Abstract
In eutherian organisms, the placenta interfaces the fetal and maternal environments. Located at the placental villous surface, in direct contact with maternal blood, is the trophoblast layer, which mediates the crucial maternal–fetal exchange of gases, nutrients, and waste products, produces hormones that support the pregnancy, and provides immunologic defense. Discovery of microRNAs (miRNAs) and their role in development, differentiation, and homeostatic resilience has increased our understanding of genomic and epigenomic networks that regulate placental function. Moreover, unique miRNA species, which are expressed by human trophoblasts and are termed “trophomiRs,” may show specialized functions during normal and pathological pregnancies. Placental miRNAs, packaged within exosomes and other vesicles or bound in protein complexes, are capable of communicating distinctive signals to maternal and/or fetal tissues. Additional research may usher in the use of circulating miRNAs as pregnancy-related disease biomarkers, providing new diagnostic and therapeutic options during pregnancy.
Unique miRNAs expressed by human trophoblasts (trophomiRs) are involved in networks that regulate placental function. Some have been associated with pathological pregnancies (e.g., preeclampsia).
The discovery of microRNAs (miRNAs) and their role in regulating the expression of mRNAs and proteins has added a new dimension to our understanding of gene regulatory networks. miRNAs are single stranded, noncoding RNA molecules of 19–24 nucleotides (nt), which act primarily by degrading mRNA transcripts or inhibiting translation of mRNA into proteins (Guo et al. 2010). Discrete genes encode miRNA precursors, which then undergo an exquisitely regulated multi-step processing before becoming fully functional as mature miRNAs. Multiple biogenesis pathways have been described. In the common one, after initial transcription by RNA polymerase II (Bartel 2004), primary (pri-) miRNAs are processed by the endonuclease Drosha and accessory proteins, resulting in a 70-nt double-stranded precursor (pre-) miRNA (Lee et al. 2003). The pre-miRNA is exported from the nucleus into the cytoplasm by the action of exportin 5, along with its accessory proteins (Perron and Provost 2009). A set of proteins, including the endonuclease Dicer and its associated proteins, process the stem-loop miRNAs into a mature form of miRNA duplex (Hutvagner et al. 2001; Jaskiewicz and Filipowicz 2008). These duplexes are incorporated into the RNA-induced silencing complex (RISC) (Matranga et al. 2005), which includes Argonaute2 (Ago2) and a set of regulatory proteins. In the RISC, the miRNA duplex unwinds and miRNA-specific sequences orchestrate RNA degradation and the inhibition of mRNA translation into protein.
Whereas sequence complementarity between the target mRNA and the seed region (nt 2–8) within the miRNA plays a key role in regulating RISC activity, other characteristics of the mRNA and miRNA are also relevant, including duplex stability in regions outside the seed, the presence of bulges at the mRNA–miRNA interaction interfaces, and the presence of DNA Alu elements in the vicinity of the interaction interface, and the like (Nykanen et al. 2001; Martinez et al. 2002; Tsutsumi et al. 2011; Chi et al. 2012; Fukunaga et al. 2012; Hibio et al. 2012; Hoffman et al. 2013). Moreover, other changes in cell physiology may impact miRNA biogenesis and processing. For example, epidermal growth factor receptor signaling leads to phosphorylation of Ago2, which attenuates its interaction with Dicer and thereby reduces miRNA processing into mature miRNAs (Shen et al. 2013).
There are more than 1000 human miRNAs, which appear to regulate >50% of human RNAs (Krol et al. 2010). Whereas the expression pattern and function of many miRNA species remain unknown, certain miRNAs have been shown to be expressed in a tissue-selective manner and to respond to discrete physiological or pathological stimuli in a manner that functionally controls gene expression. In general, most miRNAs hitherto studied have a relatively small effect on mRNA or protein expression. They likely act together as a part of gene regulatory networks to buttress cell and tissue homeostatic resilience (Esteller 2011). Several miRNA species have been implicated in important developmental, physiological, and pathological functions, which are translatable to therapeutics (van Rooij et al. 2008; Machlin et al. 2012). Key functions relevant to trophoblast biology are discussed below.
EXPRESSION AND FUNCTION OF TROPHOMIRS AND OTHER PLACENTAL miRNAs
In eutherian organisms, the placenta is obligatory for supporting fetal development and growth. Within the human hemochorial placenta, the villi are lined by the trophoblast layer, composed of multinucleated, terminally differentiated syncytiotrophoblasts and the subjacent mononucleated, less differentiated cytotrophoblasts. The syncytiotrophoblasts, which are directly bathed in maternal blood, regulate key placental functions, including maternal–fetal gas exchange, nutrient supply to the fetus, removal of waste products from the intrauterine compartment, immunological protection of the fetus, and the production of hormones that are key to fetal development and to the maintenance of gestational homeostasis. The placenta shows intricate signaling mechanisms, including miRNA-based signals, to regulate these functions. To appreciate these signals in the context of placental biology, it is imperative to define the trophoblastic miRNA landscape and changes in miRNA expression in response to diverse stimuli.
Early microarray-based reports on placental miRNA expression identified miRNA species that are differentially expressed in the placenta compared with other tissues, such as miR-141, -23a, and -136 (Barad et al. 2004). Soon thereafter, it was discovered that a large fraction of trophoblastic miRNAs are expressed from a gene cluster located on chromosome 19, termed the chromosome 19 miRNA cluster (C19MC) (Bentwich et al. 2005; Donker et al. 2012). The C19MC spans ∼100 kb of genomic DNA and includes 46 intronic miRNA genes that express 58 miRNA species. As a group, miRNAs from this cluster are the most abundant miRNAs in human trophoblasts (Bentwich et al. 2005; Donker et al. 2012). The C19MC miRNAs are primate-specific and are expressed almost exclusively in the placenta (and are thus termed trophomiRs), with markedly weaker expression in the testis, brain, and a few other organs (Bentwich et al. 2005; Liang et al. 2007). Within the human placenta, the expression of C19MC miRNAs is seen as early as 5 wk of pregnancy, and the expression gradually increases as pregnancy progresses (Zhang et al. 2008).
While C19MC members are predominantly expressed in trophoblasts, the expression of C19MCs in placenta-derived mesenchymal stromal cells in vitro has also been documented (Luo et al. 2009; Flor et al. 2012). This apparent discrepancy between in vivo and in vitro expression may represent the use of different technologies (PCR vs. in situ hybridization) or the demethylation that may occur in vitro and potentially also in tumors in vivo (Ceccom et al. 2014). Interestingly, placental cell lines derived from villous trophoblasts express C19MC. Conversely, HTR8/SVneo, derived from extravillous trophoblasts, do not express C19MC trophomiRs (Donker et al. 2012; Morales-Prieto et al. 2012; Xie et al. 2014).
While the mechanisms controlling the tissue-specific regulation of C19MC expression, as well as the regulation of individual C19MC miRNA expression, remain unknown, we do know that an imprinted, paternally expressed, CpG-rich domain plays a cardinal role in C19MC expression (Noguer-Dance et al. 2010). This differentially methylated region, located 17.6 kb upstream of the first miRNA gene, is hypermethylated in cell lines that do not express C19MCs (Tsai et al. 2009). Further, expression of C19MC miRNAs in nonexpressing cell lines can be restored using the DNA methylation inhibitor 5-aza-2′-deoxycytidine (Saito et al. 2009; Tsai et al. 2009).
The C19MC region contains recurring genomic transposable elements called “Alu repeats,” which have been implicated in recombination and gene duplication events and likely contribute to the evolution of miRNA clusters (Zhang et al. 2008; Lehnert et al. 2009). The interaction between C19MC miRNAs and Alu sequences remains unknown. Interestingly, several C19MC miRNA species show sequence complementarity with Alu repeats in the sense orientation, suggesting that these neighboring C19MC miRNAs may target transcribed Alu sequences for degradation (Lehnert et al. 2009). This is consistent with sequence complementarity between transposable elements and other miRNAs in the human genome, suggesting a general role for these miRNAs in counteracting foreign nucleic acid sequences (Shalgi et al. 2010; Ahn et al. 2013). The C19MC also harbors recurring short DNA segments (120 nt). Although the function of these repeats is unknown, they may encode spliced, noncoding, short-lived RNA transcripts (Bortolin-Cavaille et al. 2009).
Several lines of evidence suggest a role for the C19MC in cell proliferation and differentiation. Expression of C19MC miRNAs has been observed in embryonic and induced stem cells. Interestingly, their expression drops considerably when these cells differentiate, which may indicate a role in the maintenance of an undifferentiated state (Bar et al. 2008; Laurent et al. 2008; Morin et al. 2008; Ren et al. 2009; Stadler et al. 2010). Furthermore, recent reports indicate that certain C19MC miRNAs are aberrantly expressed in specific tumors, possibly reflecting reactivation of the C19MC cluster as a consequence of chromosomal rearrangements or epigenetic modifications (Li et al. 2009, 2013a; Saito et al. 2009; Tsai et al. 2009; Rippe et al. 2010; Flor and Bullerdiek 2012). In contrast, miR-519, a C19MC miRNA, was shown to have a strong tumor-suppressive activity (Marasa et al. 2010; Abdelmohsen et al. 2012). Two other C19MC members, miR-517a and miR-517c, were recently implicated in the inhibition of cell proliferation of liver carcinoma cells (Liu et al. 2013).
Recent data from our laboratory also indicates that members of the C19MC cluster are expressed at much higher levels in villous trophoblasts compared with extravillous trophoblasts and that overexpression of the C19MC cluster results in reduced migration of the extravillous trophoblast line HTR8/SVneo (Xie et al. 2014).
A complete qualitative and quantitative assessment of the miRNA landscape in the human placenta remains to be determined, although the subject has been recently reviewed (Fu et al. 2013b; Buckberry et al. 2014). Such an assessment will depend on the experimental conditions, the platforms used (e.g., microarray or RNAseq), and the use of a reliable cutoff for establishing expression. Moreover, the knowledge we gain of the placental miRNA landscape depends on the timing of profiling during pregnancy. Using RNAseq, Luo et al. (2009) showed that 80% of the short RNAs in the human placenta are miRNAs, with the remaining species representing other types of short RNAs such as piRNA, snRNA, and the like. The expression of the C19MC trophomiRs generally increases as pregnancy progresses. The chromosome 14 miRNA cluster (C14MC) is another large miRNA cluster that is expressed in the placenta (Morales-Prieto et al. 2013). Unlike C19MC, the expression of C14MC miRNA members generally declines during pregnancy and can be detected in many other tissues (Liang et al. 2007). C14MC spans 52 miRNA genes, 40 of which are organized as a single large cluster, with tandem expression of other members. It is located within the 14q32 locus, between the DLK1 and DIO3 genes, and is regulated by a maternally imprinted differentially methylated region (DMR) ∼200 kb upstream of the transcription start site (Seitz et al. 2004; Morales-Prieto et al. 2013). The expression of the 371–373 cluster, also encoded by chromosome 19, generally remains unchanged during pregnancy (Gu et al. 2013; Morales-Prieto et al. 2013).
Recent data, largely based on pregnant mouse models, have established the bona fide role of miR-675 in fetoplacental development (Keniry et al. 2012). miR-675 is expressed from the first exon of H19, a large intergenic noncoding (linc) RNA. H19 is a maternally expressed imprinted gene located on human chromosome 11p15.5, downstream from IGF2, a locus that harbors some of the most highly expressed transcripts in the placenta (Hao et al. 1993; Gabory et al. 2010). Up-regulation of miR-675, which is controlled by the stress-response RNA-binding protein HuR, restricts murine placental growth. Deficiency of H19, which harbors miR-675, promotes placental growth, and miR-675 overexpression decreases cell proliferation, likely through targeting insulin-like growth factor 1 receptor (Keniry et al. 2012). Consistent with these findings, the expression of miR-675 rises toward the end of murine pregnancy, when placental growth decelerates. In addition, miR-675 restricts trophoblast proliferation in JEG3 cells, likely through binding to the nodal modulator 1 (NOMO1) protein (Gao et al. 2012).
Other miRNAs have been suggested as having defined regulatory functions during early placental development. A number of miRNAs, such as Let-7a, miR-377, and miR-145, are involved in the regulation of placental growth factors and have been implicated in placental cell proliferation (Forbes et al. 2012; also reviewed in Doridot et al. 2013). Assessing the role of miRNAs in mediating trophoblast differentiation and fusion, Kumar et al. found that members of the miR-17∼92 cluster, as well as members of the miR-106a∼363 and miR-106b∼25 clusters, are regulated by the transcription factor c-Myc and attenuate differentiation of trophoblasts through the silencing of GCM1. The level of these miRNAs is reduced during trophoblast differentiation, allowing increased expression of GCM1, CYP19A1, and hCG. Interestingly, this c-Myc-induced pathway is up-regulated in preeclampsia, implicating this miRNA network in the diminished trophoblast differentiation that characterizes placentas from preeclamptic women (Kumar et al. 2013).
Several researchers have shown that miR-155 attenuates trophoblast proliferation, acting through direct regulation of cyclin D1 in the HTR-8/SVneo extravillous trophoblast line (Dai et al. 2012). MiR-155 also targets interleukin-1 receptor-associated kinase M (IRAKM), NFκB inhibitor interacting Ras-like 1 (NKIRAS1), and phosphatase and tensin homolog (PTEN), thereby enhancing AP-1/NFκB inflammatory pathways (Xue et al. 2013). Interestingly, PTEN is a target for miR-17 and -19b, both down-regulated in first trimester miscarriages (Ventura et al. 2013). Consistent with these observations, increased expression of miR-155 has also been implicated in conditions associated with shallow placental invasion, such as severe preeclampsia (Dai et al. 2011). Another miRNA, miR-34, has been implicated in limiting placental proliferation, with reduced expression in conditions associated with abnormal trophoblast proliferation or invasion, such as placental accreta or preeclampsia (Umemura et al. 2013; Doridot et al. 2014). Lastly, some miRNAs were shown to regulate discrete targets within the placenta. For example, miR-15a and -16 regulate the expression of serotonin transporter in a trophoblast cell line (Moya et al. 2013).
PLACENTAL DISORDERS AND miRNAs
Research into the impact of physiological or pathological conditions on miRNA expression has uncovered intriguing associations among exposures, pathology, and/or altered miRNA expression landscape and has generated hypotheses regarding the roles of placental miRNAs in these processes. One of these conditions is preeclampsia, a common disease of pregnancy that is initiated by a form of placental dysfunction and characterized by maternal hypertension, proteinuria, and a systemic vasculopathy that impact many maternal organs, with a secondary effect on fetal well-being and growth.
Several miRNAs have been consistently found to be differentially regulated in preeclampsia. The most reproducible effect has been the up-regulation of miR-210 (Pineles et al. 2007; Zhu et al. 2009; Enquobahrie et al. 2011; Lee et al. 2011; Ishibashi et al. 2012; Muralimanoharan et al. 2012; Zhang et al. 2012; Kleinrouweler et al. 2013; Xu et al. 2014). Although the precise molecular function of miR-210 remains unknown, this miRNA is known to be up-regulated in hypoxia, likely mediated by the NFκB p50 subunit and by toll-like receptor (TLR)-3 (Huang et al. 2010; Zhang et al. 2012; Kopriva et al. 2013). Lee et al. showed that miR-210 targets the 3′-UTR of iron–sulfur cluster scaffold homolog (ISCU), which is down-regulated in preeclampsia and is associated with hemosiderosis in interstitial trophoblasts in certain placental pathologies (Lee et al. 2011). The inverse expression pattern of miR-210 and ISCU has been recapitulated by others groups (Muralimanoharan et al. 2012; Colleoni et al. 2013). Yet, others have implicated miR-210 in stimulated mitochondrial respiration and mitochondrial oxygen consumption, reduced mitochondrial complex 1 expression and cytochrome c oxidase assembly protein (Muralimanoharan et al. 2012; Colleoni et al. 2013), reduction of trophoblast invasion (Anton et al. 2013), and reduced expression of hydroxysteroid (17-β) dehydrogenase 1 (Ishibashi et al. 2012).
Unlike miR-210, miR-376c is down-regulated in preeclampsia, targeting activin receptor-like kinase 5 (ALK5), and ALK7 (Fu et al. 2013a). miR-378c, which was also found to be regulated in the placentas of preeclamptic women, promotes migration and proliferation of extravillous trophoblasts by targeting Nodal (Luo et al. 2012). In addition, an agomir of miR-126, an angiogenic miRNA produced by endothelial cells, was recently shown to ameliorate hypertension, improve placental perfusion, and enhance fetal and placental weight in a rat model of preeclampsia, possibly acting by stimulating the PI3K-Akt pathway (Yan et al. 2014). Other miRNAs were shown to be regulated in placentas from preeclamptic women. Examples include miR-182, -181, -1, -17, -20a, -20b, and miR-29b (Pineles et al. 2007; Hu et al. 2009; Zhu et al. 2009; Gunel et al. 2011; Mayor-Lynn et al. 2011; Yang et al. 2011; Wang et al. 2012; Wu et al. 2012; Chen and Wang 2013; Choi et al. 2013b; Kleinrouweler et al. 2013; Li et al. 2013a). Variable miRNA levels in these studies likely reflect differences in disease definition, ethnic groups, the timing of biopsies, quality of array, analysis, and the like. Intriguingly, in most cases, there was no correlation between miRNA expression change and disease severity.
Another common manifestation of placental dysfunction is substandard fetal growth, leading to the clinical condition of fetal growth restriction (FGR). Several research groups found that miR-21 is elevated in FGR-related placentas, potentially silencing the expression of PTEN or cystathionine g-lyase, an enzyme that catalyzes the production of the vasodilator hydrogen sulfide, which may lead to increased placental vascular resistance (Maccani et al. 2011; Cindrova-Davies et al. 2013). The level of miR-141, which silences the E2F transcription factor 3 (E2F3) protein and pleiomorphic adenoma gene 1 (PLAG1), is also elevated in FGR placentas (Tang et al. 2013). Several other placental miRNAs, including miR-16 and members of the C19MC (miR-518b, -1323, -516b, -515-5p, -520h, -519d, -526b) are dys-regulated in FGR placentas (Maccani et al. 2011; Guo et al. 2013; Higashijima et al. 2013; Wang et al. 2014; reviewed in Maccani and Marsit 2011). Interestingly, the influence of hypoxia on the expression of discrete miRNA species in the placenta in vivo and in hypoxic trophoblasts in vitro may be dissimilar. For example, miR-424, which silences FGFR1, is up-regulated or unchanged in FGR-related placentas in vivo (Mouillet et al. 2010b; Huang et al. 2013), but down-regulated in hypoxic trophoblasts in vitro (Mouillet et al. 2010a, 2013). These differences likely represent variability in experimental conditions, influences that occur in the context of the intact tissue in vivo that may be eliminated in dispersed, cultured cells in vitro, and/or the stringency of analysis (Mouillet et al. 2010a). Lastly, altered placental miRNA profiles have been associated with preterm delivery and with childhood neurodevelopmental outcomes (Mayor-Lynn et al. 2011; Maccani et al. 2013).
During pregnancy, the fetoplacental unit is susceptible to diverse types of microbial infections, including viral infections. It remains unclear whether or not pregnant women show a higher susceptibility to viral infections when compared with nonpregnant women. Whereas some reports attribute an “immune deficiency of pregnancy” to a high level of circulating steroid hormones, others indicate competent immunity (Arck et al. 2007; Mor and Cardenas 2010). Irrespective of immune competence, the physiological changes of pregnancy may predispose mothers and fetuses to serious consequences of viral infections. For example, changes in maternal lung volumes and function may predispose to severe pneumonia during pregnancy, caused by viruses such as influenza, coronavirus (SARS), varicella, and rubeola, with grave secondary consequences to the fetoplacental unit (Longman and Johnson 2007). Transmission of viruses into the intrauterine compartment may cause pregnancy loss, fetal death, fetal developmental or growth abnormalities (Kaplan 1993; Duff et al. 2009). Examples include cytomegalovirus (CMV), rubella, parvovirus, and other viruses (Nigro et al. 2005; Ergaz and Ornoy 2006; Best 2007; Duff et al. 2009; Mouillet et al. 2014).
Small RNA molecules have been implicated in antimicrobial defense mechanisms (Ding 2010; Wiedenheft et al. 2012; tenOever 2013). Examples include the CRISPR (clustered, regularly interspaced, short palindromic repeats) machinery, which is based on binding of short CRIPSR sequences to nucleic acids of invading microbes and subsequent degradation (Marraffini and Sontheimer 2010; Wiedenheft et al. 2012), and antiviral RNAi pathways (at least in nonmammalian cells and in human embryonic stem cells) (Cullen et al. 2013; Maillard et al. 2013; tenOever 2013). Interestingly, some miRNAs promote viral replication or pathogenesis, such as the liver-specific miR-122, which promotes hepatitis C virus replication by shielding the viral genome from nuclease-based degradation (Li et al. 2013b; Liu et al. 2013).
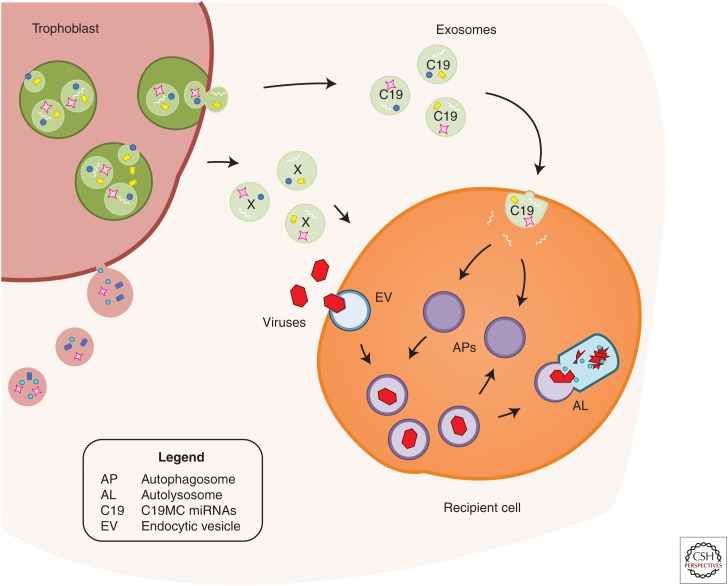
Our group has recently discovered that, when compared with nontrophoblastic cells, primary human trophoblasts (PHT cells) are resistant to infection by diverse types of DNA and RNA viruses, including coxsackievirus B, poliovirus, vesicular stomatitis virus, vaccinia virus, herpes simplex virus-1, rubella, HIV, and human CMV (Delorme-Axford et al. 2013a,b; Bayer et al. 2014; Mouillet et al. 2014). Remarkably, we found that viral resistance can be transferred to other, nonplacental primary cells and immortalized cell lines by exposing these cells to medium conditioned by PHT cells. This effect is mediated, at least in part, by the release of exosome-packaged C19MC miRNAs (Donker et al. 2012) and was not observed with nonviral pathogens such as listeria and toxoplasma (Delorme-Axford et al. 2013a; Bayer et al. 2014). Cells transfected with a BAC that harbors the C19MC locus or with individual, highly expressed C19MC miRNAs (miR-517-3p, -516b-5p, or -512-3p) also become more resistant to viral infections. Our data also indicate that PHT-conditioned media or C19MC miRNAs stimulate autophagy in recipient cells (Delorme-Axford et al. 2013a). Autophagy, a cellular prosurvival mechanism in which cellular organelles or foreign microbes are shuttled for degradation in lysosomes, (Choi et al. 2013a), can be seen to mediate the C19MC-dependent antiviral response because pharmacologic or genomic inhibition of autophagy diminishes the conferral of antiviral response to recipient cells (Delorme-Axford et al. 2013a). Although the C19MC miRNAs are likely acting in concert with other yet-to-be-identified antiviral responses, these data point to a unique and transferrable antiviral response, triggered by PHT-derived C19MC miRNAs (Fig. 1) (Delorme-Axford et al. 2013a,b; Mouillet et al. 2014).
Figure 1.
A model depicting the conferral of viral resistance to recipient cells. Primary human trophoblasts release exosomes containing C19MC (C19) miRNAs, which are taken up by recipient cells. These C19 miRNAs stimulate autophagy in the recipient cells. Incoming viruses travel within endocytic vesicles (EV) and are guided into C19-induced autophagosomes (APs). These autophagosomes fuse with lysosomes to form autolysosomes (AL), thus degrading viral endosomal vesicles. This process may cooperate with other factors (X), released into the conditioned medium. (From Delorme-Axford et al. 2013a; modified, with permission, from The National Academy of Sciences 2013.)
Various environmental exposures can influence the placental miRNA landscape. These exposures are commonly modeled in vitro, using cultured PHT cells or human trophoblast lines. We have previously showed that hypoxia regulates a set of miRNAs in trophoblasts and that one of these miRNA species, miR-205, directly regulates the expression of the transcriptional coactivator MED1 (Mouillet et al. 2010a). Others have found that exposure to bisphenol A or cigarette smoking impacts miRNA expression and that some of the responsive miRNA (e.g., miR-146a) regulate trophoblast cell proliferation (Avissar-Whiting et al. 2010; Maccani et al. 2010).
CIRCULATING miRNAs AND PLACENTAL DISEASES
Research using plasma or other body fluids such as blood, breast milk, or urine or in vitro cell culture medium has clearly established that miRNAs are released from diverse types of cells, either complexed with proteins such as Ago2, nucleophosmin1, or high-density lipoproteins (Wang et al. 2010; Arroyo et al. 2011; Vickers et al. 2011; Chen et al. 2012) or packaged within extracellular vesicles, such as exosomes, microvesicles, and/or apoptotic bodies (Fig. 2) (Valadi et al. 2007; Chen et al. 2012; Montecalvo et al. 2012; Vickers and Remaley 2012; El Andaloussi et al. 2013; Raposo and Stoorvogel 2013; Xu et al. 2013). These miRNA-containing vesicles are defined by size, protein composition, density, mechanism of biogenesis, packaging and release, and cells of origin. In general, exosomes (∼50–150 nm in diameter) are generated through a series of endocytic steps that result in the formation of intraluminal vesicles within multivesicular bodies (Scita and Di Fiore 2010; Hanson and Cashikar 2012). The sorting and loading of cargo into exosomes is largely determined by several protein complexes known as endosomal sorting complexes required for transport (ESCRT), their associated proteins (VPS4, VTA1, ALIX), and proteins such as the Rab proteins and sphingomyelinase (Trajkovic et al. 2008; Ostrowski et al. 2010; Baietti et al. 2012; Colombo et al. 2013).
Figure 2.
A schematic depicting the extracellular packages of trophoblastic miRNAs. Placental trophoblasts package miRNAs in several forms, including microvesicles, apoptotic bodies, exosomes, or protein-bound. Exosome-associated miRNAs are packaged within intraluminal vesicles contained within multivesicular bodies and are released as exosomes into the extracellular space. (From Ouyang et al. 2014; reprinted, with permission, from Elsevier 2014.)
Microvesicles (∼100–1000 nm in diameter) originate from direct shedding of the plasma membrane, where cargo is enveloped for direct exit from the cells. They contain cholesterol-rich microdomains and associated proteins (Meckes and Raab-Traub 2011). Apoptotic bodies, (apoptosomes, cell fragments, 500–4000 nm in diameter) are formed during partial or complete cell disintegration, and contain cell organelles and cytoplasmic proteins (Akers et al. 2013). Vesicles can be separated by differential ultracentrifugation with density-gradient, gel chromatography or fluid-based field-flow fractionation. The specificity and purity of each vesicle preparation can be confirmed using micro-imaging, nanoparticle tracking instruments, and determination of proteins, lipids, or nucleic acid content (El Andaloussi et al. 2013; Zheng et al. 2013). Packaging of miRNAs within vesicles or protein complexes likely enhances miRNA stability by shielding against RNase-mediated degradation and allows selective targeting of recipient cells (Valadi et al. 2007; Arroyo et al. 2011; O’Loughlin et al. 2012).
While the presence of extracellular, protein- or vesicle-bound miRNAs may provide new opportunities for biomarker discovery (discussed below), it is clear that circulating miRNAs are functionally active and have a physiological role in nonhormonal, paracrine and systemic communication among cells and tissues (Valadi et al. 2007; Arroyo et al. 2011; Vickers et al. 2011; Mittelbrunn and Sanchez-Madrid 2012; Peinado et al. 2012; Feng et al. 2013; Ramakrishnaiah et al. 2013; Raposo and Stoorvogel 2013). Nonvesicular, protein-bound miRNAs are also taken up by recipient cells, where they regulate their respective mRNA targets (Vickers et al. 2011; Vickers and Remaley 2012).
Akin to the presence of fetoplacental RNA molecules in the circulation (Poon et al. 2000; Ng et al. 2003; Maron et al. 2007; Koh et al. 2014), trophomiRs, as well as other types of miRNAs, are present in the maternal circulation during pregnancy (Gilad et al. 2008; Luo et al. 2009; Miura et al. 2010; Mouillet et al. 2010b; Donker et al. 2012), reaching median levels of 9 fmol/μg of total RNA in maternal plasma (compared with 37 fmol/μg of total placental RNA, or 25 fmol/μg of total fetal plasma RNA) (Williams et al. 2013). Williams et al. found that the most abundant miRNAs in the circulation of pregnant women were miR-451, -486, -92a, -16, let-7b, -21, -19b, -25, -22, and miR-144. In addition, the maternal plasma miRNA landscape is more closely related to that of fetal plasma miRNA than to the placental miRNA landscape (Williams et al. 2013). The rapid postpartum decline in the circulating levels of some (e.g., the C19MC, miR-127, and miR-134), but not all (miR-141, -149, -299-5p, and -135b) miRNAs suggests that they originate in the fetoplacental unit or in maternal tissues that were influenced by hormonal or other physiological changes during pregnancy (Chim et al. 2008; Williams et al. 2013).
The placenta is capable of releasing different types of vesicles, with altered dynamics of vesicle release associated with placental dysfunction in such conditions as preeclampsia or FGR (Ishihara et al. 2002; Redman and Sargent 2008; Aharon and Brenner 2013; Mayhew 2014). Recent investigation, focusing primarily on placental exosomes, indicates that these nanovesicles contain miRNAs (Luo et al. 2009; Donker et al. 2012; Redman et al. 2012; Delorme-Axford et al. 2013a; Ouyang et al. 2014; Record 2014). Whereas the function of circulating miRNAs during pregnancy remains unknown, inferring from the data discussed earlier regarding the function of extracellular C19MC miRNAs in vitro, it is plausible that C19MC trophomiRs, which are highly expressed in maternal plasma during pregnancy, can confer viral resistance to maternal endothelial cells and to other cell types.
Most research on circulating miRNAs during human pregnancy has centered on the use of these miRNAs as biomarkers for placental disorders or other pregnancy-related diseases (reviewed in Zhao et al. 2013). The accessibility of plasma analysis, along with the relative ease of measurement of abundant miRNA species, attracted many researchers to correlate the level of circulating miRNAs with diseases (Kotlabova et al. 2011). Not surprisingly, a correlation exists between plasma miR-210 and preeclampsia (Anton et al. 2013; Xu et al. 2014). Others have found associations between circulating plasma miRNA and FGR, gestational diabetes, and ectopic pregnancy (Zhao et al. 2011, 2012; Hromadnikova et al. 2012). Interestingly, searching for biomarkers of fetal hypoxia implicated in FGR, Whitehead et al. have found an association between miR-21 and miR-20b and umbilical cord lactate level (Whitehead et al. 2013). Lastly, members of the C19MC cluster (miR-520b, -520c-3p, -520f) have been suggested as indicators of response to treatment in women with hydatidiform mole (Miura et al. 2014).
Notwithstanding the interest in profiling miRNA in maternal plasma to discover biomarkers of placental disease, no single miRNA or combination of miRNAs has thus far been shown to reliably predict gestational diseases. Several studies have confirmed the relative stability of plasma miRNAs, rendering miRNA assessment an attractive diagnostic tool (Mitchell et al. 2008; Mraz et al. 2009). As discussed earlier, most miRNAs participate in maintaining homeostatic resilience, and thus do not show dramatic changes in response to physiologic or pathologic stimuli. In addition, patient-to-patient variability in plasma miRNA levels, along with temporal changes in miRNA levels during pregnancy, render this analyte less predictive. This variability in miRNA assays is amplified by the different forms of miRNA packaging in the plasma, where most analyses measure total plasma miRNA without stratification to vesicle type or protein-bound miRNA. The complexity of miRNA target prediction algorithms also hobbles the ability to envisage reliable miRNAs that match transcripts implicated in diseases. Finally, variability among researchers may be augmented by inconsistent tools used to isolate miRNAs from plasma or serum and the diversity of technology (e.g., PCR, microarrays, or RNAseq) used in the diagnostic assays.
SUMMARY
The discovery of miRNAs and their unique function in the regulation of gene expression has added a new dimension to our understanding of gene regulatory networks and their intricate roles in tissue development, differentiation, function, and the maintenance of homeostasis. Pregnancy is a unique state, in which miRNAs, as part of the genetic regulatory networks of two individuals, mediate crosstalk at the placental interface. Likewise, unique species and profiles of miRNAs that are expressed only during pregnancy, lead to intriguing questions regarding the function(s) of these miRNAs in pregnancy and the uses that might be made of these common and unique miRNA species in the diagnostics and therapeutics of gestational diseases. The identification of miRNA targets has been hampered by the suboptimal performance of miRNA target prediction algorithms. These problems are compounded by incomplete information on competitive endogenous miRNAs (e.g., circular RNAs, lncRNAs), which are known to be expressed in maternal plasma during pregnancy and may function to squelch miRNA signaling (Kosik 2013; Koh et al. 2014; Tay et al. 2014). Despite an onslaught of studies profiling the miRNA landscape in the maternal plasma, the use of miRNA-based diagnostic tools has not yet entered into the clinical arena. Basic and translational research into miRNA function and regulation during pregnancy, bolstered by analyses of miRNA communication among the maternal, placental, and fetal compartments, are greatly needed to translate the power of miRNA-based gene regulatory networks into a more complete understanding of health and disease.
ACKNOWLEDGMENTS
The authors thank Lori Rideout for assistance during preparation of this review and Bruce Campbell for editing. Research underlying our contribution to this review was supported by National Institutes of Health Grants NIH R01HD065893 and NIH R21HD071707 (to Y.S.), Pennsylvania Department of Health Research Formula Funds (to J.F.M.), NIH R01AI081759 and the Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award (to C.B.C.), and NIH R01HD075665 (to Y.S. and C.B.C.).
Footnotes
Editors: Diana W. Bianchi and Errol R. Norwitz
Additional Perspectives on Molecular Approaches to Reproductive and Newborn Medicine available at www.perspectivesinmedicine.org
REFERENCES
- Abdelmohsen K, Srikantan S, Tominaga K, Kang MJ, Yaniv Y, Martindale JL, Yang X, Park SS, Becker KG, Subramanian M, et al. 2012. Growth inhibition by miR-519 via multiple p21-inducing pathways. Mol Cell Biol 32: 2530–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharon A, Brenner B. 2013. Placenta-derived microparticles. Thromb Res 131: S22–S24. [DOI] [PubMed] [Google Scholar]
- Ahn K, Gim JA, Ha HS, Han K, Kim HS. 2013. The novel MER transposon-derived miRNAs in human genome. Gene 512: 422–428. [DOI] [PubMed] [Google Scholar]
- Akers JC, Gonda D, Kim R, Carter BS, Chen CC. 2013. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J Neurooncol 113: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton L, Olarerin-George AO, Schwartz N, Srinivas S, Bastek J, Hogenesch JB, Elovitz MA. 2013. miR-210 inhibits trophoblast invasion and is a serum biomarker for preeclampsia. Am J Pathol 183: 1437–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arck P, Hansen PJ, Mulac Jericevic B, Piccinni MP, Szekeres-Bartho J. 2007. Progesterone during pregnancy: Endocrine-immune cross talk in mammalian species and the role of stress. Am J Reprod Immunol 58: 268–279. [DOI] [PubMed] [Google Scholar]
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, et al. 2011. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci 108: 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avissar-Whiting M, Veiga KR, Uhl KM, Maccani MA, Gagne LA, Moen EL, Marsit CJ. 2010. Bisphenol A exposure leads to specific microRNA alterations in placental cells. Reprod Toxicol 29: 401–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, et al. 2012. Syndecan–syntenin–ALIX regulates the biogenesis of exosomes. Nat Cell Biol 14: 677–685. [DOI] [PubMed] [Google Scholar]
- Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK, Kroh EM, Bendoraite A, Mitchell PS, Nelson AM, et al. 2008. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells 26: 2496–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A, Bentwich I, Einav U, Gilad S, Hurban P, Karov Y, et al. 2004. MicroRNA expression detected by oligonucleotide microarrays: System establishment and expression profiling in human tissues. Genome Res 14: 2486–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bayer A, Delorme-Axford E, Sleigher C, Frey TK, Trobaugh DW, Klimstra WB, Emert-Sedlak LA, Smithgall TE, Kinchington PR, Vadia S, et al. 2014. Human trophoblasts confer resistance to viruses implicated in perinatal infection. Am J Obstet Gynecol 211: 10.1016/j.ajog.2014.1007.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. 2005. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 37: 766–770. [DOI] [PubMed] [Google Scholar]
- Best JM. 2007. Rubella. Semin Fetal Neonatal Med 12: 182–192. [DOI] [PubMed] [Google Scholar]
- Bortolin-Cavaille ML, Dance M, Weber M, Cavaille J. 2009. C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res 37: 3464–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckberry S, Bianco-Miotto T, Roberts CT. 2014. Imprinted and X-linked non-coding RNAs as potential regulators of human placental function. Epigenetics 9: 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccom J, Bourdeaut F, Loukh N, Rigau V, Milin S, Takin R, Richer W, Uro-Coste E, Couturier J, Bertozzi AI, et al. 2014. Embryonal tumor with multilayered rosettes: Diagnostic tools update and review of the literature. Clin Neuropathol 33: 15–22. [DOI] [PubMed] [Google Scholar]
- Chen DB, Wang W. 2013. Human placental microRNAs and preeclampsia. Biol Reprod 88: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Liang H, Zhang J, Zen K, Zhang CY. 2012. Secreted microRNAs: A new form of intercellular communication. Trends Cell Biol 22: 125–132. [DOI] [PubMed] [Google Scholar]
- Chi SW, Hannon GJ, Darnell RB. 2012. An alternative mode of microRNA target recognition. Nat Struct Mol Biol 19: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, Lo YM. 2008. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem 54: 482–490. [DOI] [PubMed] [Google Scholar]
- Choi AM, Ryter SW, Levine B. 2013a. Autophagy in human health and disease. N Engl J Med 368: 651–662. [DOI] [PubMed] [Google Scholar]
- Choi SY, Yun J, Lee OJ, Han HS, Yeo MK, Lee MA, Suh KS. 2013b. MicroRNA expression profiles in placenta with severe preeclampsia using a PNA-based microarray. Placenta 34: 799–804. [DOI] [PubMed] [Google Scholar]
- Cindrova-Davies T, Herrera EA, Niu Y, Kingdom J, Giussani DA, Burton GJ. 2013. Reduced cystathionine γ-Lyase and increased miR-21 expression are associated with increased vascular resistance in growth-restricted pregnancies: Hydrogen sulfide as a placental vasodilator. Am J Pathol 182: 1448–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleoni F, Padmanabhan N, Yung HW, Watson ED, Cetin I, Tissot van Patot MC, Burton GJ, Murray AJ. 2013. Suppression of mitochondrial electron transport chain function in the hypoxic human placenta: A role for miRNA-210 and protein synthesis inhibition. PLoS ONE 8: e55194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Thery C, Raposo G. 2013. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci 126: 5553–5565. [DOI] [PubMed] [Google Scholar]
- Cullen BR, Cherry S, tenOever BR. 2013. Is RNA interference a physiologically relevant innate antiviral immune response in mammals? Cell Host Microbe 14: 374–378. [DOI] [PubMed] [Google Scholar]
- Dai Y, Diao Z, Sun H, Li R, Qiu Z, Hu Y. 2011. MicroRNA-155 is involved in the remodelling of human-trophoblast-derived HTR-8/SVneo cells induced by lipopolysaccharides. Hum Reprod 26: 1882–1891. [DOI] [PubMed] [Google Scholar]
- Dai Y, Qiu Z, Diao Z, Shen L, Xue P, Sun H, Hu Y. 2012. MicroRNA-155 inhibits proliferation and migration of human extravillous trophoblast derived HTR-8/SVneo cells via down-regulating cyclin D1. Placenta 33: 824–829. [DOI] [PubMed] [Google Scholar]
- Delorme-Axford E, Donker RB, Mouillet JF, Chu T, Bayer A, Ouyang Y, Wang T, Stolz DB, Sarkar SN, Morelli AE, et al. 2013a. Human placental trophoblasts confer viral resistance to recipient cells. Proc Natl Acad Sci 110: 12048–12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme-Axford E, Bayer A, Sadovsky Y, Coyne CB. 2013b. Autophagy as a mechanism of antiviral defense at the maternal fetal interface. Autophagy 9: 2173–2174. [DOI] [PubMed] [Google Scholar]
- Ding SW. 2010. RNA-based antiviral immunity. Nat Rev Immunol 10: 632–644. [DOI] [PubMed] [Google Scholar]
- Donker RB, Mouillet JF, Chu T, Hubel CA, Stolz DB, Morelli AE, Sadovsky Y. 2012. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol Hum Reprod 18: 417–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doridot L, Miralles F, Barbaux S, Vaiman D. 2013. Trophoblasts, invasion, and microRNA. Front Genet 4: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doridot L, Houry D, Gaillard H, Chelbi ST, Barbaux S, Vaiman D. 2014. miR-34a expression, epigenetic regulation, and function in human placental diseases. Epigenetics 9: 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff P, Sweet RL, Edwards RK. 2009. Maternal and fetal infections. In Creasy and Resnik’s maternal fetal medicine (ed. Creasy RK, Resnik R, Iams JD, Lockwood CJ, Moore TR), pp. 739–798. Saunders-Elsevier, Philadelphia. [Google Scholar]
- El Andaloussi S, Mager I, Breakefield XO, Wood MJ. 2013. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat Rev Drug Discov 12: 347–357. [DOI] [PubMed] [Google Scholar]
- Enquobahrie DA, Abetew DF, Sorensen TK, Willoughby D, Chidambaram K, Williams MA. 2011. Placental microRNA expression in pregnancies complicated by preeclampsia. Am J Obstet Gynecol 204: 178.e112–178.e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergaz Z, Ornoy A. 2006. Parvovirus B19 in pregnancy. Reprod Toxicol 21: 421–435. [DOI] [PubMed] [Google Scholar]
- Esteller M. 2011. Non-coding RNAs in human disease. Nat Rev Genet 12: 861–874. [DOI] [PubMed] [Google Scholar]
- Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496: 367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor I, Bullerdiek J. 2012. The dark side of a success story: MicroRNAs of the C19MC cluster in human tumours. J Pathol 227: 270–274. [DOI] [PubMed] [Google Scholar]
- Flor I, Neumann A, Freter C, Helmke BM, Langenbuch M, Rippe V, Bullerdiek J. 2012. Abundant expression and hemimethylation of C19MC in cell cultures from placenta-derived stromal cells. Biochem Biophys Res Commun 422: 411–416. [DOI] [PubMed] [Google Scholar]
- Forbes K, Farrokhnia F, Aplin JD, Westwood M. 2012. Dicer-dependent miRNAs provide an endogenous restraint on cytotrophoblast proliferation. Placenta 33: 581–585. [DOI] [PubMed] [Google Scholar]
- Fu G, Ye G, Nadeem L, Ji L, Manchanda T, Wang Y, Zhao Y, Qiao J, Wang YL, Lye S, et al. 2013a. MicroRNA-376c impairs transforming growth factor-β and nodal signaling to promote trophoblast cell proliferation and invasion. Hypertension 61: 864–872. [DOI] [PubMed] [Google Scholar]
- Fu G, Brkic J, Hayder H, Peng C. 2013b. MicroRNAs in human placental development and pregnancy complications. Int J Mol Sci 14: 5519–5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga R, Han BW, Hung JH, Xu J, Weng Z, Zamore PD. 2012. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell 151: 533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabory A, Jammes H, Dandolo L. 2010. The H19 locus: Role of an imprinted non-coding RNA in growth and development. Bioessays 32: 473–480. [DOI] [PubMed] [Google Scholar]
- Gao WL, Liu M, Yang Y, Yang H, Liao Q, Bai Y, Li YX, Li D, Peng C, Wang YL. 2012. The imprinted H19 gene regulates human placental trophoblast cell proliferation via encoding miR-675 that targets Nodal Modulator 1 (NOMO1). RNA Biol 9: 1002–1010. [DOI] [PubMed] [Google Scholar]
- Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, et al. 2008. Serum microRNAs are promising novel biomarkers. PLoS ONE 3: e3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Sun J, Groome LJ, Wang Y. 2013. Differential miRNA expression profiles between the first and third trimester human placentas. Am J Physiol Endocrinol Metab 304: E836–E843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunel T, Zeybek YG, Akcakaya P, Kalelioglu I, Benian A, Ermis H, Aydinli K. 2011. Serum microRNA expression in pregnancies with preeclampsia. Genet Mol Res 10: 4034–4040. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Tsai SQ, Hardison NE, James AH, Motsinger-Reif AA, Thames B, Stone EA, Deng C, Piedrahita JA. 2013. Differentially expressed microRNAs and affected biological pathways revealed by modulated modularity clustering (MMC) analysis of human preeclamptic and IUGR placentas. Placenta 34: 599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Cashikar A. 2012. Multivesicular body morphogenesis. Annu Rev Cell Dev Biol 28: 337–362. [DOI] [PubMed] [Google Scholar]
- Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B. 1993. Tumour-suppressor activity of H19 RNA. Nature 365: 764–767. [DOI] [PubMed] [Google Scholar]
- Hibio N, Hino K, Shimizu E, Nagata Y, Ui-Tei K. 2012. Stability of miRNA 5′ terminal and seed regions is correlated with experimentally observed miRNA-mediated silencing efficacy. Sci Rep 2: 996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima A, Miura K, Mishima H, Kinoshita A, Jo O, Abe S, Hasegawa Y, Miura S, Yamasaki K, Yoshida A, et al. 2013. Characterization of placenta-specific microRNAs in fetal growth restriction pregnancy. Prenat Diagn 33: 214–222. [DOI] [PubMed] [Google Scholar]
- Hoffman Y, Dahary D, Bublik DR, Oren M, Pilpel Y. 2013. The majority of endogenous microRNA targets within Alu elements avoid the microRNA machinery. Bioinformatics 29: 894–902. [DOI] [PubMed] [Google Scholar]
- Hromadnikova I, Kotlabova K, Doucha J, Dlouha K, Krofta L. 2012. Absolute and relative quantification of placenta-specific microRNAs in maternal circulation with placental insufficiency-related complications. J Mol Diagn 14: 160–167. [DOI] [PubMed] [Google Scholar]
- Hu Y, Li P, Hao S, Liu L, Zhao J, Hou Y. 2009. Differential expression of microRNAs in the placentae of Chinese patients with severe pre-eclampsia. Clin Chem Lab Med 47: 923–929. [DOI] [PubMed] [Google Scholar]
- Huang X, Le QT, Giaccia AJ. 2010. MiR-210—Micromanager of the hypoxia pathway. Trends Mol Med 16: 230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Shen Z, Xu Q, Huang X, Chen Q, Li D. 2013. Increased levels of microRNA-424 are associated with the pathogenesis of fetal growth restriction. Placenta 34: 624–627. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838. [DOI] [PubMed] [Google Scholar]
- Ishibashi O, Ohkuchi A, Ali MM, Kurashina R, Luo SS, Ishikawa T, Takizawa T, Hirashima C, Takahashi K, Migita M, et al. 2012. Hydroxysteroid (17-β) dehydrogenase 1 is dysregulated by MiR-210 and MiR-518c that are aberrantly expressed in preeclamptic placentas: A novel marker for predicting preeclampsia. Hypertension 59: 265–273. [DOI] [PubMed] [Google Scholar]
- Ishihara N, Matsuo H, Murakoshi H, Laoag-Fernandez JB, Samoto T, Maruo T. 2002. Increased apoptosis in the syncytiotrophoblast in human term placentas complicated by either preeclampsia or intrauterine growth retardation. Am J Obstet Gynecol 186: 158–166. [DOI] [PubMed] [Google Scholar]
- Jaskiewicz L, Filipowicz W. 2008. Role of Dicer in posttranscriptional RNA silencing. Curr Top Microbiol Immunol 320: 77–97. [DOI] [PubMed] [Google Scholar]
- Kaplan C. 1993. The placenta and viral infections. Semin Diagn Pathol 10: 232–250. [PubMed] [Google Scholar]
- Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. 2012. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol 14: 659–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinrouweler CE, van Uitert M, Moerland PD, Ris-Stalpers C, van der Post JA, Afink GB. 2013. Differentially expressed genes in the pre-eclamptic placenta: A systematic review and meta-analysis. PLoS ONE 8: e68991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh W, Pan W, Gawad C, Fan HC, Kerchner GA, Wyss-Coray T, Blumenfeld YJ, El-Sayed YY, Quake SR. 2014. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc Natl Acad Sci 111: 7361–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva SE, Chiasson VL, Mitchell BM, Chatterjee P. 2013. TLR3-induced placental miR-210 down-regulates the STAT6/interleukin-4 pathway. PLoS ONE 8: e67760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS. 2013. Molecular biology: Circles reshape the RNA world. Nature 495: 322–324. [DOI] [PubMed] [Google Scholar]
- Kotlabova K, Doucha J, Hromadnikova I. 2011. Placental-specific microRNA in maternal circulation—Identification of appropriate pregnancy-associated microRNAs with diagnostic potential. J Reprod Immunol 89: 185–191. [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11: 597–610. [DOI] [PubMed] [Google Scholar]
- Kumar P, Luo Y, Tudela C, Alexander JM, Mendelson CR. 2013. The c-Myc-regulated microRNA-17∼92 (miR-17∼92) and miR-106a∼363 clusters target hCYP19A1 and hGCM1 to inhibit human trophoblast differentiation. Mol Cell Biol 33: 1782–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent LC, Chen J, Ulitsky I, Mueller FJ, Lu C, Shamir R, Fan JB, Loring JF. 2008. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells 26: 1506–1516. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419. [DOI] [PubMed] [Google Scholar]
- Lee DC, Romero R, Kim JS, Tarca AL, Montenegro D, Pineles BL, Kim E, Lee J, Kim SY, Draghici S, et al. 2011. miR-210 targets iron-sulfur cluster scaffold homologue in human trophoblast cell lines: Siderosis of interstitial trophoblasts as a novel pathology of preterm preeclampsia and small-for-gestational-age pregnancies. Am J Pathol 179: 590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert S, Van Loo P, Thilakarathne PJ, Marynen P, Verbeke G, Schuit FC. 2009. Evidence for co-evolution between human microRNAs and Alu-repeats. PLoS ONE 4: e4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Lee KF, Lu Y, Clarke I, Shih D, Eberhart C, Collins VP, Van Meter T, Picard D, Zhou L, et al. 2009. Frequent amplification of a chr19q13.41 microRNA polycistron (C19MC) in aggressive primitive neuroectodermal brain tumors. Cancer Cell 16: 533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Guo W, Du L, Zhao J, Wang Y, Liu L, Hu Y, Hou Y. 2013a. microRNA-29b contributes to pre-eclampsia through its effects on apoptosis, invasion and angiogenesis of trophoblast cells. Clin Sci (Lond) 124: 27–40. [DOI] [PubMed] [Google Scholar]
- Li Y, Lu J, Han Y, Fan X, Ding SW. 2013b. RNA interference functions as an antiviral immunity mechanism in mammals. Science 342: 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Ridzon D, Wong L, Chen C. 2007. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu RF, Xu X, Huang J, Fei QL, Chen F, Li YD, Han ZG. 2013. Down-regulation of miR-517a and miR-517c promotes proliferation of hepatocellular carcinoma cells via targeting Pyk2. Cancer Lett 329: 164–173. [DOI] [PubMed] [Google Scholar]
- Longman RE, Johnson TR. 2007. Viral respiratory disease in pregnancy. Curr Opin Obstet Gynecol 19: 120–125. [DOI] [PubMed] [Google Scholar]
- Luo SS, Ishibashi O, Ishikawa G, Ishikawa T, Katayama A, Mishima T, Takizawa T, Shigihara T, Goto T, Izumi A, et al. 2009. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol Reprod 81: 717–729. [DOI] [PubMed] [Google Scholar]
- Luo L, Ye G, Nadeem L, Fu G, Yang BB, Honarparvar E, Dunk C, Lye S, Peng C. 2012. MicroRNA-378a-5p promotes trophoblast cell survival, migration and invasion by targeting Nodal. J Cell Sci 125: 3124–3132. [DOI] [PubMed] [Google Scholar]
- Maccani MA, Marsit CJ. 2011. Exposure and fetal growth-associated miRNA alterations in the human placenta. Clin Epigenetics 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccani MA, Avissar-Whiting M, Banister CE, McGonnigal B, Padbury JF, Marsit CJ. 2010. Maternal cigarette smoking during pregnancy is associated with downregulation of miR-16, miR-21, and miR-146a in the placenta. Epigenetics 5: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccani MA, Padbury JF, Marsit CJ. 2011. miR-16 and miR-21 expression in the placenta is associated with fetal growth. PLoS ONE 6: e21210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccani MA, Padbury JF, Lester BM, Knopik VS, Marsit CJ. 2013. Placental miRNA expression profiles are associated with measures of infant neurobehavioral outcomes. Pediatr Res 74: 272–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlin ES, Sarnow P, Sagan SM. 2012. Combating hepatitis C virus by targeting microRNA-122 using locked nucleic acids. Curr Gene Ther 12: 301–306. [DOI] [PubMed] [Google Scholar]
- Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O. 2013. Antiviral RNA interference in mammalian cells. Science 342: 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasa BS, Srikantan S, Martindale JL, Kim MM, Lee EK, Gorospe M, Abdelmohsen K. 2010. MicroRNA profiling in human diploid fibroblasts uncovers miR-519 role in replicative senescence. Aging (Albany NY) 2: 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron JL, Johnson KL, Slonim D, Lai CQ, Ramoni M, Alterovitz G, Jarrah Z, Yang Z, Bianchi DW. 2007. Gene expression analysis in pregnant women and their infants identifies unique fetal biomarkers that circulate in maternal blood. J Clin Invest 117: 3007–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 11: 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574. [DOI] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. 2005. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123: 607–620. [DOI] [PubMed] [Google Scholar]
- Mayhew TM. 2014. Turnover of human villous trophoblast in normal pregnancy: What do we know and what do we need to know? Placenta 35: 229–240. [DOI] [PubMed] [Google Scholar]
- Mayor-Lynn K, Toloubeydokhti T, Cruz AC, Chegini N. 2011. Expression profile of microRNAs and mRNAs in human placentas from pregnancies complicated by preeclampsia and preterm labor. Reprod Sci 18: 46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckes DG Jr, Raab-Traub N. 2011. Microvesicles and viral infection. J Virol 85: 12844–12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, et al. 2008. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci 105: 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelbrunn M, Sanchez-Madrid F. 2012. Intercellular communication: Diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol 13: 328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Miura S, Yamasaki K, Higashijima A, Kinoshita A, Yoshiura K, Masuzaki H. 2010. Identification of pregnancy-associated microRNAs in maternal plasma. Clin Chem 56: 1767–1771. [DOI] [PubMed] [Google Scholar]
- Miura K, Hasegawa Y, Abe S, Higashijima A, Miura S, Mishima H, Kinoshita A, Kaneuchi M, Yoshiura K, Masuzaki H. 2014. Clinical applications of analysis of plasma circulating complete hydatidiform mole pregnancy-associated miRNAs in gestational trophoblastic neoplasia: A preliminary investigation. Placenta. 35: 787–789. [DOI] [PubMed] [Google Scholar]
- Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan ML, Karlsson JM, Baty CJ, Gibson GA, Erdos G, Wang Z, et al. 2012. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 119: 756–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor G, Cardenas I. 2010. The immune system in pregnancy: A unique complexity. Am J Reprod Immunol 63: 425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Prieto DM, Chaiwangyen W, Ospina-Prieto S, Schneider U, Herrmann J, Gruhn B, Markert UR. 2012. MicroRNA expression profiles of trophoblastic cells. Placenta 33: 725–734. [DOI] [PubMed] [Google Scholar]
- Morales-Prieto DM, Ospina-Prieto S, Chaiwangyen W, Schoenleben M, Markert UR. 2013. Pregnancy-associated miRNA-clusters. J Reprod Immunol 97: 51–61. [DOI] [PubMed] [Google Scholar]
- Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. 2008. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18: 610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillet JF, Chu T, Nelson DM, Mishima T, Sadovsky Y. 2010a. MiR-205 silences MED1 in hypoxic primary human trophoblasts. FASEB J 24: 2030–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillet JF, Chu T, Hubel CA, Nelson DM, Parks WT, Sadovsky Y. 2010b. The levels of hypoxia-regulated microRNAs in plasma of pregnant women with fetal growth restriction. Placenta 31: 781–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillet JF, Donker RB, Mishima T, Cronqvist T, Chu T, Sadovsky Y. 2013. The unique expression and function of miR-424 in human placental trophoblasts. Biol Reprod 89: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillet JF, Ouyang Y, Bayer A, Coyne CB, Sadovsky Y. 2014. The role of trophoblastic microRNAs in placental viral infection. Int J Dev Biol 58: 281–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya PR, Wendland JR, Salemme J, Fried RL, Murphy DL. 2013. miR-15a and miR-16 regulate serotonin transporter expression in human placental and rat brain raphe cells. Int J Neuropsychopharmacol 16: 621–629. [DOI] [PubMed] [Google Scholar]
- Mraz M, Malinova K, Mayer J, Pospisilova S. 2009. MicroRNA isolation and stability in stored RNA samples. Biochem Biophys Res Commun 390: 1–4. [DOI] [PubMed] [Google Scholar]
- Muralimanoharan S, Maloyan A, Mele J, Guo C, Myatt LG, Myatt L. 2012. MIR-210 modulates mitochondrial respiration in placenta with preeclampsia. Placenta 33: 816–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng EK, Tsui NB, Lau TK, Leung TN, Chiu RW, Panesar NS, Lit LC, Chan KW, Lo YM. 2003. mRNA of placental origin is readily detectable in maternal plasma. Proc Natl Acad Sci 100: 4748–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro G, Adler SP, La Torre R, Best AM. 2005. Passive immunization during pregnancy for congenital cytomegalovirus infection. N Engl J Med 353: 1350–1362. [DOI] [PubMed] [Google Scholar]
- Noguer-Dance M, Abu-Amero S, Al-Khtib M, Lefevre A, Coullin P, Moore GE, Cavaille J. 2010. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet 19: 3566–3582. [DOI] [PubMed] [Google Scholar]
- Nykanen A, Haley B, Zamore PD. 2001. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107: 309–321. [DOI] [PubMed] [Google Scholar]
- O’Loughlin AJ, Woffindale CA, Wood MJ. 2012. Exosomes and the emerging field of exosome-based gene therapy. Curr Gene Ther 12: 262–274. [DOI] [PubMed] [Google Scholar]
- Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, et al. 2010. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol 12: 19–30; sup pp. 11–13. [DOI] [PubMed] [Google Scholar]
- Ouyang Y, Mouillet JF, Coyne CB, Sadovsky Y. 2014. Review: Placenta-specific microRNAs in exosomes—Good things come in nano-packages. Placenta 35: S69–S73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar C, et al. 2012. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 18: 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron MP, Provost P. 2009. Protein components of the microRNA pathway and human diseases. Methods Mol Biol 487: 369–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineles BL, Romero R, Montenegro D, Tarca AL, Han YM, Kim YM, Draghici S, Espinoza J, Kusanovic JP, Mittal P, et al. 2007. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am J Obstet Gynecol 196: 261.e1–261.e6. [DOI] [PubMed] [Google Scholar]
- Poon LL, Leung TN, Lau TK, Lo YM. 2000. Presence of fetal RNA in maternal plasma. Clin Chem 46: 1832–1834. [PubMed] [Google Scholar]
- Ramakrishnaiah V, Thumann C, Fofana I, Habersetzer F, Pan Q, de Ruiter PE, Willemsen R, Demmers JA, Stalin Raj V, Jenster G, et al. 2013. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci 110: 13109–13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Stoorvogel W. 2013. Extracellular vesicles: Exosomes, microvesicles, and friends. J Cell Biol 200: 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record M. 2014. Intercellular communication by exosomes in placenta: A possible role in cell fusion? Placenta 35: 297–302. [DOI] [PubMed] [Google Scholar]
- Redman CW, Sargent IL. 2008. Circulating microparticles in normal pregnancy and pre-eclampsia. Placenta 29: S73–S77. [DOI] [PubMed] [Google Scholar]
- Redman CW, Tannetta DS, Dragovic RA, Gardiner C, Southcombe JH, Collett GP, Sargent IL. 2012. Review: Does size matter? Placental debris and the pathophysiology of pre-eclampsia. Placenta 33: S48–S54. [DOI] [PubMed] [Google Scholar]
- Ren J, Jin P, Wang E, Marincola FM, Stroncek DF. 2009. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J Transl Med 7: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe V, Dittberner L, Lorenz VN, Drieschner N, Nimzyk R, Sendt W, Junker K, Belge G, Bullerdiek J. 2010. The two stem cell microRNA gene clusters C19MC and miR-371–3 are activated by specific chromosomal rearrangements in a subgroup of thyroid adenomas. PLoS ONE 5: e9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Suzuki H, Tsugawa H, Nakagawa I, Matsuzaki J, Kanai Y, Hibi T. 2009. Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA-512-5p with downregulation of Mcl-1 in human gastric cancer cells. Oncogene 28: 2738–2744. [DOI] [PubMed] [Google Scholar]
- Scita G, Di Fiore PP. 2010. The endocytic matrix. Nature 463: 464–473. [DOI] [PubMed] [Google Scholar]
- Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaille J. 2004. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res 14: 1741–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R, Pilpel Y, Oren M. 2010. Repression of transposable-elements—A microRNA anti-cancer defense mechanism? Trends Genet 26: 253–259. [DOI] [PubMed] [Google Scholar]
- Shen J, Xia W, Khotskaya YB, Huo L, Nakanishi K, Lim SO, Du Y, Wang Y, Chang WC, Chen CH, et al. 2013. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 497: 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler B, Ivanovska I, Mehta K, Song S, Nelson A, Tan Y, Mathieu J, Darby C, Blau CA, Ware C, et al. 2010. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev 19: 935–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Wu W, Xu X, Huang L, Gao Q, Chen H, Sun H, Xia Y, Sha J, Wang X, et al. 2013. miR-141 contributes to fetal growth restriction by regulating PLAG1 expression. PLoS ONE 8: e58737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Rinn J, Pandolfi PP. 2014. The multilayered complexity of ceRNA crosstalk and competition. Nature 505: 344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- tenOever BR. 2013. RNA viruses and the host microRNA machinery. Nat Rev Microbiol 11: 169–180. [DOI] [PubMed] [Google Scholar]
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. 2008. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319: 1244–1247. [DOI] [PubMed] [Google Scholar]
- Tsai KW, Kao HW, Chen HC, Chen SJ, Lin WC. 2009. Epigenetic control of the expression of a primate-specific microRNA cluster in human cancer cells. Epigenetics 4: 587–592. [DOI] [PubMed] [Google Scholar]
- Tsutsumi A, Kawamata T, Izumi N, Seitz H, Tomari Y. 2011. Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nat Struct Mol Biol 18: 1153–1158. [DOI] [PubMed] [Google Scholar]
- Umemura K, Ishioka S, Endo T, Ezaka Y, Takahashi M, Saito T. 2013. Roles of microRNA-34a in the pathogenesis of placenta accreta. J Obstet Gynaecol Res 39: 67–74. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9: 654–659. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Marshall WS, Olson EN. 2008. Toward microRNA-based therapeutics for heart disease: The sense in antisense. Circ Res 103: 919–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura W, Koide K, Hori K, Yotsumoto J, Sekizawa A, Saito H, Okai T. 2013. Placental expression of microRNA-17 and -19b is down-regulated in early pregnancy loss. Eur J Obstet Gynecol Reprod Biol 169: 28–32. [DOI] [PubMed] [Google Scholar]
- Vickers KC, Remaley AT. 2012. Lipid-based carriers of microRNAs and intercellular communication. Curr Opin Lipidol 23: 91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. 2011. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 13: 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang S, Weber J, Baxter D, Galas DJ. 2010. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res 38: 7248–7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Feng L, Zhang H, Hachy S, Satohisa S, Laurent LC, Parast M, Zheng J, Chen DB. 2012. Preeclampsia up-regulates angiogenesis-associated microRNA (i.e., miR-17, -20a, and -20b) that target ephrin-B2 and EPHB4 in human placenta. J Clin Endocrinol Metab 97: E1051–E1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Na Q, Song WW, Song GY. 2014. Altered expression of miR-518b and miR-519a in the placenta is associated with low fetal birth weight. Am J Perinatol 31: 729–734. [DOI] [PubMed] [Google Scholar]
- Whitehead CL, Teh WT, Walker SP, Leung C, Larmour L, Tong S. 2013. Circulating microRNAs in maternal blood as potential biomarkers for fetal hypoxia in-utero. PLoS ONE 8: e78487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482: 331–338. [DOI] [PubMed] [Google Scholar]
- Williams Z, Ben-Dov IZ, Elias R, Mihailovic A, Brown M, Rosenwaks Z, Tuschl T. 2013. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc Natl Acad Sci 110: 4255–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhou H, Lin H, Qi J, Zhu C, Gao Z, Wang H. 2012. Circulating microRNAs are elevated in plasma from severe preeclamptic pregnancies. Reproduction 143: 389–397. [DOI] [PubMed] [Google Scholar]
- Xie L, Mouillet JF, Chu T, Parks WT, Sadovsky E, Knofler M, Sadovsky Y. 2014. C19MC microRNAs regulate the migration of human trophoblasts. Endocrinology 155: 4975–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Yang BF, Ai J. 2013. MicroRNA transport: A new way in cell communication. J Cell Physiol 228: 1713–1719. [DOI] [PubMed] [Google Scholar]
- Xu P, Zhao Y, Liu M, Wang Y, Wang H, Li YX, Zhu X, Yao Y, Wang H, Qiao J, et al. 2014. Variations of microRNAs in human placentas and plasma from preeclamptic pregnancy. Hypertension 63: 1276–1284. [DOI] [PubMed] [Google Scholar]
- Xue P, Zheng M, Diao Z, Shen L, Liu M, Gong P, Sun H, Hu Y. 2013. miR-155* mediates suppressive effect of PTEN 3′-untranslated region on AP-1/NF-κB pathway in HTR-8/SVneo cells. Placenta 34: 650–656. [DOI] [PubMed] [Google Scholar]
- Yan T, Cui K, Huang X, Ding S, Zheng Y, Luo Q, Liu X, Zou L. 2014. Assessment of therapeutic efficacy of miR-126 with contrast-enhanced ultrasound in preeclampsia rats. Placenta 35: 23–29. [DOI] [PubMed] [Google Scholar]
- Yang Q, Lu J, Wang S, Li H, Ge Q, Lu Z. 2011. Application of next-generation sequencing technology to profile the circulating microRNAs in the serum of preeclampsia versus normal pregnant women. Clin Chim Acta 412: 2167–2173. [DOI] [PubMed] [Google Scholar]
- Zhang R, Wang YQ, Su B. 2008. Molecular evolution of a primate-specific microRNA family. Mol Biol Evol 25: 1493–1502. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Fei M, Xue G, Zhou Q, Jia Y, Li L, Xin H, Sun S. 2012. Elevated levels of hypoxia-inducible microRNA-210 in pre-eclampsia: New insights into molecular mechanisms for the disease. J Cell Mol Med 16: 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Dong J, Jiang T, Shi Z, Yu B, Zhu Y, Chen D, Xu J, Huo R, Dai J, et al. 2011. Early second-trimester serum miRNA profiling predicts gestational diabetes mellitus. PLoS ONE 6: e23925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Zhao Q, Warrick J, Lockwood CM, Woodworth A, Moley KH, Gronowski AM. 2012. Circulating microRNA miR-323-3p as a biomarker of ectopic pregnancy. Clin Chem 58: 896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Moley KH, Gronowski AM. 2013. Diagnostic potential for miRNAs as biomarkers for pregnancy-specific diseases. Clin Biochem 46: 953–960. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Campbell EC, Lucocq J, Riches A, Powis SJ. 2013. Monitoring the Rab27 associated exosome pathway using nanoparticle tracking analysis. Exp Cell Res 319: 1706–1713. [DOI] [PubMed] [Google Scholar]
- Zhu XM, Han T, Sargent IL, Yin GW, Yao YQ. 2009. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am J Obstet Gynecol 200: e661–e667. [DOI] [PubMed] [Google Scholar]