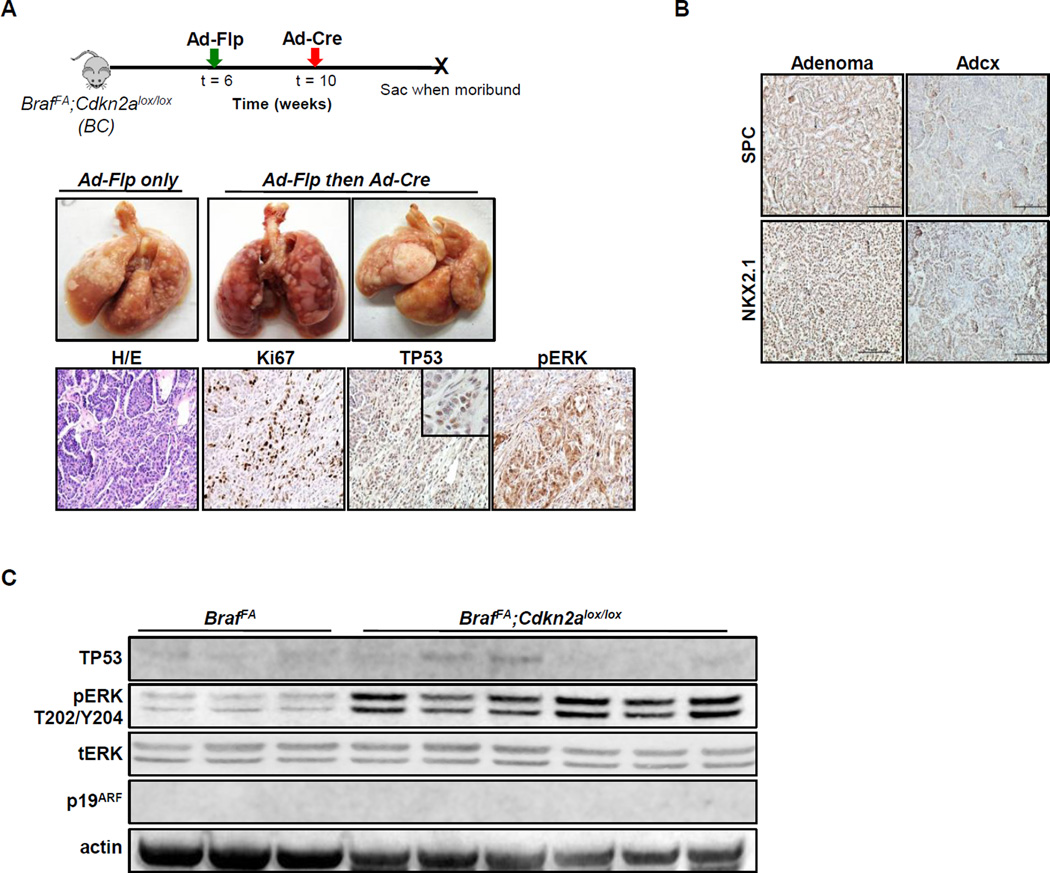
We previously demonstrated that expression of BRAFV600E combined with silencing of Cdkn2a (encoding p16INK4A, a cyclin-dependent protein kinase inhibitor and p19ARF, a regulator of TP53 activity) cooperates to induce lung adenocarcinomas (8). We therefore wished to investigate the consequences of Cdkn2a silencing in established BRAFV600E-induced benign lung tumors (BrafFA;Cdkn2alox/lox, BC mice) since both proteins are linked to senescence induction by oncogenic BRAF (24–26). Activated RAF signaling is reported to induce p16INK4A, but not p19ARF, as a trigger of cell cycle arrest in the induction of senescence in IMR-90 human fibroblasts (24,27). Similarly, RAF-induced p19ARF is thought to trigger TP53-dependent cell cycle arrest in rodent fibroblasts (25,28). Hence, we investigated the effects of activating BRAFV600E prior to INK4A/ARF silencing in the lungs of BC mice. BRAFV600E expression was initiated in six-week old BC mice with Ad-Flp and subsequently infected with Ad-Cre 4–6 weeks after tumor initiation. Similar to Trp53, silencing of Cdkn2a alone did not elicit any overt pathology in the lung. Histopathological analysis confirmed that the large lung tumors that formed in suitably manipulated BC mice were indeed adenocarcinomas as evidenced by the presence of invasive fronts, high levels of nuclear atypia and loss of differentiation. These tumors displayed readily detectable levels of pERK and were highly proliferative (Ki67). Some of these adenocarcinomas also expressed detectable levels of TP53 (Fig. 3A). Perhaps surprisingly, lung adenocarcinomas arising in BC mice appeared less differentiated than those arising in BT mice as they displayed regions of diminished expression of NKX2.1 and SP-C (Fig. 3B). Immunoblot analysis of lung tumor-derived lysates from BC mice confirmed effective recombination of Cdkn2a as evidenced by the lack of p19ARF expression. Consistent with IHC staining, tumors arising in BC mice displayed readily detectable pERK and TP53 expression when compared to BRAFV600E benign adenomas in BrafFA mice (Fig. 3C).
Figure 3. Temporal dissociation of INK4A-ARF silencing cooperates with BRAFV600E to promote lung carcinogenesis.
A. (Upper panels) Photographs of the macroscopic lungs from mice of the indicated genotypes. (Lower panels) FFPE lung sections from BrafFA;Cdkn2aflox/flox (BC) mice stained with H&E, or anti-Ki67, -TP53 or pERK by IHC.
B. Lung adenomas and adenocarcinomas from BC mice stained with antisera to detect expression of SPC or NKX2.1.
C. Immunoblot analysis of lung tumor lysates from BrafFA or BC mice as indicated.
Although Ad-Cre can infect cells in BRAFV600E-initiated tumors in BrafFA mice, the efficiency of secondary infection is generally low and shows high tumor-to-tumor variability (Fig. 2A). To enhance the efficiency of Cre-mediated recombination in established BRAFV600E lung tumors, we employed a knock-in allele (SPCCreER) in which conditionally active CreERT2 is expressed downstream of the coding sequence of SP-C, a gene uniformly expressed in BRAFV600E-induced lung tumors (8,19,29).
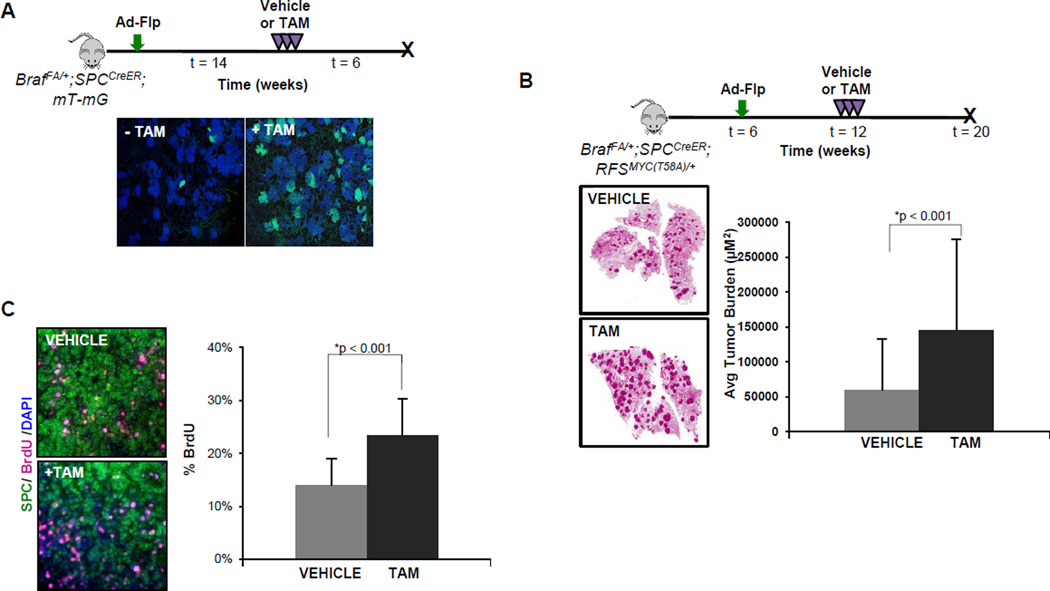
To test the efficiency of the SPCCreER allele, we utilized Rosa26mT-mG (R26mT-mG) mice that constitutively express membrane-tethered tdTomato (mT) in cells prior to the action of Cre recombinase, after which they express membrane-tethered EGFP (mG) (30). BRAFV600E-induced lung tumorigenesis was initiated with Ad-Flp in BrafFA;SPCCreER;R26mT-mG mice and were either untreated or treated with tamoxifen (TAM) 14 weeks p.i. to activate CreERT2 in SPC positive cells and euthanized 6 weeks later. Occasional EGFP positive tumors (<5%) were detected in the absence of TAM, which suggests some leakiness of the SPCCreER transgene (Fig. 4A). However, in TAM treated mice, almost all of the BRAFV600E-initiated lung tumors contained EGFP positive cells (Fig. 4A). Moreover, the percentage of EGFP positive cells in each tumor was generally high indicating the high efficiency with which the SPCCreER transgene can elicit Cre-dependent genetic alterations in Ad-Flp induced BRAFV600E driven lung tumors in mice.
Figure 4. High efficiency Cre-mediated recombination in Ad-Flp initiated BRAFV600E-driven tumors using a SPCCreER transgene.
A. Lung tumorigenesis was initiated in BrafFA;SPCCreER;mT-mG mice using Ad-Flp. 14 weeks p.i. mice were either untreated (-TAM) or treated with tamoxifen (+TAM). Six weeks later, mice were euthanized with Cre-mediated expression EGFP in lung tumors assessed by IF.
B. Lung tumorigenesis was initiated in 6 week old BrafFA;SPCCreER;RFSMYC(T58A) mice with Ad-Flp. Mice were either treated with vehicle or with tamoxifen (+TAM) 6 weeks p.i. and euthanized with lung tumor burden assessed.
C. Sections of tumor bearing lungs from BrafFA;SPCCreER;RFSMYC(T58A) mice treated with vehicle or TAM and administered BrdU prior to sacrifice. Sections were stained as indicated and BrdU positive cells were quantified below.
Recently, we demonstrated that Cre-mediated co-expression of c-MYCT58A can promote bypass of the senescence-like arrest that occurs in BRAFV600E-induced lung tumors (9, 18). To test whether expression of c-MYCT58A can reactivate proliferation in growth arrested BRAFV600E-induced lung tumors, we generated BrafFA;SPCCreER;RFSMYC(T58A) mice. Benign BRAFV600E-induced lung tumorigenesis was initiated with Ad-Flp (106 pfu) and twelve weeks p.i., mice were treated with either vehicle or TAM to induce expression of c-MYCT58A in SPC-positive, BRAFV600E-initiated lung tumor cells (Fig. 4B). Lung tumor burden was assessed in both groups of mice eight weeks later. As anticipated, SPCCreER mediated induction of c-MYCT58A expression allele led to a significant increase in overall tumor burden (p<0.05) and proliferation as gauged by Ki67 expression (not shown) or BrdU incorporation (Fig. 4C). These results confirm the utility of combining our BrafFA allele with an AT2 pneumocyte directed, SPCCreER transgene for high efficiency Cre-mediated manipulation of additional genetic events in BRAFV600E-induced lung tumor studies.
Activation of TP53 in BRAFV600E/TP53Null or BRAFV600E/INK4A-ARFNull lung cancer cells results in G1 cell cycle arrest
Since the TP53 pathway is frequently dysregulated in a wide variety of human cancers (31–34), the consequences of restoring TP53 function to cancer cells are of scientific interest and have possible therapeutic implications (23,35). To explore the consequences of reactivating TP53 in BRAFV600E-driven lung cancer cells, we generated and characterized several BRAFV600E/TP53Null (BP) or BRAFV600E/INK4A-ARFNull (BIA) lung cancer derived cell lines from suitably manipulated BT or BC mice respectively. To allow restoration of TP53 function in BP cells, they were transduced with a retrovirus encoding TP53:ER (BRAFV600E/TP53Null/TP53:ER (BPER), a conditionally active form of TP53 that requires the presence of 4-hydroxytamoxifen (4-HT) for activity or with a suitable negative control vector (pBabePuro:hbER, BPC) (36). Since BIA cells express TP53, we enhanced TP53 activity in these cells by treatment with nutlin-3a, a pharmacological inhibitor of MDM2 that leads to the accumulation of TP53 in cells (37).
As proof of concept, BPER cells (data not shown) were treated with either ethanol (ETOH) as a solvent control or 4-HT to activate TP53:ER for 48 hours with TP53:ER expression monitored by either immunofluorescence (IF) or immunoblotting (IB). Analysis by IF and IB allowed detection of TP53:ER in BPER cells that was detected with either vehicle or 4-HT with no expression detected in BPC control cells (Fig. 5A). However, at later times following 4-HT, the level of TP53:ER expression was diminished in BPER cells (data not shown). We suspect that restoration of TP53 activity would lead to induced expression of MDM2 leading to the destabilization of TP53:ER. Consistent with this hypothesis, when BPER cells were treated with 4-HT in the presence of nutlin-3a, we noted that expression of TP53:ER was sustained. We also generated subclones of BPER cells to ensure uniform levels of TP53:ER expression and performed subsequent experiments with BPER-clone D (BPER-D), which had the highest level of TP53 expression upon treatment with 4-HT.
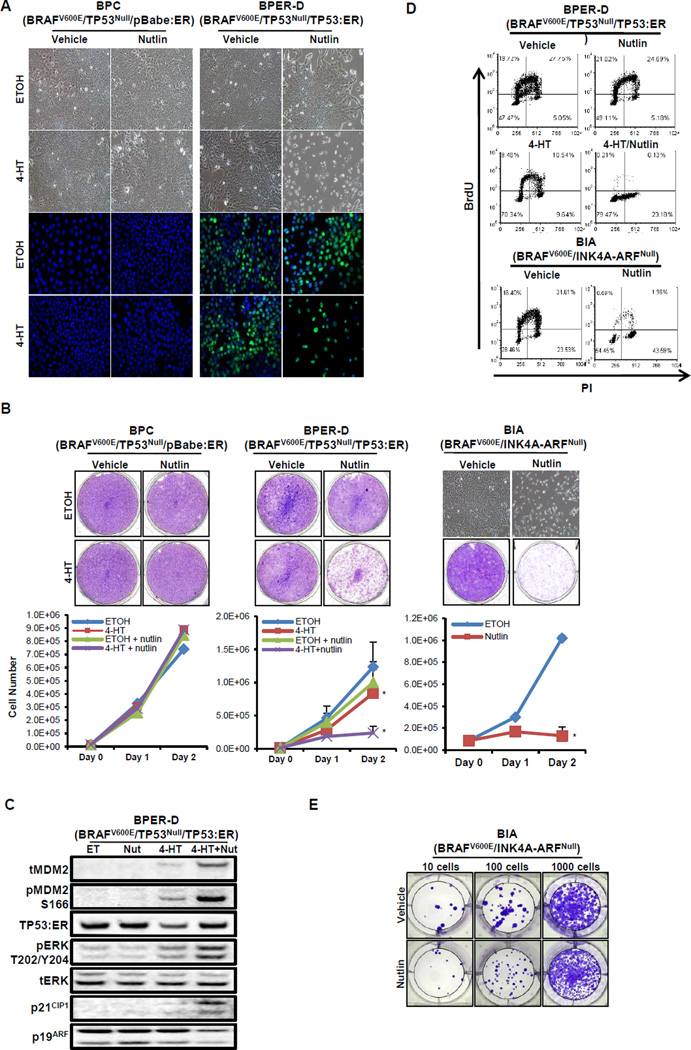
Figure 5. Restoration of TP53 function in BRAFV600E/TP53Null lung tumor cells results in a G1 cell cycle arrest.
A. Phase contrast photomicrographs of BRAFV600E/TP53Null cells infected with either a control (BPC) or a TP53:ER (BPER-D) encoding pBabePuro retrovirus vector. Cells were then treated with 4-hydroxytamoxifen (4-HT) to activate TP53:ER in the absence or presence of Nutlin-3a for 48 hours as indicated. The expression of TP53:ER (green) was assessed by IF.
B. BPC or BPER-D cells were treated with vehicle or 4-HT in the absence or presence of nutlin-3a as indicated with cell proliferation assessed using Crystal Violet staining or by cell counting (lower panels). BRAFV600E/INK4A-ARFNull (BIA) lung tumor cells were treated with vehicle or nutlin-3a as indicated with cell proliferation assessed using Crystal Violet staining or by cell counting. A 2-sided t-test was performed to assess significance (* and ** indicates p < 0.05 and p<0.001 respectively).
C. Immunoblot analysis of BPER-D cell lysates treated with ethanol (ET) as vehicle control, nutlin-3a (Nut), 4-HT or the combination as indicated.
D. Analysis of BrdU incorporation by flow cytometry in BPER-D or BIA cells after 48 hours of treatment with the indicated agents.
E. Crystal violet staining of BIA cells replated and cultured for 5 days following treatment with either vehicle or nutlin-3a for 48 hours.
We first assessed the effects of TP53:ER activation on the proliferation of BPER-D cells. Addition of 4-HT or nutlin-3a to control cells, either alone or in combination, had no effect on cell proliferation or morphology (Figs. 5A & 5B). Nutlin-3a treatment of BPER-D cells did not alter cell growth relative to ethanol treatment (1.0 ×106 vs. 1.24 ×106 cells, p = 0.17). By contrast, addition of 4-HT to activate TP53:ER reduced cell proliferation significantly compared to vehicle control (p<0.05, Fig. 5B). Moreover when TP53:ER was activated in the presence of nutlin-3a, cell growth was reduced to 19% of that observed in vehicle treated cells (p<0.001, Fig. 5B). Combination treatment also elicited alterations in cell morphology such that treated cells adopted a rounded, refractile appearance (Fig. 5A). Similar to BPER-D cells, activation of TP53 activity in BIA cells with nutlin-3a significantly reduced cell growth compared to vehicle (1.53 × 105 vs. 1.02 × 106 cells, p<0.001) and also resulted in a rounded, refractile morphology (Fig. 5B).
BPER-D cells treated with vehicle, 4-HT, nutlin-3a or the combination were analyzed by IB to assess TP53:ER activation and expression of a subset of TP53-regulated genes. Expression of TP53:ER was detected in either vehicle or nutlin-3a treated cells, but there was no expression of the TP53 targets MDM2 and p21CIP1 (Fig. 5C). Treatment of BPER-D cells with 4-HT activated TP53:ER and expression of MDM2 and p21CIP1. However, due to activation of the regulatory-feedback loop through MDM2, TP53:ER expression was reduced at later time points. By contrast, combined treatment with 4-HT and nutlin-3a led to robust total and serine 166 phosphorylated MDM2 and p21CIP1 expression, in addition to sustained TP53 levels. In addition, we observed a modest reduction in p19ARF expression, which is thought to be an indirect target of TP53 activity. These results suggest that maximal activation of TP53 in BPER-D cells requires combined treatment with 4-HT and nutlin-3a for MDM2 inhibition.
We next assessed the effects of TP53 activation on the division cycle of BPER-D or BIA cells. Cells were treated as above and labeled with BrdU and propidium iodide to assess transit through S phase and cell cycle distribution. Consistent with the proliferation assays, activation of TP53:ER (with 4-HT) led to a reduction in BrdU incorporation from 47.5% to 20% (Fig. 5D). Although nutlin-3a had no effect on its own, it dramatically potentiated the effects of TP53:ER activation reducing BrdU positive BPER-D cells to 0.34%. In addition, treatment of TP53 proficient BIA cells led to a reduction in BrdU positive cells from 48% to 1.95% (Fig. 5D). As expected, the reduction of BrdU positive BPER-D or BIA cells was accompanied with a concomitant increase in G1. Most importantly, addition of 4-HT or nutlin-3a, either alone or in combination, had no effect on the cell cycle profile in parental BP cells (data not shown).
Overall, we interpret these data to mean that reactivation of TP53 activity leads to the arrest of the BPER-D and BIA cell cycle. Since removal of the inducers of BIA or BPER-D growth arrest led to commencement of cell growth, the growth arrest is reversible and therefore not a senescence-like growth arrest (Fig. 5E and data not shown). The magnitude of the anti-proliferative effect is limited in BPER-D cells by the ability of TP53:ER to induce MDM2 that serves to inhibit the expression/activity of TP53:ER. However, when TP53:ER is activated in the presence of nutlin-3a in BPER-D cells, TP53:ER is unopposed by MDM2 and free to elicit its maximum growth suppressive properties. It is noteworthy that, even with maximal activation of TP53, we detected little to no evidence of TP53-induced apoptosis in BIA or BPER-D cells respectively. Of interest, the ability of TP53 to induce cell cycle arrest does not require p19ARF expression, which is absent from BIA cells due to Cre-mediated silencing of the Cdkn2a locus (Fig. 3C).
TP53 activation does not enhance death of BRAFV600E/INK4A-ARFNull cancer cells when combined with MEK inhibition
The ERK1/2 MAP kinase pathway is reported to promote transcriptional activation of MDM2 and consequently dampen the effects of TP53 in cells (25). Therefore, we tested the combined effects of MEK1/2 inhibition (PD325901, PD901 hereafter) and TP53 activation to determine if this combination would yield more striking anti-proliferative effects on cells. MEK inhibition dramatically reduced proliferation of BIA cells (1.02 × 106 vs. 1.23 ×105 cells, p<0.001) after 48 hours (Fig. 6A). Nutlin-3a treatment also suppressed BIA cell proliferation (1.02 × 106 vs 1.53 × 105 cells, p<0.001), while combined treatment of nutlin-3a plus PD901 further suppressed cell growth (8.6 × 104, p<0.001). Analysis of BrdU incorporation revealed that, compared to control (48% BrdU positive), nutlin-3a (~2%) more effectively inhibited S phase progression than PD901 (16.7%) with the combination most effective at inhibiting BrdU incorporation (0.2%).
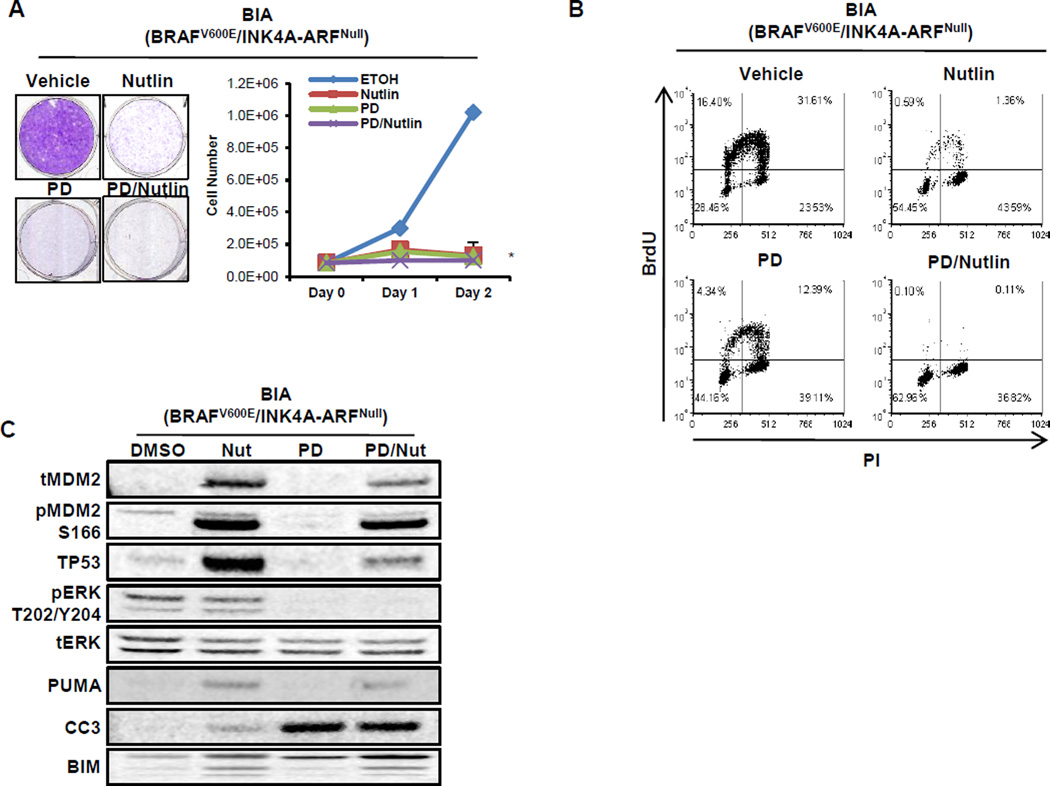
Figure 6. Inhibition of BRAFV600E/INK4A-ARFNull lung cancer cell growth by either MEK or MDM2 inhibition.
A. BRAFV600E/INK4A-ARFNull (BIA) lung cancer cells were treated with vehicle, nutlin-3a, MEK1/2 inhibitor (PD325901, PD) or the combination with cell proliferation assessed by staining with Crystal Violet or cell counting. A 2-sided t-test was performed to assess significance (* indicates p <0.001). All three conditions are significant versus vehicle, as well as combination treatment vs single agent.
B. BIA lung cancer cells were treated with vehicle, nutlin-3a, MEK1/2 inhibitor (PD) or the combination with transit through S phase of the cell division cycle assessed by labeling with BrdU. BrdU incorporation and position in the cell cycle after 24 hours of drug treatment was assessed by co-staining with an anti-BrdU antisera and propidium iodide and then detected by iF and flow cytometry.
C. Immunoblot analysis of BIA lung cancer cells treated with vehicle, nutlin-3a, MEK1/2 inhibitor (PD) or the combination as indicated.
Immunoblot analysis indicated that nutlin-3a treated BIA cells displayed increased TP53 expression with elevated expression of TP53 target genes, MDM2 and PUMA (Fig. 6C). Despite PUMA induction, a mediator of TP53-induced apoptosis (38), there was only a modest increase of cleaved-Caspase 3 (CC3), a marker of apoptosis, in nutlin-3a treated BIA cells. MEK1/2 inhibition with PD901 had no effect on MDM2 or PUMA expression but led to decreased TP53, elevated BIM expression and CC3 consistent with previous reports (39–41). However, when PD901 was combined with nutlin-3a, there was a modest diminution of both MDM2 and TP53, the significance of which is currently unclear. These results suggest that the MAP kinase pathway contributes to the TP53 regulatory loop through transcriptional activation of MDM2 as previously reported (21). Furthermore, the results demonstrate that BIA lung cancer cells express functional normal TP53 and are dependent on the BRAFV600E→MEK→ERK pathway for cell proliferation. However, they also suggest that TP53 activation does not enhance apoptosis observed following MEK inhibition.
Serum factors and differential TP53 threshold levels provide cell survival during reactivation of p53
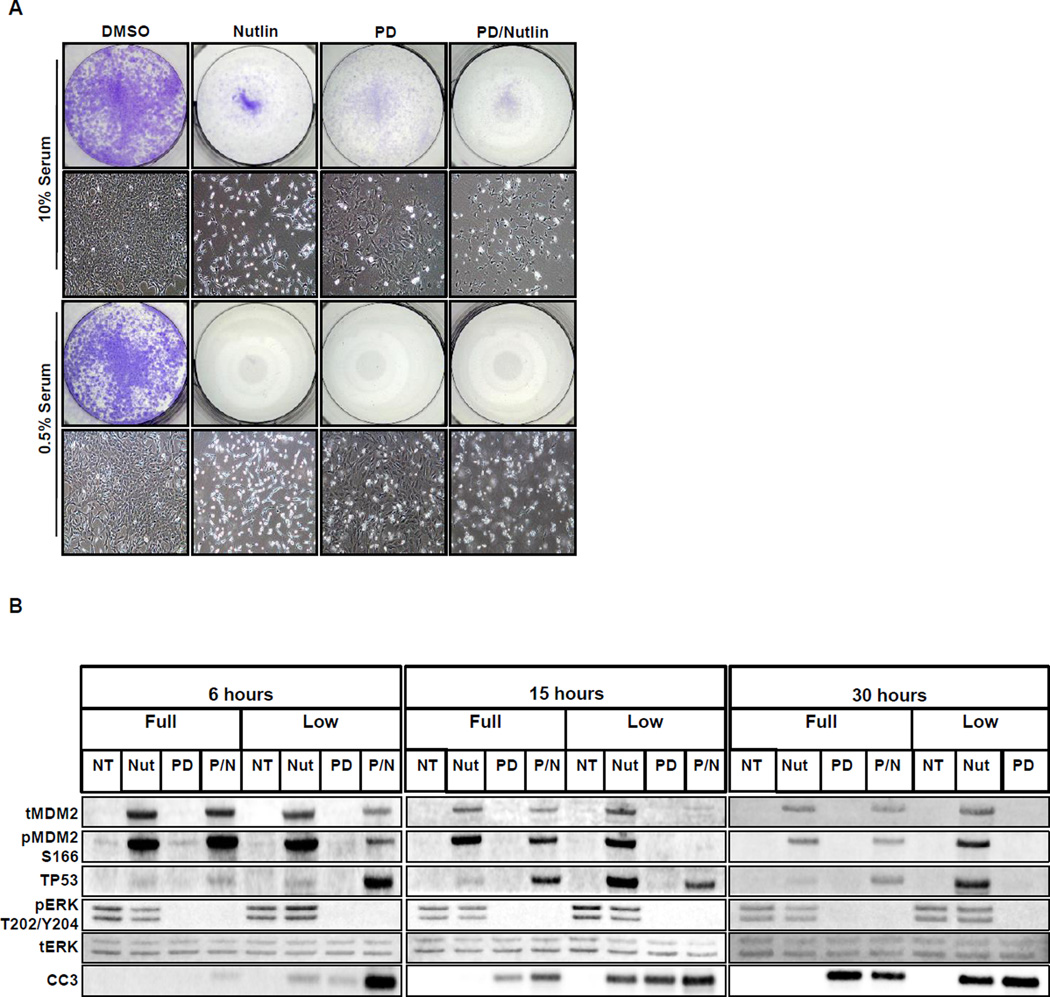
Interestingly, activation of TP53 in either BPER-D or BIA lung cancer cells resulted only in a G1 cell cycle arrest and did not elicit senescence or apoptosis. We hypothesized that perhaps factors present in fetal calf serum (FCS) may provide survival signals as well as contribute to the regulation of TP53 and/or its downstream targets. Therefore we cultured BIA cells in either full (10%) or low (0.5%) serum conditions and then activated TP53 with nutlin-3a in the absence or presence of PD901. As expected, TP53 activation in full serum led to inhibition of cell proliferation. By contrast, TP53 activation in cells cultured in low serum displayed signs of cell death as evidenced by the presence of numerous floating cells (Fig. 7A). PD901 treatment in the absence or presence of nutlin-3a enhanced cell death under low FCS conditions (Fig. 7A).
Figure 7. Serum growth factors influence the response of cells to inhibition of MDM2.
A. BIA lung cancer cells were treated with vehicle, nutlin-3a, MEK1/2 inhibitor (PD) or the combination in either 0.5%(v/v) or 10%(v/v) fetal calf serum with viable cells stained with Crystal Violet. Photomicrographs of cells in each treatment condition are also provided.
B. Immunoblot analysis of BIA cells grown in either high (10%) or low (0.5%) fetal calf serum and then treated with either control, nutlin, PD or the combination for 6, 15 or 30 hours. (NB: No sample could be harvested from BIA cells treated with the combination of nutlin-3a plus PD for 30 hours in low serum).
Cell lysates were prepared from BIA cells, cultured in 10% versus 0.5% FCS, that were treated for 6, 15 or 30 hours with nutlin-3a, PD901 or the combination of both agents (Fig. 7B). Whereas TP53 expression was elevated in response to nutlin-3a in both conditions, the magnitude of TP53 expression was higher under low serum at 15 and 30 hours post nutlin-3a addition (Fig. 7B). Again, PD901 led to substantial inhibition of pERK as early as 6 hr under both conditions. Interestingly, whereas nutlin-3a treatment of BIA cells in 10% FCS failed to elicit CC3 activation at any time point analyzed, CC3 was readily detectable in response to nutlin-3a in BIA cells cultured in low serum. Combined treatment with PD901 plus nutlin-3a led to the expected induction of CC3, which was again further increased under low FCS conditions. Indeed, we were unable to isolate sufficient cell lysates from BIA cells cultured in low serum and subjected to combination PD901 plus nutlin-3a treatment to analyze by immunoblotting. These results are consistent with the hypothesis that the presence of serum factors can gate signals elicited by activated TP53 that, in turn, contribute to whether a cell will commit to apoptosis. Furthermore, it has been suggested that high levels of TP53 are required to induce apoptosis, which may not be achieved under full serum conditions (42).
DISCUSSION
GEM models of cancer have generated a wealth of knowledge regarding the genetics, cell biology and biochemistry of cancer initiation and progression (43). They have also shed new insights on therapeutic intervention for cognate human diseases (15,44). Whereas the earliest GEM models relied on constitutive expression of activated oncogenes, recent models place an emphasis on temporal and spatial control of physiologically relevant levels of oncogene expression and appropriate cell of origin (15,45). Moreover, although tumor initiation may be driven by a single mutational event, malignant progression generally requires multiple cooperating genetic or epigenetic events, in particular oncogene activation combined with silencing of tumor suppressors. Thus more complex and sophisticated models in which multiple cooperating genes can be modified in adult mice using sequence-specific DNA recombinases such as Cre, Flp or Sleeping Beauty have become more common (46,47). A frequent drawback of using such models is that the genetic events that fuel tumor initiation and cancer progression are regulated by the same recombinase such that oncogene activation and TSG silencing occur simultaneously, a rare situation in the genesis of human cancers.
We, and others have previously described the utility of mice carrying Cre-activated alleles of Braf in various GEM models (8–10,48–51). To complement this approach, we now describe the generation of mice carrying a Braf allele (BrafFA) that encodes normal BRAF prior to recombination with Ad-Flp to encode BRAFV600E. Moreover, as previously described for a Flp-regulated KrasFSF-G12D allele, we describe the utility of BrafFA mice to generate “two-switch” models to study the progression of BRAFV600E-induced benign lung tumorigenesis to lung cancer (52). Similar to BrafCA mice, expression of BRAFV600E in the lungs of BrafFA mice gave rise to benign adenomas that failed to progress to cancer, likely due to the onset of a senescence-like proliferative arrest (8). Temporal dissociation of BRAFV600E expression from conditional silencing of either TP53 or INK4A-ARF expression promoted bypass of the senescence-like proliferative arrest observed in BRAFV600E-induced benign tumors and resulted in lung adenocarcinomas. These results suggest that the ARF-TP53 pathway is important for both the initiation and maintenance of the senescence-like proliferative arrest that occurs in BRAFV600E adenomas and is also consistent with the effects of restoration of TP53 expression in KRASG12D/TP53Null lung cancers (22,23). However, one of the limitations of using adenoviral infection as the second genetic event is the relative inefficiency of secondary infection of BRAFV600E-induced benign tumors with Ad-Cre. However, this problem is substantially overcome, by the use of an inducible SPCCre:ER transgene that is expressed in the majority of BRAFV600E benign tumor cells (8,19).
These data suggest that BRAFV600E tumors are not irreversibly genetically committed to a senescence-like proliferative arrest, consistent with the effects of expression of SV40 LT in senescent cells in tissue culture. Finally, it remains unclear what, if any role is played by the expression of p16INK4A in the senescence-like proliferative arrest displayed by BRAFV600E-induced benign tumors. Although RAF-induced p16INK4A expression is associated with oncogene-induced senescence of human IMR-90 fibroblasts, its expression is not readily detected in BRAFV600E-induced benign tumors (8). Ultimately, we will have to test the cancer promoting effects of selective silencing of either INK4A or ARF using suitably manipulated GEM models (53,54).
The BrafFA mouse model also provides an opportunity to model the effects of TP53 silencing prior to BRAFV600E expression, a situation that may occur in Li-Fraumeni patients (12,13). TP53 silencing prior to BRAFV600E expression appeared to modestly increase the multiplicity of large lung adenocarcinomas. Although TP53 silencing has no overt effect on lung epithelial cells on its own, it may potentiate BRAFV600E-induced lung carcinogenesis through effects on oncogene-induced senescence or through effects on putative stem cells of origin of lung cancer, effects on genomic instability or cell metabolism. We must also consider the less interesting possibility that TP53 deficient lung epithelial cells are more susceptible to Flp-mediated recombination of the BrafFA allele and thereby better elicit its oncogenic effects.
Previous work employing GEM models demonstrated that restoration of TP53 expression in KRASG12D/TP53Null lung cancer led to modest regression of high-grade lesions that required the expression of p19ARF. These data suggest that sustained RAF→MEK→ERK signaling driven by oncogenic KRASG12D in high-grade lesions promoted p19ARF expression that, in the absence of TP53, had no strong tumor suppressive effect. Although most of the tumor cells expressed restored TP53, inhibition of cell proliferation only occurred in cells with elevated RAF→MEK→ERK signaling and high ARF expression. This was supported by observations that restoration of TP53 expression in cultured KRASG12D/TP53Null lung cancer-derived cell lines elicits a G1 cell cycle arrest that was also p19ARF dependent (22,23). Based on these findings, we investigated whether similar observations may be made in lung tumors initiated by oncogenic BRAFV600E. In our hands, activation of TP53 in cultured BRAFV600E/TP53Null or BRAFV600E/INK4A-ARFNull lung cancer cells resulted in a reversible G1 cell cycle arrest that occurred regardless of the expression of p19ARF. Although restored TP53 expression induced pro-apoptotic proteins such as PUMA, there was no evidence of apoptosis per se unless TP53 was activated in cells cultured in low serum. These observations are consistent with the fact that FCS contains mitogenic factors such as lysophosphatidic acid and sphingosine-1-phosphate that appear to protect cells from TP53 induced apoptosis. However, these factors are insufficient to sustain the cell division cycle following TP53 activation. Indeed, the induction of G1 cell cycle arrest by TP53 may actually provide some protection against cell death in this context. It has also been reported that the levels of TP53 may play a role in determining cell fate (42) and our observation of low TP53 induction in full FCS may explain cell survival through growth arrest, rather than apoptosis. Restoration of TP53 in the face of growth factors in humans may provide a survival mechanism for tumor cells by inducing a protective growth arrest instead of cell death.
In summary, we describe a new GEM model for exploring the consequences of the temporal dissociation of BRAFV600E expression from additional cooperating events such as sustained expression of c-MYC expression or silencing of the TP53 or INK4A-ARF tumor suppressors. This model is particularly useful in the lung where the availability of viral vectors for expression of Flp recombinase can be combined with a knock-in SPCCre:ER transgene to allow ready temporal dissociation of genetic events. Moreover, the BrafFA mice will likely prove useful for extending our knowledge of the mechanisms of tumorigenesis in the wide variety of GEM models of human cancer based on conditional BRAFV600E expression (9,10,48,49,55).
Supplementary Material
ACKNOWLEDGEMENTS
We thank current and former members of the McMahon Lab for active discussions regarding this project. We thank Nate Young and Tyler Jacks for initial discussions on Adenovirus vectors encoding Flp recombinase, Rosalie Sears (OHSU) for the RFSMYC mice and Jason Rock and Hal Chapman for the SPCCreER mice. The authors also wish to thank: Jane Gordon from the HDFCCC Laboratory of Cell Analysis for assistance with microscopy and FACS analysis; Byron Hann from the HDFCCC Preclinical Therapeutics Core for advice and the Laboratory Animal Resource Center for animal husbandry. We thank Gerard Evan and Lamorna Brown-Swigart for provision and advice on the use of the p53:ER construct. This research was supported by grants from the National Cancer Institute (CA131261) and Uniting Against Lung Cancer to MM and from the American Cancer Society to AS.
REFERENCES
- 1.Hayat MJ, Howlader N, Reichman ME, Edwards BK. Cancer statistics, trends, and multiple primary cancer analyses from the Surveillance, Epidemiology, and End Results (SEER) Program. Oncologist. 2007;12(1):20–37. doi: 10.1634/theoncologist.12-1-20. [DOI] [PubMed] [Google Scholar]
- 2.Imielinski M, Greulich H, Kaplan B, Araujo L, Amann J, Horn L, et al. Oncogenic and sorafenib-sensitive ARAF mutations in lung adenocarcinoma. J Clin Invest. 2014;124(4):1582–1586. doi: 10.1172/JCI72763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heist RS, Engelman JA. SnapShot: non-small cell lung cancer. Cancer Cell. 2012;21(3):448 e2. doi: 10.1016/j.ccr.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Network CGAR. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fasbender A, Lee JH, Walters RW, Moninger TO, Zabner J, Welsh MJ. Incorporation of adenovirus in calcium phosphate precipitates enhances gene transfer to airway epithelia in vitro and in vivo. J Clin Invest. 1998;102(1):184–193. doi: 10.1172/JCI2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dankort D, Filenova E, Collado M, Serrano M, Jones K, McMahon M. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 2007;21(4):379–384. doi: 10.1101/gad.1516407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charles RP, Iezza G, Amendola E, Dankort D, McMahon M. Mutationally activated BRAF(V600E) elicits papillary thyroid cancer in the adult mouse. Cancer Res. 2011;71(11):3863–3871. doi: 10.1158/0008-5472.CAN-10-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Kobayashi T, Floc'h N, Kinkade CW, Aytes A, Dankort D, et al. B-Raf activation cooperates with PTEN loss to drive c-Myc expression in advanced prostate cancer. Cancer Res. 2012;72(18):4765–4776. doi: 10.1158/0008-5472.CAN-12-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hruban RH, Goggins M, Parsons J, Kern SE. Progression model for pancreatic cancer. Clin Cancer Res. 2000;6(8):2969–2972. [PubMed] [Google Scholar]
- 12.McBride KA, Ballinger ML, Killick E, Kirk J, Tattersall MH, Eeles RA, et al. Li-Fraumeni syndrome: cancer risk assessment and clinical management. Nat Rev Clin Oncol. 11(5):260–271. doi: 10.1038/nrclinonc.2014.41. [DOI] [PubMed] [Google Scholar]
- 13.Nichols KE, Malkin D, Garber JE, Fraumeni JF, Jr, Li FP. Germ-line p53 mutations predispose to a wide spectrum of early-onset cancers. Cancer Epidemiol Biomarkers Prev. 2001;10(2):83–87. [PubMed] [Google Scholar]
- 14.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29(4):418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 15.Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17(24):3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65(22):10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 17.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Cunningham M, Zhang X, Tokarz S, Laraway B, Troxell M, et al. Phosphorylation regulates c-Myc's oncogenic activity in the mammary gland. Cancer Res. 2011;71(3):925–936. doi: 10.1158/0008-5472.CAN-10-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman HA, Li X, Alexander JP, Brumwell A, Lorizio W, Tan K, et al. Integrin alpha6beta4 identifies an adult distal lung epithelial population with regenerative potential in mice. J Clin Invest. 2011;121(7):2855–2862. doi: 10.1172/JCI57673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juan J, Muraguchi T, Iezza G, Sears RC, McMahon M. Diminished WNT ->β-catenin ->c-MYC signaling is a barrier for malignant progression of BRAFV600E-induced lung tumors. Genes Dev. 2014;28:561–575. doi: 10.1101/gad.233627.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raymond CS, Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS ONE. 2007;2(1):e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Junttila MR, Karnezis AN, Garcia D, Madriles F, Kortlever RM, Rostker F, et al. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature. 468(7323):567–571. doi: 10.1038/nature09526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feldser DM, Kostova KK, Winslow MM, Taylor SE, Cashman C, Whittaker CA, et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010;468(7323):572–575. doi: 10.1038/nature09535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu J, Woods D, McMahon M, Bishop JM. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 1998;12(19):2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ries S, Biederer C, Woods D, Shifman O, Shirasawa S, Sasazuki T, et al. Opposing effects of Ras on p53: transcriptional activation of mdm2 and induction of p19ARF. Cell. 2000;103(2):321–330. doi: 10.1016/s0092-8674(00)00123-9. [DOI] [PubMed] [Google Scholar]
- 26.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432(7015):307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 27.Minden A, Lin A, McMahon M, Lange-Carter C, Derijard B, Davis RJ, et al. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science. 1994;266(5191):1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 28.Lomax M, Fried M. Polyoma virus disrupts ARF signaling to p53. Oncogene. 2001;20(36):4951–4960. doi: 10.1038/sj.onc.1204717. [DOI] [PubMed] [Google Scholar]
- 29.Trejo CL, Juan J, Vicent S, Sweet-Cordero A, McMahon M. MEK1/2 inhibition elicits regression of autochthonous lung tumors induced by KRASG12D or BRAFV600E. Cancer Res. 2012;72(12):3048–3059. doi: 10.1158/0008-5472.CAN-11-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 31.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137(3):413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 32.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 15(1):2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 33.Goh AM, Coffill CR, Lane DP. The role of mutant p53 in human cancer. J Pathol. 223(2):116–126. doi: 10.1002/path.2784. [DOI] [PubMed] [Google Scholar]
- 34.Junttila MR, Evan GI. p53--a Jack of all trades but master of none. Nat Rev Cancer. 2009;9(11):821–829. doi: 10.1038/nrc2728. [DOI] [PubMed] [Google Scholar]
- 35.Junttila MR, Karnezis AN, Garcia D, Madriles F, Kortlever RM, Rostker F, et al. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature. 2010;468(7323):567–571. doi: 10.1038/nature09526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christophorou MA, Martin-Zanca D, Soucek L, Lawlor ER, Brown-Swigart L, Verschuren EW, et al. Temporal dissection of p53 function in vitro and in vivo. Nat Genet. 2005;37(7):718–726. doi: 10.1038/ng1572. [DOI] [PubMed] [Google Scholar]
- 37.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 38.Hikisz P, Kilianska ZM. PUMA, a critical mediator of cell death--one decade on from its discovery. Cell Mol Biol Lett. 17(4):646–669. doi: 10.2478/s11658-012-0032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang YF, Jiang CC, Kiejda KA, Gillespie S, Zhang XD, Hersey P. Apoptosis induction in human melanoma cells by inhibition of MEK is caspase-independent and mediated by the Bcl-2 family members PUMA, Bim, Mcl-1. Clin Cancer Res. 2007;13(16):4934–4942. doi: 10.1158/1078-0432.CCR-07-0665. [DOI] [PubMed] [Google Scholar]
- 40.Cartlidge RA, Thomas GR, Cagnol S, Jong K, Molton SA, Finch AJ, et al. Oncogenic BRAF(V600E) inhibits BIM expression to promote melanoma cell survival. Pigment Cell & Melanoma Research. 2008 doi: 10.1111/j.1755-148X.2008.00491.x. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ley R, Ewings KE, Hadfield K, Cook SJ. Regulatory phosphorylation of Bim: sorting out the ERK from the JNK. Cell Death Differ. 2005;12(8):1008–1014. doi: 10.1038/sj.cdd.4401688. [DOI] [PubMed] [Google Scholar]
- 42.Kracikova M, Akiri G, George A, Sachidanandam R, Aaronson SA. A threshold mechanism mediates p53 cell fate decision between growth arrest and apoptosis. Cell Death Differ. 20(4):576–588. doi: 10.1038/cdd.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanahan D, Wagner EF, Palmiter RD. The origins of oncomice: a history of the first transgenic mice genetically engineered to develop cancer. Genes Dev. 2007;21(18):2258–2270. doi: 10.1101/gad.1583307. [DOI] [PubMed] [Google Scholar]
- 44.Casanovas O, Hicklin DJ, Bergers G, Hanahan D. Drug resistance by evasion of antiangiogenic targeting of VEGF signaling in late-stage pancreatic islet tumors. Cancer Cell. 2005;8(4):299–309. doi: 10.1016/j.ccr.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 45.Inoue M, Hager JH, Ferrara N, Gerber HP, Hanahan D. VEGF-A has a critical, nonredundant role in angiogenic switching and pancreatic beta cell carcinogenesis. Cancer Cell. 2002;1(2):193–202. doi: 10.1016/s1535-6108(02)00031-4. [DOI] [PubMed] [Google Scholar]
- 46.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15(24):3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dupuy AJ, Rogers LM, Kim J, Nannapaneni K, Starr TK, Liu P, et al. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res. 2009;69(20):8150–8156. doi: 10.1158/0008-5472.CAN-09-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collisson EA, Trejo CL, Silva JM, Gu S, Korkola JE, Heiser LM, et al. A central role for RAF-->MEK-->ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov. 2012;2(8):685–693. doi: 10.1158/2159-8290.CD-11-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41(5):544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamata T, Dankort D, Kang J, Giblett S, Pritchard CA, McMahon M, et al. Hematopoietic Expression of Oncogenic BRAF Promotes Aberrant Growth of Monocyte-Lineage Cells Resistant to PLX4720. Mol Cancer Res. 2013 doi: 10.1158/1541-7786.MCR-13-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mito JK, Min HD, Ma Y, Carter JE, Brigman BE, Dodd L, et al. Oncogene-dependent control of miRNA biogenesis and metastatic progression in a model of undifferentiated pleomorphic sarcoma. J Pathol. 2012;229(1):132–140. doi: 10.1002/path.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Young NP, Crowley D, Jacks T. Uncoupling cancer mutations reveals critical timing of p53 loss in sarcomagenesis. Cancer Res. 71(11):4040–4047. doi: 10.1158/0008-5472.CAN-10-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bertwistle D, Sherr CJ. Regulation of the Arf tumor suppressor in Emicro-Myc transgenic mice: longitudinal study of Myc-induced lymphomagenesis. Blood. 2007;109(2):792–794. doi: 10.1182/blood-2006-07-033985. [DOI] [PubMed] [Google Scholar]
- 54.Burd CE, Sorrentino JA, Clark KS, Darr DB, Krishnamurthy J, Deal AM, et al. Monitoring tumorigenesis and senescence in vivo with a p16(INK4a)-luciferase model. Cell. 152(1–2):340–351. doi: 10.1016/j.cell.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huillard E, Hashizume R, Phillips JJ, Griveau A, Ihrie RA, Aoki Y, et al. Cooperative interactions of BRAFV600E kinase and CDKN2A locus deficiency in pediatric malignant astrocytoma as a basis for rational therapy. Proc Natl Acad Sci U S A. 2012;109(22):8710–8715. doi: 10.1073/pnas.1117255109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.