Abstract
Modern tuberculosis (TB) chemotherapy is widely viewed as a crowning triumph of anti-infectives research. However, only one new TB drug has entered clinical practice in the past 40 years while drug resistance threatens to further destabilize the pandemic. Here, we review a brief history of TB drug development, focusing on the evolution of mechanism(s)-of-action studies and key conceptual barriers to rational, mechanism-based drugs.
Frontline tuberculosis (TB) drugs include isoniazid, pyrazinamide, ethambutol, and rifampicin, all developed more than 40 years ago. The rational, mechanism-based development of new TB drugs is urgently needed.
History recounts streptomycin and para-aminosalicylic acid (PAS) among the first clinical antibiotics developed. Both showed activity against Mycobacterium tuberculosis (Mtb) and were followed in rapid succession by isoniazid, pyrazinamide, cycloserine, ethionamide, ethambutol, and rifampicin, among others (Barry 2011; Zumla et al. 2013). Together, these advances transformed tuberculosis (TB) from a predictably fatal disease to a curable disease. However, current TB chemotherapies remain far from optimal. TB remains the leading bacterial cause of deaths worldwide, and drug resistance has emerged as a problem of singular importance (Nathan 2009; Russell et al. 2010; Raviglione et al. 2012).
Somewhat unexpectedly, efforts to restock this once celebrated medicine chest have faltered. The causes for this are multifactorial (IDSA 2004; Payne et al. 2007; Zumla et al. 2014). However, mounting evidence has implicated a growing inadequacy of existing tools and approaches (Nathan 2004; Nathan et al. 2008). Although technologically advanced, anti-infectives research remains rooted in the same empirical paradigm that first gave rise to penicillin. While this paradigm was initially reinforced by its delivery of all major classes of clinical antibiotics, its productivity has since waned. We believe that this early success fostered an unintended dissociation between our ability to develop antibiotics and understanding of their mechanisms. Rational, or mechanism-based, paradigms have thus lagged behind.
Here, we review existing knowledge of the mechanisms of current TB drugs and emerging experimental agents. We focus specifically on (1) the frontline agents isoniazid, pyrazinamide, ethambutol, and rifampicin, because of their clinical importance and legacy in the history of anti-infectives development and (2) bedaquiline because of its role as the first new clinically approved TB drug developed in the past 40 years. However, rather than providing an exhaustive recitation of the literature, which has been extensively cataloged elsewhere (Zhang 2005; Barry 2011; Zumla et al. 2013), we review key findings that informed our current understanding of drug mechanisms and the historical contexts in which they were discovered. By doing so, we highlight recurrent themes that have guided past and current TB drug development efforts and yet unaddressed barriers in the path toward more rational mechanism-based approaches.
HISTORICAL PERSPECTIVES ON TB DRUG DEVELOPMENT
Isoniazid (INH)
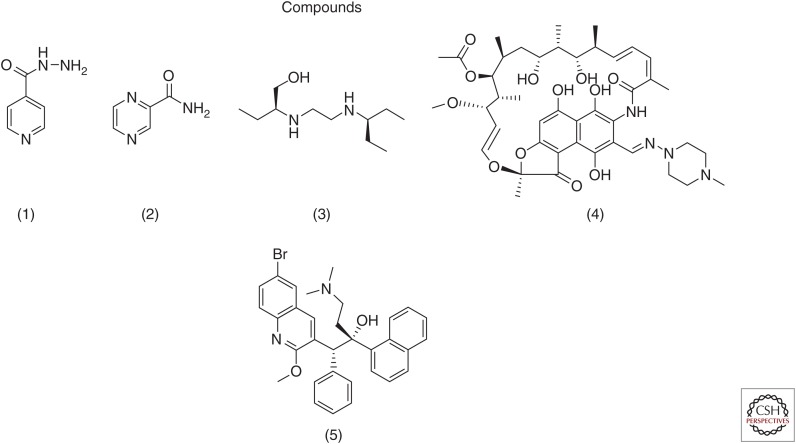
Isoniazid (INH) (isonicotinic acid hydrazide) (Fig. 1(1)) is arguably among the most clinically successful and extensively studied TB drugs ever developed (Vilcheze and Jacobs 2007). However, although once synonymous with TB chemotherapy and prophylaxis, INH has paradoxically become a defining feature of the current multidrug-resistant (MDR) and extensively drug-resistant (XDR) epidemic. Historical details of its discovery and studies around its antimycobacterial activities, however, are probably far less well appreciated.
Figure 1.
Antitubercular compounds.
INH was first synthesized in 1912. However, its antitubercular activity was only later discovered en route to the synthesis of amithiozone or thioacetazone (p-acetaminobenzaldehyde thiosemicarbazone), a pyridine-based thiosemicarbazone that was being developed in an effort to improve on the tuberculostatic activity of sulfathiazole, first reported by Domagk (Fox 1952; Long 1958). Whereas amithiozone was found to be tuberculostatic in vitro and therapeutically abandoned due to untoward gastrointestinal, allergic, and hematologic toxicities in patients, INH was found to be vastly superior to all other antitubercular compounds tested in vitro and in animals at the time, including closely related nicotinic acid and hydrazine derivatives (Fig. 1). In addition to its potency, INH was found to exhibit remarkable selectivity for mycobacteria, with activity against strains resistant to streptomycin, PAS, and amithiozone (Long 1958).
Numerous in vitro studies soon showed that INH exhibited activity against Mtb only when replicating and that this activity was preceded by a highly reproducible, although still unexplained, delay of approximately a generation and a half (1–4 d duration). During this period (the initial 16–24 h of exposure), it was noted that INH-susceptible bacilli accumulated 14C-labeled INH that could not be washed away while resistant or dead bacilli did not, and this fixation was accompanied by a loss of its acid fastness and morphologic changes such as surface wrinkling and bulging (vide supra).
Concurrent studies by Chorine (1945) serendipitously reported that the structurally related pyridine, nicotinamide, exhibited activity against Mtb in vitro and in infected guinea pigs, prompting a litany of studies examining the potential antimetabolic activity of INH (Chorine 1945; Fox 1952; Long 1958; Vilcheze and Jacobs 2007). These studies revealed reductions in NAD levels in INH-treated bacilli but failed to deliver evidence of its direct misincorporation into NAD, whereas the more closely related species nicotinic acid was found to exhibit only weak in vitro activity and no activity in guinea pigs. Notwithstanding, INH was found to induce numerous changes in the biochemical composition and activity of Mtb. These included selective effects on its extractable lipids, accumulations of soluble disaccharide trehalose and hexose phosphates, and reductions in respiratory activity. Pope further showed that the activity of INH could be antagonized by the addition of pyridoxine (a precursor of the pyridoxal phosphate cofactor required for many decarboxylation and transamination reactions) and the α-ketoacids, α-ketoglutarate and pyruvate. Although still unexplained, this antagonism was particularly remarkable as pyridoxine also appeared to paradoxically resensitize resistant strains to INH (Boone and Woodward 1953; Pope 1953, 1956; Long 1958).
Serendipity aside, progress toward a functional understanding of the mechanism of action of INH was catalyzed by INH-resistant isolates that were recovered in vitro and in vivo, either following treatment of experimentally infected animals or from the sputum of TB patients, almost immediately after the discovery of INH itself. Work by Middlebrook specifically showed that most INH resistance was accompanied by defects in catalase activity (Middlebrook 1954). Saroja and Gopinathan subsequently showed that INH sensitivity could be restored on transfer of a genetic locus from an INH-sensitive strain of Mycobacterium smegmatis encoding a catalase-like activity (manifest, at the time, as resistance to hydrogen peroxide) (Saroja and Gopinathan 1973). Together, these findings introduced the concept that INH might function as a prodrug. Work by Zhang and colleagues later extended these findings to Mtb, with the discovery of the katG-encoded catalase-peroxidase, which was found to harbor point mutations and deletions in INH-resistant clinical isolates and shown to be sufficient to restore INH susceptibility in resistant strains. Follow-up biochemical studies showed that KatG activates INH by conversion to a range of chemically reactive intermediates, including an isonicotinoyl radical (Johnsson and Schultz 1994; Wilming and Johnsson 1999; Lei et al. 2000), first hypothesized by Winder (1960). Of interest, INH was biochemically also shown to be a potent inhibitor of KatG (Marcinkeviciene et al. 1995).
Adopting a similar approach, Jacobs and colleagues isolated and genetically characterized mutants of M. smegmatis resistant to INH and ethionamide (a closely related analog) but exhibiting wild-type levels of catalase activity and susceptibility to other antitubercular compounds (Banerjee et al. 1994). These studies identified a gene named inhA, that showed >40% identity to proteins involved in fatty acid biosynthesis in Escherichia coli and Salmonella typhimurium. These mutants were found to harbor a missense mutation (S94A) that conferred an order of magnitude increase in resistance but could be corrected on allelic exchange with a wild-type copy of the inhA gene. High-level resistance could conversely be achieved by overexpressing the wild-type inhA gene on a multicopy plasmid. Biochemical studies showed that InhA was a NADH-dependent enoyl-ACP (acyl carrier protein) reductase of the fatty acid synthase type II (FASII) system (Dessen et al. 1995; Quemard et al. 1995; Marrakchi et al. 2000), involved in mycolic acid biosynthesis. Moreover, these studies showed that INH could inhibit InhA activity in cell-free lysates but that this inhibition was reduced in inhA mutant strains.
Using methods of specialized genetic transduction, Jacobs et al. finally showed that the S94A mutation was sufficient to confer coresistance to both the phenotypic and mycolic acid inhibitory activities of INH and ETH at clinically relevant levels of INH (Vilcheze et al. 2006). Together, these findings helped elucidate the significance of (1) decades-long observations reporting INH’s effects on acid-fastness (Barclay et al. 1954; Koch-Weser et al. 1955), which was later found to be caused by the unique dye-binding properties of the mycolic acids covalently linked to the 5′-hydroxyl groups of the arabinogalactan polymer of the mycobacterial cell wall (McNeil et al. 1991); (2) the demonstration that isoniazid inhibited the synthesis of mycolic acids in Mtb (Winder and Collins 1970; Winder et al. 1970) and its correlation with viability (Takayama et al. 1972); and (3) the specific inhibitory effect of isoniazid on the synthesis of saturated fatty acids greater than 26 carbons (Wang and Takayama 1972; Takayama et al. 1975; Davidson and Takayama 1979).
Building on this work, structural studies of the orthologous enoyl reductase of E. coli revealed the presence of a covalent NAD inhibitor species tightly bound to the same proposed region associated with INH resistance in InhA (Baldock et al. 1996). These studies paved the way for the unifying discovery by Rozwarski et al. (1998) that INH did not bind directly to InhA but, instead, as a covalent adduct with NAD. This INH-NAD species was shown to act as a potent, long residence time inhibitor of InhA (Lei et al. 2000; Nguyen et al. 2002; Rawat et al. 2003; Vilcheze et al. 2006). The current working model of the INH mechanism thus proposes that INH is a prodrug activated by the catalase-peroxidase KatG which gives rise to a diverse array of INH-derived radicals and adducts, some of which are capable of killing Mtb by potently inhibiting its ability to synthesize mycolic acids. The identities, quantities, and extent to which additional INH-derived species produced by KatG may also synergize with and/or contribute to the remarkable whole-cell potency of INH itself, however, remains to be elucidated.
Pyrazinamide (PZA)
As previously described, pyrazinamide (PZA) (Fig. 1(2)) emerged as an outgrowth of research conducted by Vital Chorine who discovered the ability of subcutaneous nicotinamide to prolong survival of Mtb-infected guinea pigs (Chorine 1945). Motivated by this finding, PZA emerged from Lederle Laboratories of American Cynamid as the most active pyrazine analog of nicotinamide tested in mice (Malone et al. 1952). Somewhat unexpectedly, however, PZA was found to lack activity under typical in vitro culture conditions, instead requiring incubation at an acidic pH (e.g., 5.5) similar to that associated with active inflammation (McDermott and Tompsett 1954). However, even under such conditions, PZA showed only bacteriostatic activity with minimum growth inhibitory concentrations (MIC) ranging from 6 to 50 µg mL−1 and minimal bactericidal concentrations (MBC) of >1000 µg mL−1 (Zhang and Mitchison 2003). This dissociation was further emphasized by the subsequent discovery of PZA’s unique treatment shortening (or sterilizing) activity in animals and patients at achieved serum concentrations generally at or below its in vitro MICs (Ellard 1969). Studies of PZA’s mechanism(s) of action have thus been hindered by fundamental limitations in the ability to faithfully model its therapeutic activities in vitro.
Limitations notwithstanding, studies of PZA’s mechanism(s) of action, like INH, were driven by the in vitro characterization of PZA-resistant strains. McDermott and colleagues showed that, like INH, PZA is a prodrug but activated by an amidase, later found to be encoded by the gene pncA, whose ortholog in the closely related and naturally PZA-resistant vaccine strain Mycobacterium bovis BCG (Bacillus Calmette–Guérin) was found to harbor an inactivating missense mutation (Konno et al. 1967; Scorpio and Zhang 1996). Moreover, transformation of BCG or pncA-defective Mtb strains was shown to restore or confer in vitro susceptibility to PZA (Scorpio et al. 1997; Zhang et al. 1999; Boshoff and Mizrahi 2000). Curiously, mutations associated with PZA resistance apart from those mapping to pncA have been only rarely described, and attempts to isolate mutants resistant to POA have been unsuccessful. Meticulous biochemical studies by Zhang and colleagues nonetheless showed that in vitro susceptibility to PZA was associated with an intracellular accumulation of its activated form, pyrazinoic acid (POA), as revealed by high rates of POA efflux and the natural resistance of the amidase-proficient mycobacterial species, M. smegmatis (Zhang et al. 1999). This accumulation was associated with a dissipation of mycobacterial membrane potential, although the causal significance of this effect remains unresolved (Zhang et al. 2003). More recent affinity-based studies of POA identified several additional high-affinity binding targets. Prominent among these was a subunit of the 30S ribosome, named RpsA, which plays a critical role in a specialized process termed trans-translation which involves the rescue of stalled ribosomes and has been shown to be critical for stress survival and virulence of other pathogens (Shi et al. 2011b). Reflecting the uncertain fidelity of current in vitro models, however, it remains unclear which, if any, of the foregoing findings pertain to the unique sterilizing activity of PZA.
Ethambutol (EMB)
Like PZA, ethambutol (2,2′ ethylenediimino-di-1-butanol) (Fig. 1(3)) was first discovered at Lederle Laboratories of American Cynamid and tested immediately in animals following the discovery of its remarkable stereospecific activity (Thomas et al. 1961; Shepherd et al. 1966). Comparison of all stereoisomers specifically showed that the dextro (S,S) form was 12 times more active than the meso form, whereas the levo form was entirely inactive, raising the possibility of a discrete macromolecular target (Thomas et al. 1961). However, its small size and simple structure provided few clues.
Early biochemical studies nonetheless showed that ethambutol (EMB) was rapidly taken up by both replicating and nonreplicating mycobacteria but active only against replicating bacilli, where it was found to impair glycerol metabolism as well as RNA synthesis (Forbes et al. 1962, 1965; Kuck et al. 1963). Subsequent biochemical studies (isotopic-labeling studies and analysis of cellular sugar content) showed that EMB induced an almost immediate accumulation of the major intermediate of arabinogalactan biosynthesis, β-d-arabinofuranosyl-1-monophosphodecaprenol, followed by the sequential accumulation of trehalose mono- and dimycolates and inhibition of mycolic acid incorporation into the cell wall (Takayama et al. 1979; Kilburn et al. 1981; Takayama and Kilburn 1989; Mikusova et al. 1995; Blanchard 1996; Goude et al. 2009). These findings suggested that the primary inhibitory effect of EMB lay at the polymerization of the mycobacterial cell wall arabinan.
As for the case with INH and PZA, this prediction was genetically confirmed with the in vitro generation of EMB-resistant strains and characterization of EMB-resistant clinical isolates, both of which were found to map to a cluster of genes (embCAB) associated with arabinogalactan biosynthesis (Belanger et al. 1996; Telenti et al. 1997). Curiously, however, evidence of a specific molecular target remains lacking as even the most common mutations associated with EMB resistance (such as EmbB306) have been also found in clinically susceptible isolates, whereas both EmbB and EmbC encoded arabinosyltransferases are predicted integral membrane proteins that have proven refractory to biochemical study (Safi et al. 2008; Srivastava et al. 2009; Shi et al. 2011a; Guerrero et al. 2013). Studies of EMB resistance have proven similarly challenging to interpret in that high-level resistance, which had been traditionally used as a marker of potential primary targets, was only observed in the setting of multiple mutations (Safi et al. 2013).
Primary target aside, early work by Peets and colleagues showed that the growth inhibitory activity of, and recovery from, EMB treatment could be specifically ameliorated (and often accompanied by MIC shifts of as much as 16-fold) by addition of either the polyamine spermidine or magnesium (but not amino acids, nucleotides, Zn, or Mn) (Forbes et al. 1965). More precise understanding of both its primary and downstream effects thus awaits further investigation.
Rifampicin
Rifampicin (Fig. 1(4)) is a semisynthetic derivative of a natural product ansamycin, which was originally discovered as an antimicrobial activity in the conditioned media of the soil bacterium Nocardia mediterranei termed rifamycins (named after a popular French movie about jewel heists). Remarkably, the only component of this extract (termed rifamycin B) that could be isolated in pure crystalline form was a minor (5%−10%) species with comparatively weak activity that was serendipitously noted to gain activity on incubation in oxygenated aqueous solution. Detailed chemical studies soon showed that this activation was due to a reversible oxidation of a “quinonoid-like” core followed by hydrolytic loss of a glycolic acid moiety, giving rise to the far more active rifamycins S and SV (the latter of which was found to show potent activity against Mtb). The discovery of this structure-activity relationship (SAR) provided the first evidence for a potentially specific biologic mode of action that, in conjunction with the poor bioavailability of rifamycin SV, motivated a heroic chemical campaign to develop a more potent and orally bioavailable compound. These efforts culminated in the introduction of rifampicin in the 1960s with detailed knowledge of key molecular determinants of its activity (e.g., ansa chain C21 and C23 hydroxyl groups and C1 and C8 phenols among others) (Sensi 1983; Marriner et al. 2011).
Armed with this knowledge, concurrent studies of protein and nucleic acid biosynthesis revealed that rifampicin specifically inhibited RNA synthesis and, with the identification of RNA polymerase, its molecular mode of inhibition (Wehrli and Staehelin 1971). The study of rifampicin-resistant mutants later confirmed that this inhibition, in fact, accounted for its antitubercular activity as >96% of all resistance mutations could be mapped to an 81-nucleotide region in the coding sequence of the β subunit of RNA polymerase responsible for rifampicin binding (Levin and Hatfull 1993; Telenti et al. 1993).
Bedaquiline (BDQ)
Apart from being the only clinical drug to be approved for the treatment of TB in >40 yr, bedaquiline (BDQ) (previously known as TMC207 and R207910) (Fig. 1(5)) is also the first FDA-approved drug to have been developed in the modern era of molecular sciences (Cohen 2013). BDQ was discovered in a high-throughput phenotypic screen for compounds active against the saprophytic mycobacteria, M. smegmatis, and subsequently shown to demonstrate activity against M. bovis BCG and Mtb (Andries et al. 2005). Early SAR studies of closely related congeners revealed its activity was stereoselective, such that the (R,S) stereoisomer was the most potent with MICs against Mtb ranging from 4 to 70 ng/mL, whereas the (S,R) isomer exhibited MICs of 0.35–8.8 µg/mL (Andries et al. 2005; Koul et al. 2007). This stereoselectivity suggested the existence of a specific binding target. Remarkably, BDQ was also discovered to also show activity against hypoxic, nonreplicating Mtb (Rao et al. 2008). Apart from its potency, BDQ was also found to show an unusually slow time-dependent killing such that only bacteriostatic activity was observed during the first 3–4 d but was then followed by a bactericidal phase with a 4 log10 reduction in measurable colony-forming units by day 14 (Koul et al. 2014).
Insight into the functional target of BDQ first emerged with the generation of resistant mutants followed by whole-genome resequencing. These studies identified mutations in the atpE gene encoding the membrane bound c subunit of F0ATP synthase, which were independently confirmed through comparative sequence analyses of naturally BDQ-susceptible and -resistant mycobacteria, and BDQ-selected resistant Mtb strains (Andries et al. 2005; Huitric et al. 2007, 2010). Interestingly, sequence comparisons to the orthologous mitochondrial ATP synthase revealed an alanine-to-methionine substitution at position 63 that was hypothesized to explain the apparent 20,000-fold selectivity of binding to the Mtb over mammalian enzyme (Haagsma et al. 2009). Biochemical studies nonetheless showed accompanying reductions in bacterial ATP levels, lending orthogonal evidence for the role of ATP synthase in its mechanism (Andries et al. 2005; Koul et al. 2007, 2014; Rao et al. 2008). Affinity purification-based efforts to identify binding partners (using an amine analog of BDQ immobilized to a sepharose resin) resulted in the identification of α and β subunits of ATP synthase from the total membrane fraction of M. smegmatis, although the c subunit was not recovered, perhaps because of its high hydrophobicity (Koul et al. 2007). Interestingly, however, a recent study of resistant mutants from clinical isolates showed that ∼38% of mutations were unrelated to the ATP synthase operon (Huitric et al. 2010). Thus, although the available data suggests that BDQ targets ATP synthase, it is possible that additional targets remain to be identified.
RETROSPECTIVE VIEWS ON MECHANISM-BASED STUDIES AND CONCLUSIONS
Looking back, it is interesting to note that as the tools used to develop and study antibiotics have steadily increased in scale and precision, our understanding of their mechanisms has somewhat paradoxically shrunken in scope. Indeed, early studies of antibiotic mechanisms largely consisted in broad phenotypic and biochemical characterizations of antimicrobial activity. The advent of genetic tools and approaches, however, fostered a systematic shift toward organism-wide, but gene-specific, views of drug activity. This shift was largely driven by the successful use of drug resistance as a functional “loss-of-function” window into the targets and mechanisms of INH (InhA), rifampicin (RNA polymerase β subunit), and BDQ (ATP synthase). This same experimental shift, however, was also accompanied by a limited appreciation of the only partial intersection between drug activity and resistance, and operational shift toward more empirical definitions of drug targets and mechanisms based on the genes and/or functions whose dysregulation affected activity. Although functionally powerful for its genome-wide scope, this shift had the unintended consequence of conceptually constraining drug activity to a single best target and mechanism of action because of its experimental proclivity to select for and focus on isolated genes and/or proteins.
Defined by their ability to suppress or kill bacteria, most antibiotics act through the inhibition of biochemical networks, and elicit maladaptive phenotypes that, although experimentally simple to assay, are no less complex than their adaptive physiologic counterparts. Their mechanisms of action are thus likely, if not certain, to be mediated through an equally complex array of factors, whose identities, quantitative contributions, and interactions have only been partially elucidated.
Looking ahead, it is encouraging to see growing recognition of the complexity of antibiotic action and the advent of new level technologies and disciplines to help respond to this challenge. One current area of potential promise is the rapidly emerging field of metabolomics. Metabolomics is the youngest of systems-level disciplines, defined as the global study of metabolites in a biological system under a given set of conditions (Saghatelian and Cravatt 2005; van der Werf et al. 2005, 2007; Patti et al. 2012). Because metabolites are the biochemical fuel of all physiologic processes, metabolomic approaches offer a global biochemical window into the physiologic composition and state of a given cell. From a pharmacologic point of view, metabolomic readouts offer direct biochemical insights into the intrabacterial “pharmacokinetic” fates and “pharmacodynamic” actions of a given compound within Mtb (de Carvalho et al. 2010, 2011; Pethe et al. 2010; Chakraborty et al. 2013). Although technologically advanced, current antibiotic development efforts rest heavily, if not exclusively, on the use of indirect bacteriologic or genetic readouts or in vitro measurements of enzyme activity as the primary measures of compound activity. Although valuable, these same tools and readouts have left critical ambiguities. For example, in elucidating the structure-activity relationship of a given compound, it is difficult, if not impossible, to determine the extent to which the activity of a given compound is mediated by its ability to accumulate within a bacterial cell, its biochemical affinity for a specific target, and/or its biotransformation into one or more bioactive species. Direct biochemical readouts of a compound’s intrabacterial “pharmacokinetic” fates and “pharmacodynamic” actions thus represent a major unmet scientific need of direct pharmacologic relevance. In a recent study, Chakraborty and colleagues reported one such application of metabolomic technologies using PAS, a close structural analog and competitive in vitro inhibitor of the folate biosynthetic enzyme dihydropteroate synthase (DHPS) (Chakraborty et al. 2013). Using this platform, the investigators showed that, contrary to longstanding inferences, PAS inhibited Mtb by functioning as a replacement substrate, rather than inhibitor, that, in turn, gave rise to dysfunctional folate analogs. In contrast, far more potent sulfonamide-based competitive inhibitors of Mtb’s DHPS (sulfamethoxazole, sulfaniliamide, and diaminodiphenylsulfone) were shown to lack growth-inhibitory activity because of their rapid intrabacterial inactivation, rather than failure to penetrate its notoriously thick and hydrophobic envelope. These studies thus not only showed the highly unpredictable intrabacterial fates of two well-studied antibiotics, but more importantly revised our fundamental understanding of their accompanying activities (or lack thereof). Metabolomics not withstanding, it remains important to recognize that no one approach is likely to prove sufficient to predictably determine the target and/or mechanism of any antibiotic. It is thus likely that rational mechanism-based drug development will emerge only with a continued expansion and integration of systems-level approaches.
ACKNOWLEDGMENTS
We apologize that many papers could not be discussed owing to the lack of space. The authors gratefully acknowledge the support of the Bill and Melinda Gates Foundation TB Drug Accelerator Program and National Institutes of Health for funding.
Footnotes
Editors: Stefan H.E. Kaufmann, Eric J. Rubin, and Alimuddin Zumla
Additional Perspectives on Tuberculosis available at www.perspectivesinmedicine.org
REFERENCES
- Andries K, Verhasselt P, Guillemont J, Gohlmann HW, Neefs JM, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, et al. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307: 223–227. [DOI] [PubMed] [Google Scholar]
- Baldock C, Rafferty JB, Sedelnikova SE, Baker PJ, Stuitje AR, Slabas AR, Hawkes TR, Rice DW. 1996. A mechanism of drug action revealed by structural studies of enoyl reductase. Science 274: 2107–2110. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, Collins D, de Lisle G, Jacobs WR Jr. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263: 227–230. [DOI] [PubMed] [Google Scholar]
- Barclay WR, Koch-Weser D, Ebert RH. 1954. Mode of action of isoniazid. II. Am Rev Tuberc 70: 784–792. [DOI] [PubMed] [Google Scholar]
- Barry CE. 2011. Lessons from seven decades of antituberculosis drug discovery. Curr Top Med Chem 11: 1216–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger AE, Besra GS, Ford ME, Mikusova K, Belisle JT, Brennan PJ, Inamine JM. 1996. The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc Natl Acad Sci 93: 11919–11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard JS. 1996. Molecular mechanisms of drug resistance in Mycobacterium tuberculosis. Annu Rev Biochem 65: 215–239. [DOI] [PubMed] [Google Scholar]
- Boone IU, Woodward KT. 1953. Relationship of pyridoxine and its derivatives to the mechanism of action of isoniazid. Proc Soc Exp Biol Med 84: 292–296. [DOI] [PubMed] [Google Scholar]
- Boshoff HI, Mizrahi V. 2000. Expression of Mycobacterium smegmatis pyrazinamidase in Mycobacterium tuberculosis confers hypersensitivity to pyrazinamide and related amides. J Bacteriol 182: 5479–5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Gruber T, Barry CE III, Boshoff HI, Rhee KY. 2013. Para-aminosalicylic acid acts as an alternative substrate of folate metabolism in Mycobacterium tuberculosis. Science 339: 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorine V. 1945. Action de l’amide nicotinique sur les bacilles du genre mycobcaterium. C R Hebd Seances Acad Sci 220: 150–151. [Google Scholar]
- Cohen J. 2013. Infectious disease. Approval of novel TB drug celebrated—With restraint. Science 339: 130. [DOI] [PubMed] [Google Scholar]
- Davidson LA, Takayama K. 1979. Isoniazid inhibition of the synthesis of monounsaturated long-chain fatty acids in Mycobacterium tuberculosis H37Ra. Antimicrob Agents Chemother 16: 104–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho LP, Fischer SM, Marrero J, Nathan C, Ehrt S, Rhee KY. 2010. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem Biol 17: 1122–1131. [DOI] [PubMed] [Google Scholar]
- de Carvalho LP, Darby CM, Rhee KY, Nathan C. 2011. Nitazoxanide disrupts membrane potential and intrabacterial pH homeostasis of Mycobacterium tuberculosis. ACS Med Chem Lett 2: 849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessen A, Quemard A, Blanchard JS, Jacobs WR Jr, Sacchettini JC. 1995. Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis. Science 267: 1638–1641. [DOI] [PubMed] [Google Scholar]
- Ellard GA. 1969. Absorption, metabolism and excretion of pyrazinamide in man. Tubercle 50: 144–158. [DOI] [PubMed] [Google Scholar]
- Forbes M, Kuck NA, Peets EA. 1962. Mode of action of ethambutol. J Bacteriol 84: 1099–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes M, Kuck NA, Peets EA. 1965. Effect of ethambutol on nucleic acid metabolism in Mycobacterium smegmatis and its reversal by polyamines and divalent cations. J Bacteriol 89: 1299–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox HH. 1952. The chemical approach to the control of tuberculosis. Science 116: 129–134. [DOI] [PubMed] [Google Scholar]
- Goude R, Amin AG, Chatterjee D, Parish T. 2009. The arabinosyltransferase EmbC is inhibited by ethambutol in Mycobacterium tuberculosis. Antimicrob Agents Chemother 53: 4138–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero E, Lemus D, Yzquierdo S, Vilchez G, Munoz M, Montoro E, Takiff H. 2013. Association between embB mutations and ethambutol resistance in Mycobacterium tuberculosis isolates from Cuba and the Dominican Republic: Reproducible patterns and problems. Rev Argent Microbiol 45: 21–26. [PubMed] [Google Scholar]
- Haagsma AC, Abdillahi-Ibrahim R, Wagner MJ, Krab K, Vergauwen K, Guillemont J, Andries K, Lill H, Koul A, Bald D. 2009. Selectivity of TMC207 towards mycobacterial ATP synthase compared with that towards the eukaryotic homologue. Antimicrob Agents Chemother 53: 1290–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huitric E, Verhasselt P, Andries K, Hoffner SE. 2007. In vitro antimycobacterial spectrum of a diarylquinoline ATP synthase inhibitor. Antimicrob Agents Chemother 51: 4202–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huitric E, Verhasselt P, Koul A, Andries K, Hoffner S, Andersson DI. 2010. Rates and mechanisms of resistance development in Mycobacterium tuberculosis to a novel diarylquinoline ATP synthase inhibitor. Antimicrob Agents Chemother 54: 1022–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infectious Diseases Society of America (IDSA). 2004. Bad bugs, no drugs: As antibiotic R&D stagnates, a public health crisis brews. [Google Scholar]
- Johnsson K, Schultz PG. 1994. Mechanistic studies of the oxidation of isoniazid by the catalase-peroxidase from Mycobacterium tuberculosis. J Am Chem Soc 116: 7425–7426. [Google Scholar]
- Kilburn JO, Takayama K, Armstrong EL, Greenberg J. 1981. Effects of ethambutol on phospholipid metabolism in Mycobacterium smegmatis. Antimicrob Agents Chemother 19: 346–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch-Weser D, Barclay WR, Ebert RH. 1955. The influence of isoniazid and streptomycin on acid-fastness, tetrazolium reduction, growth, and survival of tubercle bacilli. Am Rev Tuberc 71: 556–565. [DOI] [PubMed] [Google Scholar]
- Konno K, Feldmann FM, McDermott W. 1967. Pyrazinamide susceptibility and amidase activity of tubercle bacilli. Am Rev Respir Dis 95: 461–469. [DOI] [PubMed] [Google Scholar]
- Koul A, Dendouga N, Vergauwen K, Molenberghs B, Vranckx L, Willebrords R, Ristic Z, Lill H, Dorange I, Guillemont J, et al. 2007. Diarylquinolines target subunit c of mycobacterial ATP synthase. Nat Chem Biol 3: 323–324. [DOI] [PubMed] [Google Scholar]
- Koul A, Vranckx L, Dhar N, Gohlmann HW, Ozdemir E, Neefs JM, Schulz M, Lu P, Mortz E, McKinney JD, et al. 2014. Delayed bactericidal response of Mycobacterium tuberculosis to bedaquiline involves remodelling of bacterial metabolism. Nat Commun 5: 3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuck NA, Peets EA, Forbes M. 1963. Mode of action of ethambutol on Mycobacterium tuberculosis, strain H37R V. Am Rev Respir Dis 87: 905–906. [DOI] [PubMed] [Google Scholar]
- Lei B, Wei CJ, Tu SC. 2000. Action mechanism of antitubercular isoniazid. Activation by Mycobacterium tuberculosis KatG, isolation, and characterization of inha inhibitor. J Biol Chem 275: 2520–2526. [DOI] [PubMed] [Google Scholar]
- Levin ME, Hatfull GF. 1993. Mycobacterium smegmatis RNA polymerase: DNA supercoiling, action of rifampicin and mechanism of rifampicin resistance. Mol Microbiol 8: 277–285. [DOI] [PubMed] [Google Scholar]
- Long ER, ed. 1958. The chemistry and chemotherapy of tuberculosis, 3rd ed. Williams & Wilkins, Baltimore. [Google Scholar]
- Malone L, Schurr A, Lindh H, Mc KD, Kiser JS, Williams JH. 1952. The effect of pyrazinamide (aldinamide) on experimental tuberculosis in mice. Am Rev Tuberc 65: 511–518. [PubMed] [Google Scholar]
- Marcinkeviciene JA, Magliozzo RS, Blanchard JS. 1995. Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid activation. J Biol Chem 270: 22290–22295. [DOI] [PubMed] [Google Scholar]
- Marrakchi H, Laneelle G, Quemard A. 2000. InhA, a target of the antituberculous drug isoniazid, is involved in a mycobacterial fatty acid elongation system, FAS-II. Microbiology 146: 289–296. [DOI] [PubMed] [Google Scholar]
- Marriner GA, Nayyar A, Uh E, Wong SY, Mukherjee T, Via LE, Carroll M, Edwards RL, Gruber TD, Choi I, et al. 2011. The medicinal chemistry of tuberculosis chemotherapy. Top Med Chem 7: 47–124. [Google Scholar]
- McDermott W, Tompsett R. 1954. Activation of pyrazinamide and nicotinamide in acidic environments in vitro. Am Rev Tuberc 70: 748–754. [DOI] [PubMed] [Google Scholar]
- McNeil M, Daffe M, Brennan PJ. 1991. Location of the mycolyl ester substituents in the cell walls of mycobacteria. J Biol Chem 266: 13217–13223. [PubMed] [Google Scholar]
- Middlebrook G. 1954. Isoniazid-resistance and catalase activity of tubercle bacilli; a preliminary report. Am Rev Tuberc 69: 471–472. [DOI] [PubMed] [Google Scholar]
- Mikusova K, Slayden RA, Besra GS, Brennan PJ. 1995. Biogenesis of the mycobacterial cell wall and the site of action of ethambutol. Antimicrob Agents Chemother 39: 2484–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C. 2004. Antibiotics at the crossroads. Nature 431: 899–902. [DOI] [PubMed] [Google Scholar]
- Nathan C. 2009. Taming tuberculosis: A challenge for science and society. Cell Host Microbe 5: 220–224. [DOI] [PubMed] [Google Scholar]
- Nathan C, Gold B, Lin G, Stegman M, de Carvalho LP, Vandal O, Venugopal A, Bryk R. 2008. A philosophy of anti-infectives as a guide in the search for new drugs for tuberculosis. Tuberculosis (Edinb) 88: S25–S33. [DOI] [PubMed] [Google Scholar]
- Nguyen M, Quemard A, Broussy S, Bernadou J, Meunier B. 2002. Mn(III) pyrophosphate as an efficient tool for studying the mode of action of isoniazid on the InhA protein of Mycobacterium tuberculosis. Antimicrob Agents Chemother 46: 2137–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti GJ, Yanes O, Siuzdak G. 2012. Innovation: Metabolomics: The apogee of the omics trilogy. Nat Rev Mol Cell Biol 13: 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. 2007. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat Rev Drug Discov 6: 29–40. [DOI] [PubMed] [Google Scholar]
- Pethe K, Sequeira PC, Agarwalla S, Rhee K, Kuhen K, Phong WY, Patel V, Beer D, Walker JR, Duraiswamy J, et al. 2010. A chemical genetic screen in Mycobacterium tuberculosis identifies carbon-source-dependent growth inhibitors devoid of in vivo efficacy. Nat Commun 1: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope H. 1953. Antagonism of isoniazid by certain metabolites. Am Rev Tuberc 68: 938–939. [DOI] [PubMed] [Google Scholar]
- Pope H. 1956. The neutralization of isoniazid activity in Mycobacterium tuberculosis by certain metabolites. Am Rev Tuberc 73: 735–747. [DOI] [PubMed] [Google Scholar]
- Quemard A, Sacchettini JC, Dessen A, Vilcheze C, Bittman R, Jacobs WR Jr, Blanchard JS. 1995. Enzymatic characterization of the target for isoniazid in Mycobacterium tuberculosis. Biochemistry 34: 8235–8241. [DOI] [PubMed] [Google Scholar]
- Rao SP, Alonso S, Rand L, Dick T, Pethe K. 2008. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc Natl Acad Sci 105: 11945–11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raviglione M, Marais B, Floyd K, Lonnroth K, Getahun H, Migliori GB, Harries AD, Nunn P, Lienhardt C, Graham S, et al. 2012. Scaling up interventions to achieve global tuberculosis control: Progress and new developments. Lancet 379: 1902–1913. [DOI] [PubMed] [Google Scholar]
- Rawat R, Whitty A, Tonge PJ. 2003. The isoniazid-NAD adduct is a slow, tight-binding inhibitor of InhA, the Mycobacterium tuberculosis enoyl reductase: Adduct affinity and drug resistance. Proc Natl Acad Sci 100: 13881–13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozwarski DA, Grant GA, Barton DH, Jacobs WR Jr, Sacchettini JC. 1998. Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science 279: 98–102. [DOI] [PubMed] [Google Scholar]
- Russell DG, Barry CE III, Flynn JL. 2010. Tuberculosis: What we don’t know can, and does, hurt us. Science 328: 852–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safi H, Sayers B, Hazbon MH, Alland D. 2008. Transfer of embB codon 306 mutations into clinical Mycobacterium tuberculosis strains alters susceptibility to ethambutol, isoniazid, and rifampin. Antimicrob Agents Chemother 52: 2027–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safi H, Lingaraju S, Amin A, Kim S, Jones M, Holmes M, McNeil M, Peterson SN, Chatterjee D, Fleischmann R, et al. 2013. Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-β-d-arabinose biosynthetic and utilization pathway genes. Nat Genet 45: 1190–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghatelian A, Cravatt BF. 2005. Global strategies to integrate the proteome and metabolome. Curr Opin Chem Biol 9: 62–68. [DOI] [PubMed] [Google Scholar]
- Saroja D, Gopinathan KP. 1973. Transduction of isoniazid susceptibility-resistance and streptomycin resistance in mycobacteria. Antimicrob Agents Chemother 4: 643–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorpio A, Zhang Y. 1996. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med 2: 662–667. [DOI] [PubMed] [Google Scholar]
- Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M, Zhang Y. 1997. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother 41: 540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sensi P. 1983. History of the development of rifampin. Rev Infect Dis 5: S402–S406. [DOI] [PubMed] [Google Scholar]
- Shepherd RG, Baughn C, Cantrall ML, Goodstein B, Thomas JP, Wilkinson RG. 1966. Structure-activity studies leading to ethambutol, a new type of antituberculous compound. Ann N Y Acad Sci 135: 686–710. [DOI] [PubMed] [Google Scholar]
- Shi D, Li L, Zhao Y, Jia Q, Li H, Coulter C, Jin Q, Zhu G. 2011a. Characteristics of embB mutations in multidrug-resistant Mycobacterium tuberculosis isolates in Henan, China. J Antimicrob Chemother 66: 2240–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Zhang X, Jiang X, Yuan H, Lee JS, Barry CE III, Wang H, Zhang W, Zhang Y. 2011b. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science 333: 1630–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Ayyagari A, Dhole TN, Nyati KK, Dwivedi SK. 2009. emb Nucleotide polymorphisms and the role of embB306 mutations in Mycobacterium tuberculosis resistance to ethambutol. Int J Med Microbiol 299: 269–280. [DOI] [PubMed] [Google Scholar]
- Takayama K, Kilburn JO. 1989. Inhibition of synthesis of arabinogalactan by ethambutol in Mycobacterium smegmatis. Antimicrob Agents Chemother 33: 1493–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K, Wang L, David HL. 1972. Effect of isoniazid on the in vivo mycolic acid synthesis, cell growth, and viability of Mycobacterium tuberculosis. Antimicrob Agents Chemother 2: 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K, Schnoes HK, Armstrong EL, Boyle RW. 1975. Site of inhibitory action of isoniazid in the synthesis of mycolic acids in Mycobacterium tuberculosis. J Lipid Res 16: 308–317. [PubMed] [Google Scholar]
- Takayama K, Armstrong EL, Kunugi KA, Kilburn JO. 1979. Inhibition by ethambutol of mycolic acid transfer into the cell wall of Mycobacterium smegmatis. Antimicrob Agents Chemother 16: 240–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston MJ, Matter L, Schopfer K, Bodmer T. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341: 647–650. [DOI] [PubMed] [Google Scholar]
- Telenti A, Philipp WJ, Sreevatsan S, Bernasconi C, Stockbauer KE, Wieles B, Musser JM, Jacobs WR Jr. 1997. The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat Med 3: 567–570. [DOI] [PubMed] [Google Scholar]
- Thomas JP, Baughn CO, Wilkinson RG, Shepherd RG. 1961. A new synthetic compound with antituberculous activity in mice: Ethambutol (dextro-2,2′-(ethylenediimino)-di-l-butanol). Am Rev Respir Dis 83: 891–893. [DOI] [PubMed] [Google Scholar]
- van der Werf MJ, Jellema RH, Hankemeier T. 2005. Microbial metabolomics: Replacing trial-and-error by the unbiased selection and ranking of targets. J Ind Microbiol Biotechnol 32: 234–252. [DOI] [PubMed] [Google Scholar]
- van der Werf MJ, Overkamp KM, Muilwijk B, Coulier L, Hankemeier T. 2007. Microbial metabolomics: Toward a platform with full metabolome coverage. Anal Biochem 370: 17–25. [DOI] [PubMed] [Google Scholar]
- Vilcheze C, Jacobs WR Jr. 2007. The mechanism of isoniazid killing: Clarity through the scope of genetics. Annu Rev Microbiol 61: 35–50. [DOI] [PubMed] [Google Scholar]
- Vilcheze C, Wang F, Arai M, Hazbon MH, Colangeli R, Kremer L, Weisbrod TR, Alland D, Sacchettini JC, Jacobs WR Jr. 2006. Transfer of a point mutation in Mycobacterium tuberculosis inhA resolves the target of isoniazid. Nat Med 12: 1027–1029. [DOI] [PubMed] [Google Scholar]
- Wang L, Takayama K. 1972. Relationship between the uptake of isoniazid and its action on in vivo mycolic acid synthesis in Mycobacterium tuberculosis. Antimicrob Agents Chemother 2: 438–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrli W, Staehelin M. 1971. Actions of the rifamycins. Bacteriol Rev 35: 290–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilming M, Johnsson K. 1999. Spontaneous formation of the bioactive form of the tuberculosis drug isoniazid. Angew Chem Int Ed Engl 38: 2588–2590. [DOI] [PubMed] [Google Scholar]
- Winder F. 1960. Catalase and peroxidase in mycobacteria. Possible relationship to the mode of action of isoniazid. Am Rev Respir Dis 81: 68–78. [DOI] [PubMed] [Google Scholar]
- Winder FG, Collins PB. 1970. Inhibition by isoniazid of synthesis of mycolic acids in Mycobacterium tuberculosis. J Gen Microbiol 63: 41–48. [DOI] [PubMed] [Google Scholar]
- Winder FG, Collins P, Rooney SA. 1970. Effects of isoniazid on mycolic acid synthesis in Mycobacterium tuberculosis and on its cell envelope. Biochem J 117: 27P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. 2005. The magic bullets and tuberculosis drug targets. Annu Rev Pharmacol Toxicol 45: 529–564. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mitchison D. 2003. The curious characteristics of pyrazinamide: A review. Int J Tuberc Lung Dis 7: 6–21. [PubMed] [Google Scholar]
- Zhang Y, Scorpio A, Nikaido H, Sun Z. 1999. Role of acid pH and deficient efflux of pyrazinoic acid in unique susceptibility of Mycobacterium tuberculosis to pyrazinamide. J Bacteriol 181: 2044–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wade MM, Scorpio A, Zhang H, Sun Z. 2003. Mode of action of pyrazinamide: Disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. J Antimicrob Chemother 52: 790–795. [DOI] [PubMed] [Google Scholar]
- Zumla A, Nahid P, Cole ST. 2013. Advances in the development of new tuberculosis drugs and treatment regimens. Nat Rev Drug Discov 12: 388–404. [DOI] [PubMed] [Google Scholar]
- Zumla AI, Gillespie SH, Hoelscher M, Philips PP, Cole ST, Abubakar I, McHugh TD, Schito M, Maeurer M, Nunn AJ. 2014. New antituberculosis drugs, regimens, and adjunct therapies: Needs, advances, and future prospects. Lancet Infect Dis 14: 327–340. [DOI] [PubMed] [Google Scholar]