Abstract
DNA helicases are molecular motors that harness the energy of nucleoside triphosphate hydrolysis to unwinding structured DNA molecules that must be resolved during cellular replication, DNA repair, recombination, and transcription. In vivo, DNA helicases are expected to encounter a wide spectrum of covalent DNA modifications to the sugar phosphate backbone or the nitrogenous bases; these modifications can be induced by endogenous biochemical processes or exposure to environmental agents. The frequency of lesion abundance can vary depending on the lesion type. Certain adducts such as oxidative base modifications can be quite numerous, and their effects can be helix-distorting or subtle perturbations to DNA structure. Helicase encounters with specific DNA lesions and more novel forms of DNA damage will be discussed. We will also review the battery of assays that have been used to characterize helicase-catalyzed unwinding of damaged DNA substrates. Characterization of the effects of specific DNA adducts on unwinding by various DNA repair and replication helicases has proven to be insightful for understanding mechanistic and biological aspects of helicase function in cellular DNA metabolism.
Keywords: helicase, DNA damage, genomic instability, DNA repair, genetic disease
Graphical abstract
1. Introduction
Helicases involved in cellular replication, DNA repair, and transcription are likely to be among the first proteins to encounter a DNA lesion; moreover, helicase-dependent mechanisms are vital to how cells cope with endogenous or exogenous stress. Therefore, understanding how modifications to the base, sugar, or phosphate backbone of the DNA double helix affect helicase-catalyzed unwinding of the complementary strands will be insightful from both biochemical and biological viewpoints. In this review we will provide the readers a sense of the recent developments in understanding the consequences of helicase encounters with damaged DNA from an experimental perspective. An emphasis is placed on laboratory approaches that have been used for in vitro helicase studies with damaged DNA substrates so that the reader can appreciate what has been learned and some novel aspects of helicase interactions with unique forms of DNA damage that remain understudied. Although the focus of the published work has largely been on DNA unwinding measurements, future studies will likely address the effects of DNA adducts on translocation, branch-migration, and even strand annealing catalyzed by certain helicases or helicase-like proteins. With new discoveries pertaining to structural and biophysical properties of DNA helicases and their mechanisms of DNA unwinding, it seems probable that a comprehensive understanding of how helicases tolerate DNA damage will provide insights for how this specialized class of molecular motors behave in biological situations that evoke cellular responses to DNA damage and replication stress.
2. DNA Lesions and Their Effects on Helicases
Over the years, a number of laboratories including ours have investigated the effects of DNA damage on catalytic DNA unwinding performed by various purified recombinant DNA repair and replication helicases. These experiments assessed the ability of DNA helicase proteins to unwind a variety of site- and strand-specific DNA lesions located at defined positions in partial duplex DNA substrates. Such lesions are broadly categorized into two classes: 1) base modifications; 2) backbone modifications. This general classification of DNA lesions can be useful for investigating the importance of helicase contacts with the DNA substrate during single-stranded DNA translocation or duplex DNA unwinding. As discussed recently in a review from the Keck laboratory [1], crystal structures of DNA helicases from the conserved Superfamily (SF)1 and SF2 along with mechanistic studies of helicase proteins have provide important insights to how they bind and unwind DNA. This has enabled researchers to conceptually visualize how structural elements of the helicase domain and other protein domains make dynamic contacts with the bases and sugar phosphate backbone to enact base-pair (bp) separation in a manner dependent on nucleoside triphosphate hydrolysis. Studying DNA modifications that affect the bases versus the backbone of DNA can be a useful approach for probing mechanistic aspects of helicase function. Moreover, a number of the DNA lesions tested for their effects on unwinding by helicases are biologically relevant as they potentially interfere with normal maintenance of chromosomal stability and genome homeostasis. In addition, recent developments in the area of helicase-catalyzed protein-DNA remodeling are likely to be relevant to events that occur in the metabolism of DNA-protein covalent complexes (DPC) or interstrand cross-links (ICL). In the following sections, we will highlight some insights gained in understanding the effects of DNA damage on helicase function, placing an emphasis on mechanistic aspects and biological pathways characterized by recent developments within the past several years. For a more historical perspective of earlier discoveries describing effects of DNA damage on helicase function, the reader is referred to several comprehensive review articles [2–4].
3. Backbone Modifications
The large majority of biochemical studies with helicases and damaged nucleic acid substrates characterized by sugar phosphate backbone modifications have focused exclusively on synthetic non-natural types of linkages such as peptide-nucleic acid [5], polyvinyl [6,7], or polyglycol [8,9] insertions, or a methylphosphonate modification [10]. However, an emerging area of study is the biochemical and biological effects of backbone alterations arising from chemical genotoxins that can be found in the environment or are used in chemotherapy. Phosphotriester (PTE) adducts are recognized as a prominent class of such DNA lesions [11]. Thus from not only a mechanistic standpoint but also a health perspective, it is important to evaluate how helicases and other DNA metabolizing proteins are affected by chemical alterations to the DNA backbone.
3.1. Polyglycol Phosphotriester Linkage
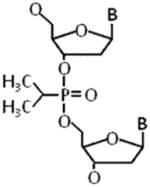
A sugar phosphate backbone modification that has been particularly useful for probing the structural elements of a DNA substrate important for efficient helicase unwinding is the polyglycol linkage that can be synthetically incorporated into the backbone. A typical polyglycol linkage which we have used in our helicase studies is an 18-atom triethylene glycol linker that spans a distance of 3 bp in duplex DNA and 6 nucleotides (nt) in single-stranded DNA (Table 1). The polyglycol linkage neutralizes the negative charge characteristic of the phosphate groups within the backbone; therefore, DNA helicase assays with this type of substrate can provide insight to the importance of ionic interactions between surface amino acid residues of the helicase protein and the DNA phosphodiester bonds. The synthetic polyglycol linkage can be placed in the single-stranded tail of the partial duplex substrate to investigate mechanistic aspects of single-stranded DNA translocation and the initiation of DNA unwinding by a helicase. To our knowledge, the first application of a polyglycol linkage in a DNA substrate to characterize a helicase was employed by the Lohman laboratory in which they provided evidence that Escherichia coli Rep helicase unwinds duplex DNA by an active mechanism [8]. More recently, we observed that the Fanconi Anemia Group J helicase (FANCJ, also called BACH1 or BRIP1) that is genetically implicated in Fanconi Anemia (FA) as well as breast and ovarian cancer [12,13] is inhibited by a polyglycol modification positioned in the 5′ single-stranded tail, but not the 3′ single-stranded tail, when it is located immediately adjacent to the duplex region of the forked helicase substrate [14]. This finding is consistent with the known 5′ to 3′ directionality of FANCJ translocation on single-stranded DNA [15].
Table 1.
Representative Backbone Modifications
| NAME | STRUCTURE |
|---|---|
| Triethylene Glycol |
|
| Alkyl Phosphotriester |
|
Information about helicase protein-DNA substrate interactions during the elongation phase of DNA unwinding can be acquired from experiments in which the polyglycol linkage is inserted in one of the two strands within the duplex segment of the helicase substrate. In this case, knowledge regarding the importance of helicase contacts with the sugar phosphate backbone of the DNA substrate can be inferred from helicase measurements. We evaluated the effect of a polyglycol linkage positioned within the duplex region 16 bp away from the fork entry and 12 bp away from the blunt duplex end of the DNA substrate [16]. This substrate was designed to assess if helicase contacts with the translocating or non-translocating strand during the elongation phase of the helicase-catalyzed DNA unwinding reaction were important. To our surprise, we observed that under multi-turnover conditions FANCJ helicase was profoundly sensitive to the polyglycol linkage in either the translocating or non-translocating strands of the duplex substrate [16]. This behavior of FANCJ was similar to certain other DNA helicases (e.g., Escherichia coli DnaB [17], hepatitis C virus NS3 [5]), but is not generalizable to other DNA helicases which display strand-specific inhibition (e.g., vaccina virus NPH-II [9], bacteriophage T7 gene 4 [18], bacteriophage T4 Dda [10,19]). We observed that the human RecQ helicase BLM defective in Bloom’s syndrome was strongly inhibited by the polyglycol linkage residing within the duplex in the helicase translocating strand but much less so in the non-translocating strand [20]. Interestingly, we found that FANCJ helicase could more efficiently unwind the DNA substrates with the polyglycol linkage in either strand nested within the duplex when the length of the 5′ single-stranded tail on which FANCJ loads was increased from 16 nt to 35 nt [16]. This result suggested that a longer single-stranded tail can enable multiple FANCJ molecules to load and generate the force production necessary to drive forward progression and translocate past the backbone modification, leading to favorable unwinding.
A subtle but informative point should be made from the FANCJ helicase studies with DNA substrates containing a polyglycol linkage in the preexisting single-stranded tail of a partial duplex forked DNA substrate versus substrates that have the polyglycol linkage nested within the duplex region of the forked substrate. When the polyglycol linkage is in the single-stranded tail, unwinding of the forked duplex substrate is only adversely effected when the backbone modification resides in the helicase translocating strand [14]. In contrast, FANCJ helicase activity is negatively impacted by the polyglycol linkage in either the translocating or non-translocating strand when the backbone modification is positioned within the duplex [16]. These results suggest that FANCJ helicase contacts with the sugar phosphate backbone in the single-stranded DNA translocation phase as the helicase loads onto the substrate during initiation of unwinding are distinguishable from those backbone contacts made by FANCJ in the duplex DNA unwinding event itself that occur during the elongation phase. A take-home message of the FANCJ helicase studies with the polyglycol linkage substrates is that the dynamics of single-stranded translocation are quite distinct from those of duplex unwinding. This topic requires further study and insight may be gleaned from combinatorial approaches including structural studies and single-molecule assays that employ uniquely positioned molecular beacons in the single-stranded or double-stranded regions of the DNA substrate. Furthermore, it is of interest if the characteristic backbone interactions of FANCJ are conserved with other Fe-S cluster helicases and if they are applicable for helicase interactions with naturally occurring backbone lesions such as alkyl phosphotriester linkages (see below).
Oligomeric assembly state may play a role in the ability of a helicase to overcome sugar backbone modifications as well as base lesions. The dimeric form of FANCJ displays superior activity over the monomeric form for helicase activity on an undamaged forked duplex DNA substrate and ATPase activity in the presence of the covalently closed M13 single-stranded DNA as the effector [21]. Therefore, it would be informative to test if the monomeric versus dimeric forms of FANCJ are differentially active in reactions containing DNA molecules which harbor sugar phosphate backbone lesions in the single-stranded or double-stranded elements. The role that helicase multimerization plays in tolerance of specific DNA adducts is very much understudied and requires immediate attention, given that helicases display a spectrum of oligomerization states which profoundly influence DNA substrate specificity and DNA unwinding efficiency on undamaged DNA substrates. In addition, the structural elements of non-helicase domains that determine the relative ability of a helicase to tolerate a backbone lesion have largely not been addressed, and will likely shed new insight to mechanisms whereby helicase-catalyzed unwinding of damaged DNA can be modulated.
DNA substrates with a polyglycol linkage nested within the duplex provided a useful tool to interrogate the putative functional interaction of the FANCJ and BLM helicases, which were found to physically bind to each other [20]. It was observed that FANCJ and BLM synergistically unwound a duplex DNA substrate with a polyglycol linkage nested within the duplex in the FANCJ translocating strand. Furthermore, the ability of a helicase-inactive FANCJ-K52R mutant to stimulate BLM helicase activity on the forked duplex substrate with the polyglycol linkage in the FANCJ translocating strand is consistent with the idea that the physical interaction between FANCJ and BLM contributes to the functional interaction. Future work should address what pathway(s) FANCJ and BLM collaborate to unwind DNA substrates with covalent damage or alternative DNA structure (e.g., G-quadruplex). One hypothesis is that FANCJ and BLM cooperate with each other to remodel stalled replication forks or with end-processing nucleases (e.g., DNA2, EXO-1) during strand resection phase of homologous recombination (HR) repair [22].
3.2. Alkyl Phosphotriester Linkage
In addition to covalent modifications imposed upon bases within genomic DNA, alkylating drugs can react with oxygen atoms of the phosphate groups residing within the sugar-phosphate backbone (for review, see [11]). This can result in the attachment of a hydrophobic alkyl group to a non-carbon-bonded oxygen and neutralization of the negative charge. The alkyl PTE modification can project out from the helical axis or project into the major groove, depending on its stereochemistry and covalent attachment site. PTE adducts are chemically stable and have been found to affect activities catalyzed by a variety of DNA metabolizing enzymes including cellular DNA polymerases and nucleases. Importantly, PTE lesions can arise from mutagenic and carcinogenic alkylating agents present in the environment. Characterizing the effects of PTE adducts on DNA helicases which encounter such backbone lesions during replication or DNA repair will help us to better understand their deleterious effects in vivo.
In our recent work, we tested a panel of DNA repair and replication helicases to unwind a forked duplex DNA substrate in which a single isopropyl PTE lesion (Table 1) was centrally positioned in either strand of the forked duplex DNA substrate, 12 bp downstream of the helicase entry site at the fork and 12 bp upstream of the blunt duplex end [23]. The design of the DNA substrate enabled the detection of helicase inhibition by the backbone modification even under conditions that the helicase partially unwound the duplex prior to the site of the PTE lesion because the remaining 12 bp between the adduct and the blunt duplex end provide enough stability for the duplex to remain intact under the helicase reaction conditions. Scoring fully unwound single-stranded DNA product by its fast migration on a native polyacrylamide gel in the classic all-or-none radiometric helicase assay is suitable for this application; however, partial unwinding of the DNA substrate prior to the site of the lesion can be inferred from a fluorometric dye displacement kinetics assay (see Experimental Approaches section below). Using reaction conditions optimal for each helicase under investigation, we found that all SF2 helicases of the Fe-S family and RecQ family (with the exception of the Werner syndrome protein (WRN)) were inhibited by the PTE lesion in a strand-specific manner, i.e., by the backbone modification in the helicase translocating strand under multi-turnover conditions. WRN was significantly inhibited by the PTE in either strand. In contrast, the Escherichia coli SF1 DNA repair helicase UvrD was unaffected by the PTE in either strand. Similarly, the ring-like replicative helicases DnaB and MCM were insensitive to the PTE lesion in either strand. Thus, the SF2 DNA repair helicases behave strikingly different from the replicative helicases or SF1 UvrD helicase, suggesting distinct mechanisms of DNA unwinding. It seems probable that the replicative helicases would be able to efficiently accommodate the translocating strand isopropyl PTE lesion within the central ring as it occludes the opposing strand outside the donut hole in a steric fashion. The fact that replicative helicases were resistant to inhibition of unwinding by the PTE lesion raises the possibility that uncoupled fork unwinding from blockage of DNA synthesis catalyzed by a DNA polymerase may pose a source of single-stranded DNA which would provide a signal for SOS induction in prokaryotes or checkpoint activation in eukaryotes (Fig. 1). Cellular studies to address this hypothesis are warranted.
Figure 1. Eukaryotic replicative helicase and DNA damage checkpoint activation.
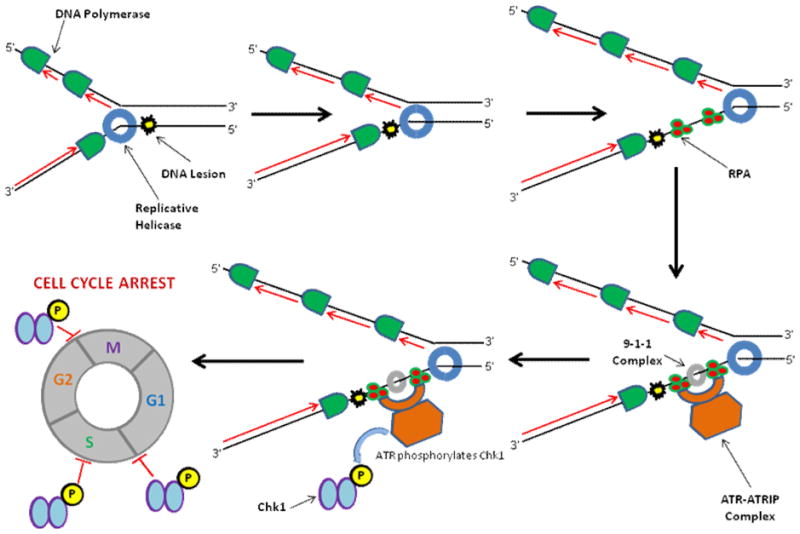
The eukaryotic replicative MCM helicase complex is able to unwind past a DNA lesion at the replication fork. The adduct blocks DNA synthesis catalyzed by a trailing replicative DNA polymerase, leading to the generation of single-stranded DNA which is coated by RPA. The ATRIP/ATR complex and other factors are recruited to the site of replication stress, where ATR is activated and can phosphorylate Chk1, leading to cell cycle arrest.
Interpretation of helicase-catalyzed unwinding on a damaged DNA substrate compared to an undamaged control substrate can be complex under ensemble conditions, particularly when multiple helicase molecules can load on a DNA substrate during the time period of incubation (multi-turnover conditions). Kinetic analysis under single-turnover conditions in which a helicase protein trap, typically a large excess of a long homo-polynucleotide (e.g., dT120) is added simultaneously with ATP to initiate the reaction is useful to measure DNA unwinding because in principle it precludes successive loading of helicase molecules on the DNA substrate during the time course (see Experimental Approaches section). Therefore, the single-turnover assay can provide a more sensitive measure of helicase inhibition by a given DNA lesion. Under single-turnover conditions, we determined that RECQ1 unwinds the DNA substrate with the isopropyl PTE lesion in the translocating strand at a significantly lower rate compared to the control (undamaged) substrate or the substrate harboring the PTE adduct in the non-translocating strand [23]. In the future, single-molecule fluorescence-based assays, discussed in a Methods paper from the Spies lab [24], will be a useful approach to study mechanistic aspects of helicase interaction with damaged DNA substrates.
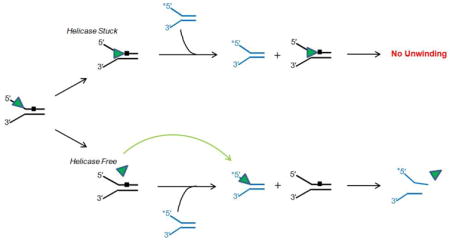
An additional mechanistic aspect of RECQ1 helicase inhibition by the PTE lesion that was addressed in vitro was sequestration. Helicase sequestration refers to the concept that a DNA lesion not only prevents further unwinding by the helicase but also causes the helicase to become trapped by the lesion harbored within the duplex rather than to immediately dissociate from the partially unwound damaged DNA substrate (see Experimental Approaches section). The action of a pinball in a pinball machine provides a good analogy for helicase sequestration versus dissociation (Fig. 2). A pinball that rapidly bounces off a cylindrical bumper provides a visual aid for non-sequestration, i.e., rapid dissociation of the helicase from the damaged DNA. In contrast, the concept of sequestration is analogous to a captive pinball that falls into a depression in the pinball table and becomes trapped in this small area of the playfield. One can imagine the ramifications of a helicase sequestered by DNA damage versus a helicase that upon collision with the obstacle becomes immediately available for other DNA unwinding tasks.
Figure 2. A pinball machine and RECQ1 helicase sequestration by a phosphotriester backbone DNA modification.
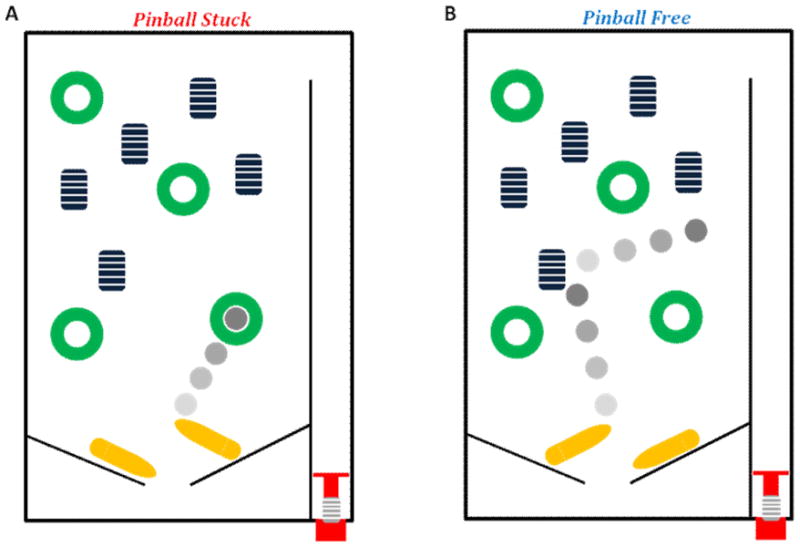
RECQ1-catalyzed DNA unwinding is inhibited by an alkyltriester linkage in the translocating strand of the DNA substrate and the helicase becomes trapped by the backbone lesion. Like a pinball that becomes captured when it falls into a depressed pocket, the helicase is unable to escape when it becomes snared by the backbone damage (Panel A). In contrast, depending on the type of lesion, certain helicases readily dissociate from the lesion, much like a pinball that bounces off a bumper (Panel B). See text for details.
Helicase sequestration assays were set up by pre-incubating RECQ1 in the presence of ATP with increasing amounts of unlabeled partial duplex DNA molecules containing the PTE lesion in the translocating or non-translocating strands versus unlabeled undamaged partial duplex DNA molecules. A radiolabeled DNA substrate of unique sequence that is efficiently unwound by the helicase (tracker substrate) is subsequently added to the reaction mixture and assayed for DNA unwinding. If the helicase under investigation rapidly dissociates from the unlabeled damaged DNA substrate (i.e., helicase bounces off the bumper), it readily becomes available for unwinding the tracker substrate. This experimental approach is very useful for assessing the relative sequestration effects exerted by a site-specific DNA lesion. Interestingly, RECQ1 was sequestered by the translocating strand PTE lesion [23], the first demonstration to our knowledge that a backbone modification induced by an alkylating agent can cause a helicase protein to become trapped on the DNA substrate it unsuccessfully attempted to unwind. Further studies should address the relative importance of helicase inhibition and sequestration by alkylated DNA damage induced by environmental agents or medicinal drugs for their mutagenic and carcinogenic effects.
4. Base Modifications
DNA base residues are susceptible to a wide range of chemical modifications arising from endogenous cellular biochemical processes (e.g., oxidation) or environmental agents (e.g., ultraviolet (UV) light, or chemical carcinogens) [25–27]. These modifications alter the chemical nature of the base by introducing functional groups to the pyrimidine or purine moiety or altering the covalent linkage of the base to the sugar (e.g., cyclopurine lesions). Hydrolysis of the sugar-phosphate bond altogether gives rise to an abasic site. Depending on the base lesion, there can be localized distortion of the DNA double helix resulting in perturbation of base stacking and bending or twisting of the helical axis. We will discuss some recent research advances characterizing the effects of unique base modifications on DNA helicase activity.
4.1. UV-Induced Cyclobutane Pyrimidine Dimer
UV light from the earth’s sun can damage genomic DNA in vivo. Among the UV-induced lesions, cyclobutane pyrimidine dimers (CPD) (Table 2) are recognized by the nucleotide excision repair (NER) machinery [28]. Recognition of DNA damage in the context of the NER pathway is quite complex. An early player in CPD recognition is the XPD helicase, a component of the eukaryotic TFIIH complex implicated in not only NER but also transcription initiation. XPB, a second DNA helicase of the TFIIH complex, also plays a dynamic role with XPD in DNA damage scanning and verification (for review, see [29–31] and references therein). These experiments have proven to be informative for understanding the structural mechanics of DNA damage sensing by XPD and the basis for cellular and clinical phenotypes of the XPB and XPD missense mutation-associated diseases Xeroderma pigmentosum (XP), Cockayne syndrome (CS), and Trichothiodystrophy.
Table 2.
Representative Base Modifications
| NAME | STRUCTURE |
|---|---|
| Thymine Glycol |
|
| 8-Oxo-Guanine |
|
| 8,5-cyclopurine-2-deoxynucleoside |
|
| Cyclobutane Pyrimidine Dimer |
|
| Benzo[c]phenanthrene |
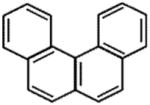
|
| Benzo[a]pyrene Diol Epoxide |
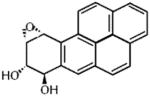
|
Yeast XPD and various archaeal XPD helicases have been tested for their sensitivity to CPD and other bulky lesions, and the results from these studies are quite mixed, suggesting differences in the ability of sequence-related XPD proteins to detect DNA damage [32,33]. Classic studies from the Friedberg lab showed that Saccharomyces cerevisiae XPD is inhibited by UV-induced DNA photoproducts and cisplatin-induced bulky adducts specifically positioned in the helicase-translocating strand [34–36]. Unique insight was gleaned from molecular characterization of two NER-defective Fe-S domain missense mutations in an XPD homolog of Ferroplasma acidarmanus (faXPD) [37]. Using a 81 bp forked duplex substrate harboring a CPD positioned 57 bp downstream of the fork entry site, the investigators were able to show that DNA unwinding of the CPD substrate was unaffected for the faXPD Fe-S missense mutants irrespective of strand, in contrast to the wild-type faXPD protein which was inhibited by the CPD in the helicase translocating strand. Moreover, neither Fe-S cluster mutant protein formed a stable protein-DNA substrate complex with the forked duplex containing the CPD lesion, suggesting they were defective in the DNA damage verification step. Altogether the cellular and biochemical data provided evidence that the DNA damage sensing ability of faXPD is required for its function in NER.
Thermoplasma acidophilum XPD (taXPD) was studied using a single-molecule approach by atomic force microscopy and it was observed that binding to a DNA lesion within duplex DNA was dependent on the type of adduct [38]. taXPD preferentially recognized a fluorescein adduct, representative of a bulky DNA lesion, that resided in the helicase translocating strand, whereas it more readily bound a CPD lesion in the non-translocating strand. It is conceivable that a less bulky lesion is recognized by taXPD in the non-translocating strand because it contacts the damage sensor, whereas a translocating strand CPD lesion passes smoothly through the channel of the helicase [38].
4.2. Polycyclic Aromatic Hydrocarbons
Polycyclic aromatic hydrocarbons (PAH) such as Benzo[c]phenanthrene (BcPh) and Benzo[a]pyrene (BaP) Diol Epoxide (DE) adducts (Table 2) in DNA arise from the concerted effects of cytochrome P-450 and epoxide hydrolase [39]. PAH are known to be mutagenic and carcinogenic, and arise from metabolites found in the environment, including diet and the combustion of fossil fuel and tobacco. Seminal studies in the metabolism, replication and repair of PAH were performed in several laboratories, including that of the late Dr. Donald M. Jerina of the National Institute of Diabetes, Digestive and Kidney Diseases, NIH. We had the opportunity to collaborate with Dr. Jerina’s laboratory to characterize the effects of defined stereoisomers of BcPh and BaP DE adducts on DNA unwinding by the WRN helicase [40,41]. From a mechanistic standpoint, these particular lesions are of great interest in DNA metabolism studies with directionally specific enzymes like ATP-dependent helicases because the hydrocarbon moieties of certain adducts intercalate into DNA with defined polarity, on the 3′ or the 5′ side, relative to the adducted base, depending on their stereochemistry. Therefore, it would be anticipated that the effects of PAH DE adducts on DNA unwinding catalyzed by a helicase may be strand- and stereoisomer-dependent. That is in fact what we observed for a defined BcPh lesion and the WRN helicase (see below). Forked duplex DNA substrates were constructed from oligonucleotides (25-mers) containing diastereomerically pure cis- and trans-opened BcPh DE-dA adducts synthesized as their 5′ phosphates by a semi-automated procedure with a manual step for coupling of the BcPh DE-dA-adducted phosphoramidites as a mixture of their 1R/1S diastereomer [41]. Absolute configurations of the chromatographically separated diastereomeric R/S pairs of cis- and trans- adducted oligonucleotides were assigned by circular dichroism.
For the DNA substrates harboring the BcPh DE-dA lesion in the strand that WRN translocates, both the cis-opened and trans-opened adducts inhibited WRN-catalyzed duplex unwinding; however, the cis-opened isomers inhibited more than the trans-opened isomers [41]. Inhibition of WRN helicase activity was strictly dependent on the stereochemistry of the trans-opened BcPh DE-dA adduct located in the strand opposite to the one WRN translocates. Only an adduct oriented toward the advancing helicase was able to inhibit WRN helicase activity. We envision this as being analogous to a gladiator ready for combat with his sword confronting the advancing warrior (Fig. 3). When the gladiator is facing him, the warrior risks being injured, whereas when the gladiator is facing the opposite direction, the warrior can deftly sneak by him. Based on our experimental findings, we concluded that the sensitivity of WRN helicase to a BcPh DE-dA adduct in the non-translocating strand projecting toward the advancing helicase is at least in part dependent on the major groove occupation of the intercalated BcPh or perturbation of the DNA double helix itself.
Figure 3. Gladiators and WRN helicase inhibition by a polyaromatic hydrocarbon DNA adduct.
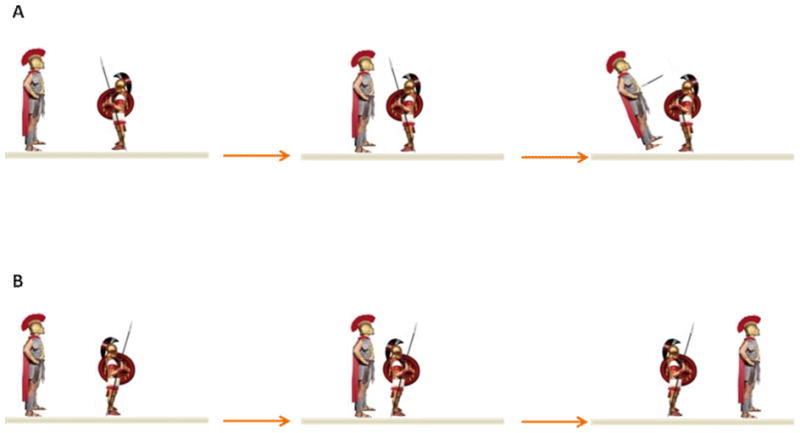
Inhibition of WRN helicase activity by a trans-opened BcPh DE adduct residing in the strand opposite to the one WRN translocates is dependent on its stereochemistry and the orientation it projects in the axis of the DNA double helix. Like a gladiator faced toward its enemy to engage in battle (Panel A), the BcPh lesion projecting toward the advancing helicase is thwarted. However, a BcPh lesion projecting in the opposite direction relative to the advancing helicase does not inhibit DNA unwinding, analogous to a rear-facing guard who allows the gladiator to pass by him without being stopped (Panel B). See text for details.
The effects of BaP DE adducts on double helical DNA are distinct from those of BcPh, but like the BcPh adduct are related to the base to which they are attached (dA, dG) and their stereochemistry ([40] and references therein). For cis-opened BaP DE –dG adducts, the modified base and its complement are flipped out from the axis of the DNA double helix regardless of stereochemistry, whereas the trans-opened BaP DE-dG adducts occupy the minor groove with defined directionality but do not significantly distort B-form DNA. In contrast, the BaP DE-dG adducts intercalate between base pairs causing them to buckle and twist in the immediate vicinity of the adduct as well as induce some local unwinding of nearby base pairs and bending of helix axis. When WRN was tested for unwinding of the BaP DE-dA DNA substrates, there was surprisingly only a very modest effect of the BaP DE-dA adducts. However, there was a strong inhibition of WRN helicase by the BaP DE-dG adduct in the helicase translocating strand, irrespective of its stereochemistry. The additional presence of the single-stranded DNA binding (SSB) protein Replication Protein A (RPA), which physically and functionally interacts with WRN [42], enabled the helicase to efficiently unwind the DNA substrates with either stereoisomer of BaP DE-dG in the helicase translocating strand [40]. This was the first evidence that RPA may serve as a critical auxiliary factor for a helicase to unwind damaged DNA. Subsequent biochemical studies with DNA substrates harboring thymine glycol (TG) also demonstrated the importance of RPA for helicase-catalyzed unwinding of substrates with that base lesion [43] (see below).
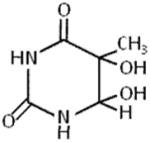
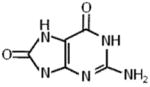
4.3. Thymine Glycol and 8-oxoguanine
Damage to DNA arising from reactive oxygen species induces a wide spectrum of base lesions on a daily basis. Two prominent oxidative lesions which exert very different effects on DNA structure are 8-oxoguanine (8-oxoG) and thymine glycol (TG) (Table 2), which are estimated to occur at a frequency of hundreds of lesions per cell per day [44]. TG causes a significant localized perturbation to the DNA double helix, resulting in a bend and extra-helical residence of the TG [45]; consequently, TG is known to pose a formidable block to cellular DNA replication. In contrast, 8-oxoG exerts only a very mild perturbation of the duplex but is mutagenic by causing G:C to T:A substitutions. We reasoned that the dramatic differences in the effect of TG versus 8-oxoG on DNA structure would be apparent when we tested the effects of these lesions on DNA unwinding catalyzed by helicase proteins. Indeed, a single TG residing in either strand at a position 10 bp downstream of the fork entry site and 15 bp upstream of the blunt duplex end caused a strong inhibition of FANCJ helicase activity under multi-turnover conditions, whereas 8-oxoG did not have any detectable effect [43]. However, the effect of TG was not universal; human RecQ helicases WRN or BLM displayed inhibition of helicase activity only by the translocating strand TG. The bacterial DNA helicases DinG, DnaB, and UvrD were largely unaffected by TG in either strand, suggesting apparent differences in mechanisms of unwinding in terms of DNA damage tolerance by the various helicases studied.
The physical and functional interaction of FANCJ with RPA and their strong co-localization in human cells subjected to agents that induce replicative stress [46] led us to ask if RPA might stimulate FANCJ to unwind damaged DNA such as the TG lesion, which is known to impede DNA synthesis catalyzed by DNA polymerases [47]. Our biochemical results demonstrated that RPA significantly stimulated FANCJ helicase activity on the forked duplex substrate that contained the TG in the strand FANCJ displaces, but no stimulation was observed when the TG resided in the translocating strand [43]. The stimulatory effect of RPA on FANCJ helicase activity was specific because Escherichia coli SSB failed to stimulate FANCJ and RPA failed to stimulate bacterial DinG helicase. The strand-specificity of RPA’s stimulatory effect on FANCJ helicase activity with the TG DNA substrate series suggested to us that the nature of the interaction of FANCJ with the TG positioned in the helicase-translocating strand versus non-translocating strand was fundamentally different. In support of this, we determined from sequestration experiments that FANCJ preferentially dissociates from the forked duplex DNA substrate it is attempting to unwind when the TG resides in the strand that FANCJ translocates in a 5′ to 3′ direction. In terms of RPA, we discovered that RPA preferentially binds to single-stranded DNA, but not double-stranded DNA, harboring a TG with a 14-fold greater affinity than undamaged single-stranded DNA of the same sequence [43]. Two other relevant observations were made: 1) RPA heterotrimer with an RPA70 missense mutation that causes a DNA binding defect failed to stimulate FANCJ helicase activity; 2) RPA stimulated RECQ1 helicase activity on the TG substrate in a strand-specific manner, i.e., only when the TG resided in the strand opposite to the one that RECQ1 translocates in a 3′ to 5′ direction. Collectively, these findings led us to propose a model in which RPA stimulates FANCJ (or RECQ1) unwinding past the TG strand by binding to the exposed TG in the non-translocating strand as the helicase attempts to unwind the nearby duplex, leading to the progression of the helicase past the TG lesion. In contrast, when FANCJ encounters the lesion in the translocating strand, the helicase dissociates from the substrate, and the partially unwound strands reanneal without affording RPA the opportunity to bind the TG in its single-stranded state. Further studies that address the biological significance of high affinity binding of RPA to TG or other types of oxidative lesions and the role this plays in RPA strand-specific stimulation of DNA processing activities are warranted. For example, involvement of RPA in glycosylase incision of TG or transcription of TG-containing template DNA should be examined in light of RPA’s high affinity binding to single-stranded harboring a TG and its strand-specific effects on DNA metabolizing enzymes such as helicases.
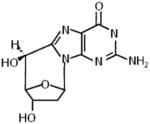
4.4. Cyclopurine
In addition to the more classical DNA base-specific effects of oxidative stress, a prominent form of oxidative DNA damage that makes a covalent modification to both the base and its connecting sugar is the 8,5′-cyclopurine-2′-deoxynucleoside (cPu) (Table 2) [48]. The cPu DNA lesion is generated by a hydroxyl radical attack of the H5-atom of the sugar moiety leading to a carbon centered radical that reacts with the C8 position of the purine (guanine (G) or adenine (A)), ultimately creating a very stable glycosidic covalent bond in the cyclization reaction. The cPu lesion in double-stranded DNA alters helix twist and base pair stacking [49,50], and is recognized by proteins of the NER pathway [51,52]. cPu lesions interfere with replication [53] and transcription [51], are proposed to be involved in XP neurological disease [54], and accumulate in congenic progeroid mice as well as aged wild-type mice [55].
Given their unusual structure in DNA and their biological profile, we investigated the effects of site- and strand-specific cyclo dA or cyclo dG adducts on duplex DNA unwinding by a panel of SF1, SF2, and SF4 DNA helicases [56]. These studies revealed wide-ranging effects on DNA unwinding catalyzed by the purified recombinant helicase proteins, even helicases within the same family such as the SF2 Fe-S cluster helicases which share significant sequence homology within the helicase core domain. Within a conserved family of DNA helicases, the RecQ enzymes behaved the most uniformly with respect to the cPu lesions in which residence of the lesion specifically in the translocating strand caused inhibition of DNA unwinding under multi-turnover conditions for BLM, WRN, RECQ1, and Escherichia coli RecQ. Inhibition of RECQ1 helicase activity by the translocating strand cPu was confirmed to also occur under single-turnover conditions. Interestingly, the effect of cPu on DNA unwinding by SF1 Escherichia coli UvrD was dependent on the conditions of the assay. UvrD helicase activity was unaffected by the cPu adduct under multi-turnover conditions, whereas it was inhibited by the translocating strand cPu under single-turnover conditions, suggesting that loading of multiple UvrD molecules on the DNA substrate under multi-turnover conditions allows lesion bypass during unwinding.
For the Fe-S cluster helicases, human FANCJ helicase was strongly inhibited by a translocating strand cPu under multi-turnover or single-turnover conditions; furthermore, sequestration experiments demonstrated that FANCJ was preferentially trapped by the translocating strand cPu. In contrast, the sequence-related human DDX11 helicase defective in the chromosomal instability disorder Warsaw Breakage syndrome was only very modestly affected by the translocating strand cPu under multi-turnover or single-turnover conditions. Archaeal Thermoplasma acidophilum XPD (taXPD), homologue to the human XPD helicase implicated in NER DNA damage verification, was only inhibited by the cPu in the non-translocating strand, whereas Escherichia coli DinG was unaffected by cPu in either the translocating or non-translocating strand. These findings strongly challenge the assumption that DNA helicases sharing sequence homology within the helicase core domain operate by the same mechanism of DNA unwinding. Thus, although recent evidence from studies of Sulfolobus acidocaldarius XPD has implicated its redox active Fe-S cluster in electron transport through double helical DNA as a mechanism for DNA lesion detection [57], it seems likely that DNA helicases of the Fe-S family operate by distinct mechanisms that are at least in part lesion- and context- dependent.
5. Novel DNA Lesions from a Helicase Perspective
Although much progress has been made in characterizing the effects of certain types of DNA damage on helicase function, there are some fairly common as well as more unusual lesions that have attracted considerable interest but remain understudied from a helicase perspective. We will highlight some of these, and suggest avenues of investigation that may be undertaken using approaches described or related to ones in this review.
5.1. Mono-Ribonucleotide
Ribonucleotides are frequently misincorporated into DNA during cellular replication (see [58–61] and references therein). The additional presence of the 2′ OH group in the sugar moiety of a mono-ribonucleotide alters the structural and mechanical properties of duplex DNA at the site of ribonucleotide as well as the nucleotide on the adjacent 3′ side. Thus, base angles and backbone shape are perturbed by a single internal ribonucleotide within duplex DNA, which can cause replication stress by impeding replisome machinery, leading to genomic instability. Defective removal of mono-ribonucleotides from DNA causes the genetic disorder Aicardi-Goutieres syndrome [62]. Despite the significant amount of research activity to understand the molecular and cellular defects stemming from the presence of ribonucleotides in DNA, the effect of mono-ribonucleotide incorporated into duplex DNA on helicase-catalyzed DNA unwinding has not been examined. A general rule is that helicases are largely RNA-specific or DNA-specific, and although some are known to unwind DNA-RNA duplexes depending on the nature of the loading strand, it seems likely that their specialization reflects basic differences in how the helicases interact with nucleic acid and impose their nucleotide-driven base pair separation. Given the abundance of ribonucleotides in DNA and their emerging prominence, it will be informative to characterize the effect of this misincorporated nucleotide on helicase catalytic function and helicase-dependent pathways in vivo.
5.2. DNA-Protein Cross-Link
Work pioneered by the Raney lab exploited the high affinity noncovalent interaction of streptavidin bound to biotinylated oligonucleotides to study the directionality of single-stranded DNA translocation by a helicase [63]. While the biotin-streptavidin complex is a potent block to single-stranded DNA translocation or duplex unwinding by many helicases, a few (e.g., FANCJ [64], gp41, Dda [65]) can effectively use their ATPase motor function to displace streptavidin off a biotinylated DNA molecule. Using streptavidin-bound biotinylated oligonucleotide-based DNA substrates that mimic roadblocks on the leading or lagging strand template, it was determined that the eukaryotic replicative DNA helicase MCM with its 3′ to 5′ directionality selectively bypasses a lagging strand roadblock, suggesting a steric exclusion model for DNA unwinding [66], a model similar to that proposed for the bacterial replicative helicase DnaB, only the latter translocating on the opposite strand with a 5′ to 3′ directionality [67].
Whereas biotin-streptavidin DNA substrates are useful for mechanistic studies, there has been emerging interest in the metabolism of covalent DNA-protein complexes (DPC) that are believed to arise biochemically in cells from endogenous processes, environmental agents, or certain chemotherapy drugs. The first study to report the effect of a site-specific and structurally well-defined DPC on helicase function was from the McCullough lab [68]. A partial duplex forked DNA substrate with a unique deoxyuridine in the translocating or non-translocating strand within the duplex was reacted with uracil DNA glycosylase to create an apurinc/apyrimidinic (AP) site followed by incubation with T4 pyrimidine dimer glycosylase/AP site lyase (T4-pdg) protein in the presence of the reducing agent NaBH4. This causes the covalent linkage of a 16-kDa protein to the DNA strand originally containing the deoxyurdine. In addition to the large steric effect, the DPC exerts a major bend of 70° in the helical axis. The purified recombinant bacterial UvrD helicase was tested for unwinding of the DPC substrates and inhibition was observed only in the case that the DPC resided in the helicase translocating strand. Interestingly, no sequestration of UvrD was detected by the translocating strand DPC, suggesting that UvrD readily dissociated from the DNA substrate upon encountering the obstacle. Elevated concentrations of UvrD were able to overcome the helicase inhibition exerted by the DPC in the translocating strand. It is possible that the ability of UvrD to initiate DNA unwinding from blunt duplex ends at higher protein contributions may contribute to unwinding the substrate with DPC in the translocating strand.
In addition to DNA repair, DPCs are likely to affect cellular replication. In Escherichia coli, it is proposed that DPCs induced by the action of DNA methyltransferase cause replication fork stalling by blocking progression of DnaB [69]; however, this remains to be formally shown. The Walter lab investigated the metabolism of a DPC using a cell-free Xenopus egg extract system and plasmid containing a site-specific DPC in which a DNA methyltransferase was covalently linked to its recognition site [70]. They determined that a leading strand template DPC arrests replication progression by causing the eukaryotic replicative helicase complex of Cdc45, MCM2-7 and GINS, designated CMG, to stall. The DPC is subsequently degraded, leaving a peptide-DNA remnant that can be bypassed by CMG. While leading strand synthesis can then resume, a translesion polymerase is required to replicate past the peptide-DNA lesion. A DPC on the lagging strand template causes transient stalling of the replisome as well as degradation of the DPC which permits copying of the Okazaki fragment. Importantly, DPC degradation is required for a primary pathway of lesion bypass, and it appears that CMG collision with the DPC is necessary for replication-coupled DPC proteolysis. A second proteolysis-independent bypass mechanism may involve either transitory opening of the CMG ring to accommodate the lesion in the leading strand template without helicase stalling or a yet-to-be-identified 5′ to 3′ accessory helicase may be recruited to dislodge CMG as the helicase complex separates strands past the lesion [70]. Mounting evidence from studies in bacteria and yeast strongly suggests that accessory helicases facilitate replication of DNA bound noncovalently by proteins (for review, see [71]), supporting their possible involvement in CMG displacement from the site of a covalent DPC. In eukaryotes, RPA may very well be involved in helicase-catalyzed CMG displacement, supported by findings that RPA-interacting human DNA repair helicases efficiently disrupt high affinity protein-DNA interactions in an RPA-dependent manner [72].
5.3. DNA Interstrand Cross-Link
A DNA ICL covalently links opposite strands of the DNA double helix, and can arise from lipid peroxidation (e.g., malondialdehyle) or common therapeutic drugs (e.g., psoralen/UVA, mitomycin C, cisplatin) [44]. It has been historically thought that the highly toxic nature of an ICL is attributed to the fact that it poses a strong blocks to cellular DNA replication or transcription; consequently, elaborate DNA repair mechanisms exist to correct ICLs. Among the most prominent of DNA ICL repair mechanisms is that provided by proteins defective in the hereditary disease Fanconi Anemia (FA), a conduit which orchestrates proteins from several different DNA repair pathways to remove the ICL and replace it with normal base-pairing sequence (for review, see [73]).
Covalent ICL DNA may serve as a useful tool to study DNA-interacting proteins that behave as base-flipping enzymes. Single base flip-out is utilized by proteins that covalently modify a base (e.g., methyltransferase [74]) or facilitate the excision of a damaged base (e.g., DNA glycosylase [75] or photolyase [76]). The ICL DNA substrate may be useful to study the thermodynamics, protein interactions, and mechanistic aspects of base-flipping by such nucleobase-modifying or nucleobase-removal enzymes. Work from the Wigley lab suggests that some SF1 helicases like Escherichia coli PcrA flip DNA bases out as hydrophobic contacts between aromatic amino acids of the helicase are made with the displaced planar bases [77]. Thus, ICL DNA substrates may be useful to study the mechanism of action of DNA helicases by halting progression of translocation and providing a snapshot of DNA intermediates and their interactions with the helicase during duplex unwinding. The ICL substrate may be particularly useful for study of helicases such as UvrD which unwind one base pair at a time [78]. Technical developments have been made in approaches to make chemically defined DNA cross-links [79]. Up to this point, such ICL DNA substrates have not been used to study DNA helicase unwinding mechanisms, but it seems likely this experimental strategy is on the horizon with the rapid development of single-molecule studies. An interesting question is if the advancing helicase can sense a DNA lesion or perturbation of the duplex caused by the lesion ahead of its arrival to the site.
From a biological perspective, DNA helicases or helicase-like motor ATPases may play unexpected roles in ICL repair. Research from the Seidman laboratory using a single-molecule approach for detection of replication fork encounters with ICLs in living cells showed that the DNA translocase complex FANCM/MHF promotes replication traverse of DNA ICL dependent on intact FANCM ATPase activity [80]. Thus surprisingly, an ICL was demonstrated to not be an absolute block to replication. Whether FANCM translocates the MCM helicase complex past the ICL or serves to activate replication restart on the opposite side of the ICL remains to be seen. In other work, the CSB ATPase/translocase defective in the rare premature aging disorder CS was found to play a role in replication-independent processing of DNA ICLs [81].
Similar to the situation mentioned above for a DPC, an accessory helicase may promote the unloading of the replicative CMG helicase complex from chromatin when the replication fork encounters an ICL. DNA repair studies using Xenopus egg extract demonstrated that the tumor suppressor protein BRCA1 plays a role in the eviction of CMG helicase when a replication fork is stalled by an ICL, allowing cross-link unhooking by dual incisions in an early event of ICL repair [82]. Reconstitution experiments suggested that other proteins or specific protein modifications in addition to BRCA1 are also required for CMG helicase eviction, raising the possibility that accessory helicases may play a role; however, this topic requires further investigation. This could be addressed experimentally with purified recombinant proteins in vitro using a modified approach of the helicase-catalyzed protein displacement procedure (described in the Experimental Approaches section) to determine if accessory helicases can dissociate CMG helicase stalled at an ICL. Among the candidate accessory helicases which may function in this capacity is the BRCA1-interacting helicase FANCJ. Aphidicolin exposure was observed to delay CMG unloading at an ICL, suggesting the involvement of a DNA polymerase to cast away CMG helicase [82]. Characterizing the precise mechanism whereby BRCA1, DNA polymerase, or other DNA repair factors such as accessory helicases promote CMG eviction during an early step of DNA repair is relevant to understanding their importance for chromosomal stability and resistance of cancer cells to DNA cross-linking agents.
6. Experimental Approaches to Monitor Helicase Activity on Damaged DNA Substrates
Advances in the synthesis of phosphoramidites representing a plethora of DNA adducts has provided new opportunities for researchers to assess how site- and strand-specific lesions affect the DNA binding and catalytic functions of various proteins (including DNA helicases) implicated in different pathways of nucleic acid metabolism. The design of DNA substrates with uniquely positioned adducts and the analysis of helicase-catalyzed unwinding of these substrates is described in the following sections. In the future, single-molecule studies and specialized DNA substrates with unique lesions and strategically placed molecular beacons are important areas of technical development to study helicase interactions with damaged DNA.
6.1. Design of DNA Substrates with Site- and Strand-Specific Adducts
A wide range of chemically synthesized phosphoramidites containing a great number of structurally diverse and biologically relevant forms of DNA damage are available for purchase from companies such as Glen Research Corporation (Sterling, VA) and Berry and Associates, Inc. (Dexter, MI). These phosphoramidities can be purchased and sent to an oligonucleotide synthesis company such as Lofstrand Labs Limited (Gaithersburg, MD), Midland Certified Reagent Company, Inc. (Midand, TX), or Integrated DNA Technologies (Coralville, IA) to be incorporated into a custom oligonucleotide. Alternatively, some commonly used DNA damage phosphoramidites (e.g., 8-oxoG) can be purchased directly from the synthesis company for incorporation into the oligonucleotide.
There are several considerations for the design of modified oligonucleotides. Some modified phosphoramidites require non-standard synthesis conditions to prevent unwanted reactions during synthesis which adds to the cost of manufacturing. For example, the TG phosphoramidite requires UltraMild synthesis conditions; however, part of this cost can be alleviated by using a deoxythymidine (dT) at the 3′ position of the oligonucleotide adjacent to the TG which eliminates the need for an UltraMild column for synthesis. In deciding on placement of the damaged phosphoramidite, the researcher should consider several issues. Placing the phosphoramidite in one of the single-stranded tails of a forked DNA substrate or near the single-stranded/double-stranded junction of a partial duplex DNA substrate could affect loading of the helicase, whereas placing it deep within the double-stranded portion of the substrate would be expected to affect the elongation phase of the unwinding reaction. In order to avoid interference of helicase loading on a partial duplex DNA substrate, the lesion can be placed close to a full turn of B-form DNA (10 bp) downstream of the helicase entry site at the fork.
When placing the adduct in the double stranded region of a DNA substrate, it is important to consider how it may affect the stability of the duplex. Some adducts are known to reduce the stability of duplex DNA. If the adduct is positioned too close to the blunt end of the partial duplex substrate on the opposite side as the helicase entry site at the fork, it is possible that the residual duplex of a helicase-catalyzed partially unwound substrate will destabilize and appear as a released product in the typical all-or-none radiometric helicase assay, despite helicase blockage by the lesion. To avoid this, a theoretical melting temperature (Tm) for the duplex region between the lesion and the blunt end can be calculated based on percent G/C content and monovalent cation concentration in the helicase reaction mixture. This Tm value can be compared to the incubation temperature and salt concentration of the helicase reaction to assess if significant destabilization of the partially unwound duplex substrate would be predicted to occur. Typically, we design DNA substrates with at least 12–13 bp with ≥50% G/C content on the blunt duplex side of the lesion to address this issue.
Another consideration is to make versatile oligonucleotides containing the DNA lesion that can be used to make multiple DNA substrates. If the researcher desires to test the effect of the DNA damage on multiple helicases, the substrate should be designed to test 5′ - 3′ helicases or 3′ - 5′ helicases. Typically, we make sure to construct DNA substrates with the adduct positioned in either the Watson or Crick strands (or neither strand) so that strand-specific inhibition of helicase activity can be assessed. The various DNA substrates used in our helicase studies were constructed from PAGE-purified oligonucleotides to avoid contamination of partially synthesized oligonucleotides that might be present in the crude oligonucleotide preparation. DNA substrates are typically prepared by annealing 10 pmol of 32P- 5′-end labeled oligonucleotide to 25 pmol of unlabeled partially complementary oligonucleotide in 50 mM NaCl. DNA substrates are stored at 4 °C, and are typically stable for several weeks.
6.2. Protocols to Measure Helicase-Catalyzed Unwinding of Damaged DNA
In the following sections, we will describe the rationale and basic set-up for experimental protocols designed to address the interactions of helicases with damaged DNA substrates. These procedures are focused on measuring helicase-catalyzed unwinding of duplex DNA substrates harboring site-specific lesions under multi-turnover or single-turnover conditions. In addition, we discuss a sequestration assay to measure helicase protein trapping by a DNA adduct and an unwinding assay that measures partial unwinding of DNA substrates. Finally, we discuss a procedure to measure protein displacement from a DNA substrate that may be useful for studies to examine the role of accessory helicases to dislodge helicases or DNA translocases stalled by covalent DNA modifications.
6.3. Standard Radiometric Strand Displacement Helicase Assay
The standard radiometric strand displacement assay addresses the ability of a helicase protein to unwind a duplex DNA substrate as a function of helicase protein concentration under multi-turnover conditions in a single time point [83] (Fig. 4). Historically, the standard radiometric helicase assay was performed with a DNA substrate consisting of a radiolabeled oligonucleotide or denatured restriction fragment annealed to a covalently closed single-stranded circular DNA molecule derived from a bacteriophage. From a more practical experimental standpoint, an oligonucleotide-based duplex DNA substrate with a lesion positioned in a specific strand and location within the duplex or single-strand overhang is now conventionally used to assess the effect of the adduct on helicase-catalyzed DNA unwinding. Reactions are typically conducted in 20 μl volumes containing 0.5 nM of the radioactively labeled forked duplex DNA substrate; however, the DNA substrate concentration can be modified according to application. The oligonucleotide-based forked duplex DNA substrate consists of partially complementary oligonucleotides with one of the two strands being kinase-labeled on the 5′ end using 32P-γ-ATP. The buffered helicase reaction mixtures contain DNA substrate, and the appropriate salt and ATP concentrations with a divalent cation (e.g., Mg2+ for WRN [84]) present as an essential cofactor optimized for the helicase under investigation. Subsequently, increasing concentrations of the helicase protein are added to each 20 μl reaction mixture and incubated at a specified temperature for an allotted period of time. Alternatively, ATP can be used to initiate the reaction. At the end of the reaction incubation period, 2X Stop buffer containing a molar excess of the divalent cation-chelating agent EDTA and a 10- to 100-fold excess of the unlabeled version of the radiolabeled strand in the DNA substrate (to prevent reannealing of the unwound radiolabeled strand) is added to each reaction mixture and the DNA products are resolved on a non-denaturing (typically 12% (19:1 acrylamide: bisacrylamide)) polyacrylamide gel for a 25–30 bp forked duplex substrate.
Figure 4. Schematic of radiometric strand displacement assay.
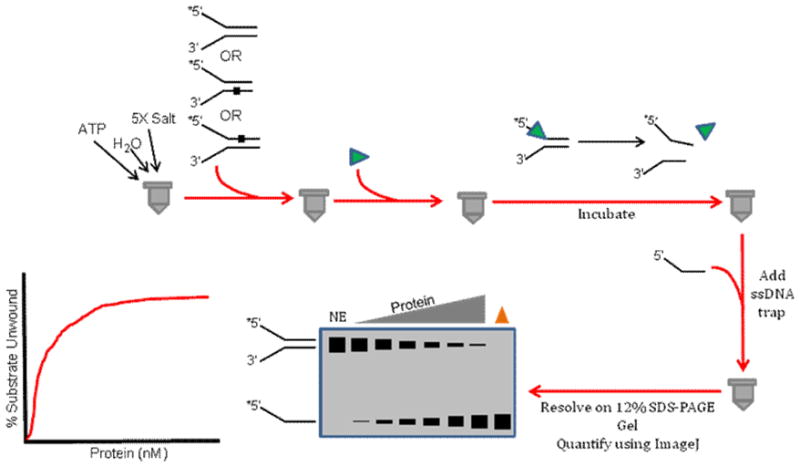
This assay is conventionally used to measure DNA unwinding as a function of protein concentration under multi-turnover conditions. See text for details.
Note that for certain DNA helicases that possess a strong annealing activity (e.g., mitochondrial Twinkle helicase [85], or Rothmund-Thomson syndrome helicase RECQL4 [86]), optimal helicase activity may require the presence of excess unlabeled oligonucleotide sharing the same base-pairing sequence as the radiolabeled strand in the helicase substrate during the helicase reaction incubation period. For helicase proteins that display high binding affinity to DNA (e.g., RECQ1 [87]), it may be necessary to incubate the helicase reaction products with 0.1 mg/ml Proteinase K at 37 °C for an additional time period (e.g., 15 min) after the EDTA quench to digest any helicase protein bound to the DNA substrate or single-stranded DNA products. After the DNA species are resolved by electrophoresis, the gel is visualized using a PhosphorImager and quantified using the ImageQuant software.
6.4. Helicase Sequestration Assay
The helicase sequestration assay is performed when the results obtained from standard strand displacement assays demonstrate helicase inhibition by a given DNA lesion. In order to assess if the helicase is trapped by the lesion or instead readily dissociates from the duplex upon encountering the site-specific damage, the helicase sequestration (protein-trapping) assay is very useful (Fig. 5). The first step in the helicase sequestration assay requires pre-incubating a defined concentration of helicase protein with increasing amounts (0–25 fmol) of unlabeled partial duplex DNA molecules containing the site- and strand-specific lesion in the presence of ATP and appropriate reaction salts for a preset period of time (typically 3–5 min) at optimal temperature to allow helicase binding and DNA unwinding. After this pre-incubation phase, 10 fmol of radioactively labeled lesion-free forked tracker DNA substrate of relatively short duplex length (e.g., 20 bp) that is unrelated to the damaged duplex molecule is added to the reaction mixture which is subsequently incubated for a defined period of time (e.g., 10 min). The reactions are then quenched, resolved on non-denaturing polyacrylamide gels, and visualized and quantified as described above. If the helicase under study is trapped by the lesion as it attempts to unwind the unlabeled DNA duplex during the pre-incubation period, the enzyme will show reduced unwinding activity on the radiolabeled tracker DNA substrate during the subsequent incubation period. However, if the helicase either rapidly dissociates from the DNA upon lesion encounter or efficiently unwinds the damaged duplex without being inhibited to any significant extent, then the helicase will be free to unwind the tracker substrate. By performing the helicase sequestration assays with increasing concentrations of unlabeled forked duplex DNA harboring the lesion in the top, bottom, or neither strand, one can compare the results from the unwinding data on the tracker DNA substrate to gain a relative indication of the degree of helicase sequestration by the damaged DNA and if the sequestration is strand-specific. We have used this assay to assess trapping of various DNA repair helicases by oxidative base lesions such as TG [43] or cPu [56] as well as backbone modifications such as an alkyltriester linkage [23].
Figure 5. Schematic of helicase sequestration assay.
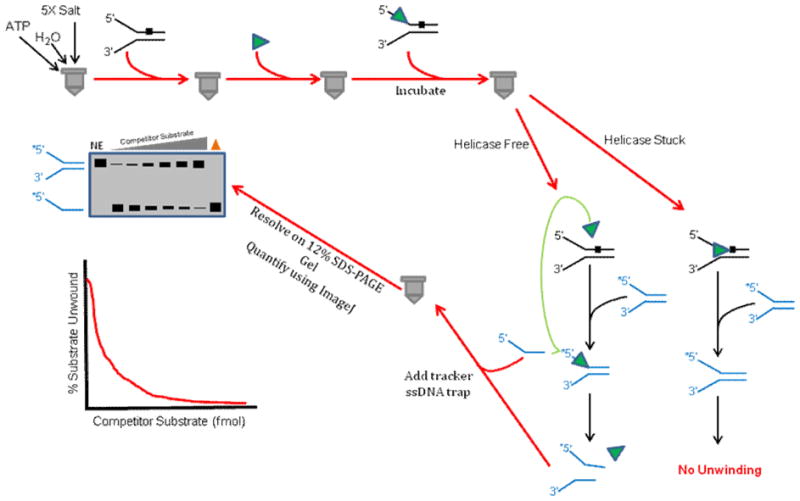
This assay is used to assess if a helicase that is blocked by a DNA lesion becomes trapped on the damaged DNA substrate or if it readily dissociates and becomes available to unwind another DNA substrate molecule. See text for details.
6.5. Single-Turnover Protein Trap Kinetics Assay
In some cases, a DNA lesion may only partially deter DNA unwinding by a helicase under multi-turnover conditions. Loading of multiple DNA helicase molecules onto the DNA substrate may allow a stalled helicase molecule to unwind past the lesion, analogous to tandem locomotives driving the lead locomotive up a steep hill. To assess experimentally if functional cooperativity among helicase molecules allows DNA unwinding past a lesion, it is informative to perform single-turnover protein trap kinetic assays (Fig. 6). This is accomplished by using a radiolabeled DNA substrate concentration (either containing the lesion or not) in which the helicase molecules are saturated with DNA substrate molecules. The helicase and DNA substrate are preincubated to allow binding, and the reaction is initiated by the simultaneous addition of ATP and a large (e.g., 100-fold) excess of dT200 to discourage helicase molecules free in solution from loading on to partially unwound forked duplex DNA substrate during the time course of the reaction. Helicase reaction mixtures are quenched and analyzed as described above for standard radiometric multi-turnover helicase assays. Typically, kinetic experiments are performed to achieve linear rates of DNA unwinding, and rates are compared for helicase-catalyzed unwinding of the damaged versus non-damaged DNA substrate. Often, single-turnover kinetic helicase assays are more stringent than multi-turnover helicase assays to assess the effect of a DNA lesion on DNA unwinding. We have employed single-turnover assays to assess the effect of an alkyltriester lesion [23] or a cyclopurine adduct [56] on helicase-catalyzed DNA unwinding.
Figure 6. Schematic of single-turnover protein trap kinetic assay.
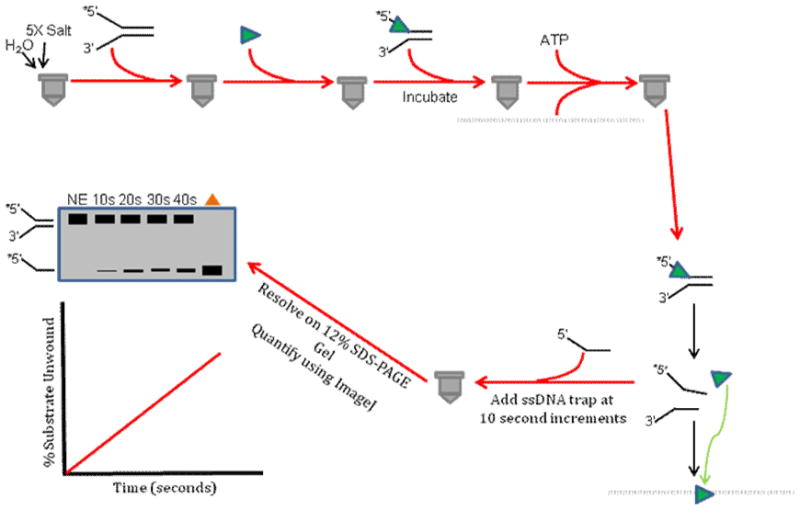
This assay is used to determine a rate of unwinding for a given helicase under conditions in which a helicase molecule (monomer or higher order oligomer) that either dissociates from the DNA substrate during the unwinding reaction or completely unwinds the substrate and subsequently dissociates from the unwound strands is prevented from binding and initiating unwinding of another DNA substrate molecule during the incubation period by the presence of excess single-stranded oligonucleotide that serves as a helicase protein trap. See text for details.
6.6. Fluorometric Dye Displacement Kinetic Assay
The fluorometric dye displacement assay allows one to measure partial unwinding of a DNA substrate (Fig. 7), which is distinct from the all-or-none DNA unwinding assays described in the previous sections. This assay is particularly relevant for monitoring helicase activity on damaged DNA substrates because it can provide evidence that a DNA adduct positioned within the duplex region of the substrate does not interfere with helicase loading or initiation of DNA unwinding on the substrate containing the DNA lesion. The dye displacement assay is built on the premise that the fluorescence property of dye molecules bound to the duplex region of a DNA substrate changes as the double-stranded DNA is unwound and the dye molecules are released into solution. For our experiments to study FANCJ helicase, we used Hoechst 33528 dye intercalated into double-stranded DNA of a forked duplex substrate, which can be excited at 344 nm and fluorescence emission can be monitored at wavelengths greater than 400 nm with a 400 nm long pass filter [16]. A stopped-flow reaction analyzer was used to measure fluorescence decrease as a function of helicase reaction time within a 200 second span. An unmodified DNA substrate fully unwound by FANCJ helicase yielded a maximal fluorescence decrease. A DNA substrate harboring a polyglycol linkage 16 bp in from the single-stranded/double-stranded junction of the fork entry site that was partially unwound by FANCJ yielded a smaller change in fluorescence because a population of the dye molecules remained intercalated with the 12 bp duplex region residing between the backbone modification and the blunt duplex end, away from the helicase entry site at the fork. The results from these assays demonstrated a reduced fluorescence change in FANCJ helicase reactions with DNA substrates containing the polyglycol modification in either the top or bottom strands of the forked duplex DNA substrate. These findings were consistent with results from standard radiometric strand displacement helicase assays in which FANCJ was found to be profoundly sensitive to the polyglycol linkage in either the translocating or non-translocating strands of the duplex [16].
Figure 7. Schematic of fluorometric dye displacement kinetic assay.
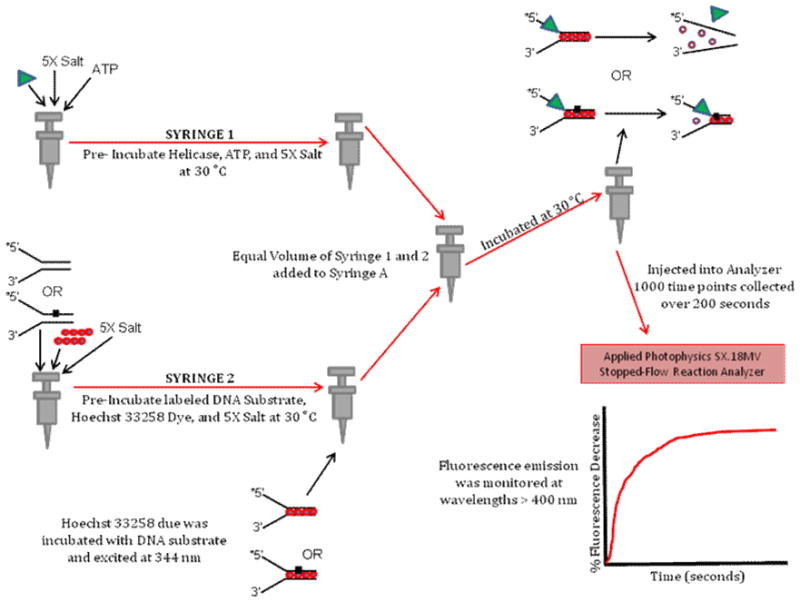
This assay utilizes a small molecule with unique fluorescence properties dependent on binding to duplex DNA to analyze partial unwinding of a duplex DNA substrate by a helicase when it encounters a lesion that blocks its progression. See text for details.
6.7. Protein Displacement Radiometric Helicase Assay
Frequently, in vitro assays that measure helicase activity are based on biochemical reactions with naked DNA substrates that may not reflect what is observed with chromatinized DNA. Proteins bound to single-stranded or double-stranded DNA affect cellular processes such as replication, transcription, or DNA repair; therefore, researchers have sought to characterize the ability of DNA helicase and helicase-like proteins to disrupt model protein-DNA complexes. This approach typically involves pre-binding a protein (e.g., stationary RNA polymerase [88], Lac repressor [89], Trp repressor [90], catalytically inactive restriction endonuclease [72], or telomere binding factor [72]) to a radiolabeled partial duplex DNA substrate and then assessing if a helicase in the presence of ATP can catalytically dislodge the protein from the substrate and unwind the underlying duplex (Fig. 8). Alternatively, helicases have been tested for their ability to disengage or bypass proteins bound to single-stranded DNA (e.g., RecA [91], Rad51 [64,92–95], RPA [96]). We have employed the radiometric assay to characterize the ability of human SF2 DNA helicases FANCJ and RECQ1 to displace the inactive restriction endonuclease BamHI-E111A from a partial duplex DNA molecule with its cognate recognition sequence which it binds with a high affinity (10−11 M) [72]. Failure to efficiently displace protein bound to DNA resulted in a poor level of DNA unwinding that was readily measured by analysis of reaction products on standard non-denaturing polyacrylamide gels as described for the standard radiometric helicase assay. We determined that the presence of the single-stranded DNA binding protein RPA which physically and functionally interacts with a number of SF2 helicases including FANCJ [46] and RECQ1 [97] could stimulate the respective helicases to displace BamHI-E111A bound to the duplex and unwind the DNA substrate. Similar observations were made for RPA-dependent FANCJ or RECQ1 helicase-catalyzed displacement of the telomere binding proteins TRF1 or TRF2 bound to a DNA substrate harboring the human telomeric G-rich repeat [72].
Figure 8. Schematic of protein displacement helicase assay.
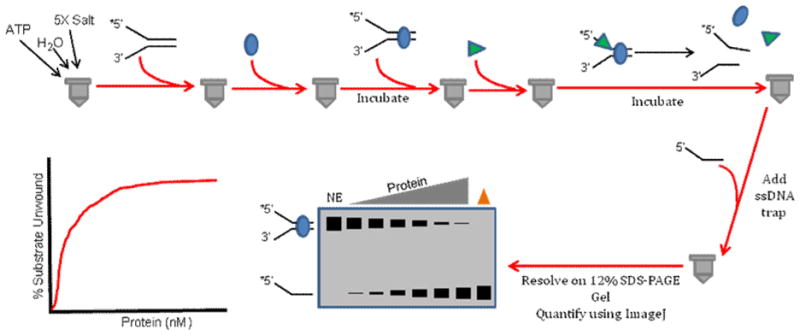
This assay is performed to monitor if a helicase can complete its ATP-dependent unwinding of a partial duplex DNA substrate when a protein is bound to the duplex region of the DNA substrate. See text for details.
The protein displacement radiometric assay may be useful to understand if helicases play a role in how DPC or ICL lesions are processed. In either case, an accessory helicase may help to dislodge the CMG helicase stalled by the covalently modified DNA lesion. Using a modified version of the protein displacement radiometric assay, one could conceivably assess by electrophoretic mobility shift assay (EMSA) using a lower percentage polyacrylamide in the native gel if a stalled CMG helicase is dissociated from an ICL or DPC DNA substrate by an accessory helicase.
7. Conclusions
Accumulation of DNA adducts over time can lead to defects in replication and transcription which undermines cellular homeostasis and chromosomal stability, potentially giving rise to neurodegeneration, cancer predisposition, and/or other clinical symptoms of aging. DNA helicases are thought to be among the first proteins that come into contact with DNA lesions in vivo and are thus suggested to be the primary recruiters of factors that play essential roles in the DNA damage response pathway. Understanding how the unwinding functions of various replication and DNA repair helicases are affected by DNA lesions that vary from base to backbone modifications as well as more unusual forms of damage such as ICLs or DPCs can provide useful information pertaining to the roles of helicases in DNA repair and the DNA damage response. Further studies in this field using classical approaches as well as newly invented technologies in more complex reconstituted systems should continue to provide insights to how helicases behave when they encounter DNA lesions and their mechanisms of action.
Acknowledgments
This work was supported by the Intramural Research Program of the NIH, National Institute on Aging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bhattacharyya B, Keck JL. Grip it and rip it: structural mechanisms of DNA helicase substrate binding and unwinding. Protein Sci. 2014;23:1498–1507. doi: 10.1002/pro.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raney KD. Chemical modifications of DNA for study of helicase mechanisms. Bioorg Med Chem. 2014;22:4399–4406. doi: 10.1016/j.bmc.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suhasini AN, Brosh RM. Mechanistic and biological aspects of helicase action on damaged DNA. Cell Cycle. 2010;9:2317–2329. doi: 10.4161/cc.9.12.11902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villani G, Tanguy LG. Interactions of DNA helicases with damaged DNA: possible biological consequences. J Biol Chem. 2000;275:33185–33188. doi: 10.1074/jbc.R000011200. [DOI] [PubMed] [Google Scholar]
- 5.Tackett AJ, Wei L, Cameron CE, Raney KD. Unwinding of nucleic acids by HCV NS3 helicase is sensitive to the structure of the duplex. Nucleic Acids Res. 2001;29:565–572. doi: 10.1093/nar/29.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertram RD, Hayes CJ, Soultanas P. Vinylphosphonate internucleotide linkages inhibit the activity of PcrA DNA helicase. Biochemistry. 2002;41:7725–7731. doi: 10.1021/bi025755s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia PL, Bradley G, Hayes CJ, Krintel S, Soultanas P, Janscak P. RPA alleviates the inhibitory effect of vinylphosphonate internucleotide linkages on DNA unwinding by BLM and WRN helicases. Nucleic Acids Res. 2004;32:3771–3778. doi: 10.1093/nar/gkh709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amaratunga M, Lohman TM. Escherichia coli rep helicase unwinds DNA by an active mechanism. Biochemistry. 1993;32:6815–6820. doi: 10.1021/bi00078a003. [DOI] [PubMed] [Google Scholar]
- 9.Kawaoka J, Jankowsky E, Pyle AM. Backbone tracking by the SF2 helicase NPH-II. Nat Struct Mol Biol. 2004;11:526–530. doi: 10.1038/nsmb771. [DOI] [PubMed] [Google Scholar]
- 10.Eoff RL, Spurling TL, Raney KD. Chemically modified DNA substrates implicate the importance of electrostatic interactions for DNA unwinding by Dda helicase. Biochemistry. 2005;44:666–674. doi: 10.1021/bi0484926. [DOI] [PubMed] [Google Scholar]
- 11.Jones GD, Le Pla RC, Farmer PB. Phosphotriester adducts (PTEs): DNA’s overlooked lesion. Mutagenesis. 2010;25:3–16. doi: 10.1093/mutage/gep038. [DOI] [PubMed] [Google Scholar]
- 12.Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, Livingston DM. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 13.Cantor SB, Guillemette S. Hereditary breast cancer and the BRCA1-associated FANCJ/BACH1/BRIP1. Future Oncol. 2011;7:253–261. doi: 10.2217/fon.10.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta R, Sharma S, Sommers JA, Jin Z, Cantor SB, Brosh RM., Jr Analysis of the DNA substrate specificity of the human BACH1 helicase associated with breast cancer. J Biol Chem. 2005;280:25450–25460. doi: 10.1074/jbc.M501995200. [DOI] [PubMed] [Google Scholar]
- 15.Brosh RM, Jr, Cantor SB. Molecular and cellular functions of the FANCJ DNA helicase defective in cancer and in Fanconi anemia. Front Genet. 2014;5:372. doi: 10.3389/fgene.2014.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta R, Sharma S, Doherty KM, Sommers JA, Cantor SB, Brosh RM., Jr Inhibition of BACH1 (FANCJ) helicase by backbone discontinuity is overcome by increased motor ATPase or length of loading strand. Nucleic Acids Res. 2006;34:6673–83. doi: 10.1093/nar/gkl964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galletto R, Jezewska MJ, Bujalowski W. Unzipping mechanism of the double-stranded DNA unwinding by a hexameric helicase: the effect of the 3′ arm and the stability of the dsDNA on the unwinding activity of the Escherichia coli DnaB helicase. J Mol Biol. 2004;343:101–114. doi: 10.1016/j.jmb.2004.07.056. [DOI] [PubMed] [Google Scholar]
- 18.Jeong YJ, Levin MK, Patel SS. The DNA-unwinding mechanism of the ring helicase of bacteriophage T7. Proc Natl Acad Sci U S A. 2004;101:7264–7269. doi: 10.1073/pnas.0400372101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tackett AJ, Morris PD, Dennis R, Goodwin TE, Raney KD. Unwinding of unnatural substrates by a DNA helicase. Biochemistry. 2001;40:543–548. doi: 10.1021/bi002122+. [DOI] [PubMed] [Google Scholar]
- 20.Suhasini AN, Rawtani NA, Wu Y, Sommers JA, Sharma S, Mosedale G, North PS, Cantor SB, Hickson ID, Brosh RM, Jr Interaction between the helicases genetically linked to Fanconi anemia group J. Bloom’s syndrome. EMBO J. 2011;30:692–705. doi: 10.1038/emboj.2010.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Y, Sommers JA, Loiland JA, Kitao H, Kuper J, Kisker C, Brosh RM. The Q Motif of FANCJ DNA Helicase Regulates its Dimerization, DNA Binding, and DNA Repair Function. J Biol Chem. 2012;22:21699–21716. doi: 10.1074/jbc.M112.351338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suhasini AN, Brosh RM., Jr Fanconi anemia and Bloom’s syndrome crosstalk through FANCJ-BLM helicase interaction. Trends Genet. 2012;28:7–13. doi: 10.1016/j.tig.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suhasini AN, Sommers JA, Yu S, Wu Y, Xu T, Kelman Z, Kaplan DL, Brosh RM., Jr DNA repair and replication fork helicases are differentially affected by alkyl phosphotriester lesion. J Biol Chem. 2012;287:19188–19198. doi: 10.1074/jbc.M112.352757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pugh RA, Honda M, Spies M. Ensemble and single-molecule fluorescence-based assays to monitor DNA binding, translocation, and unwinding by iron-sulfur cluster containing helicases. Methods. 2010;51:313–321. doi: 10.1016/j.ymeth.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 26.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 27.Wallace SS. Biological consequences of free radical-damaged DNA bases. Free Radic Biol Med. 2002;33:1–14. doi: 10.1016/s0891-5849(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 28.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 29.Egly JM, Coin F. A history of TFIIH: Two decades of molecular biology on a pivotal transcription/repair factor. DNA Repair (Amst) 2011;10:714–721. doi: 10.1016/j.dnarep.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 30.Fan L, DuPrez KT. XPB: An unconventional SF2 DNA helicase. Prog Biophys Mol Biol. 2015;117:174–181. doi: 10.1016/j.pbiomolbio.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Sugasawa K, Akagi J, Nishi R, Iwai S, Hanaoka F. Two-step recognition of DNA damage for mammalian nucleotide excision repair: Directional binding of the XPC complex and DNA strand scanning. Mol Cell. 2009;36:642–653. doi: 10.1016/j.molcel.2009.09.035. [DOI] [PubMed] [Google Scholar]
- 32.Mathieu N, Kaczmarek N, Naegeli H. Strand- and site-specific DNA lesion demarcation by the xeroderma pigmentosum group D helicase. Proc Natl Acad Sci U S A. 2010;107:17545–17550. doi: 10.1073/pnas.1004339107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudolf J, Rouillon C, Schwarz-Linek U, White MF. The helicase XPD unwinds bubble structures and is not stalled by DNA lesions removed by the nucleotide excision repair pathway. Nucleic Acids Res. 2009;38:931–941. doi: 10.1093/nar/gkp1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naegeli H, Bardwell L, Friedberg EC. The DNA helicase and adenosine triphosphatase activities of yeast Rad3 protein are inhibited by DNA damage. A potential mechanism for damage-specific recognition. J Biol Chem. 1992;267:392–398. [PubMed] [Google Scholar]
- 35.Naegeli H, Modrich P, Friedberg EC. The DNA helicase activities of Rad3 protein of Saccharomyces cerevisiae and helicase II of Escherichia coli are differentially inhibited by covalent and noncovalent DNA modifications. J Biol Chem. 1993;268:10386–10392. [PubMed] [Google Scholar]
- 36.Naegeli H, Bardwell L, Friedberg EC. Inhibition of Rad3 DNA helicase activity by DNA adducts and abasic sites: implications for the role of a DNA helicase in damage-specific incision of DNA. Biochemistry. 1993;32:613–621. doi: 10.1021/bi00053a029. [DOI] [PubMed] [Google Scholar]
- 37.Mathieu N, Kaczmarek N, Ruthemann P, Luch A, Naegeli H. DNA quality control by a lesion sensor pocket of the Xeroderma pigmentosum group D helicase subunit of TFIIH. Current Biology. 2013;23:204–212. doi: 10.1016/j.cub.2012.12.032. [DOI] [PubMed] [Google Scholar]
- 38.Buechner CN, Heil K, Michels G, Carell T, Kisker C, Tessmer I. Strand-specific recognition of DNA damages by XPD provides insights into nucleotide excision repair substrate versatility. J Biol Chem. 2014;289:3613–3624. doi: 10.1074/jbc.M113.523001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thakkar DR, Yagi H, Levin W, Wood AW, Jerina DM. Bioactivation of Foreign Compounds. 1985:117–242. [Google Scholar]
- 40.Choudhary S, Doherty KM, Handy CJ, Sayer JM, Yagi H, Jerina DM, Brosh RM., Jr Inhibition of Werner Syndrome Helicase Activity by Benzo[a]pyrene Diol Epoxide Adducts Can Be Overcome by Replication Protein A. J Biol Chem. 2006;281:6000–6009. doi: 10.1074/jbc.M510122200. [DOI] [PubMed] [Google Scholar]
- 41.Driscoll HC, Matson SW, Sayer JM, Kroth H, Jerina DM, Brosh RM., Jr Inhibition of Werner syndrome helicase activity by benzo[c]phenanthrene diol epoxide dA adducts in DNA is both strand-and stereoisomer-dependent. J Biol Chem. 2003;278:41126–41135. doi: 10.1074/jbc.M304798200. [DOI] [PubMed] [Google Scholar]
- 42.Doherty KM, Sommers JA, Gray MD, Lee JW, von Kobbe C, Thoma NH, Kureekattil RP, Kenny MK, Brosh RM., Jr Physical and functional mapping of the RPA interaction domain of the Werner and Bloom syndrome helicases. J Biol Chem. 2005;19:29494–29505. doi: 10.1074/jbc.M500653200. [DOI] [PubMed] [Google Scholar]
- 43.Suhasini AN, Sommers JA, Mason AC, Voloshin ON, Camerini-Otero RD, Wold MS, Brosh RM., Jr FANCJ Helicase Uniquely Senses Oxidative Base Damage in Either Strand of Duplex DNA and Is Stimulated by Replication Protein A to Unwind the Damaged DNA Substrate in a Strand-specific Manner. J Biol Chem. 2009;284:18458–18470. doi: 10.1074/jbc.M109.012229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berquist BR, Wilson DM., III Pathways for repairing and tolerating the spectrum of oxidative DNA lesions. Cancer Lett. 2012;327:61–72. doi: 10.1016/j.canlet.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kung HC, Bolton PH. Structure of a duplex DNA containing a thymine glycol residue in solution. J Biol Chem. 1997;272:9227–9236. doi: 10.1074/jbc.272.14.9227. [DOI] [PubMed] [Google Scholar]
- 46.Gupta R, Sharma S, Sommers JA, Kenny MK, Cantor SB, Brosh RM., Jr FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood. 2007;110:2390–2398. doi: 10.1182/blood-2006-11-057273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aller P, Rould MA, Hogg M, Wallace SS, Doublie S. A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol. Proc Natl Acad Sci U S A. 2007;104:814–818. doi: 10.1073/pnas.0606648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaruga P, Dizdaroglu M. 8,5′-Cyclopurine-2′-deoxynucleosides in DNA: mechanisms of formation, measurement, repair and biological effects. DNA Repair (Amst) 2008;7:1413–1425. doi: 10.1016/j.dnarep.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 49.Huang H, Das RS, Basu AK, Stone MP. Structure of (5′S)-8,5′-cyclo-2′-deoxyguanosine in DNA. J Am Chem Soc. 2011;133:20357–20368. doi: 10.1021/ja207407n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zaliznyak T, Lukin M, de los SC. Structure and stability of duplex DNA containing (5′S)-5′,8-cyclo-2′-deoxyadenosine: an oxidatively generated lesion repaired by NER. Chem Res Toxicol. 2012;25:2103–2111. doi: 10.1021/tx300193k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brooks PJ, Wise DS, Berry DA, Kosmoski JV, Smerdon MJ, Somers RL, Mackie H, Spoonde AY, Ackerman EJ, Coleman K, Tarone RE, Robbins JH. The oxidative DNA lesion 8,5′-(S)-cyclo-2′-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J Biol Chem. 2000;275:22355–22362. doi: 10.1074/jbc.M002259200. [DOI] [PubMed] [Google Scholar]
- 52.Kuraoka I, Bender C, Romieu A, Cadet J, Wood RD, Lindahl T. Removal of oxygen free-radical-induced 5′,8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc Natl Acad Sci U S A. 2000;97:3832–3837. doi: 10.1073/pnas.070471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jasti VP, Das RS, Hilton BA, Weerasooriya S, Zou Y, Basu AK. (5′S)-8,5′-cyclo-2′-deoxyguanosine is a strong block to replication, a potent pol V-dependent mutagenic lesion, and is inefficiently repaired in Escherichia coli. Biochemistry. 2011;50:3862–3865. doi: 10.1021/bi2004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brooks PJ. The 8,5′-cyclopurine-2′-deoxynucleosides: candidate neurodegenerative DNA lesions in xeroderma pigmentosum, and unique probes of transcription and nucleotide excision repair. DNA Repair (Amst) 2008;7:1168–1179. doi: 10.1016/j.dnarep.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J, Clauson CL, Robbins PD, Niedernhofer LJ, Wang Y. The oxidative DNA lesions 8,5′-cyclopurines accumulate with aging in a tissue-specific manner. Aging Cell. 2012;11:714–716. doi: 10.1111/j.1474-9726.2012.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khan I, Suhasini AN, Banerjee T, Sommers JA, Kaplan DL, Kuper J, Kisker C, Brosh RM., Jr Impact of age-associated cyclopurine lesions on DNA repair helicases. PLoS ONE. 2014;9:e113293. doi: 10.1371/journal.pone.0113293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sontz PA, Mui TP, Fuss JO, Tainer JA, Barton JK. DNA charge transport as a first step in coordinating the detection of lesions by repair proteins. Proc Natl Acad Sci U S A. 2012;109:1856–1861. doi: 10.1073/pnas.1120063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chiu HC, Koh KD, Evich M, Lesiak AL, Germann MW, Bongiorno A, Riedo E, Storici F. RNA intrusions change DNA elastic properties and structure. Nanoscale. 2014;6:10009–10017. doi: 10.1039/c4nr01794c. [DOI] [PubMed] [Google Scholar]
- 59.Potenski CJ, Klein HL. How the misincorporation of ribonucleotides into genomic DNA can be both harmful and helpful to cells. Nucleic Acids Res. 2014;42:10226–10234. doi: 10.1093/nar/gku773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wallace BD, Williams RS. Ribonucleotide triggered DNA damage and RNA-DNA damage responses. RNA Biol. 2014;11:1340–1346. doi: 10.4161/15476286.2014.992283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams JS, Kunkel TA. Ribonucleotides in DNA: origins, repair and consequences. DNA Repair (Amst) 2014;19:27–37. doi: 10.1016/j.dnarep.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pizzi S, Sertic S, Orcesi S, Cereda C, Bianchi M, Jackson AP, Lazzaro F, Plevani P, Muzi-Falconi M. Reduction of hRNase H2 activity in Aicardi-Goutieres syndrome cells leads to replication stress and genome instability. Hum Mol Genet. 2015;24:649–658. doi: 10.1093/hmg/ddu485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Byrd AK, Raney KD. Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat Struct Mol Biol. 2004;11:531–538. doi: 10.1038/nsmb774. [DOI] [PubMed] [Google Scholar]
- 64.Sommers JA, Rawtani N, Gupta R, Bugreev DV, Mazin AV, Cantor SB, Brosh RM., Jr FANCJ uses its motor ATPase to disrupt protein-DNA complexes, unwind triplexes, and inhibit rad51 strand exchange. J Biol Chem. 2009;284:7505–7517. doi: 10.1074/jbc.M809019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morris PD, Raney KD. DNA helicases displace streptavidin from biotin-labeled oligonucleotides. Biochemistry. 1999;38:5164–5171. doi: 10.1021/bi9822269. [DOI] [PubMed] [Google Scholar]
- 66.Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, Hurwitz J, van OA, Scharer OD, Walter JC. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaplan DL. The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J Mol Biol. 2000;301:285–299. doi: 10.1006/jmbi.2000.3965. [DOI] [PubMed] [Google Scholar]
- 68.Kumari A, Minko IG, Smith RL, Lloyd RS, McCullough AK. Modulation of UvrD helicase activity by covalent DNA-protein cross-links. J Biol Chem. 2010;285:21313–21322. doi: 10.1074/jbc.M109.078964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuo HK, Griffith JD, Kreuzer KN. 5-Azacytidine induced methyltransferase-DNA adducts block DNA replication in vivo. Cancer Res. 2007;67:8248–8254. doi: 10.1158/0008-5472.CAN-07-1038. [DOI] [PubMed] [Google Scholar]
- 70.Duxin JP, Dewar JM, Yardimci H, Walter JC. Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell. 2014;159:346–357. doi: 10.1016/j.cell.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bruning JG, Howard JL, McGlynn P. Accessory replicative helicases and the replication of protein-bound DNA. J Mol Biol. 2014;426:3917–3928. doi: 10.1016/j.jmb.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 72.Sommers JA, Banerjee T, Hinds T, Wold MS, Lei M, Brosh RM., Jr Novel Function of the Fanconi Anemia Group J or RECQ1 Helicase to Disrupt Protein-DNA Complexes in a Replication Protein A-Stimulated Manner. J Biol Chem. 2014;289:19928–19941. doi: 10.1074/jbc.M113.542456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Longerich S, Li J, Xiong Y, Sung P, Kupfer GM. Stress and DNA repair biology of the Fanconi anemia pathway. Blood. 2014;124:2812–2819. doi: 10.1182/blood-2014-04-526293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sundheim O, Talstad VA, Vagbo CB, Slupphaug G, Krokan HE. AlkB demethylases flip out in different ways. DNA Repair (Amst) 2008;7:1916–1923. doi: 10.1016/j.dnarep.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 75.Brooks SC, Adhikary S, Rubinson EH, Eichman BF. Recent advances in the structural mechanisms of DNA glycosylases. Biochim Biophys Acta. 2013;1834:247–271. doi: 10.1016/j.bbapap.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang W. Surviving the sun: repair and bypass of DNA UV lesions. Protein Sci. 2011;20:1781–1789. doi: 10.1002/pro.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dillingham MS, Soultanas P, Wiley P, Webb MR, Wigley DB. Defining the roles of individual residues in the single-stranded DNA binding site of PcrA helicase. Proc Natl Acad Sci U S A. 2001;98:8381–8387. doi: 10.1073/pnas.131009598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beuck C, Weinhold E. Reversibly locked thionucleobase pairs in DNA to study base flipping enzymes. Beilstein J Org Chem. 2014;10:2293–2306. doi: 10.3762/bjoc.10.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang J, Liu S, Bellani MA, Thazhathveetil AK, Ling C, De Winter JP, Wang Y, Wang W, Seidman MM. The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand crosslinks. Mol Cell. 2013;52:434–446. doi: 10.1016/j.molcel.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Iyama T, Lee SY, Berquist BR, Gileadi O, Bohr VA, Seidman MM, McHugh PJ, Wilson DM., III CSB interacts with SNM1A and promotes DNA interstrand crosslink processing. Nucleic Acids Res. 2015;43:247–258. doi: 10.1093/nar/gku1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Long DT, Joukov V, Budzowska M, Walter JC. BRCA1 promotes unloading of the CMG helicase from a stalled DNA replication fork. Mol Cell. 2014;56:174–185. doi: 10.1016/j.molcel.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matson SW, George JW. DNA helicase II of Escherichia coli. Characterization of the single- stranded DNA-dependent NTPase and helicase activities. J Biol Chem. 1987;262:2066–2076. [PubMed] [Google Scholar]
- 84.Choudhary S, Sommers JA, Brosh RM., Jr Biochemical and kinetic characterization of the DNA helicase and exonuclease activities of werner syndrome protein. J Biol Chem. 2004;279:34603–34613. doi: 10.1074/jbc.M401901200. [DOI] [PubMed] [Google Scholar]
- 85.Sen D, Nandakumar D, Tang GQ, Patel SS. The Human Mitochondrial DNA helicase TWINKLE is both an unwinding and an annealing helicase. J Biol Chem. 2012;287:14545–14556. doi: 10.1074/jbc.M111.309468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu X, Liu Y. Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J. 2009;28:568–577. doi: 10.1038/emboj.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, Andreoli L, Muzzolini L, Vindigni A, Brosh RM., Jr Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J Biol Chem. 2005;280:28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 88.Bedinger P, Hochstrasser M, Jongeneel CV, Alberts BM. Properties of the T4 bacteriophage DNA replication apparatus: the T4 dda DNA helicase is required to pass a bound RNA polymerase molecule. Cell. 1983;34:115–123. doi: 10.1016/0092-8674(83)90141-1. [DOI] [PubMed] [Google Scholar]
- 89.Yancey-Wrona JE, Matson SW. Bound Lac repressor protein differentially inhibits the unwinding reactions catalyzed by DNA helicases. Nucleic Acids Res. 1992;20:6713–6721. doi: 10.1093/nar/20.24.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Byrd AK, Raney KD. Displacement of a DNA binding protein by Dda helicase. Nucleic Acids Res. 2006;34:3020–3029. doi: 10.1093/nar/gkl369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Veaute X, Delmas S, Selva M, Jeusset J, Le CE, Matic I, Fabre F, Petit MA. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 2005;24:180–189. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, Jasin M, Vogel H, Sung P, Luo G. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 95.Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 96.Honda M, Park J, Pugh RA, Ha T, Spies M. Single-molecule analysis reveals differential effect of ssDNA-binding proteins on DNA translocation by XPD helicase. Mol Cell. 2009;35:694–703. doi: 10.1016/j.molcel.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cui S, Arosio D, Doherty KM, Brosh RM, Jr, Falaschi A, Vindigni A. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 2004;32:2158–2170. doi: 10.1093/nar/gkh540. [DOI] [PMC free article] [PubMed] [Google Scholar]


