Abstract
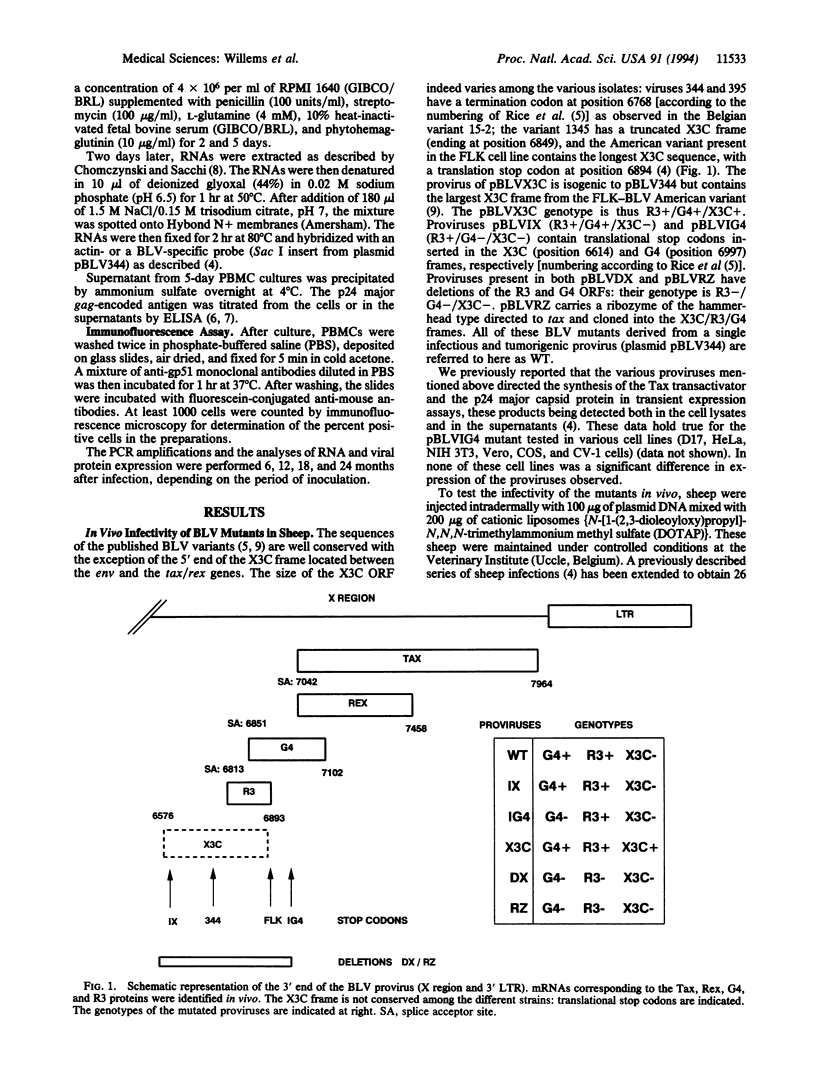
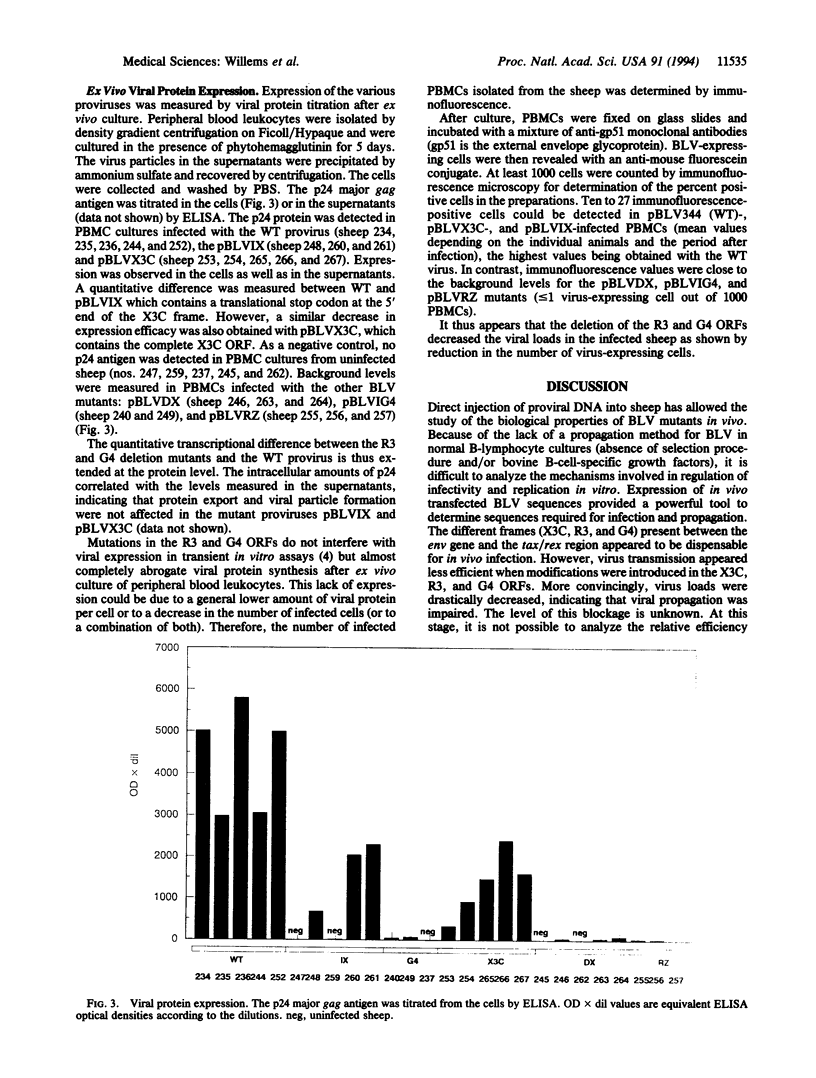
Complex oncoviruses contain, in addition to the classical retroviral genes (gag, pol, and env), a region (X) located between the envelope sequences and the 3' long terminal repeat. The X region contains two genes, tax and rex, whose protein products are involved in transcriptional and posttranscriptional regulation of viral expression. In addition to these activators, the bovine leukemia virus (BLV) and the human T-cell leukemia virus (HTLV) contain alternative open reading frames (R3 and G4 for BLV; p30, p13, and p12 for HTLV). As a virus/animal model for HTLV-induced leukemogenesis, BLV provirus can be injected intradermally into sheep, where it induced B-lymphocyte transformation. Deletion of the R3 and G4 sequences from an infectious and tumorigenic BLV provirus greatly impaired the in vivo propagation of the viruses as demonstrated by DNA polymerase chain reaction, RNA blots, structural-protein ELISA, and immunofluorescence analysis. Our results show that the alternative open reading frames are required for maintaining high virus loads during the course of persistent infection in vivo. Thus, R3 and G4 are candidates for antiviral drug development. Furthermore, viruses with a deletion in these sequences should be tested as live attenuated vaccines.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexandersen S., Carpenter S., Christensen J., Storgaard T., Viuff B., Wannemuehler Y., Belousov J., Roth J. A. Identification of alternatively spliced mRNAs encoding potential new regulatory proteins in cattle infected with bovine leukemia virus. J Virol. 1993 Jan;67(1):39–52. doi: 10.1128/jvi.67.1.39-52.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Franchini G., Mulloy J. C., Koralnik I. J., Lo Monico A., Sparkowski J. J., Andresson T., Goldstein D. J., Schlegel R. The human T-cell leukemia/lymphotropic virus type I p12I protein cooperates with the E5 oncoprotein of bovine papillomavirus in cell transformation and binds the 16-kilodalton subunit of the vacuolar H+ ATPase. J Virol. 1993 Dec;67(12):7701–7704. doi: 10.1128/jvi.67.12.7701-7704.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portetelle D., Limbach K., Burny A., Mammerickx M., Desmettre P., Riviere M., Zavada J., Paoletti E. Recombinant vaccinia virus expression of the bovine leukaemia virus envelope gene and protection of immunized sheep against infection. Vaccine. 1991 Mar;9(3):194–200. doi: 10.1016/0264-410x(91)90153-w. [DOI] [PubMed] [Google Scholar]
- Portetelle D., Mammerickx M., Burny A. Use of two monoclonal antibodies in an ELISA test for the detection of antibodies to bovine leukaemia virus envelope protein gp51. J Virol Methods. 1989 Feb;23(2):211–222. doi: 10.1016/0166-0934(89)90135-3. [DOI] [PubMed] [Google Scholar]
- Sagata N., Yasunaga T., Tsuzuku-Kawamura J., Ohishi K., Ogawa Y., Ikawa Y. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc Natl Acad Sci U S A. 1985 Feb;82(3):677–681. doi: 10.1073/pnas.82.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems L., Kettmann R., Dequiedt F., Portetelle D., Vonèche V., Cornil I., Kerkhofs P., Burny A., Mammerickx M. In vivo infection of sheep by bovine leukemia virus mutants. J Virol. 1993 Jul;67(7):4078–4085. doi: 10.1128/jvi.67.7.4078-4085.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems L., Thienpont E., Kerkhofs P., Burny A., Mammerickx M., Kettmann R. Bovine leukemia virus, an animal model for the study of intrastrain variability. J Virol. 1993 Feb;67(2):1086–1089. doi: 10.1128/jvi.67.2.1086-1089.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]





