Abstract
Entry of hepatitis B (HBV) and hepatitis D viruses (HDV) into a host cell represents the initial step of infection. This process requires multiple steps, including the low-affinity attachment of the virus to the cell surface, followed by high-affinity attachment to specific receptor(s), and subsequent endocytosis-mediated internalization. Within the viral envelope, the preS1 region is involved in receptor binding. Recently, sodium taurocholate cotransporting polypeptide (NTCP) has been identified as an entry receptor of HBV and HDV by affinity purification using a preS1 peptide. NTCP is mainly or exclusively expressed in the liver, and this membrane protein is at least one of the factors determining the narrow species specificity and hepatotropism of HBV and HDV. However, there are likely other factors that mediate the species and tissue tropism of HBV. This review summarizes the current understanding of the mechanisms of HBV/HDV entry.
The hepatitis B and D viruses enter host cells in a species- and tissue-specific manner. The process involves specific factors, such as sodium taurocholate cotransporting polypeptide, a recently identified entry receptor.
Hepatitis B virus (HBV) and hepatitis D virus (HDV), a viroid-like satellite RNA virus (see Rizzetto 2015; Taylor 2015), infect primarily hepatocytes of humans, chimpanzees, and tupaias. The HBV life cycle consists of steps including attachment, internalization, nuclear import, covalently closed circular DNA (cccDNA) formation, transcription, protein translation, encapsidation of viral RNA, reverse transcription and synthesis of genomic DNA, envelopment, and release of infectious virus particles (Fig. 1). Although HBV replication, including transcription and release, can be reproduced in many hepatocyte-derived cell lines from human and other species, HBV entry is much more restricted. So far, it occurs only in narrow host cells, including primary human (PHH) and tupaia hepatocytes (PTH) and differentiated HepaRG cells, which show a differentiated phenotype of hepatocytes (Gripon et al. 1988; Walter et al. 1996; Gripon et al. 2002). These cells have contributed to the clarification of the viral entry pathway, especially the role of viral proteins in the entry process. Recently, sodium taurocholate cotransporting polypeptide (NTCP) has been discovered as an entry receptor of HBV and HDV (Yan et al. 2012). This finding has led to the establishment of cell culture models for HBV and HDV infection and the identification of new antiviral compounds. However, recent findings suggest that there remain additional, as-yet unidentified host factor(s) that might contribute to HBV infection and its species specificity. This article reviews the current understanding of the molecular mechanisms for HBV/HDV entry and factors involved in determining their species specificity and tissue tropism.
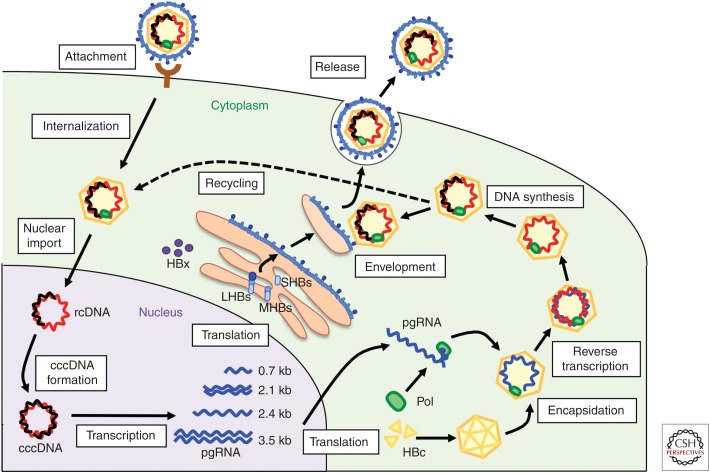
Figure 1.
The life cycle of hepatitis B virus (HBV). HBV attaches to host hepatocytes, and subsequent entry steps follow, including internalization and membrane fusion. After nuclear import, the relaxed circular HBV DNA genome is converted into covalently closed circular DNA (cccDNA). HBV replication proceeds with steps, including transcription, translation, encapsidation, reverse transcription, DNA synthesis, envelopment, release, and recycling. Only certain cell cultures can reproduce the steps from attachment to cccDNA formation, such as primary human (PHH) and tupaia (PTH) hepatocytes, differentiated HepaRG cells, and human hepatocyte-derived cell lines complemented with human sodium taurocholate cotransporting polypeptide (hNTCP). HBc, HBV core proteins; LHBs, large HBV surface proteins; MHBs, medium HBV surface proteins; pgRNA, pregenomic RNA; rcDNA, relaxed circular DNA; SHBs, small HBV surface proteins.
DOMAINS WITHIN HBV SURFACE PROTEINS THAT ARE ESSENTIAL FOR HBV AND HDV INFECTIONS
HBV and HDV share the same envelope proteins, termed HBV surface proteins (HBs), on the surface of their particles (Taylor 2013). It is, therefore, assumed that HBV and HDV enter host cells by very similar or even identical mechanisms. HBs are composed of three proteins, the large (LHBs), medium (MHBs), and small (SHBs) surface proteins, which include the preS1, preS2, and S regions, respectively (Fig. 2) (Stibbe and Gerlich 1983; Heermann et al. 1984). HBV particles include morphologically different constituents, containing abundant spherical and filamentous subviral particles (SVPs) and only a minor population of infectious HBV virions, also known as Dane particles (Patient et al. 2009). Infectious HBV virions carry all three HBs subtypes, in contrast to noninfectious spherical SVPs, which contain mainly SHBs and a small portion of MHBs, but not LHBs. Infectious HDV particles also contain LHBs, MHBs, and SHBs.
Figure 2.

Structure of hepatitis B virus (HBV) surface proteins. Large HBV surface proteins (LHBs), medium HBV surface proteins (MHBs), and small HBV surface proteins (SHBs) consist of 389–400, 281, and 226 aa (amino acids), respectively. SHBs have four transmembrane domains (I–IV). LHBs are myristoylated (myr) at its amino-terminal end, which is critically involved in HBV and HDV infection. The aa 2–48 portion of the preS1 region is essential for viral infection through receptor binding. The antigenic loop within SHBs also plays a significant role in viral infection.
Studies using neutralizing antibodies and site-directed mutagenesis have suggested that the S and preS1, but not the preS2, regions play a critical role in mediating HBV infection (Le Seyec et al. 1998; Abou-Jaoude and Sureau 2007; Salisse and Sureau 2009; Ni et al. 2010; Bremer et al. 2011). In the S region, amino acids (aa) 118–129 within the antigenic loop, especially G119, P120, C121, R122, and C124, which are located between transmembrane regions II and III and exposed to the virion surface, are critical for viral infectivity (Abou-Jaoude and Sureau 2007). Although the exact role of this region in viral infection has not been fully clarified, it has been reported that the antigenic loop mediates binding to heparan sulfate proteoglycans (HSPGs) on the host cell surface for the initial low-affinity attachment (discussed in the next paragraph) (Sureau and Salisse 2013). More important, a series of analyses using neutralizing antibodies, deletion and point mutagenesis, and peptide competition suggest that the preS1 region is essential for viral infection, probably through interaction with a specific receptor(s) on the cell surface (Le Seyec et al. 1999; Hong et al. 2004; Barrera et al. 2005; Glebe et al. 2005; Gripon et al. 2005; Engelke et al. 2006; Bremer et al. 2011). The amino-terminal end of the preS1 region is myristoylated, and this modification plays a significant role in infection (Gripon et al. 1995; Bruss et al. 1996) through the promotion of receptor binding (De Falco et al. 2001). Deletion analysis within the preS1 region suggested that aa 2–48 constitute the most essential region for infection (Le Seyec et al. 1999; Glebe et al. 2005; Engelke et al. 2006). Later, this region was shown to be a binding site for NTCP and to mediate the early viral entry process (Yan et al. 2012; Ni et al. 2014). From the above evidence, Myrcludex-B, a synthetic N-acylated preS1 lipopeptide consisting of the aa 2–48 region that shows a strong capacity to inhibit HBV entry with a subnanomolar IC50, is currently under clinical development as a new anti-HBV agent (Petersen et al. 2008; Lempp and Urban 2014).
MULTISTEP PROCESS FOR HBV AND HDV ENTRY
HBV and HDV infections of hepatocytes follow a multiple stepwise process (Fig. 3). Initially, viruses attach to host cells with low affinity in a reversible manner. This initial step involves cell surface molecules, including HSPGs (Schulze et al. 2007; Leistner et al. 2008; Lamas Longarela et al. 2013; Sureau and Salisse 2013). Subsequently, viruses interact with their specific receptor(s) with high affinity, triggering the early entry process. As described in the following paragraph, NTCP functions as an entry receptor for HBV and HDV (Yan et al. 2012; Iwamoto et al. 2014; Ni et al. 2014), although the precise mechanism for triggering viral internalization has not yet been clarified. Internalization of the viruses into host cells occurs through endocytosis. This step has been reported to involve clathrin-, caveolae-, and macropinocytosis-dependent endocytosis, which differ depending on the types of host cells, virus particles, and experimental conditions (Cooper and Shaul 2006; Macovei et al. 2010; Huang et al. 2012; Gao et al. 2013). After fusion of viral and cellular membranes inside certain vesicles, of which the molecular mechanisms have not been revealed, the nucleocapsid inside the envelope enters the cytoplasm and, ultimately, translocates into the nucleus.
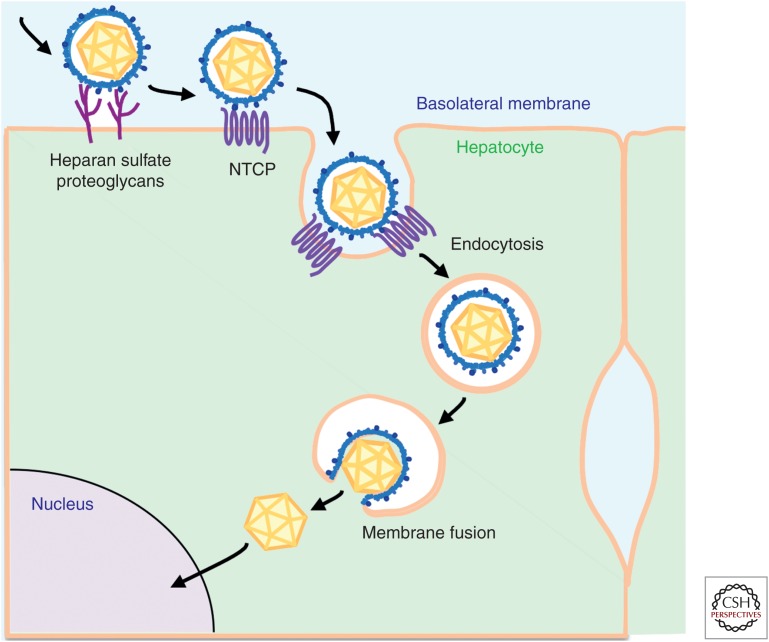
Figure 3.
Schematic representation of the HBV entry pathway. HBV particles attach to host hepatocytes through cell-surface factors, including heparan sulfate proteoglycans (HSPGs) with low affinity. The particles then interact with higher affinity with a specific receptor, sodium taurocholate cotransporting polypeptide (NTCP), to mediate the ensuing internalization. Internalization occurs in an endocytosis-dependent manner. The viral and cellular membranes are fused in certain vesicles and the nucleocapsids inside the viral particles are released into the cytoplasm, and are then destined to traffic to the nucleus for translocation via the nuclear pore complex.
NTCP AS AN ENTRY RECEPTOR FOR HBV AND HDV
After years of intense efforts, NTCP was finally revealed to be an entry receptor of HBV and HDV (Yan et al. 2012). This finding was initiated by affinity purification of a primary tupaia hepatocyte (PTH) lysate using a lipopeptide consisting of aa 2–48 of the preS1 region (preS1 peptide) as bait. Both tupaia sodium taurocholate cotransporting polypeptide (tsNTCP) and human sodium taurocholate cotransporting polypeptide (hNTCP) bind to the preS1 peptide. Knockdown of endogenous hNTCP or tsNTCP in HepaRG, PTH, and primary human hepatocytes (PHH) results in the reduction of HBV and HDV infections. Ectopic expression of hNTCP confers viral susceptibility to HepG2 and Huh-7 cells, which otherwise do not support HBV or HDV infections. These data indicate that NTCP is essential for HBV and HDV infection. Expression of NTCP correlates well with the susceptibility to viral infections; it is abundantly expressed in susceptible PHH and differentiated HepaRG cells, whereas little or no NTCP is detected in resistant cell lines, such as HepG2, Huh-7, FLC4, and HeLa cells (Watashi et al. 2014b). Importantly, physiological expression of NTCP is limited almost exclusively to the liver in humans (Hagenbuch et al. 1991; Hilgendorf et al. 2007).
NTCP, also known as a solute carrier family 10A1 (SLC10A1), is a member of the SLC10 transporter family and is a sodium-dependent transporter for bile acids (Dawson et al. 2009). NTCP on the plasma membrane cotransports one molecule of extracellular bile salt together with two sodium ions across the membrane. Its major natural substrates are conjugated bile acids, such as taurocholic acid, taurodeoxycholic acid, and taurohyodeoxycholic acid, but other xenobiotics, including statins, propranolol, furosemide, and micafungin, can be also transported by NTCP (Kramer et al. 1999; Hata et al. 2003; Yanni et al. 2010; Greupink et al. 2011; Stieger 2011). NTCP is distributed mainly on the basolateral membrane of hepatocytes and is involved in the enterohepatic circulation of bile salts. hNTCP is a 349-aa protein of ∼56 kDa in size and contains 7–9 putative transmembrane domains with its amino-terminal end in the extracellular region and carboxy-terminal end in the cytoplasm (Hagenbuch and Meier 1994; Hallen et al. 2002; Mareninova et al. 2005). However, its crystal structure has not been solved so far. Recently, the structure of a related protein, apical sodium-dependent bile salt transporter (ASBT), which is also known as SLC10A2, from Neisseria meningitis (ASBTNM) and Yersinia frederiksenii (ASBTYf) has been solved by X-ray crystallography (Hu et al. 2011; Zhou et al. 2013). However, ASBTNM contains a 10-transmembrane domain and a hydrophobic inward-facing binding cavity, which is structurally different from the hNTCP model that is predicted from structural modeling, as well as experimental data from topological analyses using antibodies and fusion tags.
MOLECULAR REQUIREMENT OF NTCP IN HBV AND HDV ENTRY
Mutation analysis of rat NTCP and the crystal structures of ASBT can be used to predict bile salt-binding sites and sodium-binding sites on hNTCP (Zahner et al. 2003; Sun et al. 2006; Hu et al. 2011). There are several studies that have attempted to determine the contribution of the region within hNTCP responsible for viral infection. Mutations of NTCP residues critical for bile salts binding or uptake (N262, S267, Q293/L294) severely reduced preS1 binding and HBV and HDV infections (Yan et al. 2014), suggesting that the putative bile acids-binding sites on NTCP overlap with the sites for binding to the preS1 region on HBV. Mutations in the sodium-binding sites (Q68, S105/N106, E257, Q261) also reduced preS1 binding and viral infections, but to a lesser extent than the bile acids–binding mutations. However, reduced transporter activity of NTCP caused by low concentration of sodium in the testing solution did not have a major effect on preS1 binding and HBV and HDV infections, indicating that the inward flow of a sodium current across cell membranes may not be required for viral entry.
Comparisons of NTCP sequences between species have shown that the region spanning aa 157–165 critically contributes to the binding to the preS1 region and thereby supports viral infection, suggesting that this region is involved in HBV binding (Yan et al. 2012). Sequence swapping between mouse and human NTCP has shown that aa 84–87 from humans can support viral infection with mouse NTCP, whereas replacement of aa 84–87 in hNTCP by its mouse counterpart drastically decreases preS1 binding and HBV and HDV infections, indicating that aa 84–87 is also involved in viral attachment and infection (Yan et al. 2013; Ni et al. 2014). Compared with the aa 157–165 region (KGIVISLV), which contains multiple hydrophobic residues, the aa 84–87 region (RLKN) in hNTCP, which is more hydrophilic and likely exposed to extracellular solvents, may be involved in direct binding to the preS1 region of HBV particles. In contrast, region 157–165 may be important for maintaining NTCP conformation, or may function after a conformational change of NTCP triggered by primary binding between preS1 and NTCP. To understand the precise regional requirements within NTCP and their roles in viral attachment and infection, the crystal structure of NTCP needs to be solved.
OTHER FACTOR(S) DETERMINING HOST SUSCEPTIBILITY TO HBV INFECTION
For reasons that are not yet understood, ectopic expression of hNTCP confers susceptibility to only a fraction of human hepatocyte cell lines, such as Huh-7, HepG2, and undifferentiated HepaRG cells (Yan et al. 2012; Iwamoto et al. 2014; Ni et al. 2014). So far, HepG2 cells show the highest infection efficiency following expression of hNTCP; however, <10% of the cells were infected with HBV according to the original studies (Yan et al. 2012), and ~20% were infected with high amounts of viral inoculum in other reports (Iwamoto et al. 2014). Intriguingly, pretreatment of hNTCP-expressing HepG2 cells with 2.5%–3% dimethyl sulfoxide (DMSO) augmented the infection efficiency to 50%–70% (Iwamoto et al. 2014; Ni et al. 2014). It remains unclear why only a portion of the cells that express hNTCP is susceptible to HBV and HDV. Moreover, HepG2 transformants expressing equivalent levels of NTCP protein on their cell surfaces differ in their susceptibility to HBV infection (Watashi et al. 2014a). These data indicate that susceptibility to HBV infection is governed by factors other than simple expression levels of NTCP. Such factors include posttranslational modification of NTCP, cell cycle status, and/or the presence of other unknown factors essential for HBV infection.
Interestingly, a new human hepatoma cell line, HLCZ01, was recently established, and these cells can be infected with HBV without ectopic expression of hNTCP (Yang et al. 2014). These cells were established by isolation of cells from a liver tumor and transplantation into nonobese diabetic/severe combined immune deficiency (NOD/SCID) mice. HLCZ01 cells express high levels of liver-specific genes, such as albumin, α1-antitrypsin, HNF4, cytochrome P450 3A4, and miR-122. The investigators detected NTCP in HLCZ01 cells, but it is curious, in their paper, that NTCP could be also detected in Huh-7 and HepG2 cells at a similar level to that in PHH. Inoculation with HBV at 20 HBV DNA genome equivalents/cell produced ∼3% HBs-positive cells at day 5 posttreatment and 36% by day 85. Treatment with lamivudine or preS1 peptide reduced the accumulation of HBsAg, HBV DNA, and HBV RNA–positive cells, suggesting that this cell line supports the spread of HBV to naïve cells in the culture. In contrast, HBV infections of HepaRG cells result in a single-cycle infection, probably because the levels of HBV production are insufficient to permit a second round of infection. Although these results require more validation, this cell line might become a useful tool to further clarify the molecular mechanisms of HBV and HDV entry, replication, and spread.
SPECIES SPECIFICITY
Hepadnaviruses, in general, have a narrow host range, restricted to species related to their natural hosts. So far, HBV has been reported to infect humans, chimpanzees, and Tupaia belangeri, but not mice and rats (Walter et al. 1996; Kock et al. 2001). The ability of the PreS1 peptide to bind hepatocytes from nonsusceptible species, such as mouse, rat, and dog, both in vitro and in vivo suggests that species specificity is determined by postattachment processes (Meier et al. 2013; Schieck et al. 2013). As rodent cells support appropriate transcription, assembly, and secretion of HBV after transfection of HBV-encoding transgenes (Araki et al. 1989; Guidotti et al. 1995), the blockade of infection in nonsusceptible species is likely because of host factor(s) related to early steps in the viral life cycle, including internalization, nuclear import, and cccDNA formation (Fig. 1). NTCP, which is likely to be involved in internalization as well as attachment, is 83.9% identical between tupaia (tsNTCP) and humans (hNTCP), 96.3% identical between crab-eating monkey (monkey sodium taurocholate cotransporting polypeptide [mkNTCP]) and humans, and 73.8% identical between mouse (mouse sodium taurocholate cotransporting polypeptide [mNTCP]) and humans. As mentioned earlier, hNTCP and tsNTCP can support HBV and HDV infections when expressed in HepG2 cells (Yan et al. 2012), whereas mNTCP does not confer HBV/HDV susceptibility (Yan et al. 2013). It is possible that mNTCP binds to HBV particles, but does not mediate viral internalization after attachment.
A second HBV-specific restriction step may occur after internalization. Complementation of hNTCP in mouse and rat hepatocyte cell lines, such as Hepa1-6, MMHD3, Hep56.1D, TC5123, AML-12, and primary mouse hepatocytes, can support HDV infection (Yan et al. 2013; Li et al. 2014; Ni et al. 2014). In contrast, these cells do not support HBV infection, although preS1 attachment is observed. Considering that HBV and HDV share the same envelope proteins and, likely, the same mechanistic pathway for the entry process, these results suggest that cellular factor(s) functioning at the postentry step are important for achieving infection of HBV, but are dispensable for HDV infections. Notably, HDV, but not HBV, can infect woodchuck hepatocytes, indicating that other human gene(s) are additionally required for HBV infections, but not for HDV infections (Taylor 2013).
HBV and HDV are assumed to attach and internalize into host hepatocytes by very similar or identical mechanisms (Taylor 2013). After release into the cytoplasm, HBV nucleocapsids and HDV ribonucleoprotein complexes may follow different pathways and require different host factors. Additionally, innate immune factors inside hepatocytes can recognize HBV and HDV differently to eliminate viruses at postentry steps. Thus, there are at least two possibilities for why infection of HBV and HDV can be distinguished in mouse hepatocytes expressing hNTCP: (1) loss of expression of host factor(s) that are required for infection of HBV but not HDV, or (2) expression of restriction factor(s) that eliminate HBV but not HDV. In an examination of the latter possibility, STING, TBK1, IRF3, and IRF7, major components of antiviral signaling pathways, did not affect HBV infection in mouse hepatocytes (Li et al. 2014). Importantly, a recent report showed that a heterokaryon established by fusing hNTCP-overexpressing mouse Hepa1-6 cells with HepG2 cells could be infected with HBV (Lempp et al. 2014). These data favor the possibility that mouse hepatocytes are deficient for cellular factor(s) required for HBV infection. Future analyses will be needed to identify these factor(s), which are necessary to better understand the molecular basis of species specificity of HBV and other hepadnaviruses. In contrast, species specificity of HDV infection is determined mainly by the sequence of NTCP. Thus, NTCP-related research has revealed unexpected differences between the mechanisms of HBV and HDV infections.
TISSUE TROPISM
HBV and HDV primarily or exclusively infect hepatocytes. A radiolabeled preS1 peptide shows specific accumulation in the liver in vivo, suggesting that viral attachment to host cells is the predominant step for determining the hepatotropism of HBV and HDV (Schieck et al. 2013). NTCP is distributed almost exclusively in the liver in humans (Hagenbuch et al. 1991; Hilgendorf et al. 2007). When hNTCP is ectopically expressed in nonhepatocyte cells, such as the human cervical cancer cell line HeLa, Chinese hamster ovary (CHO), and Africa green monkey kidney Vero cells, HDV binds and infects these cells (Yan et al. 2013), indicating that NTCP is the main barrier to distinguish and select hepatocytes as a host of HDV. In contrast, again, HBV does not achieve infection in these cells after expression of hNTCP. This suggests the existence of other restriction step(s) after attachment/internalization of HBV. During HBV replication, the transcription from cccDNA to produce viral RNA requires host transcription factors, such as peroxisome proliferator-activated receptor-α (PPARα), retinoid X receptor-α (RXRα), hepatocyte nuclear factor 4 (HNF4), and CCAAT/enhancer-binding protein (C/EBP) (Trujillo et al. 1991; Raney et al. 1997; Yu and Mertz 1997; Tang and McLachlan 2001). HNF4 shows high expression in the liver, and is also expressed in the kidney and intestine (Sladek et al. 1990), and, thus, transcription is obviously one of the restriction steps against HBV in nonhepatocyte cells.
CONCLUDING REMARKS
A series of studies, starting more than a decade ago, has revealed that the preS1 region is essential in the entry process of HBV and HDV. During these analyses, the development of culture systems using PHH, PTH, and HepaRG cells has had a significant contribution. The recent discovery of NTCP as a specific entry receptor of HBV and HDV is not only a milestone in itself in this research field, but has also accelerated the establishment of cell culture models that are useful for analyzing the molecular mechanisms underlying HBV and HDV infections. Future analyses using these systems are expected to uncover additional factors that regulate the infection and determine species tropism. These achievements might contribute to the future development of animal models for these viruses, allowing the development of new anti-HBV agents, as well as progress on the understanding of virus–host interactions.
ACKNOWLEDGMENTS
We are grateful to all of the members of the Department of Virology II, National Institute of Infectious Diseases for their helpful discussion. Our study is supported by grants-in-aid from the Ministry of Health, Labor, and Welfare, Japan, the Ministry of Education, Culture, Sports, Science, and Technology, Japan, and Japan Society for the Promotion of Science.
Footnotes
Editors: Christoph Seeger and Stephen Locarnini
Additional Perspectives on Hepatitis B and Delta Viruses available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Abou-Jaoude G, Sureau C. 2007. Entry of hepatitis delta virus requires the conserved cysteine residues of the hepatitis B virus envelope protein antigenic loop and is blocked by inhibitors of thiol-disulfide exchange. J Virol 81: 13057–13066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Miyazaki J, Hino O, Tomita N, Chisaka O, Matsubara K, Yamamura K. 1989. Expression and replication of hepatitis B virus genome in transgenic mice. Proc Natl Acad Sci 86: 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera A, Guerra B, Notvall L, Lanford RE. 2005. Mapping of the hepatitis B virus pre-S1 domain involved in receptor recognition. J Virol 79: 9786–9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer CM, Sominskaya I, Skrastina D, Pumpens P, El Wahed AA, Beutling U, Frank R, Fritz HJ, Hunsmann G, Gerlich WH, et al. 2011. N-terminal myristoylation-dependent masking of neutralizing epitopes in the preS1 attachment site of hepatitis B virus. J Hepatol 55: 29–37. [DOI] [PubMed] [Google Scholar]
- Bruss V, Hagelstein J, Gerhardt E, Galle PR. 1996. Myristylation of the large surface protein is required for hepatitis B virus in vitro infectivity. Virology 218: 396–399. [DOI] [PubMed] [Google Scholar]
- Cooper A, Shaul Y. 2006. Clathrin-mediated endocytosis and lysosomal cleavage of hepatitis B virus capsid-like core particles. J Biol Chem 281: 16563–16569. [DOI] [PubMed] [Google Scholar]
- Dawson PA, Lan T, Rao A. 2009. Bile acid transporters. J Lipid Res 50: 2340–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Falco S, Ruvo M, Verdoliva A, Scarallo A, Raimondo D, Raucci A, Fassina G. 2001. N-terminal myristylation of HBV preS1 domain enhances receptor recognition. J Pept Res 57: 390–400. [DOI] [PubMed] [Google Scholar]
- Engelke M, Mills K, Seitz S, Simon P, Gripon P, Schnolzer M, Urban S. 2006. Characterization of a hepatitis B and hepatitis delta virus receptor binding site. Hepatology 43: 750–760. [DOI] [PubMed] [Google Scholar]
- Gao Z, Li M, He W, Li W. 2013. Hepatitis B virus may enter HepG2 cells complemented with human NTCP via macropinocytosis. In 2013 International Meeting on Molecular Biology of Hepatitis B Viruses, p. O-2 Shanghai, China. [Google Scholar]
- Glebe D, Urban S, Knoop EV, Cag N, Krass P, Grun S, Bulavaite A, Sasnauskas K, Gerlich WH. 2005. Mapping of the hepatitis B virus attachment site by use of infection-inhibiting preS1 lipopeptides and tupaia hepatocytes. Gastroenterology 129: 234–245. [DOI] [PubMed] [Google Scholar]
- Greupink R, Dillen L, Monshouwer M, Huisman MT, Russel FG. 2011. Interaction of fluvastatin with the liver-specific Na+-dependent taurocholate cotransporting polypeptide (NTCP). Eur J Pharm Sci 44: 487–496. [DOI] [PubMed] [Google Scholar]
- Gripon P, Diot C, Theze N, Fourel I, Loreal O, Brechot C, Guguen-Guillouzo C. 1988. Hepatitis B virus infection of adult human hepatocytes cultured in the presence of dimethyl sulfoxide. J Virol 62: 4136–4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gripon P, Le Seyec J, Rumin S, Guguen-Guillouzo C. 1995. Myristylation of the hepatitis B virus large surface protein is essential for viral infectivity. Virology 213: 292–299. [DOI] [PubMed] [Google Scholar]
- Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, Cannie I, Guyomard C, Lucas J, Trepo C, Guguen-Guillouzo C. 2002. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci 99: 15655–15660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gripon P, Cannie I, Urban S. 2005. Efficient inhibition of hepatitis B virus infection by acylated peptides derived from the large viral surface protein. J Virol 79: 1613–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti LG, Matzke B, Schaller H, Chisari FV. 1995. High-level hepatitis B virus replication in transgenic mice. J Virol 69: 6158–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbuch B, Meier PJ. 1994. Molecular cloning, chromosomal localization, and functional characterization of a human liver Na+/bile acid cotransporter. J Clin Invest 93: 1326–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbuch B, Stieger B, Foguet M, Lubbert H, Meier PJ. 1991. Functional expression cloning and characterization of the hepatocyte Na+/bile acid cotransport system. Proc Natl Acad Sci 88: 10629–10633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallen S, Bjorquist A, Ostlund-Lindqvist AM, Sachs G. 2002. Identification of a region of the ileal-type sodium/bile acid cotransporter interacting with a competitive bile acid transport inhibitor. Biochemistry 41: 14916–14924. [DOI] [PubMed] [Google Scholar]
- Hata S, Wang P, Eftychiou N, Ananthanarayanan M, Batta A, Salen G, Pang KS, Wolkoff AW. 2003. Substrate specificities of rat oatp1 and ntcp: Implications for hepatic organic anion uptake. Am J Physiol Gastrointest Liver Physiol 285: G829–G839. [DOI] [PubMed] [Google Scholar]
- Heermann KH, Goldmann U, Schwartz W, Seyffarth T, Baumgarten H, Gerlich WH. 1984. Large surface proteins of hepatitis B virus containing the pre-s sequence. J Virol 52: 396–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgendorf C, Ahlin G, Seithel A, Artursson P, Ungell AL, Karlsson J. 2007. Expression of thirty-six drug transporter genes in human intestine, liver, kidney, and organotypic cell lines. Drug Metab Dispos 35: 1333–1340. [DOI] [PubMed] [Google Scholar]
- Hong HJ, Ryu CJ, Hur H, Kim S, Oh HK, Oh MS, Park SY. 2004. In vivo neutralization of hepatitis B virus infection by an anti-preS1 humanized antibody in chimpanzees. Virology 318: 134–141. [DOI] [PubMed] [Google Scholar]
- Hu NJ, Iwata S, Cameron AD, Drew D. 2011. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature 478: 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HC, Chen CC, Chang WC, Tao MH, Huang C. 2012. Entry of hepatitis B virus into immortalized human primary hepatocytes by clathrin-dependent endocytosis. J Virol 86: 9443–9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Watashi K, Tsukuda S, Aly HH, Fukasawa M, Fujimoto A, Suzuki R, Aizaki H, Ito T, Koiwai O, et al. 2014. Evaluation and identification of hepatitis B virus entry inhibitors using HepG2 cells overexpressing a membrane transporter NTCP. Biochem Biophys Res Commun 443: 808–813. [DOI] [PubMed] [Google Scholar]
- Kock J, Nassal M, MacNelly S, Baumert TF, Blum HE, von Weizsacker F. 2001. Efficient infection of primary tupaia hepatocytes with purified human and woolly monkey hepatitis B virus. J Virol 75: 5084–5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer W, Stengelin S, Baringhaus KH, Enhsen A, Heuer H, Becker W, Corsiero D, Girbig F, Noll R, Weyland C. 1999. Substrate specificity of the ileal and the hepatic Na+/bile acid cotransporters of the rabbit: I. Transport studies with membrane vesicles and cell lines expressing the cloned transporters. J Lipid Res 40: 1604–1617. [PubMed] [Google Scholar]
- Lamas Longarela O, Schmidt TT, Schoneweis K, Romeo R, Wedemeyer H, Urban S, Schulze A. 2013. Proteoglycans act as cellular hepatitis delta virus attachment receptors. PLoS ONE 8: e58340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leistner CM, Gruen-Bernhard S, Glebe D. 2008. Role of glycosaminoglycans for binding and infection of hepatitis B virus. Cell Microbiol 10: 122–133. [DOI] [PubMed] [Google Scholar]
- Lempp FA, Urban S. 2014. Inhibitors of hepatitis B virus attachment and entry. Intervirology 57: 151–157. [DOI] [PubMed] [Google Scholar]
- Lempp FA, Ni Y, Lipps C, Wirth D, Urban S. 2014. Hepatitis B virus infection of mouse-human heterokaryonic cells indicates the absence of host-specific dependency factor(s) in mouse liver cell lines. In 2014 International Meeting on Molecular Biology of Hepatitis B Viruses, p. O-4 Los Angeles, CA. [Google Scholar]
- Le Seyec J, Chouteau P, Cannie I, Guguen-Guillouzo C, Gripon P. 1998. Role of the pre-S2 domain of the large envelope protein in hepatitis B virus assembly and infectivity. J Virol 72: 5573–5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Seyec J, Chouteau P, Cannie I, Guguen-Guillouzo C, Gripon P. 1999. Infection process of the hepatitis B virus depends on the presence of a defined sequence in the pre-S1 domain. J Virol 73: 2052–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhuang Q, Wang Y, Zhang T, Zhao J, Zhang Y, Zhang J, Lin Y, Yuan Q, Xia N, et al. 2014. HBV life cycle is restricted in mouse hepatocytes expressing human NTCP. Cell Mol Immunol 11: 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macovei A, Radulescu C, Lazar C, Petrescu S, Durantel D, Dwek RA, Zitzmann N, Nichita NB. 2010. Hepatitis B virus requires intact caveolin-1 function for productive infection in HepaRG cells. J Virol 84: 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mareninova O, Shin JM, Vagin O, Turdikulova S, Hallen S, Sachs G. 2005. Topography of the membrane domain of the liver Na+-dependent bile acid transporter. Biochemistry 44: 13702–13712. [DOI] [PubMed] [Google Scholar]
- Meier A, Mehrle S, Weiss TS, Mier W, Urban S. 2013. Myristoylated PreS1-domain of the hepatitis B virus L-protein mediates specific binding to differentiated hepatocytes. Hepatology 58: 31–42. [DOI] [PubMed] [Google Scholar]
- Ni Y, Sonnabend J, Seitz S, Urban S. 2010. The pre-S2 domain of the hepatitis B virus is dispensable for infectivity but serves a spacer function for L-protein-connected virus assembly. J Virol 84: 3879–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Falth M, Stindt J, Koniger C, Nassal M, Kubitz R, et al. 2014. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 146: 1070–1083. [DOI] [PubMed] [Google Scholar]
- Patient R, Hourioux C, Roingeard P. 2009. Morphogenesis of hepatitis B virus and its subviral envelope particles. Cell Microbiol 11: 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Dandri M, Mier W, Lutgehetmann M, Volz T, von Weizsacker F, Haberkorn U, Fischer L, Pollok JM, Erbes B, et al. 2008. Prevention of hepatitis B virus infection in vivo by entry inhibitors derived from the large envelope protein. Nat Biotechnol 26: 335–341. [DOI] [PubMed] [Google Scholar]
- Raney AK, Johnson JL, Palmer CN, McLachlan A. 1997. Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J Virol 71: 1058–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Rizzetto M. 2015. HDV: Introduction and epidemiology. Cold Spring Harb Perspect Med 10.1101/cshperspect.a021576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisse J, Sureau C. 2009. A function essential to viral entry underlies the hepatitis B virus “a” determinant. J Virol 83: 9321–9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieck A, Schulze A, Gahler C, Muller T, Haberkorn U, Alexandrov A, Urban S, Mier W. 2013. Hepatitis B virus hepatotropism is mediated by specific receptor recognition in the liver and not restricted to susceptible hosts. Hepatology 58: 43–53. [DOI] [PubMed] [Google Scholar]
- Schulze A, Gripon P, Urban S. 2007. Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology 46: 1759–1768. [DOI] [PubMed] [Google Scholar]
- Sladek FM, Zhong WM, Lai E, Darnell JE Jr. 1990. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev 4: 2353–2365. [DOI] [PubMed] [Google Scholar]
- Stibbe W, Gerlich WH. 1983. Structural relationships between minor and major proteins of hepatitis B surface antigen. J Virol 46: 626–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieger B. 2011. The role of the sodium-taurocholate cotransporting polypeptide (NTCP) and of the bile salt export pump (BSEP) in physiology and pathophysiology of bile formation. Handb Exp Pharmacol 201: 205–259. [DOI] [PubMed] [Google Scholar]
- Sun AQ, Balasubramaniyan N, Chen H, Shahid M, Suchy FJ. 2006. Identification of functionally relevant residues of the rat ileal apical sodium-dependent bile acid cotransporter. J Biol Chem 281: 16410–16418. [DOI] [PubMed] [Google Scholar]
- Sureau C, Salisse J. 2013. A conformational heparan sulfate binding site essential to infectivity overlaps with the conserved hepatitis B virus A-determinant. Hepatology 57: 985–994. [DOI] [PubMed] [Google Scholar]
- Tang H, McLachlan A. 2001. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc Natl Acad Sci 98: 1841–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JM. 2013. Virus entry mediated by hepatitis B virus envelope proteins. World J Gastroenterol 19: 6730–6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Taylor JM. 2015. Hepatitis D virus replication. Cold Spring Harb Perspect Med 10.1101/cshperspect.a021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo MA, Letovsky J, Maguire HF, Lopez-Cabrera M, Siddiqui A. 1991. Functional analysis of a liver-specific enhancer of the hepatitis B virus. Proc Natl Acad Sci 88: 3797–3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter E, Keist R, Niederost B, Pult I, Blum HE. 1996. Hepatitis B virus infection of tupaia hepatocytes in vitro and in vivo. Hepatology 24: 1–5. [DOI] [PubMed] [Google Scholar]
- Watashi K, Iwamoto M, Sluder A, Matsunaga S, Ryo A, Morishita R, Kwon ATJ, Suzuki H, Tsukuda S, Suzuki R, et al. 2014a. Characterization of a culture system reproducing the NTCP-mediated HBV entry and its application to drug development. In 2014 International Meeting on Molecular Biology of Hepatitis B Viruses, p. O-7 Los Angeles, CA. [Google Scholar]
- Watashi K, Sluder A, Daito T, Matsunaga S, Ryo A, Nagamori S, Iwamoto M, Nakajima S, Tsukuda S, Borroto-Esoda K, et al. 2014b. Cyclosporin A and its analogs inhibit hepatitis B virus entry into cultured hepatocytes through targeting a membrane transporter, sodium taurocholate cotransporting polypeptide (NTCP). Hepatology 59: 1726–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, et al. 2012. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 1: e00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Peng B, He W, Zhong G, Qi Y, Ren B, Gao Z, Jing Z, Song M, Xu G, et al. 2013. Molecular determinants of hepatitis B and D virus entry restriction in mouse sodium taurocholate cotransporting polypeptide. J Virol 87: 7977–7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Peng B, Liu Y, Xu G, He W, Ren B, Jing Z, Sui J, Li W. 2014. Viral entry of hepatitis B and D viruses and bile salts transportation share common molecular determinants on sodium taurocholate cotransporting polypeptide. J Virol 88: 3273–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Zuo C, Wang X, Meng X, Xue B, Liu N, Yu R, Qin Y, Gao Y, Wang Q, et al. 2014. Complete replication of hepatitis B virus and hepatitis C virus in a newly developed hepatoma cell line. Proc Natl Acad Sci 111: E1264–E1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanni SB, Augustijns PF, Benjamin DK Jr, Brouwer KL, Thakker DR, Annaert PP. 2010. In vitro investigation of the hepatobiliary disposition mechanisms of the antifungal agent micafungin in humans and rats. Drug Metab Dispos 38: 1848–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Mertz JE. 1997. Differential regulation of the pre-C and pregenomic promoters of human hepatitis B virus by members of the nuclear receptor superfamily. J Virol 71: 9366–9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahner D, Eckhardt U, Petzinger E. 2003. Transport of taurocholate by mutants of negatively charged amino acids, cysteines, and threonines of the rat liver sodium-dependent taurocholate cotransporting polypeptide Ntcp. Eur J Biochem 270: 1117–1127. [DOI] [PubMed] [Google Scholar]
- Zhou X, Levin EJ, Pan Y, McCoy JG, Sharma R, Kloss B, Bruni R, Quick M, Zhou M. 2013. Structural basis of the alternating-access mechanism in a bile acid transporter. Nature 505: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]