Abstract
Neutral sphingomyelinase-2 (nSMase-2) is the major sphingomyelinase activated in response to pro-inflammatory cytokines and during oxidative stress. It is a membrane-bound 655 amino acid protein containing 22 cysteine residues. In this study, we expressed recombinant mouse nSMase-2 protein in Escherichia coli, and investigated whether nSMase-2 is a redox sensitive enzyme. Our results demonstrate that nSMase-2 exists as both monomers and multimers that are associated with high and low enzymatic activity respectively. Mutational analysis of nSMase-2 identified within its C-terminal catalytic domain several oxidant-sensitive cysteine residues that were shown to be involved in enzyme oligomerization. Changing Cys617 to Ser for example is a gain-of-function mutation associated with a decreased propensity for oligomerization. Alternatively, nSMase-2 expression in a bacterial strain that lacks endogenous thioredoxin, Rosetta-gami2, results in increased oligomer formation and lower enzyme activity. Phenotypic rescue was accomplished by treating nSMase-2 lysates with recombinant human thioredoxin. This indicates that nSMase-2 may be a novel substrate for thioredoxin. FRET analysis confirmed the presence of nSMase-2 multimers in mammalian HEK cells and their localization to the plasma membrane. In conclusion, our results identify nSMase-2 as a redox-sensitive enzyme, whose basal activity is influenced by thioredoxin-mediated changes in its oligomeric state.
Keywords: ceramide, cysteine, oligomerization, oxidative stress, sphingomyelinase, thioredoxin
INTRODUCTION
Redox signalling has emerged as a major focus area of research, as key signal transduction pathways appear to be redox-sensitive. Reactive oxygen species (ROS), particularly hydrogen peroxide (H2O2) as well as reactive nitrogen species (RNS) play a central part in these pathways as they couple intracellular changes in the redox state to biochemical functions [1]. Cysteine thiol groups react faster than the other amino acids with oxidizing species and some of the resulting oxidation products can be reversibly reduced [2], underlaying the idea that oxidant-exposed cysteine residues in proteins function as ‘switches’ that regulate protein functions during redox signalling [3,4]. Thiols are known to undergo several oxidant-mediated covalent modifications including sulfenic/sulfinic/sulfonic acid formation, sulfhydration, glutathionylation, nitrosylation and intra- or inter-molecular disulfide bond formation [5]. These modifications can lead to either activation or inactivation of a specific protein function and many of the cellular functions seem to be regulated through cysteine-dependent enzyme modifications [3-6]. The redox state of protein thiols is under the control of two major systems, namely the thioredoxin and the GSH systems. These systems are also redox-sensitive and their functions are influenced to a great extent by cellular H2O2 levels. The ever-expanding list of redox-sensitive enzymes, regulated by the thioredoxin/glutaredoxin systems includes metabolic enzymes, key regulators of cell cycle progression and apoptosis, as well as transcription factors [6]. Together they co-ordinate the highly complex cellular response/adaptation to fluctuations in ROS/RNS levels.
Ceramide is a bioactive sphingolipid that mediates cellular responses to stress [7,8]. Increased intracellular ceramide levels are observed under various clinical conditions like inflammation and oxidative stress, whereas excessive ceramide accumulation has been implicated in the induction of cell growth arrest and apoptosis [7,8]. Ceramide levels are controlled by several different metabolic enzymes, among which the sphingomyelinases are of particular importance. They are a group of phosphodiesterases that catalyse the hydrolysis of sphingomyelin to ceramide and phosphorylcholine. Importantly, practically all of the sphingomyelinases seem to be activated during oxidative stress, but the mechanisms mediating these effects are yet to be completely elucidated [9,10].
Neutral sphingomyelinase-2 (nSMase-2) has been shown to be the major sphingomyelinase activated in response to pro-inflammatory cytokines, such as tumour necrosis factor α (TNFα) [11] and interleukin-1β (IL-1β) [12,13]. It is activated by oxidative stress, like that observed in aging liver and in the lungs of smokers as well as in response to amyloid β-peptide accumulation [14-17]. nSMase-2 is a membrane-bound protein of 655 amino acids containing two N-terminal hydrophobic domains and a C-terminal catalytic domain [18]. This is a protein particularly rich in cysteine residues. Of its 22 cysteines, eight are localized in the non-catalytic domain and 14 are in the catalytic domain. nSMase-2 activity is Mg2+ -dependent and is optimal at neutral pH [11,18]. Recently, nSMase-2 was shown to contain unique anionic phospholipid lipid binding domains that aid in its localization to the plasma membrane [19]. Additionally, post-translational modifications of nSMase-2, which include cysteine-mediated palmitoylation, have been implicated in its localization and stabilization [20].
Several studies have demonstrated a marked increase in nSMase-2 activity under conditions of oxidative stress [14,15,21,22]. There is a tight, inverse correlation between nSMase-2 activity and intracellular GSH levels [15,21-23]. This antioxidant has also been shown to strongly inhibit nSMase-2 activity in vitro [21,24]. Hence, intracellular depletion of GSH has been proposed as a possible mechanism for nSMase-2 activation. nSMase-2 is also known to be nitrosylated under endoplasmic reticulum stress, through the nitration of tyrosine residues [25]. The protein is de-phosphorylated in a calcineurin-dependent manner at positions S173, S208, S289, S292 and S299 upon H2O2 treatment of human bronchial epithelial cells and A549 adenocarcinoma cells [22,26].
In this study, we expressed recombinant mouse nSMase-2 protein in Escherichia coli, and investigated whether nSMase-2 is a redox sensitive enzyme. Our results demonstrate that the redox state of nSMase-2 strongly affects its activity. Furthermore, the recombinant nSMase-2 was shown to exist in both monomeric and multimeric forms, which correspond respectively to high and low enzymatic activity. Mutational analysis of nSMase-2 identified within its C-terminal catalytic domain several oxidant-sensitive cysteine residues that were shown to be involved in enzyme oligomerization. Lastly, thioredoxin was found to affect the oligomeric state of nSMase-2, indicating that nSMase-2 might be a novel substrate for thioredoxin.
EXPERIMENTAL
Plasmids, bacterial strains, adenoviral construct, enzymes and chemicals
Plasmids pET-32a (−) and pET-26b (+) were purchased from Novagen, and pAcGFP1-C1 and +pmStrawberry were purchased from Clontech. The bacterial strains used were E. coli Rosetta2 (DE3) pLysS and Rosetta-gami2 (DE3) pLysS from Novagen, and DH5α from Invitrogen. The adenovirus coding mouse FLAG-tagged nSMase-2 (Ad-nSMase-2) was purified as previously described [12]. Restriction endonucleases and DNA modifying enzymes were purchased from New England Biolabs. All other chemicals including l-buthionine sulfoximine (BSO), diamide (DM), dithiothreitol (DTT), H2O2, isopropyl-β-d-thiogalactopyranoside (IPTG), 2-mercaptoethanol (β-Me) and sodium iodoacetamide (IAA) were purchased from Sigma–Aldrich. The Advanced Genetics Technologies Center (AGTC), at the University of Kentucky, performed sequencing analysis.
Construction of nSMase-2 bacterial expression vectors
The mouse C-terminal FLAG-tagged nSMase-2 gene (SMPD3) was PCR amplified from the previously described pBluescript-FLAG-nSMase2 plasmid [12], using the primer set 5′-CCAATCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATAT GGTTTTGTACACGACCCCCTTTCC-3′ and 5′-GATTCTCGAGCTTGTCATCGTCGTCCTTGTAGTC-3′ (underlined nucleotides denote restriction site positions). The resulting PCR product was purified and digested with XbaI and XhoI, followed by gel purification using the Wizard SV Gel and PCR cleanup kit from Promega. The FLAG-tagged nSMase-2 was then ligated into XbaI/XhoI-digested pET-32a (+) and pET-26b (+) vectors allowing for the addition of a His tag consequentially to the C-terminal FLAG tag and thereby creating pET-32a-nSMase-2-FLAG-6xHis and pET-26b-nSMase-2-FLAG-6xHis.
Two additional vectors were constructed for use in co-immunoprecipitation experiments. The pET-32a-nSMase-2-FLAG was created from the pET-32a-nSMase-2-FLAG-6xHis construct by removing the 6xHis tag. For pET-26b-nSMase-2-6xHis construction the nSMase-2 gene was PCR amplified without the FLAG tag using primers 5′-CCAATCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGGTTTTGTACACGACCCCCTTTCC-3′ and 5′-GATTCTCGAGTGCCTCCTCTTCCCCTGCAG-3′ (underlined nucleotides denote XbaI and XhoI restriction site positions) and cloned into XbaI/XhoI digested pET-26b (+) vector.
In all cases, plasmids were isolated from DH5α transformed cells. Single colonies were used to inoculate Luria-Bertani (LB) cultures for the Wizard SV pro DNA Miniprep kit (Promega). All constructs were checked for correct insert orientation using restriction enzyme digestion and sequenced to verify error-free nSMase-2 sequence.
Construction of nSMase-2 mammalian fluorescent expression vectors
For FRET experiments, the nSMase-2 gene was tagged on the N-terminal end of the enzyme with either GFP or RFP. The nSMase-2 gene was PCR amplified using the primer set 5′-CCAACTCGAGATGGTTTTGTACACGACC-3′ and 5′-GATTGGTACCTGCCTCCTCTTCCCCTGCAGACAC-3′ (underlined nucleotides denote XhoI and KpnI restriction site positions). The resulting PCR product was purified and digested with XhoI and KpnI, followed by gel purification using the Wizard SV Gel and PCR cleanup kit from Promega. The purified nSMase-2 PCR product was then ligated into XhoI/KpnI-digested pAcGFP1-C1 vector (Clontech) thereby creating the pAcGFP-C1-nSMase-2 vector.
For construction of the N-terminal tagged RFP-nSMase-2, the mStrawberry gene from the pmStrawberry plasmid (Clontech) was PCR amplified using the primer set 5′-CCAAGCTAGCGCTACCGGTCGCCACCATGGTGAGCAAGG-3′ and 5′-GATTCTCGAGATCT GAGTCCGGACTTGTACAGCTCGTCCATGCCGCCGGT-3′ (underlined nucleotides denote restriction site positions). The resulting PCR product was purified and digested with NheI and XhoI, followed by gel purification and ligation into NheI/XhoI-digested pAcGFP1-C1 vector creating the pmStrawberry-C1 vector. Subsequently, the purified XhoI/KpnI-digested nSMase-2 PCR product from above was ligated into the XhoI/KpnI-digested pmStrawberry-C1 vector creating the pmStrawberry-C1-nSMase-2 vector. All constructs were checked for correct insert orientation using restriction enzyme digestion and sequenced to verify error-free nSMase-2 sequence.
Site-directed mutagenesis of nSMase-2
The mouse nSMase-2 gene has 22 cysteine residues, with five of these (Cys53, Cys54, Cys59, Cys395 and Cys396) having previously been shown to undergo palmitoylation [20]. We mutated the other cysteines (Cys12, Cys24, Cys125, Cys132, Cys241, Cys359, Cys392, Cys400, Cys420, Cys425, Cys457, Cys471, Cys506, Cys514, Cys534, Cys561 and Cys617), either individually or sequentially, to serine using synthetic oligonucleotides (listed in Supplementary Tables S1 and S2). Mutagenesis was performed using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies) for individual cysteine-to-serine mutations, and the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies) for multiple cysteine-to-serine mutations.
Briefly, each amplification reaction consisted of 100–125 ng of each mutagenic primer (designed using the QuikChange Primer Design program of Agilent Technologies), 50 ng of plasmid template (pET-32a-nSMase-2-FLAG-6xHis) and proprietary reagents from Agilent Technologies. After an initial denaturation step at 95°C for 2 min, the mixture was subjected to 30 cycles of 95°C for 20 s, 55°C for 30 s and 65°C for 2–5 min. A final elongation step of 65°C for 5 min was performed prior to parental plasmid digestion with DpnI at 37°C for 60 min. A 1.5 μl aliquot was transformed into XL-10 Gold ultra-competent cells (Agilent Technologies). Following plating for selection, plasmids were purified using the Wizard SV Pro DNA Miniprep Kit (Promega). The desired mutation(s) were confirmed through sequencing.
Bacterial overexpression of wild-type (WT) and mutant nSMase-2
The pET-32a-nSMase-2-FLAG-6xHis, pET-26b-nSMase-2-FLAG-6xHis and mutant nSMase-2 constructs were transformed into Rosetta2 (DE3) pLysS (Novagen) using the manufacturer’s suggested protocol. The Rosetta-gami2 (DE3) pLysS cells, that have a constitutive loss-of-function mutation in the thioredoxin reductase gene (trxB), were used in experiments assessing the role of thioredoxin in nSMase-2 oligomerization. Overnight cell cultures started from single colonies were used to inoculate fresh LB medium, supplemented with either ampicillin or kanamycin (50 μg/μl) and chloramphenicol (34 μg/μl). Cultures were grown at 37°C to an D600 of 0.4–0.6 (~3 h of cell growth). nSMase-2 expression was induced by the addition of IPTG at a final concentration of 1 mM. After 4 h, 10 ml of each cell culture was centrifuged at 6000 g for 10 min at 4°C. The cell pellets were re-suspended in 500 μl of PBS and subjected to three cycles of freeze–thawing. The lysed cells were then treated with 1 μg of Benzonase (Novagen) for 30 min at 4°C and stored at −20°C.
Transfection of HEK-293-IL-1R cells
HEK 293 cells overexpressing the IL-1 receptor Type I (293-IL-1RI) were a gift from Dr. X. Li (Cleveland Clinic, Cleveland, OH) and were maintained in Dulbecco’s modified Eagles medium (DMEM) supplemented with 10% FBS and G418 (400 μg/ml). Cells were cultured on coverslips to ~70% confluency prior to transfection using Lipofectamine reagent (Invitrogen) using the manufacturers suggested protocol. Cells were then fixed in 3% paraformaldehyde and stored at −20°C.
Treatments of cell lysates from nSMase-2-expressing cells
For assessing the effect of thiol-reducing reagents, 3.5 μg of total protein was incubated for 30 min at room temperature in the presence of increasing concentrations (0–50 mM) of β-Me, DTT or GSH. The lysates were similarly incubated for 30 min at room temperature in the presence of increasing concentrations (0–20 mM) of oxidizing reagents including DM, H2O2 and GSSG. Stock solutions of GSH and GSSG were adjusted to pH 7.0 with 1 M NaOH prior to use.
Treatment of nSMase-2 with recombinant human thioredoxin was done in a coupled thioredoxin/thioredoxin reductase system. Twenty micrograms of lysates from nSMase-2 transformed Rosetta-gami2 (DE3) pLysS cells were incubated for 90 min at 37°C in 100 mM Tris/HCl (pH 7.4) containing 0.100 μmol NADPH (EMD Chemicals, Inc.), 2.0 μg of human recombinant thioredoxin (EMD Chemicals, Inc.) and 2.0 μg of yeast thioredoxin reductase (EMD Chemicals, Inc.), in a final volume of 20 μl.
Aliquots of the so treated cell lysates were subjected to nSMase activity assay and/or Western blotting analysis.
Identification of FLAG/His multimers of nSMase-2 in E. coli by co-purification
The pET-32a-nSMase-2-FLAG and pET-26b-nSMase-2-6xHis were co-transformed into Rosetta2 (DE3) pLysS cells and expression was induced with IPTG as described above. Following IPTG induction, 10 ml of cell culture was centrifuged for 10 min at 6000 g at 4°C and the pellet was resuspended in 250 μl of CelLytic B solution (Sigma). The cells were solubilized by vigorous vortexing for 15 min at room temperature and the cellular debris was pelleted by centrifugation at 16000 g for 15 min at 4°C. The supernatant was incubated with anti-FLAG M2 affinity gel (Sigma) or Ni-NTA resin (Thermo Fisher Scientific Inc.) and bound nSMase-2 protein was eluted following the manufacturer’s suggested protocols. The protein was resuspended in 2× sample buffer [125 mM Tris/HCl (pH 6.8), 4% SDS, 20% (v/v) glycerol and 0.004% Bromophenol Blue] either directly (in the case of anti FLAG M2 affinity gel) or after elution (for the Ni-NTA resin), and resolved by SDS/PAGE. Western blotting was used to identify interactions between FLAG and His-tagged nSMase-2.
SDS/PAGE and Western blotting
Protein concentration in cell lysates was measured and equal amounts of protein were resolved on a 10% polyacrylamide gel and then transferred to Immobilon-P membrane (Millipore). The wild-type and mutant nSMase-2 constructs were visualized using either a anti-FLAG (Sigma) or anti-6xHis antibody (Roche). In both cases anti-Mouse IgG alkaline phosphatase (Sigma) was used as the secondary antibody. Protein–antibody interactions were visualized on a Storm 860 fluorescence scanning instrument using enhanced chemifluorescent substrate (ECF, GE Healthcare) and were analysed using ImageQuant 5.0 software.
nSMase activity assay
nSMase activity was assayed as previously described with minor modifications [12]. Briefly, E. coli cells were pelleted and lysed with either three consecutive freeze–thaw cycles or with CelLytic B solution (Sigma). A total of 3.5 μg of protein was incubated for 60 min at 37°C in an assay mixture containing 100 mM Tris/HCl (pH 7.4), 20 μM 6-N-(7-nitro-benz-2-oxa-1,3-diazol-4-yl)amino-sphingomyelin (NBD-SM), 5 mM MgCl2, 5 mM NaF, 10 mM DTT and protease inhibitors. For pH dependence, the following buffers (100 mM) were used: acetate (pH 4.0–5.0), 2-(N-morpholino) ethanesulfonic acid (MES) buffer (pH 6.0), Tris/HCl buffer (pH 7.0–8.0) and glycine–NaOH (pH 9.0). To determine individual cation effects on activity, nSMase activity was measured in the presence of various concentrations of MgCl2, MnCl2, CaCl2, ZnCl2, CuCl2 or FeCl2. In each case, the assay was stopped by the addition of 500 μl of methanol followed by centrifugation for 15 min at 16000 g at 4°C. The supernatants were analysed by HPLC on a Nova Pac C18 reverse-phase column (Waters) using methanol:water:orthophosphoric acid (950:50:0.150, v/v) as the mobile phase with a flow rate of 2 ml/min.
FRET analysis
In our experiments, the acceptor photobleaching (APB) method was used. In the APB method, the acceptor fluorophore, in this case RFP, is selectively bleached in defined areas of interest within individual cells. The fluorescence of the donor, GFP, is monitored before (Donorpre) and after (Donorpost) the photobleaching step. The efficiency of FRET (FRETeff) is calculated using the following equation: FRETeff = (Donorpost – Donorpre)/Donorpost
For FRET measurements, 293-IL-1RI cells were co-transfected with the pAcGFP-C1-nSMase-2 and pmStrawberry-C1-nSMase-2 plasmids and examined by confocal microscopy utilizing a Leica TCS SP5 inverted microscope using a ×60 oil immersion objective and 1024 × 1024 resolution. Emission filters were 505–550 nm for 488 nm excitation (GFP detection, donor) and 594 nm for 574 nm excitation (RFP detection, acceptor). Circular regions of interest were bleached in the RFP channel using the 594 nm laser at maximum power (100% transmission). Cells that displayed comparable levels of GFP- and RFP-nSMase-2 were selected for FRET analysis. FRET efficiencies were calculated using software based on the equation given above.
RESULTS
Redox sensitivity of mammalian nSMase-2 activity
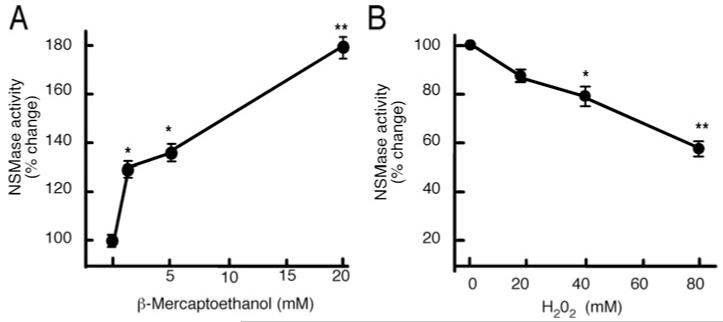
To test the sensitivity of nSMase-2 to changes in cellular redox state, 293-IL-1RI cells transiently overexpressing nSMase-2 [27] were lysed and the lysates were incubated briefly with different amounts of reducing and oxidizing agents, β-Me and H2O2 (Figure 1). Then the enzymatic activity was assayed with our standard assay buffer supplemented with the respective reducing/oxidizing agent. Increasing the concentration of β-Me led to stimulation of enzymatic activity, whereas H2O2 treatment had the opposite effect indicating that the reduced form of the enzyme has higher activity. To further study the redox sensitivity of nSMase-2, we employed a bacterial expression system, which is simpler and allows for faster results for site-directed mutagenesis studies.
Figure 1. Redox sensitivity of nSMase-2.
Lysates from 293-IL-1RI cells overexpressing nSMase-2 activity were treated with the indicated concentrations of β-Me (A) or H2O2 (B). nSMase-2 activity was determined in vitro using NBD-SM as a substrate, and the standard assay buffer supplemented with the respective concentrations of β-Me or H2O2.
Cloning and characterization of recombinant mouse nSMase-2 in E. coli
The production of functional, recombinant nSMase-2 as a FLAG and 6xHis (C-terminal) fusion protein was achieved with the help of the pET-32a (−) vector and the Rosetta2 (DE3) pLysS strain, which supplies tRNAs for rare codons not typically found in E. coli. The recombinant nSMase-2 has the anticipated properties as reported previously in mammalian cells [11,18] (Supplementary Figures S1A–S1D) and was also highly sensitive towards the presence of detergents (Supplementary Figures S1E and S1F). Deoxycholate and Triton X-100 inhibited almost 90% of the activity at concentrations of 3 mM and 0.1% respectively, whereas purification with Ni-NTA resin led to a complete loss of enzymatic activity (Supplementary Figures S1G and S1H). This loss of activity in the presence of detergents or upon purification mirrors what we saw with a mammalian expression system (results not shown) and probably reflects the need of a hydrophobic environment, which is typical for membrane-associated proteins like nSMase-2 and/or specific lipid cofactors [11,18]. All assays of enzymatic activity reported in the present study were done in bacterial lysates obtained under detergent-free conditions and are devoid of any endogenous activity.
Oligomerization of nSMase-2 and how it affects enzyme activity
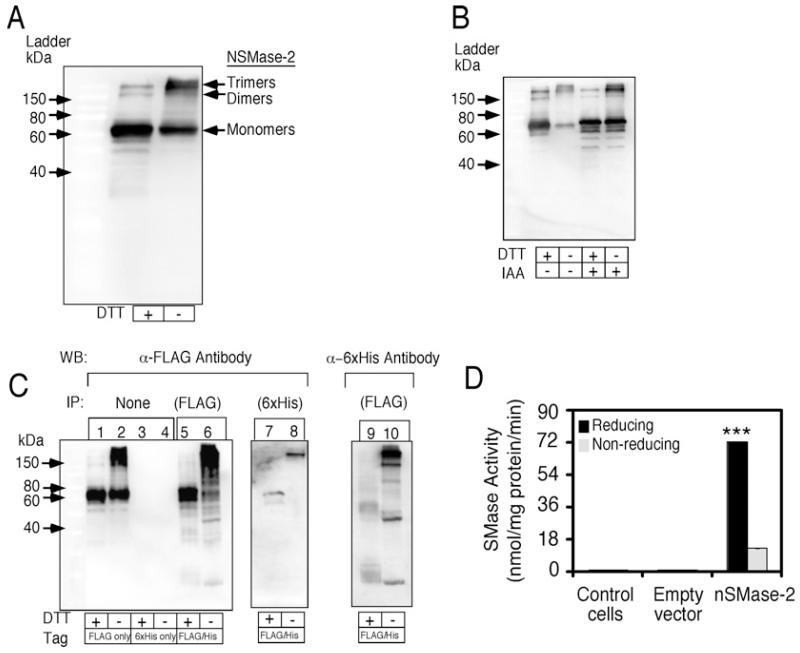
Western blot analysis revealed that nSMase-2 exists as both monomers and multimers in our bacterial expression system. Dimers and trimers of 150-kDa and 220-kDa, in addition to the 74-kDa monomers, are more evident when the lysates have not been pre-incubated with a reducing agent (10 mM DTT) prior to SDS/PAGE (Figure 2A). To ensure that these multimeric forms of nSMase-2 are not an artefact of the cell lysis process, IAA was used to irreversibly block the free cysteine thiol groups. Addition of 10 mM IAA prior to cell lysis however, did not affect the nSMase-2 multimers seen on Western blotting (Figure 2B). The slight shift higher of monomeric nSMase-2 seen on Western blotting is an indication of the successful carboxymethylation by IAA of free nSMase-2 cysteine thiol groups, proving that the treatment has the desired effect (Figure 2B).
Figure 2. Redox-sensitive nSMase-2 multimer formation.
(A) Identification of nSMase-2 multimers. Western blot analysis of lysates from nSMase-2-expressing Rosetta2 (DE3) pLysS with and without the reducing effects of DTT treatment. nSMase-2 was visualized using an antibody directed against the C-terminal FLAG. (B) Iodoacetamide (IAA) does not affect nSMase-2 oligomerization. Western blot of cells lysed in the presence or absence of 10 mM IAA and incubated in the presence and absence of 10 mM DTT for 30 min at room temperature. (C) Co-purification of nSMase-2 multimers with different tags. Rosetta2 (DE3) pLysS cells were transformed with pET-32a-nSMase-2-FLAG (lanes 1 and 2) or pET-32a-nSMase-2-6xHis (lanes 3 and 4), or co-transformed with both plasmids (lanes 7–10). Lysates from co-transformed cells were used for purification with anti-FLAG M2 affinity gel (lanes 5, 6, 9 and 10) or with Ni-NTA resin (lanes 9 and 10). nSMase-2 was visualized with anti-FLAG antibody (lanes 1–8) or with anti-6xHis antibody (lanes 7 and 8). DTT pre-treatment, where indicated, was done as described in Figure 1. (D) nSMase activity of cell lysates pre-incubated in 10 mM DTT for 30 min. Data are shown as an average ± S.D. (n = 3). ***, P < 0.001.
To further investigate nSMase-2 multimer formation, we developed an approach that allowed us to directly assess for the presence of protein–protein interactions without affecting the redox state of the protein. Two separate nSMase-2 constructs were used. One construct contained a C-terminal FLAG tag and was cloned into pET-32a (−) while the other was in pET-26b (+) and had a C-terminal 6xHis tag. This also allowed for double antibiotic selection using ampicillin and kanamycin. Both nSMase-2 constructs were co-transformed into Rosetta2 cells and interacting proteins were purified using either anti-FLAG M2 affinity gel or Ni-NTA resin. Western blot analyses illustrate the successful co-purification of His-tagged nSMase-2 with the anti-FLAG M2 affinity gel and vice versa, that of a FLAG-tagged nSMase-2 with Ni-NTA resin (Figure 2C, lanes 5–10). This provides evidence for the direct interaction of the His- and FLAG-tagged proteins.
In vitro activity assays revealed that pre-treatment of nSMase-2 cell lysates with a reducing agent resulted in a 5-fold increase in activity (Figure 2D). This suggests that the monomeric form of nSMase-2 is more active than the multimers. Taken together, these results indicate that nSMase-2 when expressed in bacterial cells exist as both monomers and multimers, and these different forms are redox sensitive and exhibit varying degrees of enzyme activity.
Dose-dependent regulation of nSMase-2 activity and oligomerization by reducing agents
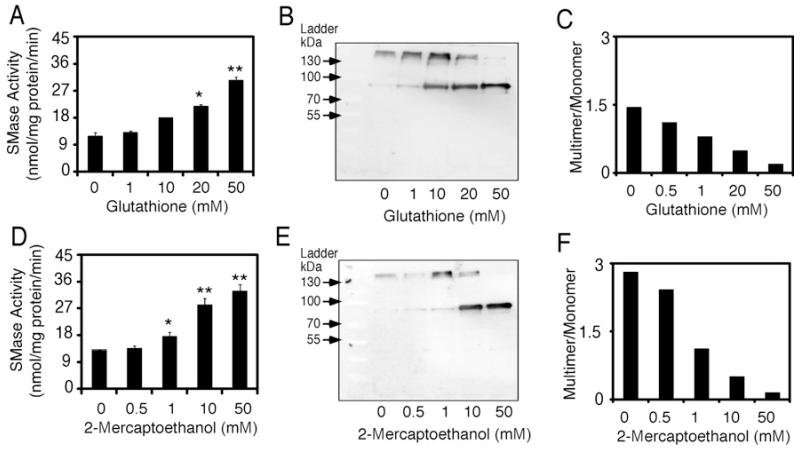
To further investigate the redox sensitivity of nSMase-2, the effects of various reducing agents on nSMase-2 activity and electrophoretic mobility were tested. In these experiments, the recombinant nSMase-2 cell lysates were pre-incubated for 30 min in the presence of increasing concentrations of GSH (Figures 3A–3C) or β-Me (Figures 3D–3F), as well as with DTT (Supplementary Figures S2A–S2C). Each sample was then subjected to activity assay and SDS/PAGE followed by Western blotting. All of the reducing agents tested caused a dose-dependent increase in NBD-SM hydrolysis that strongly correlated with the gradual conversion of nSMase-2 multimeric forms into monomers. It is of note that the maximum increase in activity was observed at reducing agent concentrations (50 mM for β-Me and GSH and 10 mM for DTT) that caused the almost complete dissociation of nSMase-2 multimers into monomers. Of the reducing agents used, DTT was the most efficient in the conversion of nSMase-2 into the monomeric form and increasing its activity. This could likely be explained by the fact that DTT provides two reducing equivalents, whereas β-Me and GSH provide only one.
Figure 3. nSMase-2 activity and electrophoretic mobility in the presence of reducing reagents.
Lysates from nSMase-2-transformed Rosetta2 (DE3) pLysS cells were pre-incubated in the presence of increasing concentrations (0–50 mM) of GSH or β-Me for 30 min at room temperature. (A, D) Effects on enzyme activity. nSMase activity assayed in 100 mM Tris, pH 7.4 supplemented with 5 mM MgCl2 and the indicated concentrations of reducing agents using NBD-SM as a substrate. (B, E) Effects on electrophoretic mobility. Western blot analysis of nSMase-2-expressing cell lysates pre-treated with reducing agents and separated on SDS/PAGE (10% gel). The nSMase-2 protein is visualized using an antibody against the FLAG tag. (C, F) Quantification of the band intensity in (B) and (E), multimer to monomer ratio is shown. Data are shown as an average ± S.D. (n = 3). *, P < 0.05, **, P < 0.01.
Oxidizing agents inhibit nSMase-2 activity in a dose-dependent manner that correlates with hyperaggregation of the nSMase-2 protein
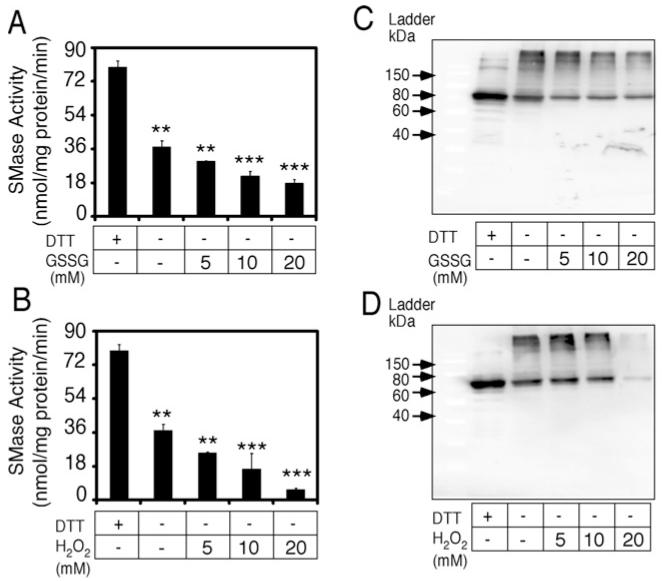
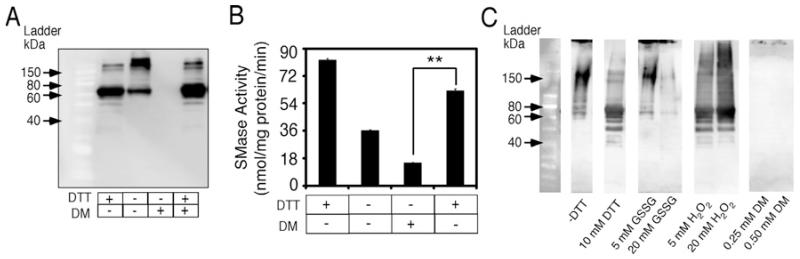
Next, we investigated how nSMase-2 was affected by oxidizing agents such as GSSG and hydrogen peroxide (Figure 4), as well as DM (Supplementary Figures S2D and S2E). Lysates from nSMase-2 expressing bacteria were pre-incubated in the presence of oxidizing reagent before being tested for activity and on a Western blot. These were compared with non-treated and DTT-treated lysates. Oxidation of nSMase-2 resulted in a dose-dependent decrease in its activity with 50% inhibition in activity being observed using 20 mM GSSG (Figure 4A). Almost complete inhibition was observed at 20 mM H2O2 and 0.5 mM DM respectively (Figure 4B and Supplementary Figure S2D). The loss in activity correlated well with a decline in the abundance of the nSMase-2 monomer caused by the oxidizing agents (Figures 4C and 4D and Supplementary Figure S2E). The abundance of the multimers also seemed to decline. However, this apparent protein loss was rescued by the addition of DTT to the oxidant-treated samples (Figure 5A). Notably, the abundance of nSMase-2 monomers in these oxidant/DTT-treated samples was identical with that in samples treated with DTT alone, indicating that the oxidant had no effect on protein stability (Figure 5A). Furthermore, DTT treatment following pre-treatment with 0.25 mM DM almost completely restored the nSMase-2 enzyme activity levels to those seen for DTT treatment only (Figure 5B). Separation of oxidant-treated samples on gradient gels revealed the presence of high molecular weight protein complexes in the oxidant-treated samples that, at a higher degree of oxidation, were incapable of entering the polyacrylamide matrix and remained at the start of the gel (Figure 5C). Collectively, these results seem to suggest that in the presence of oxidants nSMase-2 forms hyperaggregates that have no activity but that can be dissolved and the activity restored by the subsequent addition of a reducing agent. The sensitivity of these hyperaggregates to DTT would indicate that they are formed via cysteine thiol groups.
Figure 4. nSMase-2 activity and electrophoretic mobility in the presence of oxidizing agents.
Lysates from nSMase-2-transformed Rosetta2 (DE3) pLysS cells were pre-incubated in the presence of increasing concentrations (0–50 mM) of GSSG or H2O2 for 30 min at room temperature. Treatment with 10 mM DTT was used as a control. (A, B) Effects on enzyme activity. nSMase activity assayed in 100 mM Tris, pH 7.4 supplemented with 5 mM MgCl2 and the indicated concentrations of oxidizing agents using NBD-SM as a substrate. (C, D) Effects on electrophoretic mobility. Western blot analysis of nSMase-2-expressing cell lysates pre-treated with oxidizing agents and separated on SDS/PAGE (10% gel). The nSMase-2 protein is visualized using an antibody against the FLAG tag. Data are shown as an average ± S.D. (n = 3). **, P < 0.01, ***, P < 0.001.
Figure 5. Hyperaggregation of nSMase-2.
Western blot and activity analysis of wild-type nSMase-2 expressed in Rosetta2 (DE3) pLysS cells consequentially treated with DM and DTT. (A, B) Cell lysates were incubated in the presence of 0.25 mM DM for 30 min at RT (room temperature) and then in the absence or presence of 10 mM DTT. Western blot (A) and nSMase activity (B) analysis was done in parallel. (C) Western blot analysis of nSMase-2-expressing cell lysates pretreated with oxidizing reagents and separated on SDS/PAGE (4–20% gradient gel). Cell lysates were incubated in the presence of 5 and 20 mM GSSG (lanes 4–5) or H2O2 (lanes 6–7) and 0.25 mM DM (lanes 8–9) for 30 min at RT and were incubated in the absence (lane 2) or presence (lane 3) of 10 mM DTT.
Site-directed mutagenesis of nSMase-2 cysteine residues for determination of thiol groups involved in oligomerization
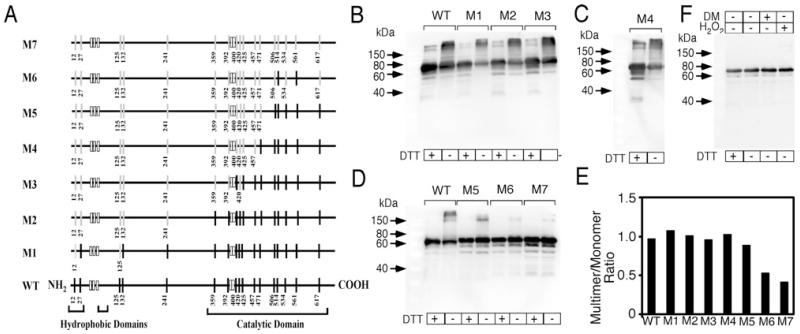
The results for the chemical modification of nSMase-2 multimers by reducing/oxidizing reagents suggest that cysteine residues might be involved in intermolecular disulfide bond formation leading to nSMase-2 oligomerization. Therefore, to ascertain the specific cysteine residues responsible, site-directed mutagenesis was employed. Cysteine (Cys) to serine (Ser) mutations were introduced sequentially in groups of two or three, generating the seven mutants shown in Figure 6(A) (the primers used are listed in Supplementary Table S1). Each individual mutant construct was then overexpressed in Rosetta2 cells. The expression levels of the mutants were similar to the wild-type nSMase-2 (Figures 6B–6D). The nSMase activity, however, progressively declined as more cysteines were replaced with serines (results not shown). The M1 and M2 mutants for example, exhibited 80% and 30% respectively of the activity seen for the wild-type. This loss of activity was not related to oligomerization of nSMase-2, as mutants M1, M2, M3 and M4 had multimer/monomer ratios similar to wild-type (Figures 6B–6C). Based on these results we can assume that cysteines in the non-catalytic region of nSMase-2 (i.e. amino acids 1–337), as well as Cys359, Cys392, Cys420, Cys425 and Cys457 in the beginning of the catalytic domain, do not participate in oligomerization. They do, however, play an essential role in the enzymatic activity of nSMase-2. The individual role of each of these cysteines in maintaining nSMase-2 activity is a subject of a separate study that is currently being prepared for publication.
Figure 6. Expression and characterization of cysteine-to-serine nSMase-2 mutants.
(A) Schematic presentation of nSMase-2 mutants. The 22 cysteines in the nSMase-2 molecule, as well as the hydrophobic and catalytic domains, are shown. Position number for each of the 17 cysteines under study is given in the diagram for the WT nSMase-2. The other five cysteines involved in palmitoylation are shown as short white rectangles. For the mutants (M1–M7), the non-mutated cysteines are shown as narrow black rectangles, and the residues mutated to serine are shown as narrow grey rectangles with the position number given. (B–D) Western blot analysis of WT and mutant (M1–M7) nSMase-2 expressed in Rosetta2 (DE3) pLysS cells. In all cases, the expressed nSMase-2 is visualized using an antibody against the C-terminal FLAG tag, and DTT treatments where indicated are for 30 min at room temperature prior to SDS/PAGE. (F) Lack of sensitivity to reducing and oxidizing agents for nSMase-2-M7 mutant. Cell lysates expressing nSMase-2-M7 were incubated with either 10 mM DTT, 0.25 mM DM or 10 mM H2O2 prior to Western blot analysis. (E) Quantification of the band intensity in (B), (C) and (D). Multimer to monomer ratio is shown.
The Western blot analysis presented in Figure 6 also shows that the nSMase-2 mutants M1 to M4 and to a lesser extent M5 retain the ability to form multimers under non-reducing conditions. Only in mutants M6 and M7, does the propensity to oligomerize seem to diminish, as judged by the equal abundance of nSMase-2 monomers under reducing and non-reducing conditions, as well as the disappearance of the high molecular weight forms in the non-DTT-treated samples (Figure 6D, lanes 6–9). Moreover, unlike the wild-type nSMase-2, the M7 mutant does not seem to form aggregates/high molecular weight complexes even when treated with oxidizing agents, such as DM and H2O2 (Figure 6F). Collectively, these results suggest that cysteines in the C-terminal region of the nSMase-2 catalytic domain, namely Cys506, Cys514, Cys534, Cys561 and Cys617 are involved in the protein oligomerization through the formation of intermolecular disulfide bridges.
Identifying specific cysteine residues important for nSMase-2 oligomerization and activity
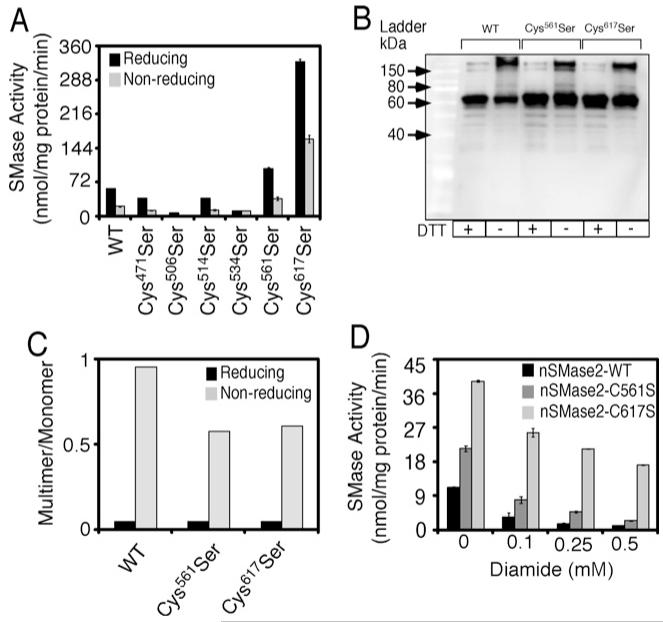
We next constructed six single-point cysteine-to-serine nSMase-2 mutants (the primers used are listed in Supplementary Table S2). The cysteine residues targeted were: Cys471, Cys506, Cys514, Cys534, Cys561 and Cys617. These are the cysteines previously mutated in the M5, M6 and M7 nSMase-2 constructs, but not in the M1, M2, M3 and M4. These single mutants were overexpressed in Rosetta2 cells, and their nSMase activity was compared with that of wild-type nSMase-2 (Figure 7A). Each of the Cys471, Cys506, Cys514 and Cys534 mutants had varying degrees of inhibited enzyme activity. The possible cause is a change in the optimal conformation of the catalytic site of the enzyme and, in the case of Cys506 and Cys534 mutants, also a change in metal binding [5]. In contrast, the Cys561Ser and Cys617Ser turned out to be gain-of-function mutations. These mutants exhibited a 2- and 7.5-fold higher activity respectively compared with the wild-type nSMase-2, both under reducing and non-reducing conditions (Figure 7C). The gain-of-function was unique for these two cysteine residues, because when individual mutations of each of the remaining 11 cysteines (from Cys12 to Cys457) in nSMase-2 were done, none resulted in increased activity (results not shown).
Figure 7. Expression and activity of single point cysteine-to-serine nSMase-2 mutants.
The single Cys471, Cys506, Cys514, Cys534, Cys561 and Cys617 mutants were expressed in Rosetta2 (DE3) pLysS cells. (A) nSMase-2 activity assay. Lysates were pre-treated in the presence or absence of DTT for 30 min at room temperature and SMase activity was measured using NBD-SM as a substrate in the standard incubation mixture. (B) Multimer formation by Cys516 and Cys617 mutants. Western blot analysis of the Cys561 and Cys617 mutants in lysates. The expressed nSMase-2 is visualized using an antibody against the C-terminal FLAG tag, and DTT treatments, where indicated, are for 30 min at room temperature. (C) Multimer-to-monomer ratios were calculated by quantification of the corresponding multimer and monomer bands from the Western blot from (B) for WT, Cys561Ser and Cys617Ser using ImageQuant 5.0 software. (D) Oxidant sensitivity of Cys561 and Cys617 mutants. Lysates were pre-treated with DM at the indicated concentrations for 30 min at room temperature and SMase activity was measured using NBD-SM as a substrate.
Diminished ability to form multimers is one possible explanation for the observed higher activity of the Cys561 and Cys617 mutants. SDS/PAGE analysis of reduced and non-reduced samples seems to support this possibility. As seen in Figures 9(B) and 9(C), in the absence of DTT, the mutants exhibited a significantly lower multimer to monomer ratio than the wild-type nSMase-2 with the Cys561 and Cys617 mutants exhibiting a ~50% decrease in their multimer to monomer ratio (Figure 7C). Moreover, in contrast with wild-type nSMase-2, for which the abundance of monomeric form was significantly elevated by DTT, the intensity of the monomeric bands of the mutants was not affected by this reducing agent (Figure 7B). It should be pointed out that the enzymatic activity of the Cys561 and Cys617 mutants retained some redox sensitivity. As seen in Figure 7(A), DTT stimulated the activity of both mutants, however, they were less sensitive to oxidizing agents than the wild-type nSMase-2 (Figure 7D). As discussed earlier and shown again in Figure 7D, the wild-type nSMase-2 loses most of its activity due to aggregation when treated with DM at concentrations as low as 0.25 mM. In contrast, the Cys617 and Cys561 mutants retained approximately 50% and 20% of their activity respectively at these concentrations of the oxidant.
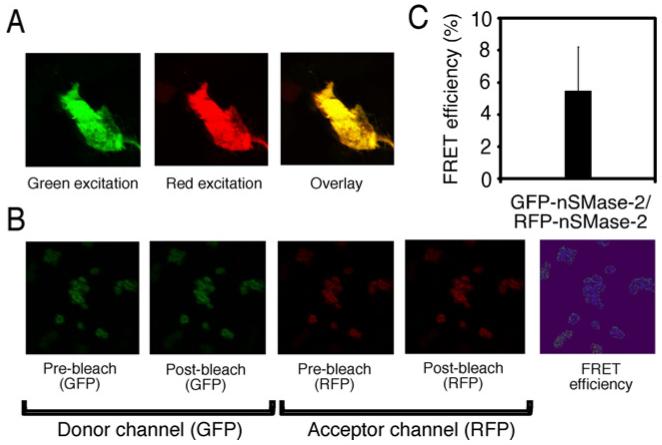
Figure 9. FRET analysis of nSMase-2.
nSMase-2 constructs with either GFP or RFP N-terminal tags were co-transfected into 293-IL-1R cells. Cells were then grown to confluency (~72 h) and fixed on coverslips prior to FRET analysis. (A) Co-expression of GFP- and RFP-nSMase-2. (B) The pmStrawberry-C1-nSMase-2 construct was co-transfected with pAcGFP1-C1-nSMase-2. Fluorescence was monitored for the donor (GFP) and acceptor (RFP) before and after subsequent photobleaching of the acceptor (RFP). (C) The FRET efficiency was calculated as the ratio between the increase in the GFP (donor) fluorescence, after photobleaching of the RFP-nSMase-2 acceptor and the post-bleaching donor fluorescence.
Thioredoxin-mediated reduction of nSMase-2 multimers to monomers
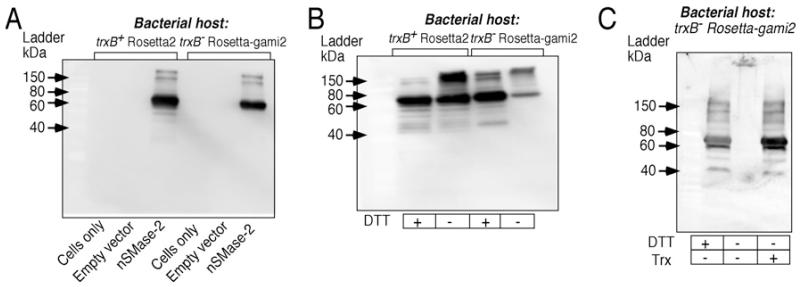
As stated earlier, the thioredoxin and glutaredoxin systems are key regulators of the intracellular redox state [28]. We hypothesized that thioredoxin may play a role in determining the balance between nSMase-2 monomers and multimers. To test this, we employed a new bacterial strain, Rosetta-gami2 (DE3) pLysS. This Rosetta2-derived strain lacks the bacterial gene thioredoxin reductase (trxB) and consequently, the capability of reducing disulfide links in proteins via thioredoxin action. Transformation of the Rosetta-gami2 cells with the nSMase-2 wild-type construct resulted in a nSMase-2 protein profile that was similar to that of the Rosetta2 strain (with a major 74-kDa band), but only when the samples are treated with a reducing agent (DTT), prior to SDS/PAGE (Figure 8A). Under non-reducing conditions the nSMase-2 produced in trxB− Rosetta-gami2 cells apparently exhibits a significantly higher propensity to form high molecular weight aggregates that cannot be separated by SDS/PAGE. This is illustrated by the fact that under non-reducing conditions, the nSMase-2 produced in trxB− Rosetta-gami2 cells appears to have less abundance of both monomeric and multimeric forms compared with the nSMase-2 in Rosetta2, but these differences mostly disappear following DTT treatment (Figure 8B). These results closely resemble the effects oxidizing agents had on the Rosetta2-produced nSMase-2 (see Figures 5 and 6).
Figure 8. nSMase-2 oligomerization is modulated by thioredoxin.
(A) Western blot of nSMase-2 expressed in Rosetta2 and Rosetta-gami2 (DE3) pLysS cells. All lysates are treated with 10 mM DTT prior to SDS/PAGE. (B) Effect of host trxB status on nSMase-2 multimer formation. Western blot analysis of nSMase-2-expressing Rosetta2 and Rosetta-gami2 (DE3) pLysS cells with and without reducing agent pre-treatment. (C) Ex vivo conversion of nSMase-2 hyperaggregates to monomers by human recombinant thioredoxin. Lysates from the nSMase-2-expressing trxB− Rosetta-gami2 (DE3) pLyS cells were incubated in either the presence of 1 μg recombinant thioredoxin (Trx), 100 nmol NADPH and 1 μg of recombinant thioredoxin reductase (TrxB), or with 10 mM DTT for 30 min at 37°C and were then separated by SDS/PAGE (4–20% gradient gel). The control (lane 2) was treated only with NADPH and recombinant thioredoxin reductase. For all Western blots, the nSMase-2 protein was detected using a primary antibody directed at the C-terminal FLAG.
To ascertain whether the trxB deletion indeed was the cause for the altered physical properties of nSMase-2 in trxB− Rosetta-gami2 cells, we treated the lysates in vitro with recombinant human thioredoxin. This was done using a coupled thioredoxin/thioredoxin reductase/NADPH system that allows thioredoxin to be maintained in a reduced state, but lacks any additional reducing equivalents. The negative control was treated identically, but without the addition of recombinant thioredoxin. As seen in Figure 8(C), the trxB− Rosetta-gami2-derived nSMase-2 that was treated with thioredoxin, had an SDS/PAGE profile that was similar to that of a sample treated with DTT instead. These results would suggest that nSMase-2 is a substrate for the human thioredoxin, and that the latter is sufficient to convert the nSMase-2 oligomers into monomers.
FRET of nSMase-2 in mammalian 293-IL-IR1 cells
FRET analysis was used to test whether the nSMase-2 protein can oligomerize in vivo in mammalian cells. For that purpose, two nSMase-2 constructs, one with GFP, and the other with RFP fused to the N-terminal end of the enzyme were coexpressed in 293-IL-IR1 cells. Initially, the subcellular localization of the two nSMase-2 proteins was examined by confocal microscopy (Figure 9A). Subsequent FRET analysis exhibited significant energy transfer, reaching an efficiency of 5.5±2.8% in cells co-transfected with GFP-nSMase-2 and RFP-nSMase-2 (Figures 9B–9C). These results suggest that the nSMase-2 molecules are in close proximity (~10 Å or 1 nm; 1 Å = 0.1 nm) for direct interaction(s) to occur. Such interactions include disulfide bond(s) formation.
DISCUSSION
Regulation of the innate activity of nSMase-2 is poorly understood. Our experiments provide evidence that the redox state of the thiol groups in nSMase-2 has a substantial impact on activity. This effect correlates, at least in part, with changes in the oligomeric state of the protein. Site-directed mutagenesis identified two cysteine residues in the catalytic domain of nSMase-2, Cys561 and Cys617, that were shown to participate in multimer formation, which in turn was linked to low activity. In contrast, high nSMase-2 activity was observed in the presence of thiol-reducing agents where nSMase-2 exists almost exclusively as monomers. Importantly, the transition between multimeric and monomeric nSMase-2 appears to be regulated by thioredoxin.
Sphingomyelinases are a family of five closely related enzymes that generate ceramide from sphingomyelin. Notably, two other members of this family, acid sphingomyelinase and nSMase-1 have been shown to be redox-sensitive [29-32]. Cysteine thiol groups within nSMase-1 are involved in the formation of intramolecular disulfide bonds that result in reversible inactivation of the enzyme [29-31]. Also, the C-terminal cysteine, Cys629, in acid sphingomyelinase has been shown to participate in the formation of intermolecular disulfide bonds, resulting in protein dimerization that, in contrast with nSMase-2, leads to increased activity [32]. This same cysteine residue seemingly could compete for the active Zn2+ required for catalysis, bringing about an inhibition of the enzymatic activity of acid sphingomyelinase.
Several lines of evidence support the notion that nSMase-2 may form oligomers, mostly dimers and trimers. Dimers (~150 kDa) and trimers (~220 kDa) are under seen on SDS/PAGE non-reducing conditions. More importantly, the abundance of these high-molecular weight complexes is not diminished by the inclusion of IAA during cell lysis. If these apparent multimers were an artefact of the lysis procedure, they would be expected to disappear when IAA was added prior to cell lysis. The existence of multimeric forms was further confirmed by an alternative approach showing the co-purification of FLAG- and His-tagged nSMase-2 from cells co-transformed with both constructs. Also, site-directed mutagenesis identified two mutants, Cys561Ser and Cys617Ser that exhibit a reduced ability to form multimers and hence confirm the specificity of the observed multimer formation. Finally, efficient FRET is observed in mammalian cells overexpressing GFP- and RFP-tagged nSMase-2.
The enzymatic activity of nSMase-2 apparently correlates with the relative abundance of monomers. This is supported by the observed need for reducing agent, for optimal activity in vitro, as well as by the inhibition of nSMase-2 enzymatic activity seen in the presence of oxidizing agents. More importantly, however, the two single mutants that had a decreased propensity for oligomerization were gain-of-function mutants, suggesting that, at least in part, the redox sensitivity of nSMase-2 could be attributed to changes in its oligomeric state. Indeed, these two single mutants also exhibited diminished sensitivity to oxidizing agents. As seen in Figure 7(D), treatment with DM substantially inhibits the innate activity of wild-type nSMase-2, possibly due to hyperaggregation. The Cys617 mutant however, is less sensitive to DM and even at the highest DM concentrations remains significantly more active than the wild-type nSMase-2. These results lead us to the conclusion that Cys617 has a key role in nSMase-2 multimerization and oxidant-induced hyperaggregation, as well as the resulting loss of enzymatic activity. At the same time, our studies also indicate that the redox state of other cysteines (mainly in the non-catalytic part of nSMase-2) is also important for enzyme activity. Although the Cys617 mutant retains some redox sensitivity and is stimulated by reducing agents (Figure 7A), mutation of certain cysteines in the non-catalytic part of the protein leads to nearly a complete loss of enzyme activity (results not shown). One possible reason for this is that these non-catalytic residues are subject to direct post-translational modification(s) by agents such as GSH/GSSG, that have been shown above to inhibit nSMase-2 activity.
The ability of human thioredoxin in vitro to convert nSMase-2 multimers into monomers suggests that nSMase-2 might be a thioredoxin substrate. Thioredoxin is a small 12-kDa cytoplasmic protein that regulates the redox state of protein thiol groups [6,28,33,34]. Under oxidative stress conditions, thioredoxin acts as an antioxidant by maintaining proteins cysteines in a reduced state and thus preventing irreversible protein oxidation. Upregulation of thioredoxin expression and activity is a hallmark of the cellular response to oxidative stress during various physiological conditions, and is also seen during chemotherapy [35] and radiation [36,37]. Thioredoxin has also been shown to be directly involved in the redox regulation of important proteins like glyceraldehyde-3-phosphate dehydrogenase (GAPDH), p53 and Ask-1 [34]. Notably, nSMase-2 is also activated under oxidative stress conditions, which has recently been shown to occur through changes in the phosphorylation of specific serine residues under the regulation of protein phosphatase 2B [22,26]. Our data suggest that thioredoxin-mediated reduction in intermolecular disulfide bonds (possibly at Cys617) and the resulting conversion of inactive nSMase-2 multimers into active monomers might be another plausible mechanism for substantial induction of nSMase-2 activity during oxidative stress. One cannot exclude the possibility that the nSMase-2 monomer/multimer balance is in a highly dynamic equilibrium that is tightly linked to the redox/oxidative state of the cells.
Supplementary Material
Acknowledgments
FUNDING
The National Institute of Health [grant numbers RO1AG026711, 2RO1AG019223 (to M.N-K.)] and the American Heart Association [grant number 10POST4300013 post-doctoral fellowship (to P.P.D. II)] supported this work.
Abbreviations
- DM
diamide
- DTT
dithiothreitol
- FRET
fluorescence resonance energy transfer
- H2O2
hydrogen peroxide
- IAA
iodoacetamide
- 293-IL-1RI
HEK 293 cells stably overexpressing the IL-1β receptor
- IPTG
isopropyl-β-d-thiogalactopyranoside
- β-Me
2-mercaptoethanol
- MES
2-(N-morpholino) ethanesulfonic acid
- NBD-SM
6-N-(7-nitro-benz-2-oxa-1,3-diazol-4-yl)amino-sphingomyelin
- nSMase-2
neutral sphingomyelinase-2
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- WT
wild-type
Footnotes
AUTHOR CONTRIBUTION
P. Patrick Dotson II planned and performed all experiments with the exception of those shown in Figure 1. He also made all plasmids for bacterial expression and FRET analysis. Alexander Karakashian cloned and characterized the original nSMase-2 cDNA constructs in the mammalian system, and performed the experiments in Figure 1(A). Mariana Nikolova-Karakashian helped in designing and planning the experiments and analysing/interpreting the results. All three co-authors participated in writing the manuscript.
REFERENCES
- 1.Bindoli A, Fukuto JM, Forman HJ. Thiol chemistry in peroxidase catalysis and redox signaling. Antioxid. Redox Signal. 2008;10:1549–1564. doi: 10.1089/ars.2008.2063. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carballal S, Alvarez B, Turell L, Botti H, Freeman BA, Radi R. Sulfenic acid in human serum albumin. Amino Acids. 2007;32:543–551. doi: 10.1007/s00726-006-0430-y. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 3.Paulsen CE, Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem. Biol. 2010;5:47–62. doi: 10.1021/cb900258z. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klomsiri C, Karplus PA, Poole LB. Cysteine-based redox switches in enzymes. Antioxid Redox Signal. 2011;14:1065–1077. doi: 10.1089/ars.2010.3376. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pace NJ, Weerapana E. Diverse functional roles of reactive cysteines. ACS Chem. Biol. 2013;8:283–296. doi: 10.1021/cb3005269. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 6.Antelmann H, Helmann JD. Thiol-based redox switches and gene regulation. Antioxid. Redox Signal. 2011;14:1049–1063. doi: 10.1089/ars.2010.3400. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nikolova-Karakashian MN, Rozenova KA. Ceramide in stress response. Adv. Exp. Med. Biol. 2010;688:86–108. doi: 10.1007/978-1-4419-6741-1_6. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Alwani M, Wu BX, Obeid LM, Hannun YA. Bioactive sphingolipids in the modulation of the inflammatory response. Pharmacol. Ther. 2006;112:171–183. doi: 10.1016/j.pharmthera.2006.04.004. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 9.Nikolova-Karakashian MN, Reid MB. Sphingolipid metabolism, oxidant signaling, and contractile function of skeletal muscle. Antioxid. Redox Signal. 2011;15:2501–2517. doi: 10.1089/ars.2011.3940. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu BX, Clarke CJ, Hannun YA. Mammalian neutral sphingomyelinases: regulation and roles in cell signaling responses. Neuromol. Med. 2010;12:320–330. doi: 10.1007/s12017-010-8120-z. CrossRef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marchesini N, Luberto C, Hannun YA. Biochemical properties of mammalian neutral sphingomyelinase 2 and its role in sphingolipid metabolism. J. Biol. Chem. 2003;278:13775–13783. doi: 10.1074/jbc.M212262200. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 12.Karakashian AA, Giltiay NV, Smith GM, Nikolova-Karakashian MN. Expression of neutral sphingomyelinase-2 (NSMase-2) in primary rat hepatocytes modulates IL-beta-induced JNK activation. FASEB J. 2004;18:968–970. doi: 10.1096/fj.03-0875fje. PubMed. [DOI] [PubMed] [Google Scholar]
- 13.Rutkute K, Karakashian AA, Giltiay NV, Dobierzewska A, Nikolova-Karakashian MN. Aging in rat causes hepatic hyperresposiveness to interleukin-1beta which is mediated by neutral sphingomyelinase-2. Hepatology. 2007;46:1166–1176. doi: 10.1002/hep.21777. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 14.Levy M, Castillo SS, Goldkorn T. nSMase2 activation and trafficking are modulated by oxidative stress to induce apoptosis. Biochem. Biophys. Res. Commun. 2006;344:900–905. doi: 10.1016/j.bbrc.2006.04.013. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rutkute K, Asmis RH, Nikolova-Karakashian MN. Regulation of neutral sphingomyelinase-2 by GSH: a new insight to the role of oxidative stress in aging-associated inflammation. J. Lipid. Res. 2007;48:2443–2452. doi: 10.1194/jlr.M700227-JLR200. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang DI, Yeh CH, Chen S, Xu J, Hsu CY. Neutral sphingomyelinase activation in endothelial and glial cell death induced by amyloid beta-peptide. Neurobiol. Dis. 2004;17:99–107. doi: 10.1016/j.nbd.2004.06.001. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 17.Lee JT, Xu J, Lee JM, Ku G, Han X, Yang DI, Chen S, Hsu CY. Amyloid-beta peptide induces oligodendrocyte death by activating the neutral sphingomyelinase-ceramide pathway. J. Cell Biol. 2004;164:123–131. doi: 10.1083/jcb.200307017. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann K, Tomiuk S, Wolff G, Stoffel W. Cloning and characterization of the mammalian brain-specific, Mg2+-dependent neutral sphingomyelinase. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5895–5900. doi: 10.1073/pnas.97.11.5895. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu BX, Clarke CJ, Matmati N, Montefusco D, Bartke N, Hannun YA. Identification of novel anionic phospholipid binding domains in neutral sphingomyelinase 2 with selective binding preference. J. Biol. Chem. 2011;286:22362–22371. doi: 10.1074/jbc.M110.156471. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tani M, Hannun YA. Neutral sphingomyelinase 2 is palmitoylated on multiple cysteine residues – role of palmitoylation in subcellular localization. J. Biol. Chem. 2007;282:10047–10056. doi: 10.1074/jbc.M611249200. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 21.Liu B, Andrieu-Abadie N, Levade T, Zhang P, Obeid LM, Hannun YA. Glutathione regulation of neutral sphingomyelinase in tumor necrosis factor-alpha-induced cell death. J. Biol. Chem. 1998;273:11313–11320. doi: 10.1074/jbc.273.18.11313. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 22.Filosto S, Fry W, Knowlton AA, Goldkorn T. Neutral sphingomyelinase 2 (nSMase2) is a phosphoprotein regulated by calcineurin (PP2B) J. Biol. Chem. 2010;285:10213–10222. doi: 10.1074/jbc.M109.069963. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavrentiadou SN, Chan C, Kawcak T, Ravid T, Tsaba A, van der Vliet A, Rasooly R, Goldkorn T. Ceramide-mediated apoptosis in lung epithelial cells is regulated by glutathione. Am. J. Respir. Cell Mol. Biol. 2001;25:676–684. doi: 10.1165/ajrcmb.25.6.4321. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu B, Hannun YA. Inhibition of the neutral magnesium-dependent sphingomyelinase by glutathione. J. Biol. Chem. 1997;272:16281–16287. doi: 10.1074/jbc.272.26.16281. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 25.Chaube R, Kallakunta VM, Espey MG, McLarty R, Faccenda A, Ananvoranich S, Mutus B. Endoplasmic reticulum stress-mediated inhibition of NSMase2 elevates plasma membrane cholesterol and attenuates NO production in endothelial cells. Biochim. Biophys. Acta. 2012;1821:313–323. doi: 10.1016/j.bbalip.2011.10.015. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 26.Filosto S, Ashfaq M, Chung S, Fry W, Goldkorn T. Neutral sphingomyelinase 2 activity and protein stability are modulated by phosphorylation of five conserved serines. J. Biol. Chem. 2012;287:514–522. doi: 10.1074/jbc.M111.315481. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobierzewska A, Shi L, Karakashian AA, Nikolova-Karakashian MN. Interleukin 1beta regulation of FoxO1 protein content and localization: evidence for a novel ceramide-dependent mechanism. J. Biol. Chem. 2012;287:44749–44760. doi: 10.1074/jbc.M112.378836. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanschmann EM, Godoy JR, Berndt C, Hudemann C, Lillig CH. Thioredoxins, glutaredoxins, and peroxiredoxins-molecular mechanisms and health significance: from cofactors to antioxidants to redox signaling. Antioxid. Redox Signal. 2013;19:1539–1605. doi: 10.1089/ars.2012.4599. CrossRef PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fensome AC, Rodrigues-Lima F, Josephs M, Paterson HF, Katan M. A neutral magnesium-dependent sphingomyelinase isoform associated with intracellular membranes and reversibly inhibited by reactive oxygen species. J. Biol. Chem. 2000;275:1128–1136. doi: 10.1074/jbc.275.2.1128. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 30.Rodrigues-Lima F, Fensome AC, Josephs M, Evans J, Veldman RJ, Katan M. Structural requirements for catalysis and membrane targeting of mammalian enzymes with neutral sphingomyelinase and lysophospholipid phospholipase C activities. Analysis by chemical modification and site-directed mutagenesis. J. Biol. Chem. 2000;275:28316–28325. doi: 10.1074/jbc.M003080200. PubMed. [DOI] [PubMed] [Google Scholar]
- 31.Josephs M, Katan M, Rodrigues-Lima F. Irreversible inactivation of magnesium-dependent neutral sphingomyelinase 1 (NSM1) by peroxynitrite, a nitric oxide-derived oxidant. FEBS Lett. 2002;531:329–334. doi: 10.1016/s0014-5793(02)03551-2. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 32.Qiu H, Edmunds T, Baker-Malcolm J, Karey KP, Estes S, Schwarz C, Hughes H, Van Patten SM. Activation of human acid sphingomyelinase through modification or deletion of C-terminal cysteine. J. Biol. Chem. 2003;278:32744–32752. doi: 10.1074/jbc.M303022200. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 33.Lu J, Holmgren A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2014;66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 34.Mahmood DF, Abderrazak A, El Hadri K, Simmet T, Rouis M. The thioredoxin system as a therapeutic target in human health and disease. Antioxid. Redox Signal. 2013;19:1266–1303. doi: 10.1089/ars.2012.4757. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 35.Kim SJ, Miyoshi Y, Taguchi T, Tamaki Y, Nakamura H, Yodoi J, Kato K, Noguchi S. High thioredoxin expression is associated with resistance to docetaxel in primary breast cancer. Clin. Cancer Res. 2005;11:8425–8430. doi: 10.1158/1078-0432.CCR-05-0449. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
- 36.Wei SJ, Botero A, Hirota K, Bradbury CM, Markovina S, Laszlo A, Spitz DR, Goswami PC, Yodoi J, Gius D. Thioredoxin nuclear translocation and interaction with redox factor-1 activates the activator protein-1 transcription factor in response to ionizing radiation. Cancer Res. 2000;60:6688–6695. PubMed. [PubMed] [Google Scholar]
- 37.Didier C, Kerblat I, Drouet C, Favier A, Beani JC, Richard MJ. Induction of thioredoxin by ultraviolet-A radiation prevents oxidative-mediated cell death in human skin fibroblasts. Free Radic. Biol. Med. 2001;31:585–598. doi: 10.1016/s0891-5849(01)00617-7. CrossRef PubMed. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.