Abstract
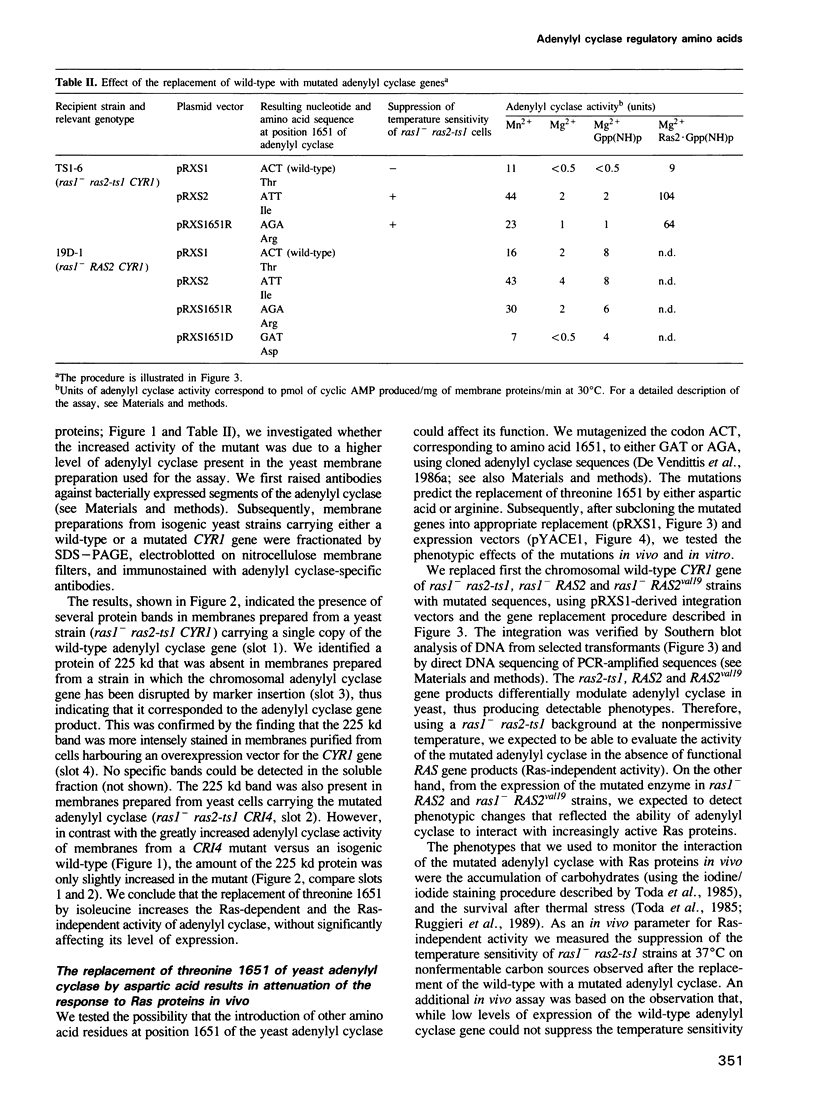
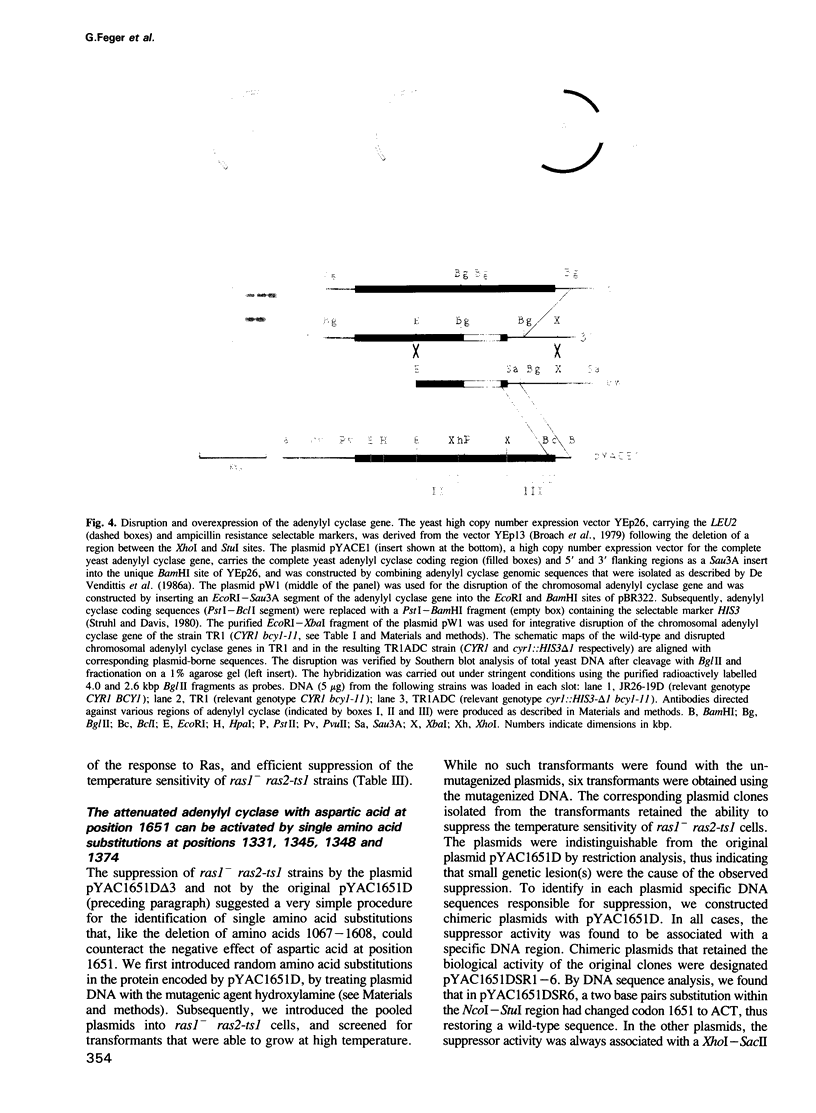
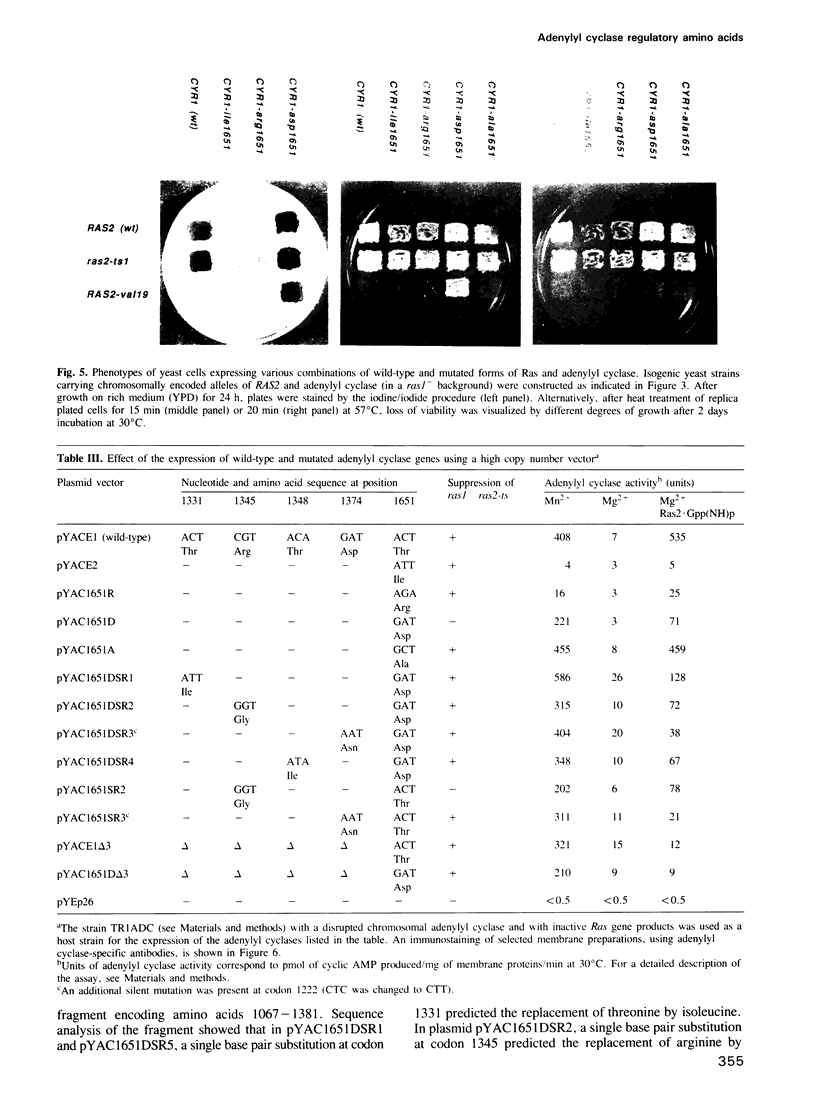
We have attempted to identify amino acid residues of the yeast adenylyl cyclase that are involved in the regulation of its activity, by isolating adenylyl cyclase-linked spontaneous mutations capable of suppressing the temperature-sensitive phenotype of ras1- ras2-ts1 strains. We previously identified a mutated adenylyl cyclase in which a single point mutation, called CR14, led to the replacement of threonine 1651 with isoleucine. We have now investigated the biological effects of CR14, and of other mutations that cause the replacement of threonine 1651 by distinct amino acids. We have observed that the response of adenylyl cyclase to Ras can be either enhanced or attenuated, without significant effects on the steady-state level of the former enzyme in vivo, depending on the amino acid side chain at position 1651. Therefore, this residue identifies a regulatory region on the adenylyl cyclase molecule. We have also taken advantage of the attenuation of adenylyl cyclase function caused by the replacement of threonine 1651 with aspartic acid to isolate intragenic suppressor mutations. We have identified several point mutations, leading to single amino acid substitutions, individually capable of reactivating the attenuated adenylyl cyclase. The corresponding amino acid changes are located within a relatively small region, including residues 1331, 1345, 1348 and 1374. This region could be physiologically involved in the negative control of the carboxy-terminal catalytic domain.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Broach J. R., Deschenes R. J. The function of ras genes in Saccharomyces cerevisiae. Adv Cancer Res. 1990;54:79–139. doi: 10.1016/s0065-230x(08)60809-x. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Broek D., Samiy N., Fasano O., Fujiyama A., Tamanoi F., Northup J., Wigler M. Differential activation of yeast adenylate cyclase by wild-type and mutant RAS proteins. Cell. 1985 Jul;41(3):763–769. doi: 10.1016/s0092-8674(85)80057-x. [DOI] [PubMed] [Google Scholar]
- Casperson G. F., Walker N., Brasier A. R., Bourne H. R. A guanine nucleotide-sensitive adenylate cyclase in the yeast Saccharomyces cerevisiae. J Biol Chem. 1983 Jul 10;258(13):7911–7914. [PubMed] [Google Scholar]
- Colicelli J., Field J., Ballester R., Chester N., Young D., Wigler M. Mutational mapping of RAS-responsive domains of the Saccharomyces cerevisiae adenylyl cyclase. Mol Cell Biol. 1990 Jun;10(6):2539–2543. doi: 10.1128/mcb.10.6.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vendittis E., Vitelli A., Zahn R., Fasano O. Suppression of defective RAS1 and RAS2 functions in yeast by an adenylate cyclase activated by a single amino acid change. EMBO J. 1986 Dec 20;5(13):3657–3663. doi: 10.1002/j.1460-2075.1986.tb04696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vendittis E., Zahn R., Fasano O. Regeneration of the GTP-bound from the GDP-bound form of human and yeast ras proteins by nucleotide exchange. Stimulatory effect of organic and inorganic polyphosphates. Eur J Biochem. 1986 Dec 1;161(2):473–478. doi: 10.1111/j.1432-1033.1986.tb10468.x. [DOI] [PubMed] [Google Scholar]
- Engelberg D., Poradosu E., Simchen G., Levitzki A. Adenylyl cyclase activity of the fission yeast Schizosaccharomyces pombe is not regulated by guanyl nucleotides. FEBS Lett. 1990 Feb 26;261(2):413–418. doi: 10.1016/0014-5793(90)80605-i. [DOI] [PubMed] [Google Scholar]
- Fasano O., Crechet J. B., De Vendittis E., Zahn R., Feger G., Vitelli A., Parmeggiani A. Yeast mutants temperature-sensitive for growth after random mutagenesis of the chromosomal RAS2 gene and deletion of the RAS1 gene. EMBO J. 1988 Nov;7(11):3375–3383. doi: 10.1002/j.1460-2075.1988.tb03210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor-Chaiken M., Deschenes R. J., Broach J. R. SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell. 1990 Apr 20;61(2):329–340. doi: 10.1016/0092-8674(90)90813-t. [DOI] [PubMed] [Google Scholar]
- Field J., Broek D., Kataoka T., Wigler M. Guanine nucleotide activation of, and competition between, RAS proteins from Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jun;7(6):2128–2133. doi: 10.1128/mcb.7.6.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J., Nikawa J., Broek D., MacDonald B., Rodgers L., Wilson I. A., Lerner R. A., Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988 May;8(5):2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J., Vojtek A., Ballester R., Bolger G., Colicelli J., Ferguson K., Gerst J., Kataoka T., Michaeli T., Powers S. Cloning and characterization of CAP, the S. cerevisiae gene encoding the 70 kd adenylyl cyclase-associated protein. Cell. 1990 Apr 20;61(2):319–327. doi: 10.1016/0092-8674(90)90812-s. [DOI] [PubMed] [Google Scholar]
- Field J., Xu H. P., Michaeli T., Ballester R., Sass P., Wigler M., Colicelli J. Mutations of the adenylyl cyclase gene that block RAS function in Saccharomyces cerevisiae. Science. 1990 Jan 26;247(4941):464–467. doi: 10.1126/science.2405488. [DOI] [PubMed] [Google Scholar]
- Gibbs J. B., Marshall M. S. The ras oncogene--an important regulatory element in lower eucaryotic organisms. Microbiol Rev. 1989 Jun;53(2):171–185. doi: 10.1128/mr.53.2.171-185.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman M. E., Adhya S., Das A. Transcription antitermination by bacteriophage lambda N gene product. J Mol Biol. 1980 Jun 15;140(1):57–75. doi: 10.1016/0022-2836(80)90356-3. [DOI] [PubMed] [Google Scholar]
- Hall A. The cellular functions of small GTP-binding proteins. Science. 1990 Aug 10;249(4969):635–640. doi: 10.1126/science.2116664. [DOI] [PubMed] [Google Scholar]
- Heideman W., Casperson G. F., Bourne H. R. Adenylyl cyclase in yeast: antibodies and mutations identify a regulatory domain. J Cell Biochem. 1990 Apr;42(4):229–242. doi: 10.1002/jcb.240420406. [DOI] [PubMed] [Google Scholar]
- Hortsch M., Avossa D., Meyer D. I. A structural and functional analysis of the docking protein. Characterization of active domains by proteolysis and specific antibodies. J Biol Chem. 1985 Aug 5;260(16):9137–9145. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T., Broek D., Wigler M. DNA sequence and characterization of the S. cerevisiae gene encoding adenylate cyclase. Cell. 1985 Dec;43(2 Pt 1):493–505. doi: 10.1016/0092-8674(85)90179-5. [DOI] [PubMed] [Google Scholar]
- Kataoka T., Powers S., McGill C., Fasano O., Strathern J., Broach J., Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984 Jun;37(2):437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- Krupinski J., Coussen F., Bakalyar H. A., Tang W. J., Feinstein P. G., Orth K., Slaughter C., Reed R. R., Gilman A. G. Adenylyl cyclase amino acid sequence: possible channel- or transporter-like structure. Science. 1989 Jun 30;244(4912):1558–1564. doi: 10.1126/science.2472670. [DOI] [PubMed] [Google Scholar]
- Löwenadler B., Nilsson B., Abrahmsén L., Moks T., Ljungqvist L., Holmgren E., Paleus S., Josephson S., Philipson L., Uhlén M. Production of specific antibodies against protein A fusion proteins. EMBO J. 1986 Sep;5(9):2393–2398. doi: 10.1002/j.1460-2075.1986.tb04509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall M. S., Gibbs J. B., Scolnick E. M., Sigal I. S. An adenylate cyclase from Saccharomyces cerevisiae that is stimulated by RAS proteins with effector mutations. Mol Cell Biol. 1988 Jan;8(1):52–61. doi: 10.1128/mcb.8.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Uno I., Oshima Y., Ishikawa T. Isolation and characterization of yeast mutants deficient in adenylate cyclase and cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2355–2359. doi: 10.1073/pnas.79.7.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A., Reed S. I. Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2119–2123. doi: 10.1073/pnas.77.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J., Cameron S., Toda T., Ferguson K. M., Wigler M. Rigorous feedback control of cAMP levels in Saccharomyces cerevisiae. Genes Dev. 1987 Nov;1(9):931–937. doi: 10.1101/gad.1.9.931. [DOI] [PubMed] [Google Scholar]
- Ruggieri R., Tanaka K., Nakafuku M., Kaziro Y., Toh-e A., Matsumoto K. MSI1, a negative regulator of the RAS-cAMP pathway in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8778–8782. doi: 10.1073/pnas.86.22.8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon Y., Londos C., Rodbell M. A highly sensitive adenylate cyclase assay. Anal Biochem. 1974 Apr;58(2):541–548. doi: 10.1016/0003-2697(74)90222-x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Davis R. W. A physical, genetic and transcriptional map of the cloned his3 gene region of Saccharomyces cerevisiae. J Mol Biol. 1980 Jan 25;136(3):309–332. doi: 10.1016/0022-2836(80)90376-9. [DOI] [PubMed] [Google Scholar]
- Tamanoi F. Yeast RAS genes. Biochim Biophys Acta. 1988 Aug 3;948(1):1–15. doi: 10.1016/0304-419x(88)90002-9. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Weisblum B. Construction of a colicin E1-R factor composite plasmid in vitro: means for amplification of deoxyribonucleic acid. J Bacteriol. 1975 Jan;121(1):354–362. doi: 10.1128/jb.121.1.354-362.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Uno I., Ishikawa T., Powers S., Kataoka T., Broek D., Cameron S., Broach J., Matsumoto K., Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985 Jan;40(1):27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- Uno I., Mitsuzawa H., Tanaka K., Oshima T., Ishikawa T. Identification of the domain of Saccharomyces cerevisiae adenylate cyclase associated with the regulatory function of RAS products. Mol Gen Genet. 1987 Dec;210(2):187–194. doi: 10.1007/BF00325683. [DOI] [PubMed] [Google Scholar]
- Voss H., Schwager C., Wirkner U., Sproat B., Zimmermann J., Rosenthal A., Erfle H., Stegemann J., Ansorge W. Direct genomic fluorescent on-line sequencing and analysis using in vitro amplification of DNA. Nucleic Acids Res. 1989 Apr 11;17(7):2517–2527. doi: 10.1093/nar/17.7.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamawaki-Kataoka Y., Tamaoki T., Choe H. R., Tanaka H., Kataoka T. Adenylate cyclases in yeast: a comparison of the genes from Schizosaccharomyces pombe and Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5693–5697. doi: 10.1073/pnas.86.15.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]