Abstract
The budding yeast Saccharomyces cerevisiae is a platform organism for bioethanol production from various feedstocks and robust strains are desirable for efficient fermentation because yeast cells inevitably encounter stressors during the process. Recently, diverse S. cerevisiae lineages were identified, which provided novel resources for understanding stress tolerance variations and related shaping factors in the yeast. This study characterized the tolerance of diverse S. cerevisiae strains to the stressors of high ethanol concentrations, temperature shocks, and osmotic stress. The results showed that the isolates from human-associated environments overall presented a higher level of stress tolerance compared with those from forests spared anthropogenic influences. Statistical analyses indicated that the variations of stress tolerance were significantly correlated with both ecological sources and geographical locations of the strains. This study provides guidelines for selection of robust S. cerevisiae strains for bioethanol production from nature.
Introduction
The budding yeast Saccharomyces cerevisiae is a vitally important microorganism for human being, which has been instrumental to baking, brewing, distilling, and wine making since ancient times [1]. More recently, S. cerevisiae has been used as a platform organism for production of bioethanol from various biomass feedstocks, such as corn starch, sugarcane, and lignocellulose [2,3]. Regardless of what kind of biomass feedstock is used, stressors inhibiting yeast growth and fermentation are generated during fermentation processes. High ethanol concentrations, temperature shocks, and osmotic stress are among the most common stressors during fermentations [4,5]. Strains of S. cerevisiae resistant to such stressors are desirable for bioethanol production [6].
The yeast S. cerevisiae is among the most thoroughly studied eukaryotic model organisms at the cellular, molecular, and genetic levels. However, the ecology of S. cerevisiae has not attracted enough attention until recently. Natural isolates of S. cerevisiae inhabiting tree exudates in forests were discovered fifteen years ago [7,8]. Subsequent studies proposed the existing of both domesticated populations and wild populations [9]. In recent years, it has been recognized that S. cerevisiae is a ubiquitous species in nature and it occupies numerous habitats from human-associated environments as well as habitats remote from human activity [10,11]. Five world lineages and eight Chinese lineages have been identified as well as many genetic mosaics strains through investigation of diverse isolates from industrial environments, hospitals, breweries, tree barks, leaves, soil, etc. The identification of clear lineages and mosaics strains offered new insights into genetic structure of S. cerevisiae [11–13]. Meanwhile, phenotypic variations of these S. cerevisiae lineages attracted increasing attention [12,14–16]. Wild S. cerevisiae strains collected from oak exudates present apparent differences from vineyard or sake isolates in phenotypic similarity, growth rate, and stress tolerance to a variety of stressors, such as copper sulfate and freeze [16,17]. The finding of eight Chinese S. cerevisiae lineages provided novel materials for investigating phenotypic variations and related driving forces in diverse S. cerevisiae populations.
This study aims to investigate the variations of stress tolerance in novel S. cerevisiae lineages and the influence of ecology and geography on the varied phenotypes. Considering that high ethanol concentrations, temperature shocks, and osmotic stress are the most common stressors during ethanol fermentation, we evaluated the tolerance of S. cerevisiae strains to these stressors. The results showed that both ecological sources and geographical locations presented significant correlation with variations of stress tolerance. Overall, the isolates from human-associated environments represented a higher level of stress tolerance compared with those from forests spared anthropogenic influences, which provides guidelines for selection of robust S. cerevisiae strains for bioethanol production from nature.
Materials and Methods
Strains
The panel of S. cerevisiae strains used in stress tolerance analysis consisted of 34 Chinese forest isolates and 37 Chinese orchard isolates, as well as 12 industrial strains deposited in China General Microbiological Culture Collection Center (CGMCC) (Table 1). The forest and orchard isolates were collected and identified according to the methods described previously [11]. All necessary permits were obtained for the described field studies. The owners of the orchards and the authorities responsible for the forests agreed with the collection of samples and the sampling plan. The geographic locations of strains isolated in China covered Jilin province (latitude and longitude across the sampling sites, 41°35'~42°25', 127°40'~128°16'), Beijing areas (39°26'~41°03', 115°25'~117°30'), Shandong province (35°36'~36°25', 116°58'~119°12'), Xinjing province (41°68'~42°95', 86°06'~90°25'), Shannxi province (34°17', 108°20'), Yunnan province (22°00'~25°34', 100°52'~102°73'), and Hainan province (19°52'~20°02', 109°57'~110°35').
Table 1. Strains of S. cerevisiae used in stress tolerance analysis.
| Lineage | Strain | Location | Source | Date |
|---|---|---|---|---|
| Orchard group | ||||
| CHN IV | BJPTE | Shunyi, Beijing | Vineyard grape | 2008 |
| BJ3 | Shunyi, Beijing | Vineyard grape | 2008 | |
| CHN VI | AQPT45 | Anqiu, Shandong | Vineyard grape | 2008 |
| AQT2 | Anqiu, Shandong | Grapevine | 2008 | |
| CHN VIII | SH28-1 | Shanshan, Xinjiang | Vineyard grape | 2006 |
| BJ2 | Shunyi, Beijing | Apple in orchard | 2008 | |
| SD4 | Qufu, Shandong | Vineyard grape | 2008 | |
| QFPT43 | Qufu, Shandong | Vineyard grape | 2008 | |
| Clade 1 | BJPG4 | Shunyi, Beijing | Apple in orchard | 2008 |
| BJPG14 | Shunyi, Beijing | Apple in orchard | 2008 | |
| BJL8 | Shunyi, Beijing | Pear in orchard | 2008 | |
| BJL10 | Shunyi, Beijing | Pear in orchard | 2008 | |
| Clade 2 | XL3-1 | Korla, Xinjiang | Pear in orchard | 2006 |
| XL4 | Korla, Xinjiang | Pear in orchard | 2006 | |
| XL11-2 | Korla, Xinjiang | Pear in orchard | 2006 | |
| XL13-2 | Korla, Xinjiang | Pear in orchard | 2006 | |
| XL14-1 | Korla, Xinjiang | Pear in orchard | 2006 | |
| XL15-2 | Korla, Xinjiang | Pear in orchard | 2006 | |
| P10-1 | Korla, Xinjiang | Apple in orchard | 2006 | |
| P16 | Korla, Xinjiang | Apple in orchard | 2006 | |
| P16-2 | Korla, Xinjiang | Apple in orchard | 2006 | |
| Sake | YN2 | Jinghong, Yunnan | Orange in orchard | 2006 |
| BJL4 | Shunyi, Beijing | Pear in orchard | 2008 | |
| BJL7 | Shunyi, Beijing | Pear in orchard | 2008 | |
| Wine/European | XJ5 | Shanshan, Xinjiang | Vineyard grape | 2006 |
| WA3-4 | Shunyi, Beijing | Vineyard grape | 2008 | |
| Mosaic | XJ4 | Shanshan, Xinjiang | Vineyard grape | 2006 |
| XJ6 | Korla, Xinjiang | Pear in orchard | 2006 | |
| P7-1 | Korla, Xinjiang | Apple in orchard | 2006 | |
| XJ2 | Korla, Xinjiang | Apple in orchard | 2006 | |
| XJ3 | Korla, Xinjiang | Apple in orchard | 2006 | |
| BL6 | Jinghong, Yunnan | Pineapple in plantation | 2006 | |
| YN1 | Jinghong, Yunnan | Pineapple in plantation | 2006 | |
| BL11C | Jinghong, Yunnan | Pineapple in plantation | 2006 | |
| BJL3 | Shunyi, Beijing | Pear in orchard | 2008 | |
| JL3 | Changbai Mt, Jilin | Peach | 2007 | |
| YC77 | Changbai Mt, Jilin | Oak bark | 2008 | |
| Wild group | ||||
| CHN I | HN6 | Bawangling Mt, Hainan | Rotten wood | 2007 |
| HN1 | Diaoluo Mt, Hainan | Rotten wood | 2007 | |
| CHN II | SX2 | Qinling Mt, Shaanxi | Oak bark | 2006 |
| SX4 | Qinling Mt, Shaanxi | Oak bark | 2006 | |
| SX5 | Qinling Mt, Shaanxi | Oak bark | 2006 | |
| SX6 | Qinling Mt, Shaanxi | Oak bark | 2006 | |
| SX7 | Qinling Mt, Shaanxi | Oak bark | 2006 | |
| SX8 | Qinling Mt, Shaanxi | Oak bark | 2006 | |
| CHN III | HN19 | Wuzhi Mt, Hainan | Oak bark | 2006 |
| CHN IV | BJ18 | Dongling Mt, Beijing | Oak bark | 2008 |
| LS26 | Dongling Mt, Beijing | Oak bark | 2008 | |
| LS541 | Dongling Mt, Beijing | Oak bark | 2008 | |
| LS542 | Dongling Mt, Beijing | Oak bark | 2008 | |
| BJ23 | Dongling Mt, Beijing | Oak bark | 2008 | |
| BJ24 | Dongling Mt, Beijing | Oak bark | 2008 | |
| BJ20 | Dongling Mt, Beijing | Oak bark | 2008 | |
| CHN V | HN17 | Wuzhi Mt, Hainan | Oak bark | 2006 |
| HN18 | Wuzhi Mt, Hainan | Oak bark | 2006 | |
| HN12 | Hainan, China | Rotten wood | 2007 | |
| HN15 | Hainan, China | Soil | 2007 | |
| HN16 | Hainan, China | Soil | 2007 | |
| CHN VI | AQSZ8 | Anqiu, Shandong | Oak bark | 2008 |
| AQSZ30 | Anqiu, Shandong | Oak bark | 2008 | |
| SD2 | Tai’an, Shandong | Oak bark | 2008 | |
| Sake | TABL2 | Tai’an, Shandong | Oak bark | 2008 |
| JL2 | Changbai Mt, Jilin | Oak bark | 2008 | |
| Mosaic | SX9 | Qinling Mt, Shaanxi | Wild tomato | 2006 |
| SX11 | Qinling Mt, Shaanxi | Oak bark | 2006 | |
| YN6 | Zixi Mt, Yunnan | Oak bark | 2006 | |
| YDX4 | Dali, Yunnan | Soil | 2008 | |
| YN5 | Dali, Yunnan | Soil | 2008 | |
| BJ3P | Haidian, Beijing | A peach flower | 2006 | |
| TAHT7 | Tai’an, Shandong | Oak bark | 2008 | |
| SD3 | Tai’an, Shandong | Oak bark | 2008 | |
| Industrial group | ||||
| Clade 3 | AS2.2 | England | Beer | Pre-1952 |
| AS2.3 | England | Beer | Pre-1952 | |
| AS2.4 | England | Beer | Pre-1952 | |
| Sake | AS2.666 | Guizhou, China | Distilled spirit | Pre-1960 |
| Wine/European | AS2.24 | England | Brewary | Pre-1952 |
| AS2.521 | Unknown | Ratafee | Pre-1960 | |
| AS2.526 | Unknown | Distilled spirit | Pre-1960 | |
| AS2.1422 | Japan | Distilled spirit | 1955 | |
| Mosaic | AS2.381 | Russia | Paper mill | Pre-1960 |
| AS2.383 | Russia | Paper mill | Pre-1960 | |
| AS2.395 | Russia | Paper mill | Pre-1960 | |
| AS2.1427 | Japan | Distilled spirit | Pre-1943 | |
Abbreviation: Mt, mountain.
Sequence analysis
Four housekeeping genes (ACT1, RPB1, RPN2, and TBP) and three stress response genes (HSP104, HXK1 and TPS1) were sequenced in this study [18–21]. Three of the four housekeeping genes (ACT1, RPB1 and RPN2) were previously used to infer the diversity of S. cerevisiae [22, 23]. Gene sequences were amplified using the primer pairs as described in S1 Table. The gene amplification conditions were as follows: 4 min initial denaturation at 94°C, 30 cycles each of 30 sec at 94°C, 45 sec at 52°C or 55°C, and 90 sec at 72°C, and a final extension period of 8 min at 72°C. Population genetic analysis was completed using DnaSP software [24]. McDonald and Kreitman test was performed to evaluate the neutral evolution for the genes [25]. Neighbor-joining tree was constructed from the evolutionary distance data calculated from Kimura’s two-parameter model [26].
Population structure analysis
Population structure was inferred using the program Structure 2.3.2 [27] based on 143 SNPs, for which neutral evolution was not rejected based on McDonald & Kreitman test. Admixture model with correlated allele frequencies was used. The result was visualized using Distruct 1.1 [28].
Culture conditions
All the strains were initially grown on YPD slants (10 g L-1 of yeast extract, 20 g L-1 of peptone, 20 g L-1 of dextrose and 20 g L-1 of agarose, pH 6.0) for 3 days at 30°C. Afterwards, a loop of each of the colonies was inoculated into 3 mL of YPD broth and cultured for 48 h at 30°C with shaking at 200 rpm. In order to invigorate all the strains, 20 μL of each YPD broth culture was transferred into a new YPD broth tube and cultured for an additional 24 h at 30°C with shaking at 200 rpm. The optical density at 600 nm (OD600) of the pre-cultured broth was determined by using a Biotek Synergy HT spectrophotometer, and then the pre-cultured broth was inoculated into tubes containing 3 mL of yeast minimal medium (YMM) and brought to an OD600 reading of 0.02. YMM consisted of 6.7 g L-1 yeast nitrogen base without amino acids (Difco, Beijing), 20 g L-1 dextrose and one type of stressor. The stressors applied were either 10% (v/v) of ethanol, 42°C (heat) or 2 M of KCl (osmotic stress), respectively. Strains grown in YMM broth without stressors were used as controls and incubated at 30°C with shaking at 200 rpm for 72 h. All experiments were performed in duplicate and repeated at least three times.
Biomass dry weight determination
To determine the biomass dry weight (in mg) of stress cultures, 3 mL of sample suspensions was transferred into weighed dry Eppendorf tubes and centrifuged for 5 min at 13400 g. The pellets were washed twice with distilled water. Then, the pellets were dried for 8 h at 105°C after which they were weighed by using a Mettler Toledo XS105 Dual Range Analytical Balance (Schweiz, Readability = 0.01 mg).
Stress tolerance analysis
To investigate stress tolerance variations of strains, we initially evaluated three levels for each of the stressors (ethanol: 8% (v/v), 10%, or 12%; heat: 40°C, 42°C, or 45°C; osmotic stress: 1 M, 2 M, or 3 M of KCl). The highest divergence was represented at middle stress levels for all stressors and thus the comparison of stress tolerance was performed by using these conditions (10% ethanol, 42°C, and 2 M of KCl). The stress tolerance of strains was evaluated by biomass dry weight after growth in stressful media. The comparison analyses among different groups were performed by using the Analysis of Variance (ANOVA) method and Nonparametric Tests. For independent samples (k, k>2), One-way ANOVA including Homogeneity of Variance Test and Least-Significant Difference (LSD) was valid when equal variance was assumed. Kruskal-Wallis test as an efficient Nonparametric Tests method was used when equal variance was not assumed. The correlations between the variability of stress tolerance and ecological sources or geographic locations were estimated by the Pearson correlation coefficient r. Strains were categorized into sub-groups according to ecological sources or geographic locations. The stress tolerance (biomass) of strains between sub-groups was compared and the Pearson correlation coefficient was calculated by ANOVA analysis. The hierarchical clustering of phenotypes was performed using Cluster 3.0 software [29].
Results and Discussion
Population genetic variation
Genetic variation was surveyed in 121 strains of S. cerevisiae using a dataset including 6519 bp of DNA sequence. The S. cerevisiae populations consist of 71 Chinese natural isolates, 12 industrial strains from CGMCC, and 38 SGRP isolates (Table 1) [11,12]. A total of 227 SNPs were identified, constituting 93 haplotypes. Heterozygous sites were found in 8 of 83 strains (SGRP isolates excluded) and at 45 SNPs. The natural isolates displayed a lower heterozygote to homozygote ratio of 5.6% (4/71) compared with the industrial strains representing that of 33.3% (4/12). Most natural isolates are homothallic diploids and haploid spores are capable of switching mating type and selfing, which may facilitate the remove of heterozygosity [14].
Neutral evolution was estimated in the 7 gene loci as described in the Material and Method section by the McDonald & Kreitman (MK) test [25]. Fixations were inferred from comparison of S. cerevisiae and the closest outgroup, S. paradoxus. The MK test did not reject neutral evolution for ACT1, HSP104, RPB1, RPN2, and TBP, while rejected neutral evolution for HXK1 and TPS1 due to an excess of replacement polymorphism than expected under neutral evolution (Table 2). To determine whether synonymous sites in HXK1 and TPS1 were affected by selection, codon bias was calculated and the number of unpreferred and preferred changes was compared along the lineage leading to S. cerevisiae and within strains of S. cerevisiae. In S. cerevisiae, those genes with expected neutral synonymous sites had a high effective number of codons (ENC > 45) and an equal number of preferred to unpreferred (P to U) and unpreferred to preferred (U to P) changes [30]. An ENC of 49.8 and equal P to U and U to P were exhibited in TPS1 for both polymorphic and fixed synonymous changes, in line with the expectation for neutral sites. HXK1 showed an ENC of 35.3 and a significant nonequilibrium on polymorphic changes (18:3 for P to U versus U to P), indicating that the synonymous sites in HXK1 were not effectively neutral. The HXK1 gene encodes the hexokinase isoenzyme I. HXK1 gene was previously reported locating in quantitative trait loci related to ethanol resistance and it was upregulated after ethanol shock [20,21]. TPS1 is a necessary gene for trehalose biosynthesis in S. cerevisiae, and trehalose is a major reserve carbohydrate involved in responses to thermal, osmotic, oxidative, and ethanol stresses [18]. The function of HXK1 and TPS1 in stress response may correlate with the evolution of them.
Table 2. McDonald–Kreitman Tests of Neutrality.
| Gene | P-value | Number of alleles | Polymorphic | Fixed | |||
|---|---|---|---|---|---|---|---|
| S.cer | S. par | syn | nonsyn | syn | nonsyn | ||
| ACT1 | \ | 121 | 36 | 19 | 0 | 9 | 0 |
| HSP104 | 0.154 | 121 | 36 | 59 | 5 | 40 | 0 |
| HXK1 | 0.00777 | 121 | 36 | 99 | 25 | 58 | 3 |
| RPB1 | 0.127 | 121 | 22 | 26 | 5 | 40 | 2 |
| RPN2 | 0.14 | 121 | 36 | 50 | 9 | 47 | 3 |
| TBP | 0.0768 | 121 | 36 | 41 | 6 | 29 | 0 |
| TPS1 | 0.00227 | 121 | 36 | 72 | 13 | 88 | 2 |
Population structure
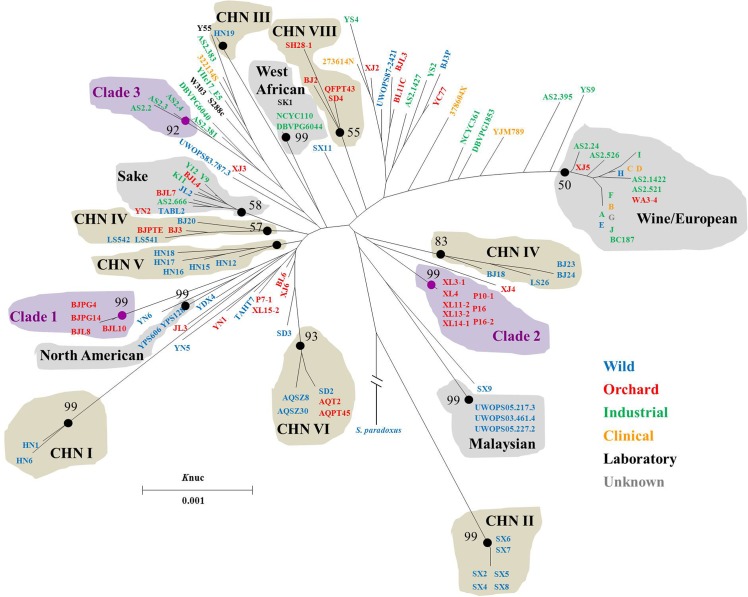
Phylogenetic relationships among strains of S. cerevisiae were inferred from a neighbor-joining (NJ) tree constructed from concatenated sequences of gene loci, for which neutral evolution was not rejected, namely ACT1, HSP104, RPB1, RPN2, TBP, and the synonymous sites in TPS1. For the strains containing heterozygous sites, one randomly selected haplotype was used in the analysis. Seven of the eight Chinese lineages and the five world lineages previously proposed were depicted in the NJ tree (Fig 1). The strains in Chinese lineage CHN IV were separated into two clades in this study, which should be due to the used gene loci different from previous study for phylogenetic analysis [11]. In addition to the previous reported lineages, three other clades with strong bootstrap values (>92%) were identified and designated as Clade 1, Clade2, and Clade 3 (Fig 1). The Clade 1 and Clade2 strains were isolated from fruits in northern China and western China, respectively, while the Clade3 strains constituted beer yeasts from England (Table 1). Structure analysis supported the general groupings of the strains (S1 Fig). Additional gene loci or genome sequences are needed to estimate if the three Clades are novel S. cerevisiae lineages.
Fig 1. Neighbor-joining tree constructed from concatenated sequences of gene loci, for which neutral evolution was not rejected.
Bootstrap percentages over 50% from 1000 bootstrap replicates were shown. Saccharomyces paradoxus was used as the out-group. Light grey shadow denotes the world lineages, tawny shadow denotes the Chinese lineages, and purple shadow denotes the Clades proposed in this study.
Population stress tolerance
The strains used for stress tolerance analysis include 71 Chinese natural isolates and 12 industrial strains (Table 1). Growth lag, growth rate and growth efficiency are the key variables of cellular growth dynamics [31]. Growth dynamics of representative strains showed a longer growth lag, a smaller growth rate, and lower growth efficiency under stress conditions compared with the controls without stress during the tests for 116 hours, indicating the feasibility of these variables used to reflect stress tolerance (S2 Fig). The growth efficiency (biomass) for 72 h was used to infer stress tolerance in this study. Because ethanol and heat stress lead to cell aggregation and flocculation, which might cause a biased and unreliable measurement of cell density, biomass dry weight of cultures instead of optical density was determined. Bivariate correlation analysis between the data in duplicate represented a high correlation at p<0.001 for each stressors (Heat, Pearson r = 0.726; ethanol, Pearson r = 0.83; osmotic stress, Pearson r = 0.905) supporting the reliability of the measurement.
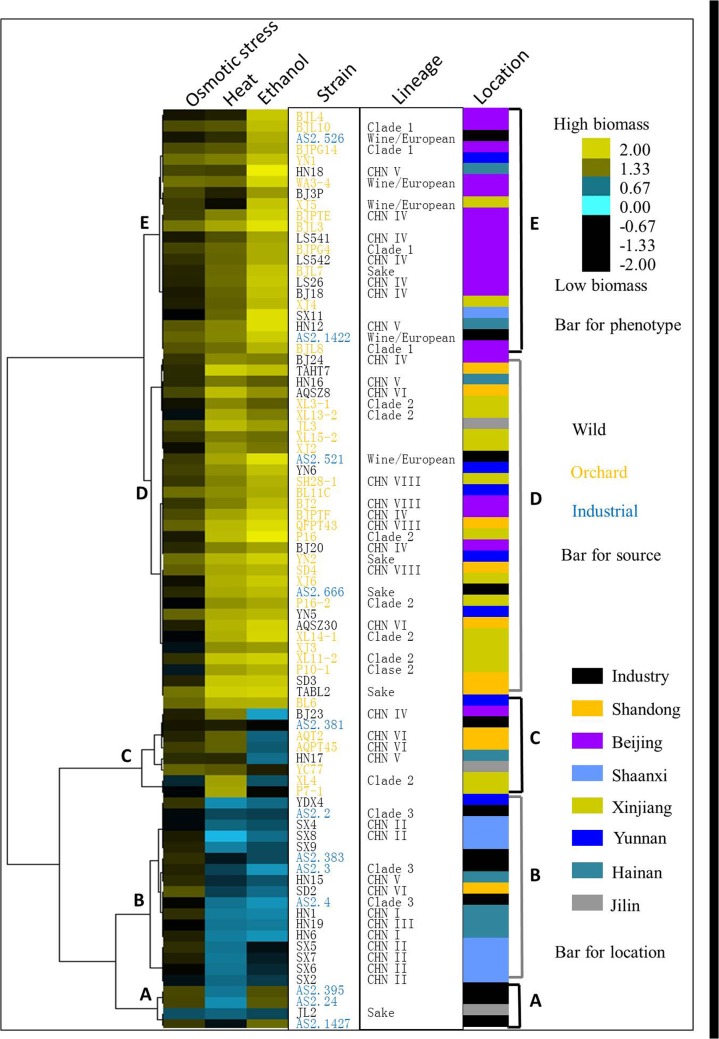
Hierarchical clustering of the 83 strains of S. cerevisiae was performed according to biomass dry weight data (Fig 2). Five clusters were recognized and designated as A to E. Most strains (40/60) in an identical genetic lineage were grouped in an identical phenotypic cluster. All isolates in lineages CHN I, CHN II, CHN III, and Clade 3 gathered in cluster B; all isolates in lineage CHN VIII and Clade 1 gathered in cluster D and E, respectively; The Clade 2 isolates excluding XL4 were placed in cluster D. Varied stress tolerance was shown in lineages CHN IV, CHN V and CHN VI, in which the isolates were placed in clusters B to E (Fig 2). These findings indicated correspondence between genotype and phenotype in lineages of S. cerevisiae.
Fig 2. Stress tolerance variations in the S. cerevisiae isolates.
Hierarchical clustering of phenotypes was performed using a centered Pearson correlation metric and average linkage mapping. Biomass of strains cultured for 72h in stressful conditions was used to infer stress tolerance. Locations of the isolates were also depicted. The industrial strains from multiple locations were not subdivided.
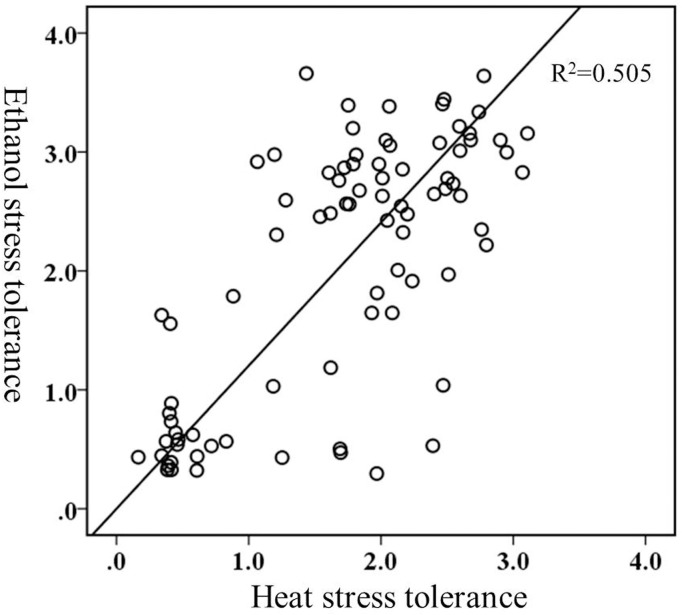
The range of biomass variations under stresses of ethanol, heat, and osmotic stress were 0.3 to 3.7 mg, 0.17 to 3.1 mg, and 0.54 to 1.9 mg, respectively. These data displayed a larger variation on tolerance to ethanol or heat compared with osmotic stress in S. cerevisiae populations (Fig 2). Bivariate correlation analysis represented a high correlation (Pearson r = 0.71 at p<0.0001) between ethanol and heat stress tolerance (Fig 3). This finding was similar (Pearson r = 0.51 at p<0.0006) to that deduced from testing a panel of strains from vineyard, sake and clinical environments [15]. Overlapping stress responses could be induced by heat and ethanol shock, such as causing similar changes to plasma membrane protein composition and inducing heat shock proteins that contribute to both thermal and ethanol tolerance [32]. These phenomena can be explained by environmental modification of S. cerevisiae via fermentation and the subsequent selection pressures imposed on them by the production of both ethanol and heat together [33,34].
Fig 3. Pearson correlation between ethanol and heat stress tolerance determined by bivariate correlation analysis.
Ethanol and heat stress tolerance was inferred from the biomass (gram) of 3 ml cultures grown for 72h under ethanol or heat stress.
Influences of environment on stress tolerance variations
To evaluate the impact of ecological source on stress tolerance variations of S. cerevisiae strains, the 83 strains were categorized into three groups based on ecological sources, namely Orchard, Wild, and Industrial (Table 1). Stress tolerance was compared between the three groups. Significantly higher levels of tolerance to ethanol (ANOVA, at the 0.05 level) and heat stress (Kruskal-Wallis Test, at the 0.05 level) presented in the Orchard group compared with the Wild and Industrial groups, indicating the influence of ecological source on phenotypic variations of S. cerevisiae strains (Fig 4A and 4B). Tolerance to osmotic stress was comparable among the three groups (Fig 4C). Unexpectedly, the Industrial group did not overall generate a high stress tolerance level. This is because the Clade 3 and mosaic strains in the Industrial group represented stress sensitivity, though the Sake and Wine/European strains from distilled spirit in the Industrial group showed tolerance to ethanol and heat stress (Table 1 and Fig 2).
Fig 4. Boxplots depicting stress tolerance variations between the ecological groups.
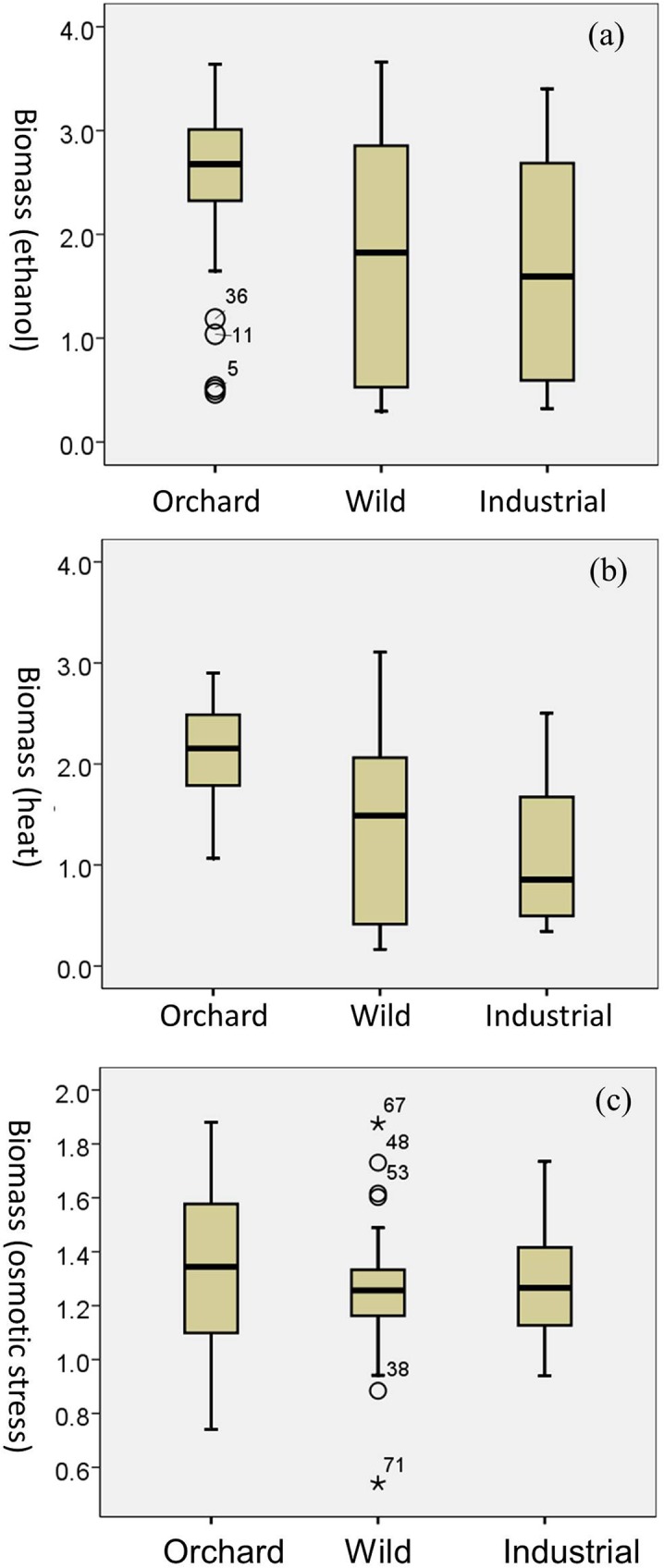
(a) ethanol stress; (b) heat stress; (c) osmotic stress. The number of strains in the Orchard, Wild, and Industrial group was 37, 34, and 12, respectively. The box contains the middle 50% of the data. Horizontal line inside the box: median; Upper boundary of whisker: largest observed value that is not an outlier; Lower boundary of whisker: smallest observed value that is not outlier. ○ = values >1.5 box-lengths from the box but not extremes (outliers). * Values >3 box-lengths from the box (extremes).
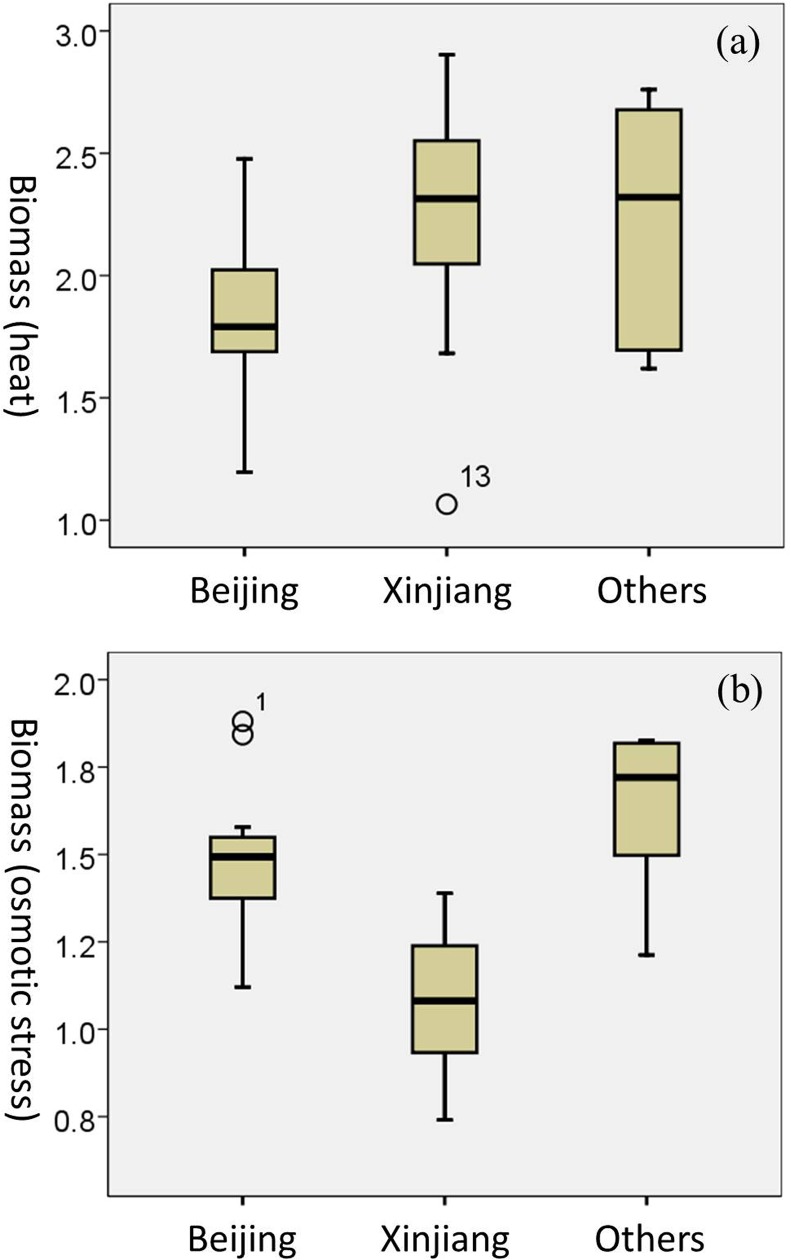
To investigate influences of geographic location on stress tolerance variations, strains in an identical ecological group were divided into sub-groups based on geographic locations (provinces) (Table 1 and S3 Fig). Significant differences (ANOVA, at the 0.05 level) presented among the sub-groups in both the Wild group and the Orchard group (Fig 5 and Fig 6). For example, the Wild strains isolated from Beijing and Shandong province displayed a significantly (ANOVA, at the 0.01 level) higher level of tolerance to ethanol and heat stress compared with those Wild strains (lineage CHN II) from Shaanxi province (Fig 5). The Pearson correlation coefficient (r) to the variability of ethanol and heat stress was 0.51 and 0.77, from which 25.67% and 59.98% of the variability could be explained by geographic locations, respectively. The Orchard isolates from Xinjiang province presented significantly (ANOVA, at the 0.05 level) higher sensitivity to osmotic stress compared with those Orchard isolates in other sub-groups (Fig 6). The Pearson correlation coefficient to the variability of osmotic stress was 0.78, from which 60.73% of the variability could be explained by geographic locations. These results indicated the correlation between phenotypic variations of S. cerevisiae strains and geographic location.
Fig 5. Boxplots depicting stress tolerance variations in the Wild group.
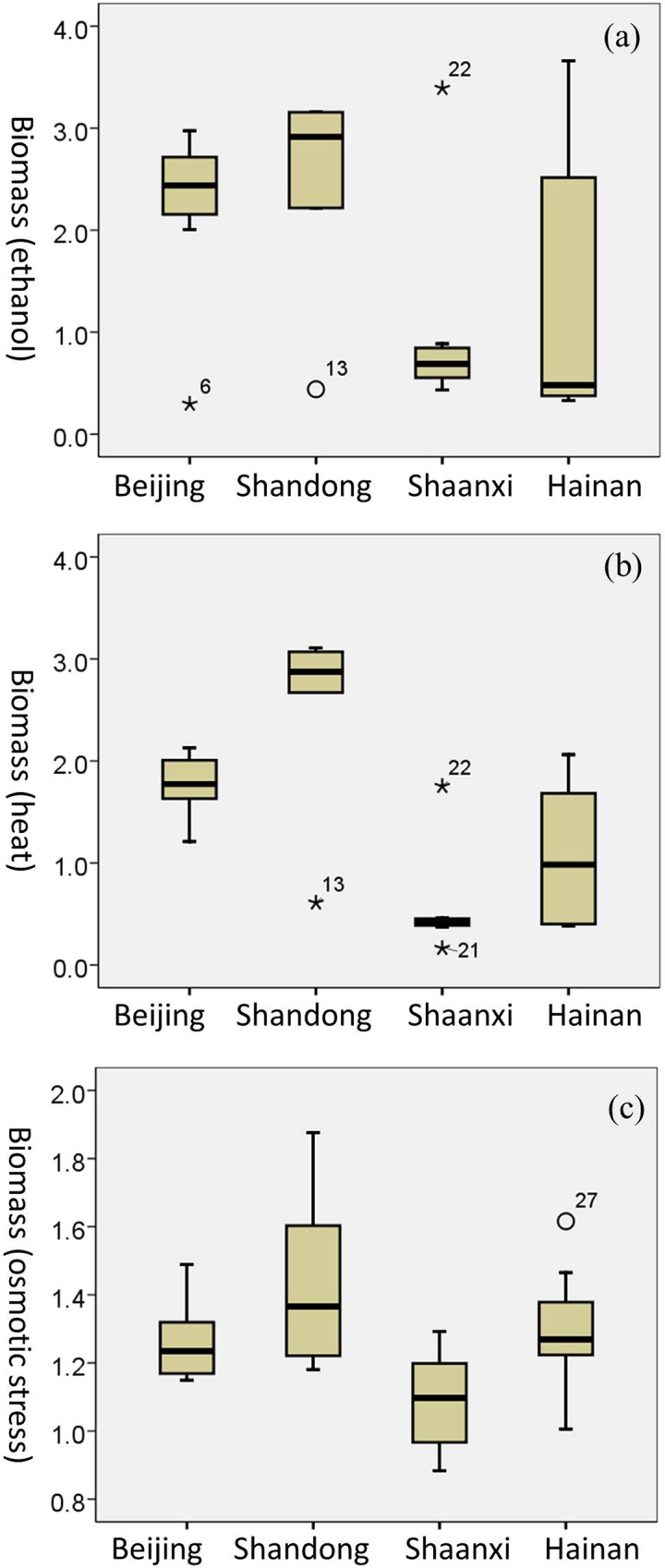
(a) ethanol stress; (b) heat stress; (c) osmotic stress. ○ = values >1.5 box-lengths from the box but not extremes (outliers). * Values >3 box-lengths from the box (extremes).
Fig 6. Boxplots depicting stress tolerance variations in the Orchard group.
(a) heat stress and (b) osmotic stress. Ethanol stress data was not shown because no significant difference was presented among sub-groups. ○ = values >1.5 box-lengths from the box but not extremes (outliers). * Values >3 box-lengths from the box (extremes).
Overall, both ecological source and geographic location showed close correlation to the phenotypic variations of S. cerevisiae populations. In the evolutionary history, S. cerevisiae has interacted with insects and/or human activities, which may help to shape the ecology and geography of the yeast [35,37]. In this study, most of the isolates from human-associated environments possess higher levels of stress tolerance compared with those from settings spared anthropogenic influences. The lineages from Shandong province and Beijing areas (CHN VIII and Clade 1) showed high levels of stress tolerance, while the lineage from Shaanxi province (CHN II) and most isolates from Hainan province (CHN V) were sensitive to stressors (Fig 2 and S3 Fig). Beijing, Shandong and Shaanxi provinces are all located in northern China and have a temperate climate (S3 Fig). Hainan province is a tropical island in southern China. This information indicated that climatic zones are not closely related to stress tolerance variations in S. cerevisiae. The strains from Shaanxi (CHN II) and Hainan province (CHN V) were all isolated in primeval forests spared anthropogenic influences, like its closest wild relative, S. paradoxus [11,37,38]. The strains from Shandong province and Beijing areas (CHN VIII and Clade 1) were isolated in orchards, which are human associated environments. It seems that human activities have exerted strong influences on stress tolerance variations in S. cerevisiae. This viewpoint achieved supports from this study and previous publications [11,36,37,39–41]. The evidence from study of Chinese lineages was described as follows. First, S. cerevisiae population migration was previously observed between different Chinese geographic locations and from primeval forests to human-associated environments [11]. Second, the Orchard group represented significantly higher level of tolerance to ethanol and heat stress than the Wild group in this study (Fig 4). Third, the Chinese lineage CHN VIII displaying a high level of stress tolerance consisted of strains isolated from orchards in three different Chinese geographic regions, while the stress sensitive Chinese lineages CHN I, II, or III consisted of strains from primeval forests (Table 1, Figs 1 and 2). Fourth, the domestic Sake and Wine/European lineages also contained Chinese isolates and most of them represented higher levels of stress tolerance than those from primeval forests (Figs 1 and 2).
Industrial bioethanol yeast strains are generally considered possessing strong tolerance to stressors. Nevertheless, there are natural S. cerevisiae strains can outcompete industrial bioethanol strains in tolerance to specific stressors [42,43]. In our previous study, we found that the strain JZ1C in Orchard group showed high tolerance to heat and this strain could efficiently converse inulin into ethanol at 40°C [44]. These findings indicated the possibility to isolate novel superior S. cerevisiae strains for ethanol fermentation from nature.
Conclusions
In this study, we compared the stress tolerance of diverse S. cerevisiae strains and estimated the influence of ecology and geography on variations of stress tolerance in the yeast. Both ecological sources and geographical locations presented significant correlations with variations of stress tolerance, which might be explained by the influence of human activities on the evolutionary history of S. cerevisiae. Overall, the isolates from human-associated environments represented a higher level of stress tolerance compared with those from forests spared anthropogenic influences. This study provides guidelines for selection of robust S. cerevisiae strains for bioethanol production from nature.
Supporting Information
Admixture model with correlated allele frequencies was used. Both the length of burnin and the MCMC repetitions after burnin were 100,000. Each strain is represented by a single vertical line broken into K colored segments, with lengths proportional to each of the K inferred clusters.
(DOCX)
YMM cultures of 3 ml were weighed at each selected time point. Stressors: ethanol, 10% (v/v); heat, 42°C; osmotic stress, 2 M of KCl. All of the experiments were performed in triplicate. Standard deviations (error bars) are shown.
(DOCX)
A, Xinjiang province in Northwest China; B, Shaanxi province in the Loess Plateau; C, Yunnan province in Southwest China; D, Hainan province in Maritime South China; E, Shandong province in North China Plain; F, The suburb of Beijing in North China Plain; G, Jilin province in Northeast China.
(DOCX)
(DOCX)
Acknowledgments
We thank Prof. Feng-Yan Bai in Institute of Microbiology, Chinese Academy of Sciences, for providing wild and orchard S. cerevisiae strains. We also thank Prof. Fu-Li Li in Qingdao Institute of BioEnergy and Bioprocess Technology, Chinese Academy of Sciences, for proofreading the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the National Natural Science Foundation of China (No. 30900007) and Scientific Research Foundation of Shandong University of Science and Technology for Recruited Talents (2015RCJJ054).
References
- 1. Cavalieri D, McGovern PE, Hartl DL, Mortimer R, Polsinelli M (2003) Evidence for S. cerevisiae fermentation in ancient wine. J Mol Evol 57: S226–232. [DOI] [PubMed] [Google Scholar]
- 2. Nevoigt E (2008) Progress in metabolic engineering of the yeast Saccharomyces cerevisiae . Microbiol Mol Biol Rev 72: 379–412. 10.1128/MMBR.00025-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lau MW, Dale BE (2009) Cellulosic ethanol production from AFEX-treated corn stover using Saccharomyces cerevisiae 424A(LNH-ST). Proc Natl Acad Sci USA 106:1368–1373. 10.1073/pnas.0812364106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Attfield PV (1997) Stress tolerance: the key to effective strains of industrial baker's yeast. Nat Biotechnol 15:1351–1357. [DOI] [PubMed] [Google Scholar]
- 5. Tao X, Zheng D, Liu T, Wang P, Zhao W, Zhu MY et al. (2012) A novel strategy to construct yeast Saccharomyces cerevisiae strains for very high gravity fermentation. PLoS ONE 7(2): e31235 10.1371/journal.pone.0031235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao XQ, Bai FW (2009) Mechanism of yeast stress tolerance and its manipulation for efficient ethanol production. J Biotechnol 144:23–30. 10.1016/j.jbiotec.2009.05.001 [DOI] [PubMed] [Google Scholar]
- 7. Naumov GI, Naumova ES, Sniegowski PD (1998) Saccharomyces paradoxus and Saccharomyces cerevisiae are associated with exudates of North American oaks. Can J Microbiol 44:1045–1050. [PubMed] [Google Scholar]
- 8. Sniegowski PD, Dombrowski PG, Fingerman E (2002) Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res 1:299–306. [DOI] [PubMed] [Google Scholar]
- 9. Fay JC, Benavides JA (2005a) Evidence for domesticated and wild populations of Saccharomyces cerevisiae . PLoS Genet 1:66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Landry CR, Townsend JP, Hartl DL, Cavalieri D (2006) Ecological and evolutionary genomics of Saccharomyces cerevisiae . Mol Ecol 15:575–591. [DOI] [PubMed] [Google Scholar]
- 11. Wang QM, Liu WQ, Liti G, Wang SA, Bai FY (2012) Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Mol Ecol 21:5404–5417. [DOI] [PubMed] [Google Scholar]
- 12. Liti G, Carter DM, Moses AM, Warringer J, Parts L, Stephen A (2009) Population genomics of domestic and wild yeasts. Nature 458:337–341. 10.1038/nature07743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. (2009) Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae . Nature 458:342–345. 10.1038/nature07670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mortimer RK, Romano P, Suzzi G, Polsinelli M (1994) Genome renewal: a new phenomenon revealed from a genetic study of 43 strains of Saccharomyces cerevisiae derived from natural fermentation of grape musts. Yeast 10: 1543–1552. [DOI] [PubMed] [Google Scholar]
- 15. Kvitek DJ, Will JL, Gasch AP (2008) Variations in stress sensitivity and genomic expression in diverse S. cerevisiae isolates. PLoS Genet 4:e1000223 10.1371/journal.pgen.1000223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fay JC, McCullough HL, Sniegowski PD, Eisen MB (2004) Population genetic variation in gene expression is associated with phenotypic variation in Saccharomyces cerevisiae . Genome Biol 5: R26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Will JL, Kim HS, Clarke J, Painter JC, Fay JC, Gasch AP (2010) Incipient balancing selection through adaptive loss of aquaporins in natural Saccharomyces cerevisiae populations. PLoS Genet 6(4): e1000893 10.1371/journal.pgen.1000893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bell W, Klaassen P, Ohnacker M, Boller T, Herweijer M, Schoppink P, et al. (1992) Characterization of the 56-kDa subunit of yeast trehalose-6-phosphate synthase and cloning of its gene reveal its identity with the product of CIF1, a regulator of carbon catabolite inactivation. Eur J Biochem 209:951–959. [DOI] [PubMed] [Google Scholar]
- 19. Sanchez Y, Taulien J, Borkovich KA, Lindquist S (1992) Hsp104 is required for tolerance to many forms of stress. EMBO J 11:2357–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alexandre H, Ansanay-Galeote V, Dequin S, Blondin B (2001) Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae . FEBS Letters 498:98–103. [DOI] [PubMed] [Google Scholar]
- 21. Hu XH, Wang MH, Tan T, Li JR, Yang H, Leach L, et al. (2007) Genetic dissection of ethanol tolerance in the budding yeast Saccharomyces cerevisiae . Genetics 175:1479–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ayoub MJ, Legras JL, Saliba R, Gaillardin C (2006) Application of Multi Locus Sequence Typing to the analysis of the biodiversity of indigenous Saccharomyces cerevisiae wine yeasts from Lebanon. J Appl Microbiol 100:699–711. [DOI] [PubMed] [Google Scholar]
- 23. Diezmann S, Dietrich FS (2009) Saccharomyces cerevisiae: Population divergence and resistance to oxidative stress in clinical, domesticated and wild isolates. PLoS ONE 4:e5317 10.1371/journal.pone.0005317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175. [DOI] [PubMed] [Google Scholar]
- 25. McDonald JH, Kreitman M (1991) Adaptive protein evolution at the Adh locus in Drosophila . Nature 351:652–654. [DOI] [PubMed] [Google Scholar]
- 26. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 27. Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rosenberg NA (2004) Distruct: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138. [Google Scholar]
- 29. Eisen M, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fay JC, Benavides JA (2005. b) Hypervariable noncoding sequences in Saccharomyces cerevisiae . Genetics 170:1575–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Warringer J, Anevski D, Liu B, Blomberg A (2008) Chemogenetic fingerprinting by analysis of cellular growth dynamics. BMC Chem Biol 8:3 10.1186/1472-6769-8-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Piper PW (1995) The heat shock and ethanol stress responses of yeast exhibit extensive similarity and functional overlap. FEMS Microbiol Lett 134: 121–127. [DOI] [PubMed] [Google Scholar]
- 33. Thomson JM, Gaucher EA, Burgan MF, DeKee DW, Li T, Aris JP, et al. (2005) Resurrecting ancestral alcohol dehydrogenases from yeast. Nat Genet 37:630–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goddard MA (2008) Quantifying the complexities of Saccharomyces cerevisiae's ecosystem engineering via fermentation. Ecology 89:2077–2082. [DOI] [PubMed] [Google Scholar]
- 35. Stefanini I, Dapporto L, Legras JL, Calabretta A, Di Paola M, De Filippo C, et al. (2012) Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc Natl Acad Sci USA 109: 13398–13403. 10.1073/pnas.1208362109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Warringer J, Zörgö E, Cubillos FA, Zia A, Gjuvsland A, Simpson JT, et al. (2011) Trait variation in yeast is defined by population history. PLoS Genet 7:e1002111 10.1371/journal.pgen.1002111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koufopanou V, Hughes J, Bell G, Burt A (2006) The spatial scale of genetic differentiation in a model organism: the wild yeast Saccharomyces paradoxus . Trans R Soc Lond B Biol Sci 361:1941–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kuehne HA, Murphy HA, Francis CA, Sniegowski PD (2007) Allopatric divergence, secondary contact, and genetic isolation in wild yeast populations. Curr Biol 17:407–411. [DOI] [PubMed] [Google Scholar]
- 39. Cromie GA, Hyma KE, Ludlow CL, Garmendia-torres C, Gilbert TL, May P, et al. (2013) Genomic sequence diversity and population structure of Saccharomyces cerevisiae assessed by RAD-seq. G3 3(12): 2163–2171. 10.1534/g3.113.007492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Legras JL, Merdinoglu D, Cornuet JM, Karst F (2007) Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol Ecol 16: 2091–2102. [DOI] [PubMed] [Google Scholar]
- 41. Strope PK, Skelly DA, Kozmin SG, Mahadevan G, Stone EA, Magwene PM, et al. (2015) The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res 25:762–774. 10.1101/gr.185538.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mukherjee V, Steensels J, Lievens B, Van de Voorde I, Verplaetse A, Aerts G, et al. (2014) Phenotypic evaluation of natural and industrial Saccharomyces yeasts for different traits desirable in industrial bioethanol production. Appl Microbiol Biotechnol 98:9483–9498. 10.1007/s00253-014-6090-z [DOI] [PubMed] [Google Scholar]
- 43. Wimalasena TT, Greetham D, Marvin ME, Liti G, Chandelia Y, Hart A, et al. (2014) Phenotypic characterisation of Saccharomyces spp. yeast for tolerance to stresses encountered during fermentation of lignocellulosic residues to produce bioethanol. Microb Cell Fact 13:47 10.1186/1475-2859-13-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hu N, Yuan B, Sun J, Wang SA, Li FL (2012) Thermotolerant Kluyveromyces marxianus and Saccharomyces cerevisiae strains representing potentials for bioethanol production from Jerusalem artichoke by consolidated bioprocessing. Appl Microbiol Biotech 95:1359–1368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Admixture model with correlated allele frequencies was used. Both the length of burnin and the MCMC repetitions after burnin were 100,000. Each strain is represented by a single vertical line broken into K colored segments, with lengths proportional to each of the K inferred clusters.
(DOCX)
YMM cultures of 3 ml were weighed at each selected time point. Stressors: ethanol, 10% (v/v); heat, 42°C; osmotic stress, 2 M of KCl. All of the experiments were performed in triplicate. Standard deviations (error bars) are shown.
(DOCX)
A, Xinjiang province in Northwest China; B, Shaanxi province in the Loess Plateau; C, Yunnan province in Southwest China; D, Hainan province in Maritime South China; E, Shandong province in North China Plain; F, The suburb of Beijing in North China Plain; G, Jilin province in Northeast China.
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.