Abstract
Reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) is a reliable technique for measuring and evaluating gene expression during variable biological processes. To facilitate gene expression studies, normalization of genes of interest relative to stable reference genes is crucial. The western flower thrips Frankliniella occidentalis (Pergande) (Thysanoptera: Thripidae), the main vector of tomato spotted wilt virus (TSWV), is a destructive invasive species. In this study, the expression profiles of 11 candidate reference genes from nonviruliferous and viruliferous F. occidentalis were investigated. Five distinct algorithms, geNorm, NormFinder, BestKeeper, the ΔC t method, and RefFinder, were used to determine the performance of these genes. geNorm, NormFinder, BestKeeper, and RefFinder identified heat shock protein 70 (HSP70), heat shock protein 60 (HSP60), elongation factor 1 α, and ribosomal protein l32 (RPL32) as the most stable reference genes, and the ΔC t method identified HSP60, HSP70, RPL32, and heat shock protein 90 as the most stable reference genes. Additionally, two reference genes were sufficient for reliable normalization in nonviruliferous and viruliferous F. occidentalis. This work provides a foundation for investigating the molecular mechanisms of TSWV and F. occidentalis interactions.
Introduction
The western flower thrips, Frankliniella occidentalis (Pergande) (Thysanoptera: Thripidae), is a destructive species that is found globally. This insect damages hundreds of plant species through direct and indirect mechanisms via feeding and transmitting tospoviruses, respectively [1–3]. The tomato spotted wilt virus (Family: Bunyaviridae; Genus: Tospovirus; TSWV) is transmitted in a circulative, propagative manner by thrips, with F. occidentalis as the most efficient vector of TSWV [4]. Currently, TSWV ranks among the top ten most economically important plant viruses worldwide [5]. TSWV infects in as many as 1,000 plant species, causing disease in many vegetables and ornamentals [6]. Global epidemics of TSWV are likely related to the worldwide distribution of F. occidentalis [7].
To better understand the molecular mechanisms of TSWV and F. occidentalis interactions, the transcriptomes of F. occidentalis were generated [8–11]. Additionally, RNA interference (RNAi) tool that enables functional genomics assays was successfully developed for F. occidentalis [12]. Given the nature of RNAi mechanisms, the impact of RNAi will likely be a method to control gene expression profiles of pest organisms. To make better use of these genomic resources, the establishment of a standardized reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) procedure in F. occidentalis according to the MIQE (Minimum Information for publication of Quantitative real-time PCR Experiments) guidelines is instrumental for functional genomics studies of this pest [13].
RT-qPCR is a high-throughput technique for measuring and evaluating gene expression [14]; however, there remain limitations that can significantly influence the normalization of gene expression, including sample amount, RNA quality and quantity, efficiency of reverse transcription, cDNA quality, PCR efficiency, and experimental operation between different samples [15, 16]. Previously, Actin was used as stable reference gene to investigate the TSWV titers in F. occidentalis [12, 17–19], whereas another reference gene, RP49, was used as an internal control gene to estimate the abundance of TSWV N RNA in F. occidentalis [10]. The 18S gene was used as an endogenous control to investigate the expression profiles of 36 selected genes in viruliferous and non-viruliferous F. occidentalis [8], however, only one reference gene for the RT-qPCR experiment was used in these studies [8–11, 17–19]. In standardized RT-qPCR experiments, two or several reference genes are commonly used to normalize gene expression data, which are stably expressed across various experimental conditions and serve as the internal controls [14–16]. Recently, seven candidate genes from F. occidentalis including Actin, 18S, H3, Tubulin, GAPDH, EF-1A, and RPL32 were evaluated for their suitability as reference genes across different developmental stages and temperatures [20]. A previous study strongly suggests the necessity of conducting custom reference gene selection designed for all experimental conditions, even when examining the same abiotic or biotic factor [21].
The objective of this study was to determine most stable reference genes in nonviruliferous and viruliferous F. occidentalis. Here, 11 candidate reference genes from F. occidentalis were tested, namely, β-actin (Actin), α-tubulin (Tubulin), elongation factor 1 α (EF1A), vacuolar-type H + -ATPase (ATPase), NADH-ubiquinone oxidoreductase (NADH), heat shock protein 60 (HSP60), heat shock protein 70 (HSP70), heat shock protein 90 (HSP90), ribosomal protein l32 (RPL32), 28S ribosomal RNA (28S), and 18S ribosomal RNA (18S). To validate the selected reference genes, these candidates were further examined by RT-qPCR analysis against a TSWV-receptor gene in both nonviruliferous and viruliferous F. occidentalis.
Materials and Methods
Ethics Statement
The western flower thrips F. occidentalis (Pergande) (Thysanoptera: Thripidae) was collected from clover plants, Trifolium repens L., at the Experimental Station of Qingdao Agricultural University. No specific permit was required for the collection described.
Insect rearing, plant cultures, and TSWV inoculation
F. occidentalis was reared on the common bean Phaseolus vulgaris and maintained in MGC-250BP-2 incubators (Shanghai Yiheng Instruments, China) at 55–60% relative humidity and under a light: dark cycle of 16: 8 h. F. occidentalis adults were allowed to lay eggs on P. vulgaris for 1 day, and the adults were subsequently removed. First instar larvae were obtained in a few days.
Pepper (Capsicum annuum L., cv. Zhongjiao 6) plants were grown in soil mixed with vermiculite and organic fertilizer in 1.5-L pots (one plant per pot) under natural light and controlled temperatures (30 ± 2°C) in a greenhouse.
TSWV was maintained on Datura stramonium L. (Solanaceae). TSWV-infected pepper plants were prepared according to the method described in our previous study [22]. Control plants were treated similarly, but were inoculated by applying ground healthy plant material.
Nonviruliferous and viruliferous F. occidentalis
Healthy and TSWV-infected pepper leaf discs (diameter, 26 mm) were obtained using a cork borer. Each leaf was kept in a ventilated vial containing 3 ml of 1.0% agar to keep them fresh. Thirty newly hatched first instar larvae (nymphs) as one replicate were maintained on healthy or TSWV-infected discs in respective vials for 24 h to obtain nonviruliferous and viruliferous F. occidentalis, respectively [23]. Each treatment consisted of four replicates. Samples were frozen in liquid nitrogen and stored at –80°C.
Total RNA extraction and cDNA synthesis
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) according to previously described methods [24]. First-strand cDNA was synthesized from 1.0 μg of total RNA using a PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa, Shiga, Japan) according to the manufacturer’s recommendations. cDNA was diluted 10-fold for subsequent RT-qPCR studies.
Reference gene selection and primer design
Eleven candidate reference genes were selected that are commonly used in RT-qPCR and that have been verified as stable genes in other species (Table 1). Primers were designed based on the sequences obtained from GenBank (Table 1). The primers used for RT-qPCR analysis were designed online (https://www.idtdna.com/Primerquest/Home/Index) using the following parameters: amplicon length 75–150 bp with the optimum amplicon at 100 bp, Tm 59–65°C with the optimum amplicon at 62°C, primer lengths 17–30 bp, optimized to 22 bp, and GC content 35–65% with the optimal content at 50%. PCR amplifications were performed in 25-μl reactions containing 2.5 μl of 10× PCR Buffer (Mg2+ Plus), 0.5 μl of dNTP mix (10 mM of each nucleotide), 0.5 μl of each primer (10 μM each), and 0.25 μl of TaKaRa Taq (5 U/μl) (TaKaRa). Target genes were amplified using the following parameters: initial denaturation at 94°C for 3 min; 35 cycles of 94°C for 30 s, 59°C for 45 s, and 72°C for 1 min; and a final elongation step of 72°C for 10 min. Primer specificity was confirmed by melting curve analysis and agarose gel electrophoresis of the amplification product.
Table 1. Summary of the 11 housekeeping genes tested in this study.
| Gene | Description | Accession No. | Primer sequences (5’-3’) | Length (bp) | E(%)* | R2, ** |
|---|---|---|---|---|---|---|
| HSP70 | heat shock protein 70 | KC148536 | F: GTCACCGTACCCGCATATTT | 104 | 0.95 | 0.9845 |
| R: GCAGTGGGCTCGTTGATAATA | ||||||
| HSP60 | heat shock protein 60 | JX967580 | F: CTGGACTGTAAGCGTGCTATAA | 80 | 0.91 | 0.9903 |
| R: GGCACGATGAACACCTATGA | ||||||
| EF1A | elongation factor 1 α | AB277244 | F: AAGGAACTGCGTCGTGGATA | 99 | 1.05 | 0.991 |
| R: AGGGTGGTTCAGGACAATGA | ||||||
| RPL32 | ribosomal protein l32 | AB572580 | F: CTGGCGTAAACCTAAGGGTATT | 96 | 0.98 | 0.9998 |
| R: GTCTTGGCATTGCTTCCATAAC | ||||||
| ATPase | vacuolar type H + -ATPase | JN835456 | F: TACCAAATGGGACTCCAATACC | 130 | 0.90 | 0.9970 |
| R:GTAAGTAAGAGGTGGCCAGATAC | ||||||
| HSP90 | heat shock protein 90 | JX967579 | F: CTCGCAACCAGGACGATATTAG | 110 | 0.96 | 0.9918 |
| R: CTGACCCTCCACAGAGAAATG | ||||||
| NADH | NADH-ubiquinone oxidoreductase | YP_006576366 | F: AGCTACTAAACCGCCTCATAAA | 99 | 0.95 | 0.9656 |
| R:GGTGGTTATGGTATTTATCGTTTGT | ||||||
| 18S | 18S ribosomal RNA | JX002704 | F: CTGCGGAAATACTGGAGCTAATA | 109 | 1.09 | 0.9960 |
| R: AAGTAGACGATGGCCGAAAC | ||||||
| Actin | β-actin | AF434716 | F:CCTCATCCCTAGTTGTCTTGTG | 96 | 0.86 | 0.9788 |
| R: TTCTCGCTCAGCTGTAATTGT | ||||||
| 28S | 28S ribosomal RNA | GU980314 | F: GGGTGGTAAACTCCATCTAAGG | 108 | 0.97 | 0.9969 |
| R:CACGTACTCTTGAACTCTCTCTTC | ||||||
| Tubulin | α-tubulin | KC513334 | F: GTGGACAACGAAGCCATCTA | 77 | 1.04 | 0.9900 |
| R: CGGTTCAGGTTGGTGTAGG |
"*": PCR efficiency (calculated from the standard curve)
"**": Regression coefficient
RT-qPCR
RT-qPCR was performed on a qTOWER 2.2 Real-Time Thermal Cycler system (Analytik Jena, Germany). PCR reactions (20 μl) contained 7.2 μl of ddH2O, 10.0 μl of 2× SYBR Premix Ex Taq (TaKaRa), 0.4 μl of each specific primer (10 μM), and 2.0 μl of first-strand cDNA. The RT-qPCR program amplified target genes using the following parameters: initial denaturation for 3 min at 95°C followed by 40 cycles of denaturation at 95°C for 15 s and annealing at 60°C for 30 s. For melting curve analysis, a dissociation step cycle (55°C for 10 s, and then an increase of 0.5°C every 10 s up to 95°C) was used. The reactions were set up in 96-well Microseal PCR plates (Sangon, Shanghai, China) in triplicate. A 5-fold dilution series of cDNA (1/5, 1/25, 1/125, 1/625, and 1/3125) was used to construct a standard curve. The RT-qPCR efficiency was calculated according to the following equation: E = (10[-1/slope] -1)×100.
Validation of selected reference genes
One TSWV-receptor gene of F. occidentalis was used to evaluate the candidate reference genes (GenBank No. AF247969). TSWV-receptor gene expression levels were investigated in nonviruliferous and viruliferous F. occidentalis. Two normalization factors (NFs) were calculated based on (1) the geometric mean of genes with the lowest Geomean values, and (2) a single reference with the lowest or highest Geomean value (as determined by RefFinder). Relative quantification of the TSWV-receptor gene in different samples was performed using the 2-ΔΔCt method [25].
Data analysis
All biological replicates were used to calculate the average C t value. The stability of candidate reference genes was evaluated by the algorithms geNorm [14], NormFinder [15], BestKeeper [26], and the ΔC t method [27]. Finally, the tested candidates were compared and ranked using the web-based comprehensive analysis tool RefFinder (http://www.leonxie.com/referencegene.php). Nonparametric tests (K independent samples) were used to compare the expression levels of the TSWV-receptor gene in viruliferous and nonviruliferous F. occidentalis. Statistical analysis was conducted using SPSS 20.0 (SPSS Inc., Chicago, IL, USA).
Results
Transcriptional profiling of candidate reference genes
All tested genes were visualized as a single amplicon of the expected size on a 2.0% agarose gel (S1 Fig). Furthermore, gene-specific amplification was confirmed by a single peak in real-time melting curve analysis (S2 Fig). The linear regression equation, correlation coefficient, and PCR efficiency for each standard curve are shown in Table 1.
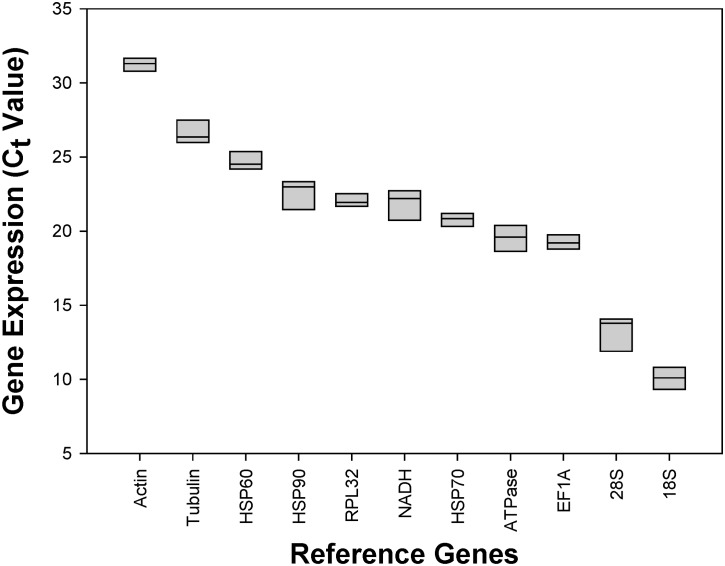
The mean and standard deviation (SD) of C t values were calculated for all samples (S1 Table). EF1A (SD = 0.68) had the least variable expression and this was reflected in its low SD values. By contrast, 28S (SD = 1.35) had the most variable expression, as shown by its high SD values. Additionally, 18S had the lowest C t values (C tavg = 10.09), suggesting that it had the highest expression level, whereas Actin was the lowest expressed gene among the candidates (C tavg = 31.62) (Fig 1; S1 Table).
Fig 1. Expression profiles of 11 candidate reference genes in Frankliniella occidentalis.
The expression levels of the candidate reference genes are documented by the C t values. The median is represented by the line in the box. The interquartile rang is bordered by the upper and lower edges, which indicate the 75th and 25th percentiles, respectively.
Stability of candidate reference genes
GeNorm determines its ranking based on the geometric mean of the SD of each transformed gene pair combination (M-value). The lower the M-value, the higher the ranking. EF1A and HSP70 were co-ranked as the most stable genes (M = 0.270). The overall order based on geNorm from the most stable to the least stable reference gene was as follows: EF1A = HSP70, RPL32, HSP60, ATPase, HSP90, NADH, 28S, 18S, Actin, and Tubulin (Table 2).
Table 2. Ranking of the 11 housekeeping genes using five different algorisms.
| RefFinder | geNorm | NormFider | ΔC t | BestKeeper | |||||
|---|---|---|---|---|---|---|---|---|---|
| Genes | GM | Genes | SV | Genes | SV | Genes | SV | Genes | SD |
| HSP70 | 1.68 | EF1A | 0.270 | HSP60 | 0.152 | HSP60 | 0.69 | EF1A | 0.499 |
| HSP60 | 2.00 | HSP70 | 0.270 | HSP70 | 0.299 | HSP70 | 0.71 | HSP70 | 0.521 |
| EF1A | 2.11 | RPL32 | 0.335 | RPL32 | 0.398 | RPL32 | 0.75 | RPL32 | 0.556 |
| RPL32 | 3.00 | HSP60 | 0.375 | EF1A | 0.507 | HSP90 | 0.80 | HSP60 | 0.560 |
| ATPase | 5.89 | ATPase | 0.537 | ATPase | 0.531 | EF1A | 0.80 | Actin | 0.566 |
| HSP90 | 6.16 | HSP90 | 0.592 | HSP90 | 0.547 | ATPase | 0.82 | 18S | 0.595 |
| NADH | 7.45 | NADH | 0.627 | NADH | 0.638 | NADH | 0.85 | Tubulin | 0.784 |
| 18S | 8.13 | 28S | 0.667 | 28S | 0.852 | 28S | 1.00 | ATPase | 0.809 |
| Actin | 8.41 | 18S | 0.754 | 18S | 0.878 | 18S | 1.06 | NADH | 0.894 |
| 28S | 8.66 | Actin | 0.815 | Actin | 0.893 | Actin | 1.08 | HSP90 | 0.915 |
| Tubulin | 9.82 | Tubulin | 0.888 | Tubulin | 1.060 | Tubulin | 1.22 | 28S | 1.069 |
The ΔC t method relies on relative pair-wise comparisons. Using raw C t values, the average SD of each gene set is inversely proportional to its stability. HSP60 (0.69) was the top-ranked gene (Table 2). The overall order from the most stable to the least stable reference gene based on the ΔC t method was as follows: HSP60, HSP70, RPL32, HSP90, EF1A, ATPase, NADH, 28S, 18S, Actin, and Tubulin (Table 2; S2 Table).
A low stability value suggests a more stable gene by NormFinder. HSP60 (0.152) was the most reliable and stable reference gene. The overall order from the most stable to the least stable reference gene based on NormFinder was as follows: HSP60, HSP70, RPL32, EF1A, ATPase, HSP90, NADH, 28S, 18S, Actin, and Tubulin (Table 2).
A low SD value suggests a more stable gene by BestKeeper. The stability of a gene is inversely proportional to the SD value. EF1A (SD = 0.499) had the least variable expression levels across all samples (Table 2). The overall order from the most stable to the least stable reference gene based on BestKeeper was as follows: EF1A, HSP70, RPL32, HSP60, Actin, 18S, Tubulin, ATPase, NADH, HSP90, and 28S (Table 2).
Quantitative analysis of candidate reference genes based on geNorm
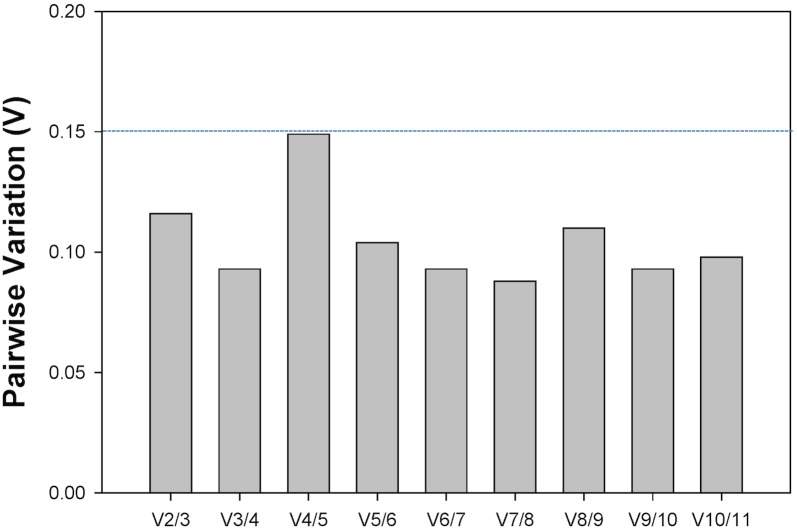
Under plant virus stress, the first V-value less than 0.15 was after V2/3 (Fig 2). This means that two reference genes were sufficient for reliable normalization regardless of the virus infection status of the insect.
Fig 2. Determination of the optimal number of reference genes under plant virus stress.
To determine the minimum number of genes required for normalization, the V-value was computed using geNorm. Starting with two genes, the software sequentially adds another gene and recalculates the NF ratio. If the added gene does not increase the NF ratio above the proposed 0.15 cut-off value, then the starting pair of genes is sufficient for normalization. If the NF ratio is adequately increased, more genes should be incorporated.
Comprehensive ranking of the best reference genes using RefFinder
According to RefFinder, which integrates the above-mentioned four software tools to compare and rank candidates, the comprehensive ranking of candidate reference genes from the most to the least stable was as follows: HSP70, HSP60, EF1A, RPL32, ATPase, HSP90, NADH, 18S, Actin, 28S, and Tubulin (Table 2). Among these, Tubulin had a geometric mean of almost 10.0, and was determined to be the least suitable candidate to serve as a reliable reference gene for normalizing gene expression.
Validation of selected reference genes
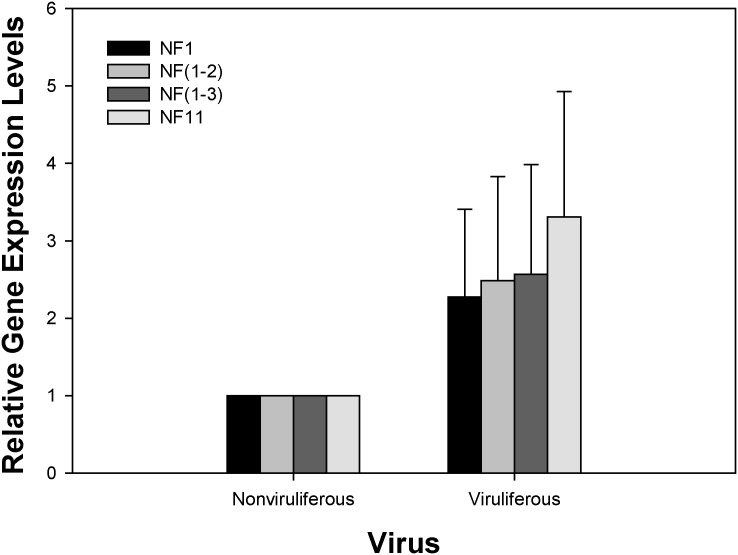
Using one, two, or three of the best reference gene combinations for normalization, expression of the TSWV-receptor gene did not differ between viruliferous and nonviruliferous F. occidentalis (P > 0.05) (Fig 3).
Fig 3. Validation of the recommended reference gene.
The expression profiles of the TSWV-receptor gene in nonviruliferous and viruliferous Frankliniella occidentalis were investigated using different combinations of reference genes. NF1, NF (1–2), NF (1–3), and NF9 indicate that expression of the TSWV-receptor gene was normalized using the best, top two, top three, and worst reference genes, respectively. The bar represents the mean and standard error of four biological replicates.
Discussion
Recent studies suggest that there is no single reference gene that is suitable for all types of normalization experiments [14, 28–30]. Therefore, candidate reference genes should be evaluated before they are used for normalization [21, 31]. Recently, a study found that the ranking of candidate reference genes in the sweet potato whitefly, Bemisia tabaci varied substantially among intra- and inter-classes of insecticides [21]. This strongly suggested the need for conducting reference gene selection specifically designed for all experimental conditions, even when examining the same abiotic or biotic factor [21]. Zheng et al. (2014) focused on the selection of reference genes across different developmental stages and temperatures in F. occidentalis [20], while this study mainly focused on the impact of virus infection on the stability of these internal controls. As expected, these selected reference genes varied across different experimental conditions. For example, our study demonstrated that Tubulin was the least appropriate reference gene in F. occidentalis, which is inconsistent with Zheng et al. (2014) where Tubulin was a suitable reference gene for F. occidentalis across different experimental conditions [20]. In addition, Actin was not appropriate in our study, which was consistent with the Zheng et al. (2014) study [20]. Interestingly, Actin has been used previously as reference gene to investigate the TSWV titer in F. occidentalis [12, 17–19]. Therefore, we suggest that custom reference gene selection should be conducted for each experimental condition.
In previous studies, only one reference gene was selected for the RT-qPCR experiment in F. occidentalis [8–11, 17–19]. To avoid biased normalization, many researchers have started to advocate the use of multiple reference genes to analyze gene expression [14, 24, 28–30]. The geNorm program first calculates an expression stability value (M) for each gene and compares the pair-wise variation (V) of this gene with the other genes. A threshold of V<0.15 was suggested for valid normalization. geNorm starts with a single gene pair, and tests whether the inclusion of a 3rd gene adds significant variation. The pair-wise variation (Vn/Vn+1) was analyzed between the normalization factors NFn and NFn+1 by geNorm to determine the optimal number of references genes required for qRT-PCR data normalization. In our study, the first pair-wise variation value less than 0.15 was after V2/3 (Fig 2), suggesting that two reference genes are sufficient for studying gene expression in nonviruliferous and viruliferous F. occidentalis (Fig 2).
In short, 11 candidate reference genes were selected for RT-qPCR analysis in nonviruliferous and viruliferous F. occidentalis assessed by five algorithms (geNorm, NormFinder, BestKeeper, ΔC t method, and RefFinder). Among them, HSP60, HSP70, and RPL32 are the three most stable reference genes under the impact of plant virus infection as found by all five algorithms. Recently, the introduction of RNAi as a tool for functional genomics assays for F. occidentalis was developed [28]. Therefore, our study not only provides a standardized protocol for the quantification of gene expression in F. occidentalis, but also provides a solid foundation for genomic and functional genomics research assessing the interactions between TSWV and F. occidentalis.
Supporting Information
M,DL 2000 bp Marker; Templates in the PCR reactions were as follows: 1) 18S; 2) 28S; 3) Actin; 4) ATPase; 5) EF1A; 6) HSP60; 7) HSP70; 8) HSP90; 9) NADH; 10) RPL32; 11) Tubulin.
(TIF)
(TIF)
(DOCX)
(DOCX)
Acknowledgments
Special thanks go to Jeffrey Edward Noland (University of Kentucky) for his help in revising the manuscript. This research was supported by a Special Fund for Agroscience Research in the Public Interest (Award Agreement No.: 201303028), the Science and Technology Development Planning Program of Qingdao (13-1-3-108-nsh), the Shandong Modern Agricultural Technology & Industry System (SDAIT-02-021-11), and the Joint Funds for Young Scientist of Hunan (14JJ6058). The granting agencies have no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Special Fund for Agroscience Research in the Public Interest (201303028), the Shandong Modern Agricultural Technology & Industry System (SDAIT-02-021-11), the Science and Technology Development Planning Program of Qingdao (13-1-3-108-nsh), and the Taishan Mountain Scholar Constructive Engineering Foundation of Shandong, China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Brødsgaard HF (1994) Effect of photoperiod on the bionomics of Frankliniella occidentalis (Pergande) (Thysanoptera, Thripidae). J Appl Entomol 117: 498–507. [Google Scholar]
- 2. Brunner PC, Frey JE (2010) Habitat‐specific population structure in native western flower thrips Frankliniella occidentalis (Insecta, Thysanoptera). J Evolution Biol 23: 797–804. [DOI] [PubMed] [Google Scholar]
- 3. Jones DR (2005) Plant viruses transmitted by thrips. Eur J Plant Pathol 113: 119–157. [Google Scholar]
- 4. Whitfield AE, Ullman DE, German TL (2005) Tospovirus-thrips interactions. Annu Rev Phytopathol 43: 459–489. [DOI] [PubMed] [Google Scholar]
- 5. Scholthof KBG, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, et al. (2011) Top 10 plant viruses in molecular plant pathology. Mol Plant Pathol 12: 938–954. 10.1111/j.1364-3703.2011.00752.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pappu HR, Jones RAC, Jain RK (2009) Global status of tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res 141: 219–236. 10.1016/j.virusres.2009.01.009 [DOI] [PubMed] [Google Scholar]
- 7. Hogenhout SA, Ammar ED, Whitfield AE, Redinbaugh MG (2008) Insect vector interactions with persistently transmitted viruses. Annu Rev Phytopathol 46: 327–359. 10.1146/annurev.phyto.022508.092135 [DOI] [PubMed] [Google Scholar]
- 8. Zhang ZJ, Zhang PJ, Li WD, Zhang JM, Huang F, Yang J, et al. (2013) De novo transcriptome sequencing in Frankliniella occidentalis to identify genes involved in plant virus transmission and insecticide resistance. Genomics 101: 296–305 10.1016/j.ygeno.2013.02.005 [DOI] [PubMed] [Google Scholar]
- 9. Rotenberg D, Whitfield AE (2010) Analysis of expressed sequence tags for Frankliniella occidentalis, the western flower thrips. Insect Mol Biol 19: 537–551. 10.1111/j.1365-2583.2010.01012.x [DOI] [PubMed] [Google Scholar]
- 10. Badillo-Vargas IE, Rotenberg D, Schneweis DJ, Hiromasa Y, Tomich JM, Whitfield AE (2012) Proteomic analysis of Frankliniella occidentalis and differentially-expressed proteins in response to Tomato spotted wilt virus infection. J Virol JVI-00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stafford-Banks CA, Rotenberg D, Johnson BR, Whitfield AE, Ullman DE (2014) Analysis of the salivary gland transcriptome of Frankliniella occidentalis . PLOS ONE 9: e94447 10.1371/journal.pone.0094447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Badillo-Vargas IE, Rotenberg D, Schneweis BA, Whitfield AE (2015) RNA interference tools for the western flower thrips, Frankliniella occidentalis . J Insect Physiol 76: 36–46. 10.1016/j.jinsphys.2015.03.009 [DOI] [PubMed] [Google Scholar]
- 13. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 14. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–5250. [DOI] [PubMed] [Google Scholar]
- 16. Guo JL, Ling H, Wu QB, Xu LP, Que YX (2014). The choice of reference genes for assessing gene expression in sugarcane under salinity and drought stresses. Sci Rep 4: 7042 10.1038/srep07042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boonham N, Smith P, Walsh K, Tame J, Morris J, Spence N, et al. (2002) The detection of Tomato spotted wilt virus (TSWV) in individual thrips using real time fluorescent RT-PCR (TaqMan). J Virol Methods 101: 37–48. [DOI] [PubMed] [Google Scholar]
- 18. Rotenberg D, Krishna Kumar NK, Ullman DE, Montero-Astúa M, Willis DK, German T L, et al. (2009) Variation in Tomato spotted wilt virus titer in Frankliniella occidentalis and its association with frequency of transmission. Phytopathology 99: 404–410. 10.1094/PHYTO-99-4-0404 [DOI] [PubMed] [Google Scholar]
- 19. Margaria P, Bosco L, Vallino M, Ciuffo M, Mautino GC, Tavella L, et al. (2014) The NSs protein of Tomato spotted wilt virus is required for persistent infection and transmission by Frankliniella occidentalis . J Virol 88: 5788–5802. 10.1128/JVI.00079-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng YT, Li HB, Lu MX, Du YZ (2014) Evaluation and validation of reference genes for qRT-PCR normalization in Frankliniella occidentalis (Thysanoptera: Thripidae). PLOS ONE 9: e111369 10.1371/journal.pone.0111369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liang P, Guo YJ, Zhou XG, Gao XW (2014) Expression profiling in Bemisia tabaci under insecticide treatment: indicating the necessity for custom reference gene selection. PLOS ONE 9: e87514 10.1371/journal.pone.0087514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pan HP, Chen G, Li F, Wu QJ, Wang SL, Xie W, et al. (2013) Tomato spotted wilt virus infection reduces the fitness of a nonvector herbivore on pepper. J Econ Entomol 106: 924–928. [DOI] [PubMed] [Google Scholar]
- 23. Mason G, Roggero P, Tavella L (2003) Detection of Tomato spotted wilt virus in its vector Frankliniella occidentalis by reverse transcription-polymerase chain reaction. J Virol Methods 109: 69–73. [DOI] [PubMed] [Google Scholar]
- 24. Yang CX, Pan HP, Liu Y, Zhou XG (2015) Stably expressed housekeeping genes across developmental stages in the two-spotted spider mite, Tetranychus urticae . PLOS ONE 10: e0120833 10.1371/journal.pone.0120833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 26. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26: 509–515. [DOI] [PubMed] [Google Scholar]
- 27. Silver N, Best S, Jiang J, Thein SL (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol 7: 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jacob F, Guertler R, Naim S, Nixdorf S, Fedier A, Hacker NF, et al. (2013) Careful selection of reference genes is required for reliable performance of RT-qPCR in human normal and cancer cell lines. PLOS ONE 8: e59180 10.1371/journal.pone.0059180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sinha DK, Smith CM (2014) Selection of reference genes for expression analysis in Diuraphis noxia (Hemiptera: Aphididae) fed on resistant and susceptible wheat plants. Sci Rep 4: 5059 10.1038/srep05059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang CX, Pan HP, Liu Y, Zhou XG (2014) Selection of reference genes for expression analysis using quantitative real-time PCR in the pea aphid, Acyrthosiphon pisum (Harris) (Hemiptera, Aphidiae). PLOS ONE 9: e110454 10.1371/journal.pone.0110454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lilly ST, Drummond RSM, Pearson MN, MacDiarmid RM (2011) Identification and validation of reference genes for normalization of transcripts from virus-infected Arabidopsis thaliana . Mol Plant Microbe In 24: 294–304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
M,DL 2000 bp Marker; Templates in the PCR reactions were as follows: 1) 18S; 2) 28S; 3) Actin; 4) ATPase; 5) EF1A; 6) HSP60; 7) HSP70; 8) HSP90; 9) NADH; 10) RPL32; 11) Tubulin.
(TIF)
(TIF)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.