Introduction
House dust mites belong to the most frequent indoor allergen sources worldwide and are potent inducers of perennial asthma and rhinitis [1-3]. Major allergens in house dust are associated with mites of the genus Dermatophagoides [4]. To date, 17 groups of allergens from Dermatophogoides sp. with diverse biological functions have been described (www.allergen.org) [5, 6]. Especially group 1 and 2 allergens from Dermatophagoides pteronyssinus (Der p) and D. farina (Der f), secreted from mite bodies or excreted with their feces, are among the strongest immunogens in humans [7, 8]. Der p 1 and Der f 1 are proteins with cysteine protease activity while Der p 2 and Der f 2 contribute to allergenicity by interacting with the innate immune system [9, 10]. Recently, a new HDM allergen, Der p 23, a perithrophin-like protein was identified. It was shown to induce Immunoglobulin E (IgE)-levels comparable to Der p 1 and 2 and hence represents another major HDM allergen [11].
IgE is of particular importance for the pathogenesis of an allergic immune response. Cross-linking of IgE molecules bound to Fcε-receptors on the surface of mast cells and basophil granulocytes by allergens leads to the release of histamine and other proinflammatory mediators and triggers immediate type clinical symptoms of allergic disease [12, 13]. T cells are also major contributors to allergic disease. T helper type 2 (Th2)-derived cytokines such as IL-4 and IL-13 induce immune class switching from IgG4 to IgE in B cells [14-16]. In addition to promoting the production of IgE, Th2 cells may also contribute to the immune response directly by releasing proinflammatory cytokines such as IL-5 that trigger eosinophilia.
While several studies have reported several T cell epitopes derived from HDM [17-23], a comprehensive characterization of the epitopes recognized by human T cells in the context of house dust mite is still lacking. The precise definition and exact mapping of the epitopes involved, antigens of origin, and patterns of associated T helper cell responses would allow the identification of immunodominant epitopes and thereby contribute to a better understanding of pathogenic immune responses. A number of studies have shown that the frequency of allergen-specific T cells is extremely low, in the range of 1 to 6/10000 cells [22, 24]. In particular, epitope sets that allow ex vivo characterization of HDM-specific T cell responses are lacking, and most if not all data is derived from protocols that utilize in vitro re-stimulation. This issue is of relevance, since it is well known that in vitro re-stimulation can alter the phenotype of the responding cells.
In this study, we analyzed patterns of immune recognition of Der p- and Der f-proteins, which are considered most dominant within mite allergens [7, 8]. We comprehensively defined T cell responses and characterized the Th1/Th2 balance in HDM-specific immune responses directed against different house dust mite allergens and epitopes. To examine whether the group 1 and 2 allergens also dominate at the T cell level, over other antigens that are subdominant in terms of IgE titers, we also included the recently described Der p 23 [11] into our studies. Based on these results we define a pool of immunodominant epitopes that allows the detection of HDM-specific T cells directly ex vivo.
Methods
Study population
PBMCs from 55 house dust mite allergic and 10 non-allergic individuals were kindly provided by ALK-Abello A/S (Horsholm, Denmark). From each volunteer 20 to 40 million PBMCs were obtained. PBMCs were isolated from whole blood by density gradient centrifugation according to manufacturers’ instructions (Ficoll-Hypaque, Amersham Biosciences, Uppsala, Sweden). A total of 55 allergic patients were analyzed in the study. However, due to limitations in cell numbers, not all patients were tested in the same experiment. The results sections indicate the number of patients tested in any particular experiment.
Der p- and Der f-specific IgE-titers were also provided by ALK-Abello and are summarized in supplementary tables 1A and B of this manuscript. Serum levels of Der p/f-specific IgE-antibodies were determined in whole blood using the ImmunoCAP-system (Thermo Fischer, Uppsala, Sweden).
Selection of 15-mer peptides from house dust mite sequences
Sequences of mite Der p allergen groups 1, 2, 23- and Der f- groups 1 and 2 were selected from 7 databases: Allergen Database for Food Safety (http://allergen.nihs.go.jp/ADFS), Food Allergy Research and Resource Program (www.farrp.org), Allergen Nomenclature (www.allergen.org/Allergen.aspx), Allergome (www.allergome.org), Swissprot (www.expasy.ch/sprot), Structural Database of Allergenic Proteins (http://fermi.utmb.edu/SDAP), and the Biotechnology Information for Food Safety Database (Gendel et al. 2006, Mol Nutr Food Res). Sets of peptides of 15 amino acids in length, overlapping by 10 residues, were generated to cover the entire allergen protein sequences. Overall, a total of 178 peptides were assembled in 10 pools and screened for their immunogenicity.
Peptide synthesis
Peptides were purchased from Mimotopes (Clayton, Victoria, Australia) and/or A and A (San Diego, CA) as crude material on a small (1-mg) scale. Individual peptides were resuspended in DMSO at a final concentration of 40 mg/ml. A pool of 75 immunodominant peptides from Der p/f allergens identified in this study or previously published [17, 20, 21, 23] was also used. This set is referred to as the Dust Mite Pool (DMP) throughout the manuscript. To generate this pool 10 μl of each individual peptide dissolved in DMSO were pooled and re-lyophilized (A and A, San Diego, CA). Following lyophilization the pool was reconstituted in DMSO so that each peptide was present at a concentration of 4 mg/ml.
Stimulation and expansion of dust-mite specific T cells
For in vitro expansion of house dust mite specific T cells, PBMCs of HDM-allergic and non-allergic individuals were stimulated with Der p- and Der f-extract (ALK-Abello A/S (Horsholm, Denmark) at 5 μg/ml, respectively. The concentration was determined by titration experiments (data not shown). Cells were cultured in RPMI1640 supplemented with 5% human AB serum in 24 well plates (BD Bioscience, San Diego, CA) at a density of 2×106/ml and incubated at 37°C. IL-2 was added every 3 days after initial stimulation. Cells were harvested on day 14 and screened for IFNγ/IL-5-production by dual ELISPOT assays.
Dual ELISPOT assays
In vitro restimulation
The production of IFNγ and IL-5 from cultured PBMCs in response to antigenic stimulation was assessed by dual ELISPOT assays as described previously [25]. Flat-bottom 96-well plates were coated with 5 μg/ml anti-human IFNγ (Clone 1-D1K; Mabtech, Cincinati, OH) and anti-human IL-5 (clone TRFK5; Mabtech). PBMCs were harvested after 14 days of expansion and incubated in 96 well ELISPOT plate at a density of 1×105 cells/well either with peptide pools (10 μg/ml) or individual peptides (10 μg/ml), HDM extract (5 μg/ml), PHA (10 μg/ml), or medium containing 1% DMSO (corresponding to the percentage of DMSO in the pools/peptides) as a control. After 24 hours, cells were removed and plates were incubated with 2 μg/ml biotinylated anti-human IL-5 ab (Mabtech) and HRP-conjugated anti-human IFNγ-ab at 37°C for 2 hours. Spots corresponding to the biotinylated IL-5-antibody were incubated with alkaline phosphatase complex (Vector Laboratories, Burlingame, CA), and developed using Vector Blue Alkaline Phosphatase Substrate Kit III according to manufacturers instruction. Spots corresponding to the HRP-conjugated IFNγ-antibody were developed with 3-amino-9-ehtylcarbazole solution (Sigma Aldrich, St. Louis, MO). Spot forming cells (SFC) were counted by computer assisted image analysis (KS-ELISPOT reader, Zeiss, Munich, Germany). Each assay was performed in triplicates. Student’s t-test using the mean of triplicate values of the response against the extract, pool or individual peptides, compared to the response against medium control was applied to calculate statistical significance. As previously described, criteria for positivity were 100 SFCs per 106 PBMCs, P < 0.05, and a stimulation index > 2 [26-29]. Positive pools were deconvoluted to identify the individual epitopes inducing the response. In the case of individual peptides, criteria for positivity were 20 SFCs per 106 PBMCs, P < 0.05, and a stimulation index > 2.
Ex vivo restimulation
To analyze cytokine production in response to ex vivo antigenic stimulation PBMCs were thawed, washed with RPMI and resuspended at 4×106 PBMC/ml. Thereafter, 2×105 PBMC/well were stimulated with HDM extract (5 μg/ml), individual peptides (10 μg/ml), DMP (1 μg/ml), PHA (10 μg/ml) or medium in 96 well plate and IL-10- and IL-17- responses were assessed in addition to IFNγ and IL-5 by ELISPOT. IFNγ-/IL-5 production was determined exactly as described above.
To determine IL-10- and IL-17-production, 96 well ELISPOT plates were coated with anti-human IL-10 (clone 9D7; Mabtech) and anti-human IL-17 antibody (clone 147178; eBioscience, San Diego, CA). After 24 hours, cells were removed and plates were incubated with 2 μg/ml biotinylated anti-human IL-10-antibody (Mabtech) and FITC-conjugated anti-human IL-17 antibody (eBioscience) at 37°C for 2 hours. Spots corresponding to the biotinylated IL-10 antibody were incubated with alkaline phosphatase complex (Vector Laboratories, Burlingame, CA), and developed using Vector Blue Alkaline Phosphatase Substrate Kit III according to manufacturers instruction. Spots corresponding to the anti-FITC-HRP conjugated IL-17 antibody were developed with 3-amino-9-ehtylcarbazole solution (Sigma Aldrich, St. Louis, MO). Spot forming cells (SFC) were counted by computer assisted image analysis (KS-ELISPOT reader, Zeiss, Munich, Germany) and analyzed as described in the in vitro restimulation section of this manuscript.
Results
Determination of T cell reactivity to Der p and Der f allergens
The initial series of experiments in this study characterized T cell reactivity of HDM allergic donors to group 1 and 2 allergens from Der p and Der f and the recently described Der p 23 allergen [11]. While 17 different groups of Der p/Der f allergens are listed in the IUIS database (www.allergen.org), the Der p1/2 from Dermatophagoides pteronyssinus and Der f 1/2 from D. farinae are amongst the most widely reported and well characterized mite allergens in humans [7, 8].
For these initial studies a cohort of 21 male and female (66%/33%) donors, age 19-66, with a clinical history of HDM allergy and IgE CAP (Der f or Der p) titers >0.35 kU/L was recruited. To assess HDM-specific responses, PBMCs from each donor were stimulated in vitro with a mixture of Der p/f extract. After 14 days, pools of 15/20-mer peptides, overlapping by 10 residues entirely spanning the various antigens, were tested using ELISPOT assays for their capacity to elicit IFNγ and/or IL-5 responses. IL-5 and IFNγ were chosen as representative of Th2 and Th1 cytokines, respectively. Positive pools were deconvoluted to identify the specific individual epitopes eliciting the responses. Overall, positive IL-5 responses to at least one antigen could be detected in all 21 donors, and 16/21 generated IFNγ responses (Supplementary Table 1A).
Breadth and immunodominance in T cell responses to group 1, 2 and 23 HDM antigens
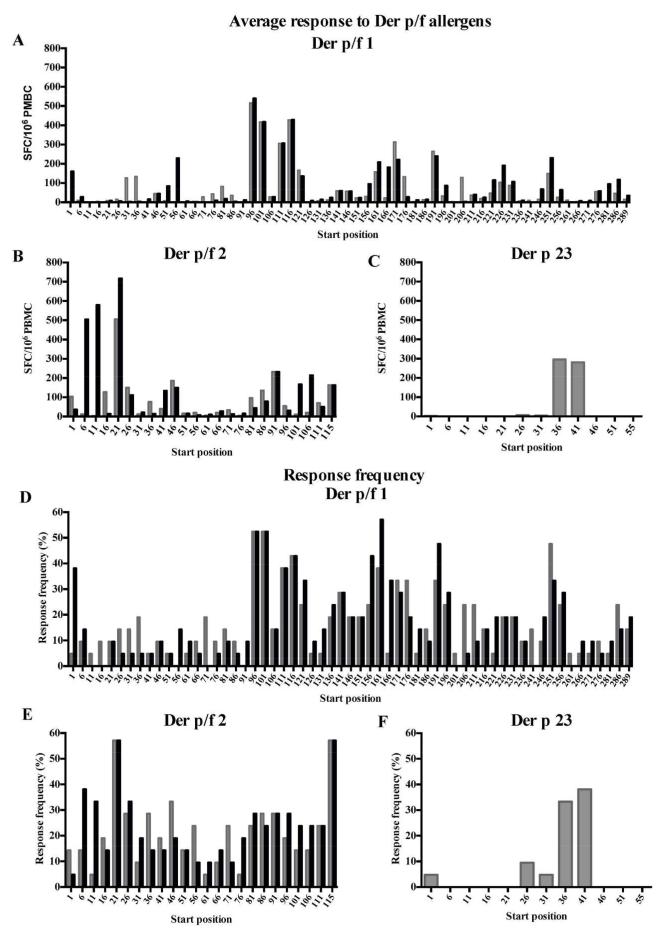
The overall reactivity detected in the course of these experiments is shown in Figures 1 A-C, which depict the sum total SFC detected for each peptide for each of the three antigen groups examined. Group 1 reactivity was strongest, exceeding reactivity to group 2 by about two-fold. Furthermore, reactivity to group 1 and 2 allergens is much stronger than the reactivity observed against Der p 23 (>10-fold). This might be due to differences in the allergen content in the HDM extract, with it likely that the Der p 1 and Der p 2 content is higher than that of Der p 23. The corresponding overall frequency of donor recognition of each peptide is shown in Figures 1 D-F. The analysis of response frequencies reveals a hierarchy of immunodominance similar to the one observed for response magnitude.
Figure 1.
A-C: Average SFC response (total SFC/no. donors tested, n=21) to individual 15-mer peptides spanning allergens from Der p (grey bars) and Der f (black bars) group 1 (A), group 2 (B) or Der p group 23 (C). D-F: Frequency of the responding donors to the respective peptides from Der p and Der f group 1 (D), group 2 (E) and Der p group 23-allergens (F) shown as percentage of donors investigated. *Starting position for the Der p 1 peptide is 95. The Der f 1 homolog starts at position 96.
A total of 819 positive responses were detected in any donor/readout/peptide combination, mapping to a total of 170 (or 95.5% of the 178) different peptides. Further analysis was undertaken to eliminate redundancies. Accordingly, in instances in which two overlapping peptides were positive in the same donor, the highest response was chosen as representative of the epitope reactivity. As a result, a total of 137 different, non-redundant, potential epitopes were recorded. These results underscore the heterogeneity of human allergen responses to HDM. A summary of donor IFNγ and IL-5 responses is provided in Supplemental Table 1A.
We also noted that relatively few regions accounted for a large fraction of the total response. Table 1 lists the main antigenic regions defined for each allergen, to be inclusive of any region accounting for 1% or more of the total reactivity to the corresponding antigen. In summary, 11-13 regions per allergen are sufficient to account for 90% or more of the reactivity to the Der p and Der f group 1 antigens, and 6 each for the two group 2 antigens. Interestingly, 90% of the total Der p 23 responses was accounted for by one single epitope region.
Table 1.
Dominant peptides from Der p/f group 1, 2 and 23 antigenic regions detected in this study.
| Sequence | Position | % donors |
SFC | IL5:IFNγ | % protein SFC |
Cumulative response |
|---|---|---|---|---|---|---|
| Der p 1 | ||||||
| DLRQMRTVTPIRMQGGCGSC | 95 | 61.9 | 14716 | 7.9 | 23.0 | 23.0 |
| GCGSCWAFSGVAATESAYLA | 111 | 47.6 | 10701 | 3.3 | 16.7 | 39.7 |
| QESYYRYVAREQSCR | 170 | 33.3 | 6563 | 8.2 | 10.3 | 49.9 |
| RFGISNYCQIYPPNA | 190 | 23.8 | 5250 | +++ | 8.2 | 58.1 |
| HAVNIVGYSNAQGVD | 250 | 42.9 | 3076 | 8.1 | 4.8 | 63.0 |
| IEYIQHNGVVQESYY | 160 | 28.6 | 3073 | 3.4 | 4.8 | 67.8 |
| VKYVQSNGGAINHLS | 36 | 14.3 | 2783 | 6.7 | 4.3 | 72.1 |
| NKIREALAQTHSAIA | 205 | 19.0 | 2647 | 8.9 | 4.1 | 76.2 |
| DLNAETNAC SINGNAPAEI | 76 | 14.3 | 2626 | 3.5 | 4.1 | 80.3 |
| KDLDAFRHYDGRTIIQRDNG | 225 | 19.0 | 2206 | 5.0 | 3.4 | 83.8 |
| QELVDCASQHGCHGDTIPRG | 141 | 38.1 | 2173 | 28.8 | 3.4 | 87.2 |
| NWGDNGYGYFAANID | 275 | 9.5 | 1160 | 19.5 | 1.8 | 89.0 |
| VIIGIKDLDAFRHYD | 220 | 4.8 | 990 | +++ | 1.5 | 90.5 |
| INHLSDLSLDEFKNR | 46 | 9.5 | 937 | 5.0 | 1.5 | 92.0 |
| AANIDLMMIEEYPYV | 285 | 19.0 | 887 | +++ | 1.4 | 93.4 |
| Sequence | Position | % donors |
SFC | IL5:IFNγ | % protein SFC |
Cumulative response |
|---|---|---|---|---|---|---|
| Derf 1 | ||||||
| DLRSLRTVTPIRMQGGCGSC | 96 | 61.9 | 15723 | 10.2 | 21.7 | 21.7 |
| GCGSCWAFSGVAATESAYLA | 111 | 47.6 | 10701 | 3.3 | 14.8 | 36.5 |
| YQPNYHAVNIVGYGSTQGVD | 246 | 38.1 | 5893 | 11.0 | 8.1 | 44.6 |
| HYGISNYCQIYPPDVKQIRE | 191 | 61.9 | 5781 | 14.5 | 8.0 | 52.6 |
| TIPRGIEYIQQNGVVEERSY | 156 | 61.9 | 5416 | 21.6 | 7.5 | 60.1 |
| EFKNRYLMSAEAFEQ | 56 | 14.3 | 4787 | 1.8 | 6.6 | 66.7 |
| EERSYPYVAREQQCR | 171 | 28.6 | 4636 | 72.2 | 6.4 | 73.1 |
| KDLRAFQHYDGRTIIQHDNG | 226 | 23.8 | 4180 | 10.1 | 5.8 | 78.9 |
| GYGYFQAGNNLMMIEQYPYV | 281 | 14.3 | 3516 | 2.2 | 4.9 | 83.7 |
| RPASIKTFEEFKKAF | 1 | 38.1 | 3370 | 13.4 | 4.7 | 88.4 |
| QELVDCASQHGCHGDTIPRG | 141 | 38.1 | 2173 | 28.8 | 3.0 | 91.4 |
| VIIGIKDLRAFQHYD | 221 | 4.8 | 833 | 0.7 | 1.2 | 92.5 |
| Sequence | Position | % donors |
SFC | IL5:IFNγ | % protein SFC |
Cumulative response |
|---|---|---|---|---|---|---|
| Der p 2 | ||||||
| CHGSEPCIIHRGKPFQLEAV | 21 | 57.1 | 10494 | 12.7 | 34.2 | 34.2 |
| YDIKYTWNVPKIAPKSENVV | 86 | 38.1 | 6679 | 16.6 | 21.8 | 56.0 |
| NTKTAKIEIKASIDG | 46 | 23.8 | 3762 | 5.2 | 12.3 | 68.2 |
| GVLACAIATHAKIRD | 115 | 57.1 | 3417 | 67.3 | 11.1 | 79.4 |
| DQVDVKDCANHEIKK | 1 | 14.3 | 2173 | 15.7 | 7.1 | 86.5 |
| QLEAVFEANQNTKTA | 36 | 19.0 | 1430 | +++ | 4.7 | 91.1 |
| VKGQQYDIKYTWNVP | 81 | 4.8 | 937 | +++ | 3.1 | 94.2 |
| NACHYMKCPLVKGQQ | 71 | 14.3 | 563 | +++ | 1.8 | 96.0 |
| Sequence | Position | % donors |
SFC | IL5:IFNγ | % protein SFC |
Cumulative response |
|---|---|---|---|---|---|---|
| Der f 2 | ||||||
| CHGSDPCIIHRGKPFTLEAL | 21 | 57.1 | 15510 | 33.2 | 31.2 | 31.2 |
| KDCANNEIKKVMVDGCHGSD | 6 | 38.1 | 12955 | 24.1 | 26.1 | 57.3 |
| YDAKYTWNVPKIAPKSENVV | 86 | 38.1 | 6056 | 23.3 | 12.2 | 69.5 |
| SENVVVT VKL VGDNGVL ACA | 101 | 23.8 | 5126 | 8.2 | 10.3 | 79.8 |
| FDANQNTKTAKIEIKASLDG | 41 | 19.0 | 3623 | 9.1 | 7.3 | 87.1 |
| GVLACAIATHAKIRD | 115 | 52.4 | 3334 | 65.7 | 6.7 | 93.8 |
| DQVDVKDCANNEIKK | 1 | 4.8 | 753 | +++ | 1.5 | 95.3 |
| Sequence | Position | % donors |
SFC | IL5:IFNγ | % protein SFC |
Cumulative response |
|---|---|---|---|---|---|---|
| Der p 23 | ||||||
| PKDPHKFYICSNWEAVHKDC | 36 | 38.1 | 7060 | +++ | 97.6 | 97.6 |
| CPSRFGYFADPKDPH | 26 | 9.5 | 130 | 0.6 | 1.8 | 99.4 |
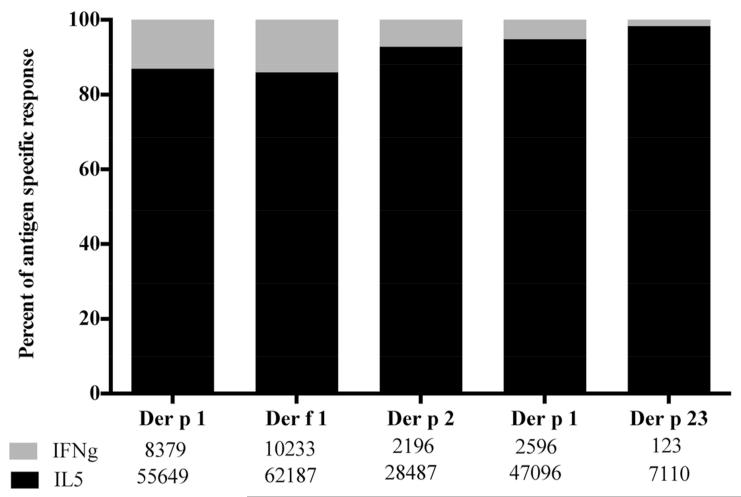
Next, the data was assessed for polarization of Th-responses. As expected, because of the allergic status of the donors, Th2 responses dominated Th1 responses by a factor of approximately 8:1. Additionally, we were interested in whether responses to group 1 and 2 allergens differed in their Th1/Th2 balance. As shown in Figure 2, regardless of the allergen, a similar pattern of IL-5 dominance was observed. Further analysis to test whether any of the main antigenic regions would be associated with a differential pattern of Th1/Th2 responses revealed similar patterns of polarization regardless of the antigenic region considered (see Table 1).
Figure 2.
IL-5/IFNγ-balance as a function of HDM antigen. Data of 21 patients is shown as percentage of total antigen specific response (SFC) attributable to IL-5 (black) and IFNγ (gray).
Generation of a comprehensive pool of HDM epitopes
In most instances, because of the low frequency of antigen-specific CD4+ T cells within PBMC, the study of allergen-specific T cell responses requires an in vitro stimulation step to expand allergen reactive T cells [22, 30]. We reasoned that simultaneous use of as many different epitopes as possible might obviate the need for in vitro restimulation, because while the frequency of T cells recognizing each individual epitope may be below the limit of detection, an epitope pool might collectively yield a signal above the limit of detection. To test this hypothesis, and_based on the results from our epitope screening studies we assembled a pool of immunodominant house dust mite epitopes as presented in Table 1. This pool included the 34 most dominant peptides cumulatively accounting for 90% of the total allergen- specific response in our screen. To take previously identified epitopes into consideration an additional set of 41 epitopes was also selected from the Immune Epitope Database (IEDB) [31]. The set included Der p/f epitopes, of canonical HLA class II length of 13 to 25 residues, and was edited to eliminate redundant and/or nested sequences. Only epitopes identified in ELISA, ELISPOT, intracellular cytokine staining (ICS) assays or by HLA-tetramer staining were considered and for epitopes tested in 10 or more donors, those recognized in less than 10% were eliminated. This set allowed us to include epitopes that were previously identified in an unbiased fashion by other studies and investigators, and in independent cohorts. Further, this analysis allowed coverage of the Der p 4 allergen, which was not specifically investigated in the current study. No epitope from any other Der p/f allergen matching the criteria above could be identified from the literature or the IEDB resource. In total, a set of 75 epitopes eliciting Th1 or Th2 responses was selected (Supplementary Table 2).
The comprehensive HDM epitopes pool allows detection of ex vivo T cell responses
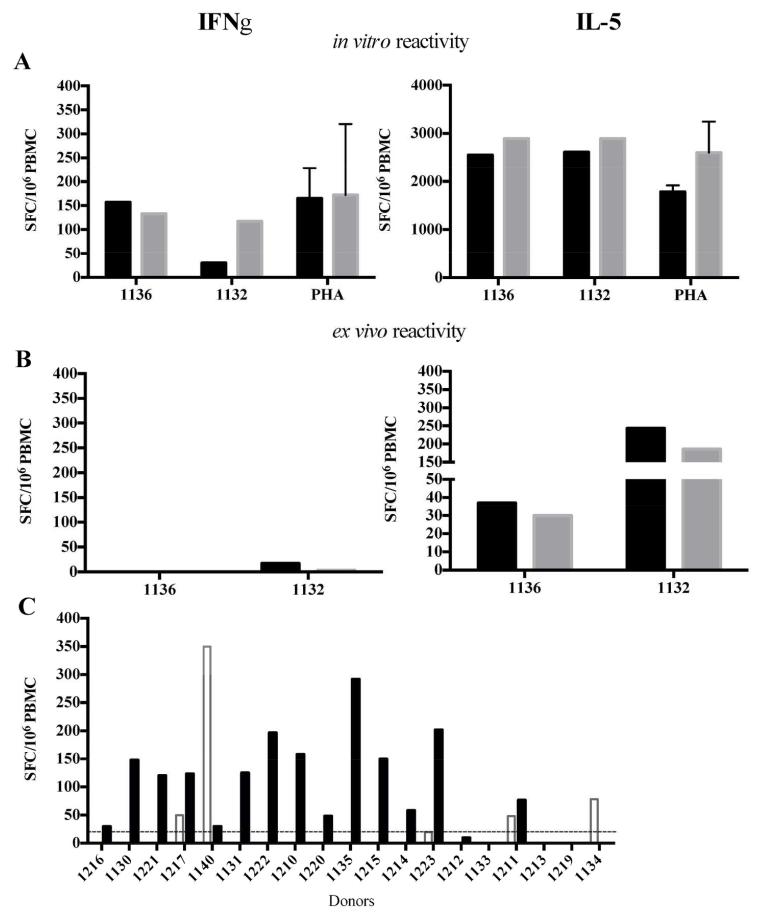
This pool of 75 immunodominant peptides from Der p/f allergens (referred to as Dust Mite Pool, or DMP hereafter) was then tested in PBMCs of two house dust mite allergic individuals in vitro following 14 days of culture as well as ex vivo (Figure 3). After in vitro expansion the response magnitude of DMP-stimulated PBMCs was comparable to PHA-stimulated samples (Figure 3A). In line with previous results from this study, IL-5 production was significantly higher than IFNγ in the in vitro assay. Moreover, significant numbers of IL-5-producing cells were detected directly ex vivo following DMP stimulation, demonstrating that this pool allows the detection of Th2 responses without any further in vitro manipulation (Figure 3B). The experiments depicted in Fig 3A and B were exploratory and performed in two representative donors. Based on the results a larger panel of donors was tested as shown in Figure 3C.
Figure 3.
A comprehensive pool of immunodominant peptides from Der p/f allergens allows ex vivo detection of immune responses. IFNγ-/IL-5-reactivity against a house dust mite pool was tested in PBMCs from two allergic individuals (1132 and 1136) (A) in vitro after 14 days of culture as well as (B) ex vivo without any further in vitro manipulation. In each case, the left panel shows IFNγ- and the right panel shows IL-5 responses. The response magnitude to two concentrations of the pool or PHA is shown (black bars, 5 μg/ml; grey bars, 0.5 μg/ml; PHA, mean and SD are plotted). (C) Ex vivo IL-5- (black) and IFNγ– (white) -reactivity detected in a panel of house dust mite allergic individuals.
To validate these findings and test whether ex vivo reactivity could be detected in a broader set of subjects we tested allergic individuals that were previously analyzed after in vitro expansion, ex vivo (see Supplementary Table 1A for results of the in vitro assay). Due to limited cell sample availability only 19 of 21 donors of the first subject cohort were analyzed. Using the peptide pool we detected IL-5 responses in 14 out of 19 donors (Figure 3C). As expected, IFNγ responses were much less frequently observed (4/19 subjects).
DMP ex vivo reactivity correlates with IgE-reactivity
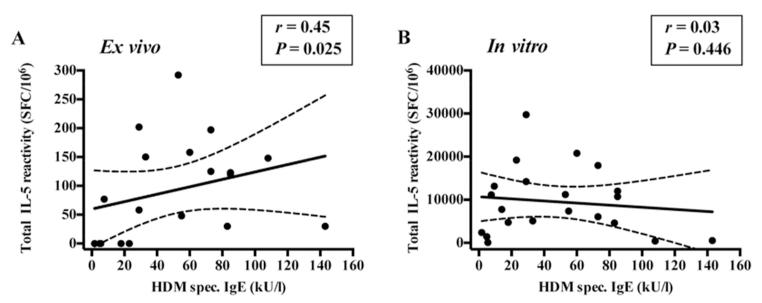
To establish whether a correlation could be detected between T cell responses and IgE-titers either following in vitro restimulation or in ex vivo assays, Der p/f-specific IgE-titers, defined as the sum total of specific IgE directed against Der p and Der f extracts, were determined (see supplementary table 1A). IL-5 reactivity was defined as the sum of total IL-5 responses for each Der allergen tested in vitro (supplementary table 1A) or ex vivo responses against the DMP as shown in Figure 3C. Ex vivo, a positive correlation between IL-5-reactivity and the titer of dust mite specific IgE was detected (Spearman r=0.45, P=0.025, one-tailed) whereas no significant correlation between these two parameters was observed following the in vitro culture (Figure 4). We also evaluated, for both group 1 and 2 allergens, the correlation between Der p or Der f1 specific IgE and the allergen-specific T cell response. As shown in supplemental figure 1, no correlation was detected.
Figure 4.
Ex vivo IL-5-production correlates with IgE, while in vitro does not. Correlation between HDM-specific IgE-titer and (A) ex vivo (n=19) and (B) in vitro (n=21) IL-5-reactivity in house dust mite allergic individuals. Spearman r and P-values (one-tailed) are shown. Best-fit line and 95% confidence band is plotted.
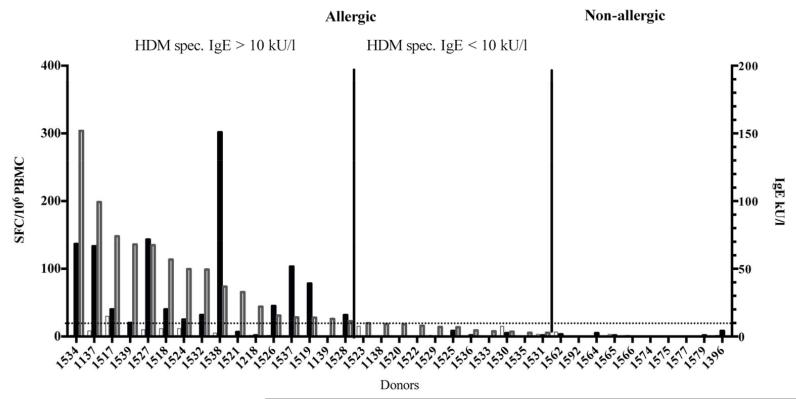
To further confirm and validate our results, we assembled an additional cohort composed of 27 allergic and 10 non-allergic individuals and tested their PBMCs for ex vivo IFNγ/IL-5 reactivity to DMP stimulation as described before (supplementary table 1B). The results are presented in Figure 5. In line with our observation from the previous cohort, the immune reactivity in allergic individuals was IL-5 dominated and associated with higher IgE-titers. When donors with HDM specific IgE-titers >10 kU/L were compared to those with low IgE (<10 kU/L) IL-5 responses were observed in 12 out of 16 donors versus 0 out of 11 in the low titer group (Figure 5). In non-allergic individuals no significant responses were detected above the threshold of 20 SFC. A compilation of data from all tested donors (Figures 3 to 5) revealed that DMP stimulation leads to detectable IL-5 responses in 88% of allergic individuals (31 of 35 donors) whereas no IL-5 signal was detected in any of the tested non-allergic subjects (Supplementary Table 3).
Figure 5.
IL-5-reactivity (black bars) and IgE-titers (grey bars) in a secondary patient cohort including 27 house dust mite allergic and 10 non-allergic individuals.
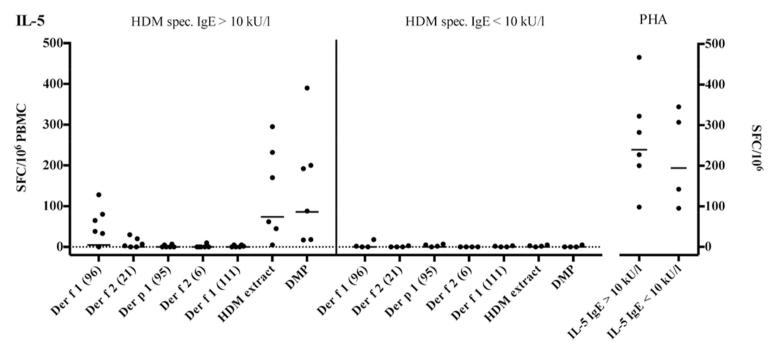
IL-5 reactivity against the DMP is similar to that against the HDM extract
As mentioned above, our hypothesis was that simultaneous use of the many different epitopes included in DMP obviates the need for in vitro restimulation, because while the frequency of T cells recognizing each individual epitope may be below the limit of detection, an epitope pool might yield a signal above the limit of detection. To further address this point, the ex vivo IFNγ/IL-5 reactivity against the DMP and HDM extracts was measured in a set of 10 HDM allergic donors (Figure 6). Six of these donors had not been previously tested, while four were included in the experiment of Figures 3-5. As a control we included five individual peptides eliciting the highest responses in our screen (Table 1).
Figure 6.
Comparison of ex vivo IL-5-reactivity against extract, individual 15/20mer peptides and DMP in house dust mite allergic donors.
Overall, the magnitude of IL-5-reactivity against the DMP was similar to the reactivity of HDM extract stimulated samples. Responses to single peptides were below the detection limit in most cases except for Der f 1961-15 and Der f 221-40 eliciting responses in five and two donors with IgE-titers above 10kU/l, respectively. These results are consistent with the notion that the various less dominant epitope specificities, including also the ones described in the literature contribute to enhancing the signal detected by the DMP pool.
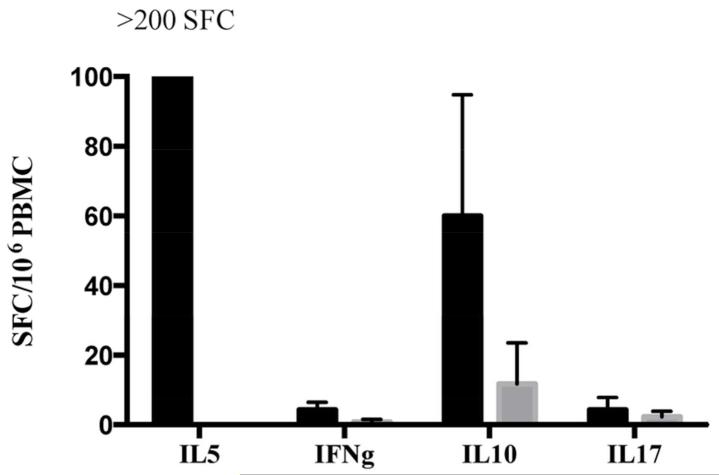
Ex vivo DMP stimulation of PBMCs from allergic donors results in a predominant Th2 response
In the experiments presented above, IL-5 responses to the DMP were measured based on previous experiments in our hands that suggested that IL-5 is the best readout for Th2 responses in the ELISPOT. Here we wanted to provide a proof of concept that the DMP could be used to also assay for additional cytokines, representative of different Th subsets, with the expectation that Th2 responses would be dominant. For this reason, we tested 2 HDM allergic donors, and 2 non-allergic controls in ELISPOT assays for IL-5, IFNγ, IL-10 and IL-17 as cytokines representative of Th1, Treg, and Th-17 subsets, respectively. The results shown in Figure 7 demonstrate that vigorous IL-5 responses are detected in the allergic but not in the non-allergic cohort. By comparison, IL-10, IFNγ and IL-17 responses are less vigorous, albeit also only detected in the allergic individuals and not in the non-allergic controls.
Figure 7.
Ex vivo DMP stimulation of PBMCs results in a predominant Th2 response in allergic donors (black bars) compared to non-allergic individuals (grey bars).
Discussion
In the present study, we analyzed the patterns of T cell recognition to several described house dust mite (HDM) allergens and epitopes, including group 1, 2 and 23 allergens from Der p and group 1 and 2 allergens from Der f. We mapped T cell responses and characterized the Th1/Th2 balance in T cell responses directed against different Der p and Der f epitopes.
The reactivity pattern observed against Der p-derived group 1 and group 2 allergens was compared with the reactivity observed against group 1 and 2 Der f-derived allergens. In general the reactivity pattern was very similar, as expected on the basis of the close phylogenetic relationship between Dermatophagoides pteronyssinus and Dermatophagoides farinae, and the significant sequence similarity between the allergens derived from the two species (84% sequence identity) [32, 33]. However, some less well conserved regions were recognized in a species-specific fashion, such as regions Der f 26-25, and Der p 126-50, where the reactivity was much stronger compared to the homologous counterpart, Der p 26-25 and Der f 126-50, respectively. Differences or similarities in patterns of recognition could not be addressed in the case of Der p 23, since the potential Der f homolog has not yet been identified.
Comparing response magnitudes, the immune reactivity in group 1 was found to be the strongest, exceeding reactivity to group 2 by about two-fold. Furthermore, reactivity to group 1 and 2 allergens was more than 10-fold higher than the reactivity observed against Der p 23. This might be due to differences in the allergen content in the HDM extracts, with it likely that Der p 1>Der p 2>>Der p 23. It is well known that Der p 1 and Der p 2 can be detected in HDM extracts, whereas the amount of Der p 23 is hardly detectable [7, 8].
In terms of immunodominance of HDM group 1, 2 and 23 allergens at the T cell level, we observed that relatively few regions accounted for a large fraction of the total response. 11-13 regions each were found to be sufficient to account for 90% or more of the reactivity to the Der p and Der f group 1 antigens, and 6 each for the two group 2 antigens. It should be noted that group 1 and group 2 allergens differ in size, with group 1 being twice as large as group 2. Indeed, when the number of epitopes per 100 residues is calculated, very similar values are observed (range 3.6-4.7, with an average of 4.3), suggesting that the difference in size between the two allergens might explain the difference in the number of epitopes. In the case of Der p 23, a single epitope accounted for the majority of the response. This lower number of epitopes could only be partially explained by the smaller size of the Der p 23 protein, as the number of epitopes/100 residue value was still 3 fold lower compared to what was observed for group 1 and 2 allergens. These data thus emphasize that the immunodominance hierarchy observed at the level of T cell response magnitude and frequency of responses is reflected in the epitope density in the various allergens. The data also demonstrate how relatively few regions elicit strong T cell responses to the antigens studied.
As stated above, few regions that accounted for >90% of the T cell reactivity. These results match previous observations in other systems such as Timothy Grass [25], the preponderance of the antigen specific response could be accounted for by HLA promiscuous binding peptides. Similar patterns have also been reported in the context of cockroach allergens [34] and several other common allergens [35]. In the present case, we have not performed HLA class II binding measurements. However, utilizing binding prediction algorithms available on the IEDB website, we note that 27 of the 44 regions defined here are predicted to bind 10 or more of the 27 most common HLA class II specificities. Together, these 27 regions account for between 60 and 98% (average 74%) of the response to the 5 antigens studied. Thus, while the issue of promiscuity was not specifically addressed here, we do suspect that as appears to be the case with other allergens, promiscuity will also be an appreciable factor in the context of the HDM response.
We also note that several of these epitopes partially match previously described epitopes in the HDM allergens ([17, 36-38] Eddie James). In most cases, thymidine incorporation assays were used to identify epitopes from Dermatophagoides pteronyssinus and D. farinae [17, 36-38]. In our study, epitopes were identified by ELISPOT following in vitro expansion of allergen-specific cells. However, simultaneous use of many different epitopes might represent a powerful approach to detecting T cell responses; while even if the frequency of T cells recognizing each individual epitope may be below the limit of detection, a pool of a large number of epitopes might pass the limit of detection. Accordingly, based on the results from our epitope screening studies we assembled a pool of immunodominant house dust mite epitopes cumulatively accounting for 90% of the Der p/f 1, 2 and Der p 23 allergen specific responses and supplemented them with additional epitopes published in the literature [17, 18, 20-23]. This pool (DMP) recapitulated the IL-5 reactivity against house dust mite allergens and allowed ex vivo detection of antigen-specific T cells. In allergy, studying antigen-specific T cell responses without any further in vitro manipulation is often hampered by the low frequency of antigen-specific CD4+ T cells within peripheral blood mononuclear cells. In vitro approaches are, therefore, commonly used [22, 30] to expand allergen reactive T cells. However, while the epitope specificity is maintained following expansion, possible alteration of phenotypic specificity during in vitro expansion cannot be excluded. Our approach utilized the higher sensitivity achieved by in vitro expansion to identify individual HDM epitopes. Then, as described pooling of the epitopes allowed generation of a reagent which broadly recruits most HDM T cell specificities, and confers enough sensitivity to detect responses ex vivo. The fact that we could not observe a good correlation between magnitude of T cell responses and IgE-reactivity following in vitro culture, but did when considering ex vivo responses could be a result of the culture system skewing the response magnitudes. The DMP could therefore serve as a useful research tool to characterize allergen-specific CD4+ T cell responses directly ex vivo. The DMP could be used to detect the HDM-T cells in allergic patients but also in patients treated by SIT [39].
Additionally, the DMP might serve as a great tool to study potential differential responses between patients suffering from allergic rhinitis and allergic asthma. Moreover, not only correlations between dust-mite specific T cell responses and severity of clinical manifestations, but also the potential influence of seasonality could be studied directly ex vivo (e. g. fall compared with summer) [22, 40, 41]. Moreover, allergen extracts are of a more complex nature and the fact that commercial extracts are not standardized is well known [42]. Indeed, it is possible that the Der p/f 1-2 content may be drastically different between commercial HDM extracts. Variations in the amount of specific allergens could be critical for the effectiveness of T-cell stimulation, and different results could be obtained with different allergen extracts [42, 43]. Furthermore, allergen extracts are often contaminated by other substances, such as LPS, adding potential variability to the results obtained. The DMP, however, as a well-defined entity might therefore be a useful tool for more detailed studies of dust mite-specific CD4+ T cells leading to a better understanding of immune pathogenesis of house dust mite allergic responses. Furthermore, it might be useful to monitor potential phenotypic changes within the course of allergen-specific immunotherapy, and even as a direct therapeutic agent in allergen specific immunotherapy.
Supplementary Material
Acknowledgements
Funding was provided by ALK-Abello A/S (Horsholm, Denmark).
This project was also funded in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, under grant number U19 AI100275.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
REFERENCES
- 1.Platts-Mills TA, Thomas WR, Aalberse RC, Vervloet D, Champman MD. Dust mite allergens and asthma: report of a second international workshop. The Journal of allergy and clinical immunology. 1992;89:1046–60. doi: 10.1016/0091-6749(92)90228-t. [DOI] [PubMed] [Google Scholar]
- 2.Platts-Mills TA, Vervloet D, Thomas WR, Aalberse RC, Chapman MD. Indoor allergens and asthma: report of the Third International Workshop. The Journal of allergy and clinical immunology. 1997;100:S2–24. doi: 10.1016/s0091-6749(97)70292-6. [DOI] [PubMed] [Google Scholar]
- 3.Salo PM, Arbes SJ, Jr., Crockett PW, Thorne PS, Cohn RD, Zeldin DC. Exposure to multiple indoor allergens in US homes and its relationship to asthma. The Journal of allergy and clinical immunology. 2008;121:678–84 e2. doi: 10.1016/j.jaci.2007.12.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voorhorst R. House dust mite and house dust allergy. Annals of allergy. 1977;38:71. [PubMed] [Google Scholar]
- 5.Chapman MD, Pomes A, Breiteneder H, Ferreira F. Nomenclature and structural biology of allergens. The Journal of allergy and clinical immunology. 2007;119:414–20. doi: 10.1016/j.jaci.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Pomes A. Allergen structures and biologic functions: the cutting edge of allergy research. Current allergy and asthma reports. 2008;8:425–32. doi: 10.1007/s11882-008-0082-y. [DOI] [PubMed] [Google Scholar]
- 7.Tovey ER, Chapman MD, Platts-Mills TA. Mite faeces are a major source of house dust allergens. Nature. 1981;289:592–3. doi: 10.1038/289592a0. [DOI] [PubMed] [Google Scholar]
- 8.Thomas WR, Smith WA, Hales BJ. The allergenic specificities of the house dust mite. Chang Gung medical journal. 2004;27:563–9. [PubMed] [Google Scholar]
- 9.Ando T, Ino Y, Haida M, Honma R, Maeda H, Yamakawa H, Iwaki M, Okudaira H. Isolation of cysteine protease in the crude mite extract, Dermatophagoides farinae. International archives of allergy and applied immunology. 1991;96:199–205. doi: 10.1159/000235495. [DOI] [PubMed] [Google Scholar]
- 10.Stewart GA, Lake FR, Thompson PJ. Faecally derived hydrolytic enzymes from Dermatophagoides pteronyssinus: physicochemical characterisation of potential allergens. International archives of allergy and applied immunology. 1991;95:248–56. doi: 10.1159/000235437. [DOI] [PubMed] [Google Scholar]
- 11.Weghofer M, Grote M, Resch Y, Casset A, Kneidinger M, Kopec J, Thomas WR, Fernandez-Caldas E, Kabesch M, Ferrara R, Mari A, Purohit A, Pauli G, Horak F, Keller W, Valent P, Valenta R, Vrtala S. Identification of Der p 23, a peritrophin-like protein, as a new major Dermatophagoides pteronyssinus allergen associated with the peritrophic matrix of mite fecal pellets. Journal of immunology. 2013;190:3059–67. doi: 10.4049/jimmunol.1202288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mudde GC, Hansel TT, von Reijsen FC, Osterhoff BF, Bruijnzeel-Koomen CA. IgE: an immunoglobulin specialized in antigen capture? Immunology today. 1990;11:440–3. doi: 10.1016/0167-5699(90)90172-6. [DOI] [PubMed] [Google Scholar]
- 13.Larche M, Akdis CA, Valenta R. Immunological mechanisms of allergen-specific immunotherapy. Nature reviews Immunology. 2006;6:761–71. doi: 10.1038/nri1934. [DOI] [PubMed] [Google Scholar]
- 14.Lebman DA, Coffman RL. Interleukin 4 causes isotype switching to IgE in T cell-stimulated clonal B cell cultures. The Journal of experimental medicine. 1988;168:853–62. doi: 10.1084/jem.168.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gauchat JF, Lebman DA, Coffman RL, Gascan H, de Vries JE. Structure and expression of germline epsilon transcripts in human B cells induced by interleukin 4 to switch to IgE production. The Journal of experimental medicine. 1990;172:463–73. doi: 10.1084/jem.172.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Punnonen J, Aversa G, Cocks BG, de Vries JE. Role of interleukin-4 and interleukin-13 in synthesis of IgE and expression of CD23 by human B cells. Allergy. 1994;49:576–86. doi: 10.1111/j.1398-9995.1994.tb00122.x. [DOI] [PubMed] [Google Scholar]
- 17.Yssel H, Johnson KE, Schneider PV, Wideman J, Terr A, Kastelein R, De Vries JE. T cell activation-inducing epitopes of the house dust mite allergen Der p I. Proliferation and lymphokine production patterns by Der p I-specific CD4+ T cell clones. Journal of immunology. 1992;148:738–45. [PubMed] [Google Scholar]
- 18.Okano M, Nagano T, Nakada M, Masuda Y, Kino K, Yasueda H, Nose Y, Nishimura Y, Ohta N. Epitope analysis of HLA-DR-restricted helper T-cell responses to Der p II, a major allergen molecule of Dermatophagoides pteronyssinus. Allergy. 1996;51:29–35. doi: 10.1111/j.1398-9995.1996.tb04546.x. [DOI] [PubMed] [Google Scholar]
- 19.Verhoef A, Higgins JA, Thorpe CJ, Marsh SG, Hayball JD, Lamb JR, O’Hehir RE. Clonal analysis of the atopic immune response to the group 2 allergen of Dermatophagoides spp.: identification of HLA-DR and -DQ restricted T cell epitopes. International immunology. 1993;5:1589–97. doi: 10.1093/intimm/5.12.1589. [DOI] [PubMed] [Google Scholar]
- 20.Joost van Neerven R, van t’Hof W, Ringrose JH, Jansen HM, Aalberse RC, Wierenga EA, Kapsenberg ML. T cell epitopes of house dust mite major allergen Der p II. Journal of immunology. 1993;151:2326–35. [PubMed] [Google Scholar]
- 21.Bonvalet M, Wambre E, Moussu H, Horiot S, Kwok WW, Louise A, Ebo D, Hoarau C, Van Overtvelt L, Baron-Bodo V, Moingeon P. Comparison between major histocompatibility complex class II tetramer staining and surface expression of activation markers for the detection of allergen-specific CD4(+) T cells. Clinical and experimental allergy: journal of the British Society for Allergy and Clinical Immunology. 2011;41:821–9. doi: 10.1111/j.1365-2222.2011.03708.x. [DOI] [PubMed] [Google Scholar]
- 22.Wambre E, Bonvalet M, Bodo VB, Maillere B, Leclert G, Moussu H, Von Hofe E, Louise A, Balazuc AM, Ebo D, Hoarau C, Garcia G, Van Overtvelt L, Moingeon P. Distinct characteristics of seasonal (Bet v 1) vs. perennial (Der p 1/Der p 2) allergen-specific CD4(+) T cell responses. Clinical and experimental allergy: journal of the British Society for Allergy and Clinical Immunology. 2011;41:192–203. doi: 10.1111/j.1365-2222.2010.03641.x. [DOI] [PubMed] [Google Scholar]
- 23.Crack LR, Chan HW, McPherson T, Ogg GS. Identification of an immunodominant region of the major house dust mite allergen Der p 2 presented by common human leucocyte antigen alleles. Clinical and experimental dermatology. 2012;37:266–76. doi: 10.1111/j.1365-2230.2011.04227.x. [DOI] [PubMed] [Google Scholar]
- 24.Akdis M, Verhagen J, Taylor A, Karamloo F, Karagiannidis C, Crameri R, Thunberg S, Deniz G, Valenta R, Fiebig H, Kegel C, Disch R, Schmidt-Weber CB, Blaser K, Akdis CA. Immune responses in healthy and allergic individuals are characterized by a fine balance between allergen-specific T regulatory 1 and T helper 2 cells. The Journal of experimental medicine. 2004;199:1567–75. doi: 10.1084/jem.20032058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oseroff C, Sidney J, Kotturi MF, Kolla R, Alam R, Broide DH, Wasserman SI, Weiskopf D, McKinney DM, Chung JL, Petersen A, Grey H, Peters B, Sette A. Molecular determinants of T cell epitope recognition to the common Timothy grass allergen. Journal of immunology. 2010;185:943–55. doi: 10.4049/jimmunol.1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui HH, Grey H, Sette A. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nature biotechnology. 2006;24:817–9. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 27.Oseroff C, Peters B, Pasquetto V, Moutaftsi M, Sidney J, Panchanathan V, Tscharke DC, Maillere B, Grey H, Sette A. Dissociation between epitope hierarchy and immunoprevalence in CD8 responses to vaccinia virus western reserve. Journal of immunology. 2008;180:7193–202. doi: 10.4049/jimmunol.180.11.7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arlehamn CS, Sidney J, Henderson R, Greenbaum JA, James EA, Moutaftsi M, Coler R, McKinney DM, Park D, Taplitz R, Kwok WW, Grey H, Peters B, Sette A. Dissecting mechanisms of immunodominance to the common tuberculosis antigens ESAT-6, CFP10, Rv2031c (hspX), Rv2654c (TB7.7), and Rv1038c (EsxJ) Journal of immunology. 2012;188:5020–31. doi: 10.4049/jimmunol.1103556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiskopf D, Yauch LE, Angelo MA, John DV, Greenbaum JA, Sidney J, Kolla RV, De Silva AD, de Silva AM, Grey H, Peters B, Shresta S, Sette A. Insights into HLA-restricted T cell responses in a novel mouse model of dengue virus infection point toward new implications for vaccine design. Journal of immunology. 2011;187:4268–79. doi: 10.4049/jimmunol.1101970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wambre E, Van Overtvelt L, Maillere B, Humphreys R, von Hofe E, Ferhat L, Ebo D, Moingeon P. Single cell assessment of allergen-specific T cell responses with MHC class II peptide tetramers: methodological aspects. International archives of allergy and immunology. 2008;146:99–112. doi: 10.1159/000113513. [DOI] [PubMed] [Google Scholar]
- 31.Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, Wheeler DK, Gabbard JL, Hix D, Sette A, Peters B. The immune epitope database (IEDB) 3.0. Nucleic acids research. 2014 doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chua KY, Stewart GA, Thomas WR, Simpson RJ, Dilworth RJ, Plozza TM, Turner KJ. Sequence analysis of cDNA coding for a major house dust mite allergen, Der p 1. Homology with cysteine proteases. The Journal of experimental medicine. 1988;167:175–82. doi: 10.1084/jem.167.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dilworth RJ, Chua KY, Thomas WR. Sequence analysis of cDNA coding for a major house dust mite allergen, Der f I. Clinical and experimental allergy: journal of the British Society for Allergy and Clinical Immunology. 1991;21:25–32. doi: 10.1111/j.1365-2222.1991.tb00800.x. [DOI] [PubMed] [Google Scholar]
- 34.Oseroff C, Sidney J, Tripple V, Grey H, Wood R, Broide DH, Greenbaum J, Kolla R, Peters B, Pomes A, Sette A. Analysis of T cell responses to the major allergens from German cockroach: epitope specificity and relationship to IgE production. Journal of immunology. 2012;189:679–88. doi: 10.4049/jimmunol.1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oseroff C, Sidney J, Vita R, Tripple V, McKinney DM, Southwood S, Brodie TM, Sallusto F, Grey H, Alam R, Broide D, Greenbaum JA, Kolla R, Peters B, Sette A. T cell responses to known allergen proteins are differently polarized and account for a variable fraction of total response to allergen extracts. Journal of immunology. 2012;189:1800–11. doi: 10.4049/jimmunol.1200850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hales BJ, Thomas WR. T-cell sensitization to epitopes from the house dust mites Dermatophagoides pteronyssinus and Euroglyphus maynei. Clinical and experimental allergy: journal of the British Society for Allergy and Clinical Immunology. 1997;27:868–75. [PubMed] [Google Scholar]
- 37.O’Brien RM, Thomas WR, Tait BD. An immunogenetic analysis of T-cell reactive regions on the major allergen from the house dust mite, Der p I, with recombinant truncated fragments. The Journal of allergy and clinical immunology. 1994;93:628–34. doi: 10.1016/s0091-6749(94)70074-5. [DOI] [PubMed] [Google Scholar]
- 38.O’Hehir RE, Verhoef A, Panagiotopoulou E, Keswani S, Hayball JD, Thomas WR, Lamb JR. Analysis of human T cell responses to the group II allergen of Dermatophagoides species: localization of major antigenic sites. The Journal of allergy and clinical immunology. 1993;92:105–13. doi: 10.1016/0091-6749(93)90044-g. [DOI] [PubMed] [Google Scholar]
- 39.Schulten V, Tripple V, Sidney J, Greenbaum J, Frazier A, Alam R, Broide D, Peters B, Sette A. Association between specific timothy grass antigens and changes in TH1- and TH2-cell responses following specific immunotherapy. The Journal of allergy and clinical immunology. 2014;134:1076–83. doi: 10.1016/j.jaci.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michaud B, Gouvis-Echraghi R, Candon S, Couderc R, Jais JP, Bach JF, Chatenoud L, Just J. Quantification of circulating house dust mite-specific IL-4- and IL-13-secreting T cells correlates with rhinitis severity in asthmatic children and varies with the seasons. Clinical and experimental allergy: journal of the British Society for Allergy and Clinical Immunology. 2014;44:222–30. doi: 10.1111/cea.12222. [DOI] [PubMed] [Google Scholar]
- 41.Pacciani V, Corrente S, Gregori S, Pierantozzi A, Silenzi R, Chianca M, Moschese V, Chini L, Angelini F. Correlation of Der p 2 T-cell responses with clinical characteristics of children allergic to house dust mite. Annals of allergy, asthma & immunology: official publication of the American College of Allergy, Asthma, & Immunology. 2012;109:442–7. doi: 10.1016/j.anai.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 42.Focke M, Marth K, Valenta R. Molecular composition and biological activity of commercial birch pollen allergen extracts. European journal of clinical investigation. 2009;39:429–36. doi: 10.1111/j.1365-2362.2009.02109.x. [DOI] [PubMed] [Google Scholar]
- 43.Esch RE. Allergen source materials and quality control of allergenic extracts. Methods. 1997;13:2–13. doi: 10.1006/meth.1997.0491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.