Abstract
The Anopheles genus is a member of the Culicidae family and consists of approximately 460 recognized species. The genus is composed of 7 subgenera with diverse geographical distributions. Despite its huge medical importance, a consensus has not been reached on the phylogenetic relationships among Anopheles subgenera. We assembled a comprehensive dataset comprising the COI, COII and 5.8S rRNA genes and used maximum likelihood and Bayesian inference to estimate the phylogeny and divergence times of six out of the seven Anopheles subgenera. Our analysis reveals a monophyletic group composed of the three exclusively Neotropical subgenera, Stethomyia, Kerteszia and Nyssorhynchus, which began to diversify in the Late Cretaceous, at approximately 90 Ma. The inferred age of the last common ancestor of the Anopheles genus was ca. 110 Ma. The monophyly of all Anopheles subgenera was supported, although we failed to recover a significant level of statistical support for the monophyly of the Anopheles genus. The ages of the last common ancestors of the Neotropical clade and the Anopheles and Cellia subgenera were inferred to be at the Late Cretaceous (ca. 90 Ma). Our analysis failed to statistically support the monophyly of the Anopheles genus because of an unresolved polytomy between Bironella and A. squamifemur.
Introduction
Malaria is a vector-borne disease that is transmitted by Anopheles mosquitoes infected with Plasmodium protozoans. Although declining in incidence, 207 million cases of this tropical malady were estimated in 2012, with more than 620,000 casualties; most of these were in the African continent (www.who.int/gho/malaria). The decline has been associated with the implementation of effective protective measures for exposed individuals and with a reduction in the longevity and density of the vector mosquito population (WHO, World Malaria Report 2012).
The mosquito genus Anopheles belongs to the subfamily Anophelinae, family Culicidae [1], a monophyletic group supported by molecular phylogenetics [2, 3] and numerous distinct morphological features [4]. The genus currently harbors 465 recognized species that are allocated across seven subgenera based on the number and position of the specialized setae on the male genitalia. Nevertheless, the diversity and the geographical distribution of the species assigned to each Anopheles subgenus vary tremendously [1].
The two largest subgenera, for instance, include 86% of the diversity of the genus Anopheles: the cosmopolitan Anopheles with 182 species and the Old-World Cellia with 220 species. Four smaller subgenera are restricted to the Neotropics: Kerteszia with 12 species, Lophopodomyia with six species, Nyssorhynchus with 39 species and Stethomyia with five species. Finally, the monotypic Baimaia subgenus has only recently been described to include the type species A. kyondawensis, which is restricted to the Oriental region [5]. The four largest subgenera contain clades of the 40 species that have been identified as dominant malaria vectors [6].
Molecular and morphological studies have generally supported the monophyletic status of the subgenera Cellia, Nyssorhynchus and Kerteszia [2–4]. Additionally, a close association has been consistently found between two neotropical subgenera, Nyssorhynchus and Kerteszia [3, 4, 7]. Nevertheless, the relationship between this neotropical clade with the other neotropical subgenera (Lophopodomyia and Stethomyia) has not been properly tested as many studies lacked the necessary taxon sampling [2, 3, 8].
One exception was the study of Sallum et al. [7], which used molecular markers that included all major Anopheles subgenera and approximately 30 Anophelinae species. Nevertheless, in that study, the monophyletic status of Nyssorhynchus was challenged for the first time as Kerteszia grouped within that clade. Additionally, the study indicated as tentative the placement of the neotropical Stethomyia (grouped to the non-tropical Cellia subgenus in the phylogeny) as this taxon was represented by one species sequenced for a single marker. Because the statistical support for the clades in the study was generally low, it remains to be determined whether a clearer picture emerges if taxon and marker sampling is incremented, particularly for the neotropical Stethomyia.
In this study, we used a comprehensive phylogeny with 50 species representing all major subgenera to unveil the neotropical diversification in the genus Anopheles. As the number and the time of colonizations to the continent are critical for a well-defined diversification picture, we also included time tree and ancestral area reconstruction analyses that allowed us to make biogeographical considerations regarding the colonization of the Neotropics by ancestral anophelines. Our most prominent result is the statistical support for a new clade composed of the subgenera Stethomyia, Kerteszia and Nyssorhynchus that is confined to the Neotropical region.
Materials and Methods
Taxon sampling and molecular markers
We included 47 Anopheles species representing six out of the seven subgenera. From each subgenus, all species that are considered to be dominant vectors for malaria were included in the dataset to refine their phylogenetic position. The classification of the species in subgenera followed [1]. As the monophyletic status of Anopheles has been previously questioned, we have also included species of the other two Anophelinae genera: Chagasia bathana and Bironella hollandi. The topology was rooted using Aedes aegypti sequences. Phylogenetic analyses were conducted using three molecular markers. Two markers were mitochondrial, the cytochrome oxidase subunits I and II (COI and COII), and one was nuclear, the ribosomal 5.8S subunit. Molecular markers were selected based on the availability of sequences for a large portion of the Anopheles diversity, avoiding an incomplete supermatrix. Sequence data were downloaded from GenBank, and accession numbers are provided in Table 1.
Table 1. GenBank accession numbers of sequences.
| Subgenus | Species | COI | COI | COII | 5.8S |
|---|---|---|---|---|---|
| Anopheles | Anopheles atroparvus | - | - | - | AY634533 |
| Anopheles | Anopheles barbirostris | AY729982 | EU797194 | AB331589 | EU812783 |
| Anopheles | Anopheles freeborni | - | AF417717 | AF417753 | - |
| Anopheles | Anopheles labranchiae | HQ860331 | - | - | AY365008 |
| Anopheles | Anopheles lesteri | EU699001 | - | EU699056 | AF384172 |
| Anopheles | Anopheles messeae | HE659586 | - | AY953352 | AF504213 |
| Anopheles | Anopheles pseudopunctipennis | HM022407 | AF417721 | AF417757 | U49735 |
| Anopheles | Anopheles quadrimaculatus | NC_000875 | NC_000875 | NC_000875 | U32503 |
| Anopheles | Anopheles sacharovi | AY135694 | - | - | AF462088 |
| Anopheles | Anopheles sinensis | AY768950 | HM488283 | AF325715 | HM590510 |
| Cellia | Anopheles aconitus | DQ000253 | - | AF194448 | DQ000252 |
| Cellia | Anopheles annularis | AY917197 | - | EU620675 | DQ478878 |
| Cellia | Anopheles arabiensis | AF252877 | AF417705 | AF417741 | DQ287723 |
| Cellia | Anopheles balabacensis | - | - | U94289 | - |
| Cellia | Anopheles culicifacies | - | AF116834 | HQ377221 | AY168883 |
| Cellia | Anopheles dirus | - | AJ877310 | AJ877309 | U60410 |
| Cellia | Anopheles farauti | HQ840792 | HQ840792 | DQ674709 | HM584396 |
| Cellia | Anopheles flavirostris | - | AY943650 | AJ512742 | GU062188 |
| Cellia | Anopheles fluviatilis | GQ906980 | AF116830 | AJ512740 | GQ857445 |
| Cellia | Anopheles funestus | NC_008070 | NC_008070 | NC_008070 | JN994135 |
| Cellia | Anopheles gambiae | NC_002084 | NC_002084 | NC_002084 | NW_163551 |
| Cellia | Anopheles koliensis | HQ840838 | HQ840838 | U94304 | EF042756 |
| Cellia | Anopheles latens | - | DQ897936 | - | - |
| Cellia | Anopheles maculatus | JN596972 | - | AF448468 | AY803346 |
| Cellia | Anopheles melas | DQ792579 | DQ792579 | DQ792579 | GQ870314 |
| Cellia | Anopheles merus | - | - | - | GQ870313 |
| Cellia | Anopheles minimus | GQ259180 | AF116832 | AF194452 | DQ336436 |
| Cellia | Anopheles moucheti | - | DQ069721 | DQ069719 | - |
| Cellia | Anopheles nili | - | DQ069722 | DQ069720 | - |
| Cellia | Anopheles punctulatus | HQ840857 | HQ840857 | U94312 | HM584446 |
| Cellia | Anopheles sergentii | - | - | - | AY533851 |
| Cellia | Anopheles stephensi | FJ210893 | AF417713 | AY949851 | EU847233 |
| Cellia | Anopheles subpictus | AF222327 | AF417711 | EF601864 | GQ870328 |
| Cellia | Anopheles sundaicus | AF222324 | AF417712 | AF417748 | AF369559 |
| Cellia | Anopheles superpictus | - | AY900633 | FJ526436 | DQ487148 |
| Kerteszia | Anopheles bellator | - | - | AF417740 | DQ364652 |
| Kerteszia | Anopheles homunculus | JQ291235 | - | - | JQ291246 |
| Kerteszia | Anopheles lepidotus | JQ041282 | - | - | JN967765 |
| Lophopodomyia | Anopheles squamifemur | - | AF417723 | AF417759 | - |
| Nyssorhnchus | Anopheles albimanus | - | AF417695 | AF417731 | L78065 |
| Nyssorhnchus | Anopheles albitarsis | HQ335344 | HQ335344 | HQ335344 | AF462385 |
| Nyssorhnchus | Anopheles aquasalis | - | AF417697 | AF417733 | DQ020123 |
| Nyssorhnchus | Anopheles darlingi | NC_014275 | NC_014275 | NC_014275 | GU477277 |
| Nyssorhnchus | Anopheles marajoara | DQ076216 | DQ076216 | AF417735 | AY028127 |
| Nyssorhnchus | Anopheles nuneztovari | AF368065 | AF270915 | AF417736 | HQ020405 |
| Stethomyia | Anopheles acanthotorynus | - | AF417724 | AF417760 | - |
| Stethomyia | Anopheles nimbus | HM022409 | HM022409 | - | - |
| Outgroup | Aedes aegypti | NC_010241 | NC_010241 | NC_010241 | M95126 |
| Outgroup | Bironella hollandi | - | - | EU477545 | EF619445 |
| Outgroup | Chagasia bathana | AF417726 | AF417726 | AF417762 | - |
Alignment and phylogenetic analysis
Each gene was aligned individually using the program MAFFT 7 [9]. Alignments were then inspected and edited in MEGA version 5.1 [10]. Individual alignments were then concatenated in the SeaView 4 program [11] assuming species-level monophyly. The final alignment matrix included 157 bp of the 5.8S marker, 525 bp of the first segment of COI and 562 bp of the second segment, and 684 bp of COII, summing to a total length of 1,928 bp. The final alignment is available online at the Dryad database and at www.edarwin.net/data/Anopheles. Two methods of phylogenetic reconstruction were implemented using the GTR+G+I substitution model as indicated by the jModelTest2 program [12]. The first was a Bayesian inference (BI) method conducted in the program MrBayes 3.2 [13]. The Markov Chain Monte Carlo (MCMC) algorithm was executed in two independent runs. Each run was sampled every 1,000th generation until 10,000 trees were obtained, with 25% excluded as burn-in. In this tree, the clade Bayesian posterior probability (BP) was used as a metric of topological support. Convergence of the chains was assessed via the potential scale reduction factor, which was close to 1.0 for all parameters, and the effective sample size (ESS), which was > 200 for all parameters. The second method was maximum likelihood (ML). In this case, the algorithm was implemented in the PhyML program package 3 [14], and the topological support test was the approximate likelihood ratio statistic, aLRT [15].
We have also investigated whether data partitioning would impact topological inference. Partitioning scheme was inferred with the PartitionFinder software [16] by searching through all substitution models and using the Bayesian information criterion (BIC) to choose between alternative models. Three data blocks were tested, namely, 5.8S, COI and COII, and the greedy search algorithm was used. The best partitioning scheme was composed of two partitions, a mitochondrial partition containing COI and COII, under the GTR+G+I model, and a single partition for 5.8S, under the K80+G model. Phylogenetic inference using the estimated partitioning scheme was conducted in MrBayes, using the same MCMC settings as above, and also in RAxML 8 [17], which implements a fast maximum likelihood topological search.
Divergence times and ancestral area reconstruction analyses
The molecular dating analysis was conducted in a Bayesian framework with the program BEAST 1.7.8 [18] that also uses a MCMC algorithm to infer the posterior distribution of the parameters. As in the phylogenetic inference, the elected model for nucleotide substitution was GTR+G+I. The prior distribution of the evolutionary rates along branches was modeled by the uncorrelated lognormal distribution, whereas the Yule process was adopted to model the tree prior. The MCMC run consisted of 100,000,000 generations with parameters sampled every 1,000th step. A burn-in period of 25,000 generations was discarded. The BEAST analysis was repeated twice to check for convergence, which was assessed by the potential scale reduction factor as implemented in the coda package of the R programming environment (www.r-project.org). ESSs were also calculated in Tracer 1.6, resulting in values > 200 for all parameters.
To decompose the branch lengths (i.e., genetic distances) into absolute times and evolutionary rates, calibration priors on node ages are required. Usually, these priors are obtained from the fossil record or from the mean evolutionary rate. As with most non-vertebrate taxa, however, the Anopheles fossil record is very scarce because only two Anopheles fossils are currently recognized. The oldest record is Anopheles (Nyssorhynchus) dominicanus from the Late Eocene (33.9–41.3 Ma) [19], and the most recent is Anopheles rottensis from the Late Oligocene (13.8–33.9 Ma) [20]. Nevertheless, the usage of these records as time priors has been deemed notably problematic. Although the A. domincanus fossil was assigned to subgenus Nyssorhynchus, the age of the fossil varies from 15 Ma to 45 Ma, depending on the dating technique applied [8]. Thus, as in many studies with mosquitoes, we have relied on the split between Aedes and Anopheles that has been estimated at 145 Ma (97.7–193.7) by Logue et al. [21], in which the timescale was calibrated using the estimate of the Drosophila-Anopheles divergence at 260 Ma obtained by Gaunt and Miles [22]. Thus, a Gaussian calibration prior with mean = 145 Ma and standard deviation = 25 Ma was adopted to accommodate the 97.7–193.7 range within the 95% highest probability density interval.
For the ancestral area reconstruction analysis, we first associated each terminal taxon to one of the following area categories: (1) Americas; (2) Africa; (3) Europe; (4) India plus West Asia; and (5) Southeast Asia plus the Pacific. Geographical areas were categorized according to Sinka et al. (2012), in which comprehensive distribution data for dominant malaria vectors was gathered and an Anopheles global map was created. Ancestral reconstruction was implemented using the ML method [23] available in the APE package [24] of the R programming environment. We also implemented the ancestral geographic range estimation method using the Lagrange software [25].
Results
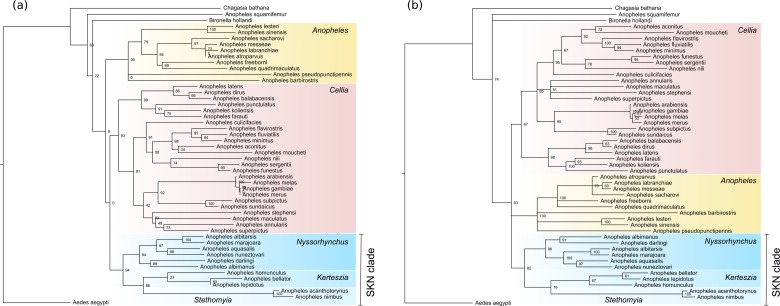
Statistical tests performed on our trees suggest a relatively high level of support for our main clades (Fig 1a and 1b). Phylogenetic relationships between major clades were, however, poorly supported. Our results indicate that five subgenera are monophyletic and most of the results are backed with statistical support: Anopheles (0.99 aLRT and 100% BP); Cellia (0.93 aLRT and 87% BP); Stethomyia (1.00 aLRT and 100% BP); Kerteszia (0.23 aLRT and 67% BP); and Nyssorhynchus (0.94 aLRT and 96% BP; Fig 1a and 1b). The monophyly of subgenus Lophopodomyia, however, remains to be tested, because only a single species was included (A. squamifemur). Data partitioning into nuclear and mitochondrial segments have not altered the topology of Anopheles evolution in both Bayesian and ML trees.
Fig 1. Phylogeny of the Anopheles genus.
(a) Maximum likelihood tree with aLRT statistical support. (b) Bayesian inference tree with clade posterior probabilities.
As well documented in previous studies, in our tree, we identified genus Chagasia as a sister lineage to the clade containing the genera Bironella and Anopheles [2, 7]. Additionally, a monophyletic classification for genus Anopheles was not supported in our trees (Fig 1a and 1b). In the BI tree, A. squamifemur was retrieved as the sister lineage of the large clade that joined the remaining Anopheles diversity plus Bironella hollandi (Fig 1a). This result has also been reported previously, and further supports the inclusion of Bironella as part of the greater Anopheles diversity (morphology Sallum et al. 2000; molecular Sallum et al. 2002, 2005). On the other hand, in the ML tree, the node was unresolved, joining A. squamifemur, B. hollandi and the remaining Anopheles spp. in a polytomy.
The paraphyletic status of Anopheles in our tree is possibly a result of misplacement of the root. This artifact could occur due to poor outgroup choice or low taxon sampling causing long-branch issues. On the other hand, a comprehensive morphological analysis also indicates the inclusion of Bironella lineages within Anopheles diversity. Furthermore, we report mixed results as our nodes were not statistically supported, and it would be important to include more sequences of Bironella and Chagasia to definitively settle this matter.
Due to its large diversity, taxonomists have also assigned the species of Anopheles into infrageneric categories such as sections, series, groups, subgroups and complexes. In our dataset, we have included many species from these to enable monophyletic tests of the categories. For instance, subgenera Anopheles and Nyssorhynchus are divided into sections. We included members of two sections of Anopheles, namely, Angusticorn and Laticorn. In this case, the status of these sections is non-resolved due to a polytomy. The polytomic node includes A. pseudopunctipennis (section Angusticorn), A. barbirotris (section Laticorn), and the other species of the Anopheles subgenus in which diversity is assembled into two monophyletic clades, each with the remaining diversity of each of these two sections. With regard to Nyssorhynchus, we also included species assigned to two sections, Albimanus and Argyritarsis, that were not recovered as monophyletic; A. albitarsis (section Albimanus) grouped with A. marajoara (section Argyritarsis) with a maximum level of support. The other two species of Albimanus (A. aquasalis and A. nuneztovari) also grouped, as did the remaining two species of Argyritarsis (A. darling and A. albimanus).
Less inclusive groups include the series in which Anopheles, Cellia and Nyssorhynchus are divided. Our dataset included members of two series of Anopheles, series Anopheles (section Angusticorn) and series Myzorhynchus (section Laticorn). Because a single series from each section was included, the same polytomy described above was also observed with regard to the series of Anopheles. The genus Cellia is represented by members of four series, Myzomyia, Neocellia, Neomyzomyia, and Pyretophorus, none of which are monophyletic. For instance, A. nili (series Neomyzomyia) grouped within the diversity of the series Myzomyia; the high bootstrap support and the fact that this grouping took place in both topologies indicate that these series are not natural groups. Additionally, A. subpictus (series Neocellia) and A. sundaicus (series Pyretophorus) were tightly grouped, rendering their series non-monophyletic groups. Apart from these examples, the remaining diversity of the series grouped for Neocellia (A. annularis, A. maculatus, A. superpictus and A. stephensi), for Neomyzomyia (A. balabacensis, A. dirus, A. farauti, A. koliensis, A. latens, and A. punctulatus) and for Pyretophorus (A. arabiensis, A. gambiae, A. melas, A. merus) were so grouped with high support. With regard to Nyssorhynchus, four series were included, Albimanus, Albitariss, Argyritarsis and Oswaldoi, but only two included more than a single species. These were series Albitarsis and Oswaldoi, in which both species grouped with high statistical support in our topology.
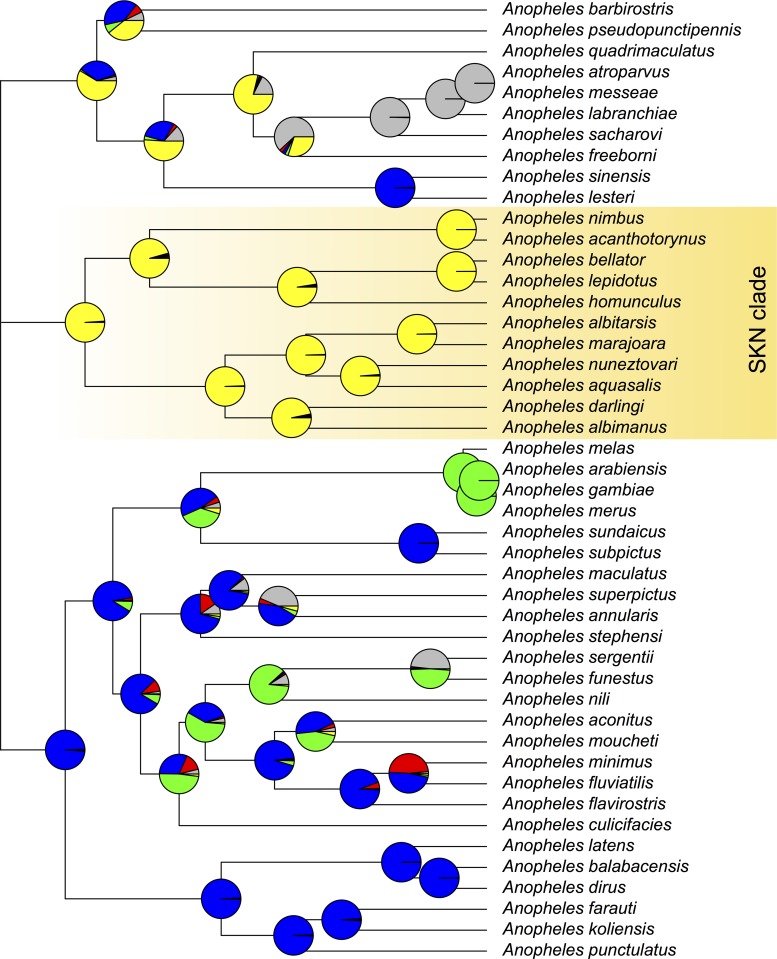
Ancestral area reconstruction presented a higher likelihood that the ancestor of the SKN clade was geographically distributed in the Americas (Fig 2), with 95.2% probability as estimated in Lagrange. An ancestral distribution in Southeast Asia plus the Pacific was favored for the Cellia subgenus, whereas the ancestor of the (A. melas, A. arabiensis, A. gambiae and A. merus) clade was distributed in Africa, with full support in both geographical analyses. Maximum support was also obtained for a Southeast Asia plus the Pacific ancestral distribution for the (A. latens, A. balabacensis, A. dirus, A. farauti, A. koliensis and A. punctulatus) and the (A. sinensis, A.lesteri) clades. Finally, the ancestral area of the (A. atroparvus, A. messeae, A. labranchiae and A. sacharovi) clade was inferred to be India plus West Asia with the highest likelihood. The ancestral geographical distribution of the remaining nodes was not fully resolved.
Fig 2. Ancestral area reconstruction conducted using the maximum likelihood method in the APE package.
Circles depict the relative probabilities of each region. Color codes are as follows: green—Africa; yellow—Americas; blue—Southeast Asia and the Pacific; gray—Europe plus Middle East; and red—India plus West Asia.
Discussion
Our analysis of ancestral geographical distribution is dubious with regard to the Anopheles clade, indicating that Neotropic and African distributions in the Early Cretaceous (113 Ma) are equally likely. The timescale inferred here is in agreement with a recent study of 16 Anopheles genomes, which also inferred the age of the last common ancestor (LCA) of Anopheles at ~100 Ma [26]. Previous studies have estimated more recent ages for the ancestor of Anopheles, suggesting a split for Anopheles and Cellia subgenera in the late Cretaceous, at 93.6 Ma [8] and 81 Ma [21], or even in the Eocene at 43.1 Ma [27]. Moreno et al. [8] and Logue et al. [21] also dated the age of the Nyssorhynchus subgenera at 79 Ma and 94 Ma, respectively. Our divergence time for the Nyssorhynchus lineage was not comparable with those studies as our analyses grouped these subgenera with Stethomyia and Kerteszia within the SKN clade.
A comparison between the evolutionary histories of the Anopheles genus and Plasmodium is meaningful. The phylogeny and timing of the evolution of Plasmodium has been the focus of several recent studies [28–31]. It is presumed that the evolution of Plasmodium species is strongly associated with host-switching events [29, 32, 33], which poses a major challenge for the elucidation of the Plasmodium phylogeny [33]. A total of five Plasmodium species are the main causes of malaria in humans: P. falciparum, P. vivax, P. ovale, P. malariae and P. knowlesi [34, 35]. Estimates of the divergence times for these species are much younger than those inferred for their Anopheles hosts in our analysis, although no consensus on Plasmodium divergence times has been reached [31, 33, 36].
In the Cellia subgenus, two major clades were clearly recognized; the first lineage separated a clade containing species from Southeast Asia and the Pacific from the remaining species. Additionally, the African group (A. gambiae, A. merus, A. melas and A. arabiensis) was conspicuously characterized by the small genetic distances between species. Within the Anopheles subgenus, ML and BI methods inferred different topological associations. In ML tree, a clade containing the predominantly European species, A. atroparvus, A. messeae, A. labranchiae and A. sacharovi, was well supported (aLTR 0.97), whereas the same clade presented 78% BP in the Bayesian tree. Moreover, a sister group relationship between A. quadrimaculatus and A. freeborni was not recovered in the BI tree. This phylogenetic relationship was recovered in studies that used the ITS2 ribosomal DNA marker [37–39], with minor discrepancies. Moreover, a sister group relationship between A. quadrimaculatus and A. freeborni was not recovered in Marinucci et al.’s [37] study.
Phylogenetic relationships within the Cellia subgenus are in general agreement with a recent genomic study by Neafsey et al. [26], who also found that the Southeast Asia plus the Pacific (A. dirus + A. farauti) clade consisted of the first lineage to diverge within this subgenus. Differences between our results and that of Neafsey et al. rely on the relationship of major subgenera. Neafsey et al. inferred the (Nyssorhynchus, (Anopheles, Cellia)) relationship, while we could not significantly resolve this higher-level evolutionary relationship within Anopheles. In this sense, Krzywinski [2, 3] also recovered the same relationship of Neafsey et al., whereas Sallum [7] estimated a different association, namely, (Cellia, (Nyssorhynchus, Anopheles)).
Specifically, our results are, however, at odds with Kamali et al. and Neafsey at al. with respect to the relationship between A. gambiae, A. stephensi, A. funestus and A. nili. Both works found a closer evolutionary affinity between A. funestus and A. stephensi, with A. gambiae as sister group, whereas we inferred a (A. funestus + A. nili) clade. Both studies, however, favored gene sampling instead of species sampling. Kamali et al. analyzed 49 genes, while Neafsey et al. studied more than 720,000 aminoacid sites. Recent analysis has shown that increasing taxonomic sampling, even with incomplete gene sampling, augments phylogenetic resolution [40]. Therefore, we expect that our analysis, although restricted in the number of loci, gained phylogenetic resolution by the increased sampling of taxa.
It is worth noting that the small genetic distances found in the African group (A. gambiae, A. merus, A. melas and A. arabiensis) are expected as a result of a complex speciation process permeated with recurrent introgressive hybridization events in the recent evolutionary past, as reported by Fontaine et al. [41]. Controlling and measuring the extent of introgression in phylogenetic reconstruction is not feasible by using a few molecular markers or by including a single representative individual per species. Such thorough investigation is only effective when a large sample of molecular markers and individuals per species is available. Unfortunately, this is not the case of Anopheles. With restricted sampling of loci and individuals, it is not feasible to distinguish between phylogenetic discordance caused by stochastic errors, incomplete lineage sorting and introgression. Thus, the extent of the phylogenetic errors due to introgression in Anopheles species groups may be unveiled in the future as the number of available genomes increases. Moreover, because of their large population sizes, inference of mosquito species phylogeny is expected to be difficult due to incomplete lineage sorting [42, 43].
The most important topological result of our study, however, is the clade composed of three subgenera of Anopheles ((Stethomyia, Kerteszia), Nyssorhynchus). This clade was recovered for the first time in our ML and BI analyses with a relatively high level of support (0.94 aLRT and 82% BP). In this study, we termed this new clade the SKN clade. The association between Nyssorynchus + Kerteszia has been previously found in molecular [2, 7] and morphological studies [4]. The relationship of the SKN clade with other Anopheles subgenera also presents a large discrepancy with other studies. Here, we have found weak support for the sister group relationship between SKN and Cellia. Previous studies have placed Cellia as a sister-group to the subgenera Anopheles, Lophopodomyia, Stethomyia and the genus Bironella [4]; Cellia was proposed to be the sister-group of the Anopheles subgenus [3], and it has also been associated with the subgenera Lophopodomyia, Nyssorhynchus, Kereszia and genus Bironella [7]. The ML topology of this work places the Old-World Cellia as a sister-group to the SKN Neotropical clade, although this grouping presented virtually no statistical support (Fig 1 a). The discrepancies found between different analyses are likely due to the choice and number of species sampled, which varied significantly among previous studies.
Within SKN, the divergence between subgenera Stethomyia and Kerteszia was estimated at ca. 70 Ma. Both methods of ancestral reconstruction of geography showed that the LCA of the SKN clade was American with a relative probability of 95.2%, which implies that the early radiation of the SKN subgenera took place in the Americas (Fig 2). The ages of the LCA of subgenera Anopheles, Cellia and the SKN clade were all inferred to be in the Late Cretaceous at ca. 90 Ma. The geographic distribution of the ancestor of the Cellia subgenera was likely in the Southeast Asia and the Pacific region. The ancestral area of the remaining subgenera-level splits could not be resolved with high probability. In conclusion, our results support the concept that the Neotropical subgenera Stethomyia + Kerteszia + Nyssorhynchus comprise a monophyletic group that begun to diversify in the Late Cretaceous. The association of the SKN clade with other Anopheles subgenera is unclear. The radiation of the Cellia subgenera as well as the Anopheles subgenera took place at approximately the same time. Furthermore, although we recovered the monophyly of the Anopheles subgenus, our analysis failed to statistically support the monophyly of the Anopheles genus.
Acknowledgments
CGS was funded by the Brazilian Research Council-CNPq grant 307982/2012-2 and the Rio de Janeiro State Science Foundation-FAPERJ grants 110.028/2011 and 111.831/2011. CMV was funded through CNPq grant 481843/2013-2. LAF was financially supported by a scholarship from the Brazilian Ministry of Education (CAPES). CAMR was funded by a fellowship from the Brazilian Research Council-CNPq and a grant from the Rio de Janeiro State Science Foundation.
Data Availability
All relevant data are uploaded to Dryad (doi:10.5061/dryad.bp6gv).
Funding Statement
CGS was funded by the Brazilian Research Council-CNPq grant 307982/2012-2 and the Rio de Janeiro State Science Foundation-FAPERJ grants 110.028/2011 and 111.831/2011. CMV was funded through CNPq grant 481843/2013-2. LAF was financially supported by a scholarship from the Brazilian Ministry of Education (CAPES). CAMR was funded by a fellowship from the Brazilian Research Council-CNPq and a grant from the Rio de Janeiro State Science Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Harbach RE. The Culicidae (Diptera): a review of taxonomy, classification and phylogeny. Zootaxa. 2007;(1668):591–638. [Google Scholar]
- 2. Krzywinski J, Wilkerson RC, Besansky NJ. Toward understanding Anophelinae (Diptera, Culicidae) phylogeny: Insights from nuclear single-copy genes and the weight of evidence. Systematic Biology. 2001;50(4):540–56. [PubMed] [Google Scholar]
- 3. Krzywinski J, Wilkerson RC, Besansky NJ. Evolution of mitochondrial and ribosomal gene sequences in Anophelinae (Diptera: Culicidae): Implications for phylogeny reconstruction. Molecular phylogenetics and evolution. 2001;18(3):479–87. [DOI] [PubMed] [Google Scholar]
- 4. Sallum MAM, Schultz TR, Wilkerson RC. Phylogeny of Anophelinae (Diptera Culicidae) based on morphological characters. Annals of the Entomological Society of America. 2000;93(4):745–75. [Google Scholar]
- 5. Harbach RE, Rattanarithikul R, Harrision BA. Baimaia, a new subgenus for Anopheles kyondawensis Abraham, a unique crabhole-breeding anopheline in Southeastern Asia. Proceedings of the Entomological Society of Washington. 2005;107(4):750–61. [Google Scholar]
- 6. Sinka ME, Bangs MJ, Manguin S, Rubio-Palis Y, Chareonviriyaphap T, Coetzee M, et al. A global map of dominant malaria vectors. Parasites & vectors. 2012;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sallum MAM, Schultz TR, Foster PG, Aronstein K, Wirtz RA, Wilkerson RC. Phylogeny of Anophelinae (Diptera: Culicidae) based on nuclear ribosomal and mitochondrial DNA sequences. Systematic Entomology. 2002;27(3):361–82. [Google Scholar]
- 8. Moreno M, Marinotti O, Krzywinski J, Tadei WP, James AA, Achee NL, et al. Complete mtDNA genomes of Anopheles darlingi and an approach to anopheline divergence time. Malaria Journal. 2010;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular biology and evolution. 2013;30(4):772–80. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular biology and evolution. 2011;28(10):2731–9. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular biology and evolution. 2010;27(2):221–4. 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- 12. Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods. 2012;9(8):772-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–4. [DOI] [PubMed] [Google Scholar]
- 14. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Systematic Biology. 2010;59(3):307–21. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 15. Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55(4):539–52. [DOI] [PubMed] [Google Scholar]
- 16. Lanfear R, Calcott B, Ho SYW, Guindon S. PartitionFinder: Combined Selection of Partitioning Schemes and Substitution Models for Phylogenetic Analyses. Molecular biology and evolution. 2012;29(6):1695–701. 10.1093/molbev/mss020 [DOI] [PubMed] [Google Scholar]
- 17. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–3. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Molecular biology and evolution. 2012;29(8):1969–73. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zavortink TJ, Poinar GO. Anopheles (Nyssorhynchus) dominicanus sp n. (Diptera: culicidae) from Dominican amber. Annals of the Entomological Society of America. 2000;93(6):1230–5. [Google Scholar]
- 20. Poinar GO, Zavortink TJ, Pike T, Johnston PA. Paleoculicis minutus (Diptera: Culicidae) n. gen., n. sp., from Cretaceous Canadian amber, with a summary of described fossil mosquitoes. Acta Geol Hisp. 2000;35:119–28. [Google Scholar]
- 21. Logue K, Chan ER, Phipps T, Small ST, Reimer L, Henry-Halldin C, et al. Mitochondrial genome sequences reveal deep divergences among Anopheles punctulatus sibling species in Papua New Guinea. Malaria Journal. 2013;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gaunt MW, Miles MA. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Molecular biology and evolution. 2002;19(5):748–61. [DOI] [PubMed] [Google Scholar]
- 23. Schluter D, Price T, Mooers AO, Ludwig D. Likelihood of ancestor states in adaptive radiation. Evolution. 1997;51(6):1699–711. [DOI] [PubMed] [Google Scholar]
- 24. Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20(2):289–90. [DOI] [PubMed] [Google Scholar]
- 25. Ree RH, Smith SA. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Systematic Biology. 2008;57(1):4–14. 10.1080/10635150701883881 [DOI] [PubMed] [Google Scholar]
- 26. Neafsey DE, Waterhouse RM, Abai MR, Aganezov SS, Alekseyev MA, Allen JE, et al. Mosquito genomics. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science. 2015;347(6217):1258522 10.1126/science.1258522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reidenbach KR, Cook S, Bertone MA, Harbach RE, Wiegmann BM, Besansky NJ. Phylogenetic analysis and temporal diversification of mosquitoes (Diptera: Culicidae) based on nuclear genes and morphology. BMC evolutionary biology. 2009;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Davalos LA, Perkins SL. Saturation and base composition bias explain phylogenomic conflict in Plasmodium. Genomics. 2008;91(5):433–42. 10.1016/j.ygeno.2008.01.006 [DOI] [PubMed] [Google Scholar]
- 29. Martinsen ES, Perkins SL, Schall JJ. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): Evolution of life-history traits and host switches. Molecular phylogenetics and evolution. 2008;47(1):261–73. 10.1016/j.ympev.2007.11.012 [DOI] [PubMed] [Google Scholar]
- 30. Duval L, Fourment M, Nerrienet E, Rousset D, Sadeuh SA, Goodman SM, et al. African apes as reservoirs of Plasmodium falciparum and the origin and diversification of the Laverania subgenus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(23):10561–6. 10.1073/pnas.1005435107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ricklefs RE, Outlaw DC. A Molecular Clock for Malaria Parasites. Science. 2010;329(5988):226–9. 10.1126/science.1188954 [DOI] [PubMed] [Google Scholar]
- 32. Hayakawa T, Culleton R, Otani H, Horii T, Tanabe K. Big bang in the evolution of extant malaria parasites. Molecular biology and evolution. 2008;25(10):2233–9. 10.1093/molbev/msn171 [DOI] [PubMed] [Google Scholar]
- 33. Pacheco MA, Battistuzzi FU, Junge RE, Cornejo OE, Williams CV, Landau I, et al. Timing the origin of human malarias: the lemur puzzle. BMC evolutionary biology. 2011;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sermwittayawong N, Singh B, Nishibuchi M, Sawangjaroen N, Vuddhakul V. Human Plasmodium knowlesi infection in Ranong province, southwestern border of Thailand. Malaria Journal. 2012;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rich SM, Ayala FJ. Evolutionary Origins of Human Malaria Parasites In: Dronamraju KR, Arese P, editors. Malaria: Genetic and Evolutionary Aspects. New ork, NY: Springer; 2005. p. 125–68. [Google Scholar]
- 36. Silva JC, Egan A, Friedman R, Munro JB, Carlton JM, Hughes AL. Genome sequences reveal divergence times of malaria parasite lineages. Parasitology. 2011;138(13):1737–49. 10.1017/S0031182010001575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marinucci M, Romi R, Mancini P, Di Luca M, Severini C. Phylogenetic relationships of seven palearctic members of the maculipennis complex inferred from ITS2 sequence analysis. Insect molecular biology. 1999;8(4):469–80. [DOI] [PubMed] [Google Scholar]
- 38. Kampen H. The ITS2 ribosomal DNA of Anopheles beklemishevi and further remarks on the phylogenetic relationships within the Anopheles maculipennis group of species (Diptera: Culicidae). Parasitology Research. 2005;97(2):118–28. [DOI] [PubMed] [Google Scholar]
- 39. Djadid ND, Gholizadeh S, Tafsiri E, Romi R, Gordeev M, Zakeri S. Molecular identification of Palearctic members of Anopheles maculipennis in northern Iran. Malaria Journal. 2007;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wiens JJ, Tiu J. Highly Incomplete Taxa Can Rescue Phylogenetic Analyses from the Negative Impacts of Limited Taxon Sampling. PloS one. 2012;7(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fontaine MC, Pease JB, Steele A, Waterhouse RM, Neafsey DE, Sharakhov IV, et al. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science. 2015;347(6217):42–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Edwards SV. Is a new and general theory of molecular systematics emerging? Evolution. 2009;63(1):1–19. 10.1111/j.1558-5646.2008.00549.x [DOI] [PubMed] [Google Scholar]
- 43. Schrago CG, Menezes AN, Furtado C, Bonvicino CR, Seuanez HN. Multispecies Coalescent Analysis of the Early Diversification of Neotropical Primates: Phylogenetic Inference under Strong Gene Trees/Species Tree Conflict. Genome Biol Evol. 2014;6(11):3105–14. 10.1093/gbe/evu244 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are uploaded to Dryad (doi:10.5061/dryad.bp6gv).