Abstract
Lung cancer is the leading cause of cancer deaths, with small cell lung cancer (SCLC) representing the most aggressive subtype. Standard treatments have not changed in decades, and the 5-year survival rate has remained <7%. Genomic analyses have identified key driver mutations of SCLC that were subsequently validated in animal models of SCLC. To provide better treatment options, a deeper understanding of the cellular and molecular mechanisms underlying SCLC initiation, progression, metastasis, and acquisition of resistance is required. In this review, we describe the genetic landscape of SCLC, features of the cell of origin, and targeted therapeutic approaches.
Keywords: SCLC, cell of origin, neuroendocrine, driver, GEMM, targeted therapy
Lung cancer accounts for more than one-tenth of all cancer cases worldwide and is one of the most common types of cancer. In addition, it has a very poor prognosis, which is reflected in a 5-year survival rate of 18% (Ferlay et al. 2010). Smoking is the major risk factor for the development of lung cancer, where extensive exposure to carcinogens from tobacco smoke is able to induce a high mutational load in cells of the lung. Accumulation of mutations in different oncogenes and tumor suppressors is ultimately the cause of tumorigenesis for almost all lung cancers (Govindan 2006). Within the large group of lung cancers, a vast diversity of morphological appearances and genetic aberrations has been observed, indicating that this is a heterogeneous disease. Lung cancer has therefore been classified into multiple subgroups, with the broadest division made between non-small-cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC is the most abundant form of lung cancer and comprises several subclasses that include adenocarcinomas, squamous cell carcinomas, and large cell carcinomas (Kamangar et al. 2006). SCLC represents ∼15% of all lung cancer cases and can be distinguished by its neuroendocrine (NE) features (van Meerbeeck et al. 2011). According to the 2004 World Health Organization classification, SCLC is one of the NE tumors of the lung. This group, which represents around a third of all lung tumors, includes low-grade typical carcinoids, intermediate-grade atypical carcinoids, and two high-grade tumor types: SCLC and large cell NE carcinoma (Linnoila 2006).
In addition to morphological and histological differences, lung cancer subtypes also have distinct disease progression patterns, with SCLC showing the most rapid growth and a tendency to metastasize to distant sites of the body early in the disease (Kato et al. 1969; Jackman and Johnson 2005). After its diagnosis, SCLC is most commonly classified as either limited stage disease (LD) or extensive stage disease (ED), depending on the absence or presence of distant metastases (Micke et al. 2002). Around two-thirds of all SCLC patients are diagnosed with ED, with metastases commonly observed in the contralateral lung, liver, brain, and bones (Kalemkerian et al. 2013). In spite of the late detection of SCLC, a good initial response to chemotherapy and radiotherapy is observed in the majority of patients. Unfortunately, following this initial response, almost all patients relapse within 6–12 mo with resistant disease. Despite numerous clinical trials aimed to improve the therapeutic management for SCLC, the results have been disappointing, and, consequently, treatment regimens have remained largely unchanged for the last 30 years.
In order to improve the durability of the responses in SCLC, an in-depth molecular characterization of this tumor type is urgently needed. Detailed analyses of the origin of the cancer cells and their genetic aberrations should lead to a better understanding of targets that can be more successfully exploited for therapeutic intervention.
History of SCLC
Lung cancer had been a rare disease before smoking became common, representing only ∼1% of all cancers in 1880 (Witschi 2001). SCLC in particular is strongly correlated with smoking and occurs almost exclusively in heavy smokers. SCLC was originally classified as mediastinal sarcoma (Bensch et al. 1968). The oval-shaped tumor cells of this cancer contain scant cytoplasm and thus resemble oat grains. In 1926, Barnard (1926) described the bronchial origin of “oat-celled sarcoma” of the mediastinum. He showed that this tumor type was actually a carcinoma of the lung and suggested changing the classification to bronchial carcinoma. According to Barnard (1926), tumors arose from germinal cells found in the basal layer of bronchial epithelium.
In 1959, Azzopardi (1959) provided a histochemical description of 100 cases of oat cell carcinoma and identified considerable cytological and structural variation among tumor samples, which he categorized into streams, ribbons, pseudorosettes, rosettes, tubules, and ductules.
In 1962, Watson and Berg (1962) helped to further define oat cell carcinoma of the lung as a specific tumor type as to its origin, symptoms, clinical presentation, and response to treatment. They noted early involvement of the main bronchi but also described cases of normal uninvolved lung in the background of massive extrapulmonary nodal disease and distant metastases. Oat cells spread through lymphatic and blood vessels earlier than any other type of lung cancer. Interestingly, oat cell carcinoma showed a favorable response to nitrogen mustard, with 90% of patients responding, pointing further to a unique biology of this cancer type. The duration of response, however, was brief. Watson and Berg (1962) proposed radiation together with nitrogen mustard treatment as a preferred form of palliation. Interestingly, the same type of approach is still being used today, almost 60 years later.
Subsequently, a new cell of origin for oat cell carcinoma was described. In 1965, Bensch et al. (1965) published the ultrastructure of bronchial carcinoid tumors. The most striking feature was the presence of electron-opaque granules of uniform size within these tumor cells, unique to this tumor type (Bensch et al. 1965). The cells also contained pseudopods, which extended over a considerable distance between neighboring cells and often invaded surrounding connective tissue. At the same time, Bensch et al. (1968) identified a very similar cell type in normal bronchial epithelium, closely resembling the secretory argentaffin (Kultschitzky) cells found in the gastrointestinal tract. These cells were scant and situated between the basal parts of the columnar epithelium and likely were the precursor cells for the tumor. Bronchial carcinoid tumors were located centrally within the lung, consistent with the location of the described normal cells. Furthermore, hormonal changes unique to patients with this cancer type further supported the neurosecretory nature of the precursor cells (Hattori et al. 1972).
The developmental lineage of these pulmonary cells was uncertain at that time. Studies in mouse models provided further evidence for a NE cell as the likely cell of origin of small cell carcinoma.
In the next section, we provide a brief overview of lung development and epithelial lineage specification, with particular attention given to pulmonary NE cells (PNECs).
Respiratory lineage specification and function
The respiratory system arises from the anterior foregut endoderm. Progenitors of the trachea and lung are first identified in mice at embryonic day 9.0 (E9.0), and, at E9.5, primordial buds of the right and left lung are formed (Cardoso 2006; Hogan et al. 2014; Kotton and Morrisey 2014). During the pseudoglandular period (E10.5–E17), the lung epithelium undergoes branching morphogenesis to form the respiratory tree (Metzger et al. 2008). At the same time, the epithelium begins to differentiate into secretory (club), ciliated, and NE cells. Respiratory bronchioles, alveolar ducts, and primitive alveoli develop during the canalicular stage (E16.5–E17.5), which is followed by the final saccular stage (E17.5 to postnatal day 5), during which alveoli, lined by alveolar epithelial type 1 (AEC1) and AEC2 cells, are formed (Cardoso 2006; Rock and Hogan 2011). These early events in lung development are controlled by a variety of signaling pathways, including Fgf, Tgfb, Wnt, Sox, Hedgehog, Notch, and retinoic acid (Rock and Hogan 2011; Kotton and Morrisey 2014).
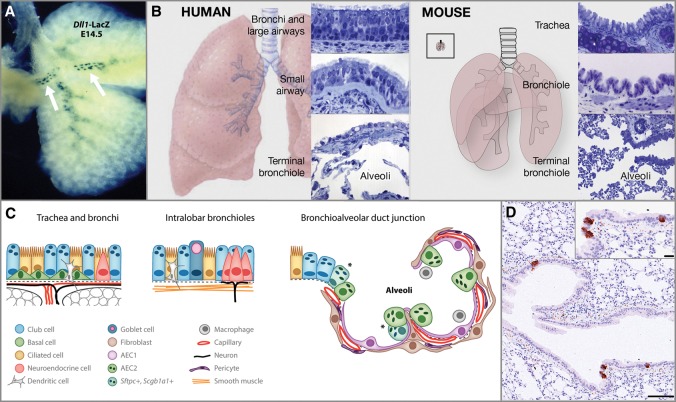
NE cells are the first epithelial cells to arise within the lung and are more abundant in fetal and neonatal lungs, suggestive of a role in pulmonary development. They are derived from a population of multipotent epithelial progenitors marked by expression of the basic helix–loop–helix (bHLH) transcription factor ID2 (Rock and Hogan 2011). Lineage tracing analysis of ID2-expressing cells, labeled at E11.5, showed their ability to give rise to all of the major respiratory epithelial cell types, including PNECs (Rawlins et al. 2009). Evidence suggests that PNEC fate specification is controlled by cross-talk between bHLH activator and repressor genes, a mechanism conserved between Drosophila and mammals (Ito et al. 2000). In the mouse lung, the bHLH factor achaete-scute homolog 1 (ASCL1) activates NE differentiation, while hairy and enhancer of split 1 (HES1) represses this pathway by inhibiting ASCL1/TCF3 complex formation and reducing Ascl1 transcription (Ito et al. 2000; Rock and Hogan 2011). Precocious PNECs developed in both embryonic and adult lungs of Hes1-deficient mice, likely as a consequence of increased expression of ASCL1 (Ito et al. 2000). In contrast, Ascl1-null mice had no detectible PNECs (Borges et al. 1997). NOTCH signaling was also shown to be important in PNEC lineage specification. NOTCH ligand delta-like-1 (DLL1) is expressed in presumptive NE cells within proximal airways as early as E13.5 (Fig. 1A), and its activation may be under the control of ASCL1 (Post et al. 2000; Linnoila 2006). Recent evidence shows the involvement of multiple NOTCH receptors in the maintenance of Hes1 expression and in NE compartment size regulation (Morimoto et al. 2012). Thus, interplay between bHLH factors and the NOTCH pathway contributes significantly to pulmonary NE lineage specification.
Figure 1.
Structure and cell types of the respiratory system. (A) Expression of Dll1-LacZ (white arrows) in presumptive NE cells within proximal airways at E14.5. (Republished with permission of Annual Reviews from Rock and Hogan 2011; permission conveyed through the Copyright Clearance Center, Inc.) (B) Schematic of human and mouse lungs with epithelial histology of regions along the proximal–distal axis. (Republished with permission of Elsevier from Hogan et al. 2014; permission conveyed through Copyright Clearance Center, Inc.) (Left panel) The human trachea, bronchi, and bronchioles >1 mm in diameter are lined by a pseudostratified epithelium with basal, multiciliated, and secretory club cells. Club cells predominate in the small airways. The alveoli are lined by squamous AEC1s and cuboidal AEC2s. (Right panel) In mice, only the trachea and main stem bronchi are lined by a pseudostratified mucociliary epithelium with basal cells. The smaller bronchi and bronchioles are lined by a simple epithelium with multiciliated and club cells. The inset illustrates a mouse lung to the same scale as the human lung in the left panel. (C) Schematic representation of individual cell types within the mouse respiratory epithelial system. (Republished with permission of Annual Reviews from Rock and Hogan 2011; permission conveyed through the Copyright Clearance Center, Inc.) (D) Section of the adult mouse lung, with rare NE cells marked by the expression of synaptophysin (brown). Bars: 100 μm; inset, 20 μm.
Within the mature respiratory system, epithelial cells differ along the proximal–distal axis (Fig. 1B,C). Trachea and bronchi are lined with pseudostratified columnar epithelium that contains ciliated and secretory cells, solitary PNECs, and basal cells. More distal intrapulmonary conducting airways in mice are lined by a simple columnar epithelium of ciliated and secretory cells (Rock et al. 2010; Hogan et al. 2014). Gas exchange is carried out within alveolar epithelium, which consists of AEC1 and AEC2 cells. Notably, most of the airways of the human lung are lined by pseudostratified epithelium.
PNECs are found in species ranging from primitive amphibians to mammals and constitute a rare respiratory cell population, representing only 0.4% of epithelial cells in the adult lung (Van Lommel et al. 1999; Van Lommel 2001; Linnoila 2006). They are located as either solitary cells within the nasal epithelium, trachea, and mainstream bronchi or clusters of cells (NE bodies [NEBs]), often within airway bifurcations (Fig. 1D). PNECs have properties of both endocrine and neuronal cells. They express neuronal markers, such as neural cell adhesion molecule (NCAM1) and ASCL1, a key determinant of neuronal fate specification (Linnoila 2006; Ali et al. 2014; Chanda et al. 2014). In addition, NEBs are associated with intraepithelial nerve fibers and can transmit signals to the CNS. At the same time, they often contain characteristic electron-dense cytoplasmic vesicles that accumulate peptides, such as calcitonin gene-related peptide CGRP (CALCA) and bombesin, as well as amines, such as serotonin. The content of the vesicles is likely being released into the extracellular space and the blood stream, thus providing both local and distant regulation. Serotonin acts as a pulmonary vasoconstrictor, while CGRP acts as a vasodilator. Suggested functions of PNECs include oxygen sensing, control of airway tone and pulmonary blood flow, and immunomodulation (Van Lommel 2001; Linnoila 2006).
Even though PNECs are specified early in lung morphogenesis, which may suggest their progenitor function, their absence in Ascl1-deficient mice does not prevent differentiation of other respiratory epithelial cell types, such as secretory and alveolar cells (Borges et al. 1997). At the same time, a number of studies point to their supportive role in both the developing lung and the injured adult lung. Temporal–spatial distribution of club cells in the developing human lung shows their close association with NEBs, suggesting that either direct contact or paracrine factors secreted by NEBs may support neighboring epithelial cells (Van Lommel et al. 1999; Van Lommel 2001). Indeed, epithelial cells located next to NEBs in the embryonic lung were strongly labeled with 3H-thymidine, indicative of division, and the amount of label decreased with increasing distance from the NEBs (Van Lommel 2001). PNEC hyperplasia was reported following both pathological and induced forms of damage, such as oxidant stress, smoking, and burn injury (Stevens et al. 1997; Linnoila 2006). Similarly, acute injury following naphthalene treatment, which causes selective elimination of club cells in mouse airways, leads to rapid expansion of PNECs (Stevens et al. 1997). Naphthalene-resistant club cells (also called variant club cells), located adjacent to NEBs, were capable of restoring injured epithelium (Reynolds et al. 2000). This result suggested that NEBs may provide a unique supportive microenvironment for progenitor cells. Interestingly, a NE-specific CGRP-CreER lineage tracing model documented a direct contribution of CGRP-positive cells toward club and ciliated cell populations following naphthalene treatment, although depletion of CGRP-positive cells before naphthalene treatment did not delay regeneration of the epithelium (Song et al. 2012). Moreover, the epithelium was not restored after genetic ablation of club cells upon administration of ganciclovir to transgenic mice expressing herpes simplex virus thymidine kinase under a club cell-specific promoter (Hong et al. 2001). Thus, the role of NE cells in the context of lung epithelial homeostasis and regeneration is not yet fully clarified.
The lung is a quiescent organ with a very slow cell turnover but a robust regenerative response following injury. Distinct regions of the lung contain different progenitor cells that are responsible for physiological homeostasis and regeneration. Unlike classical stem cells, putative progenitor populations of the lung are well differentiated. Nonetheless, recent studies point to a remarkable plasticity. Depending on the type and severity of damage, multiple respiratory cell types can acquire stem/progenitor activity. Currently, cells with regenerative capacity include basal cells, club cells, variant club cells, AEC2 cells, BASCs, and ITGA6+/ITGB4+ cells (Hogan et al. 2014; Kotton and Morrisey 2014). Recently, a novel putative stem cell compartment of K5+/p63+ distal epithelial cells was defined. This cell population was capable of restoring alveolar epithelium following H1N1 influenza virus infection (Vaughan et al. 2015; Zuo et al. 2015). Thus, the lung shows a high degree of plasticity and can use a number of different cell types for its regeneration. This suggests that the full regenerative potential and possible enhanced progenitor characteristics of PNEC may still be revealed and await further investigation.
Specific mutations can dramatically alter the cellular phenotype, and a unique combination of a cell type with genetic lesions sculpts the resulting tumor. The genetic alterations found in SCLC cells are discussed next.
Genetic landscape in SCLC
Recurrent mutations found in cancers are the likely driving force for tumor development. Comprehensive genome-wide characterization of these drivers for SCLC is currently lagging behind compared with other cancer types, mainly due to a scarcity of patient material available for research purposes. Tumor material is obtained only by fine needle aspiration and endobronchial biopsies used for regular histology (Renshaw et al. 2005). The genomic studies that have been performed show that SCLC is characterized by an extremely high frequency of gains and losses (Wistuba et al. 2000a). In addition, this cancer has one of the highest mutation rates, which is linked to the exposure to mutagens in tobacco smoke (Toyooka et al. 2003).
Some of the early techniques used to discover drivers of SCLC, such as karyotyping and comparative genomic hybridizations, gave clear insights into the overall chromosomal alterations in this tumor type (Salgia and Skarin 1998). These studies identified genomic rearrangements common to all lung cancers as well as aberrations specific for SCLC. The almost uniform loss of large regions of chromosome 3p in all types of lung cancer suggested that it contains important tumor suppressors (Otterson et al. 1992). Although many genes in this region could have a tumor-suppressive role in the lung, FHIT, ROBO1, and RASSF1 have been suggested to be the main candidates (Wistuba et al. 2000a; George et al. 2015). However, the evidence for their functional involvement in lung cancer development is currently still limited. Regions on chromosome arms 4p, 4q, 10q, 13q, 16q, and 17p show high frequencies of loss of heterozygosity (LOH) unique to SCLC, suggesting that genes in these regions may be involved in the development of this lung cancer subtype (Ried et al. 1994; Virmani et al. 1998; Shivapurkar et al. 1999).
The most striking alterations found at the individual gene level in SCLC are the nearly uniform loss of function of the tumor suppressors TP53 and RB1. The frequency of TP53 mutations in SCLC is between 75% and 90%, indicating that the loss of this gene is an important event in the onset of SCLC development (Takahashi et al. 1989). The p53 protein is normally activated when cells encounter DNA damage or hypoxia and performs an essential role in the maintenance of genomic integrity by inducing a cell cycle arrest or apoptosis upon genomic stress (Carvajal and Manfredi 2013). Loss of functional p53 would therefore allow for genomic instability, which could be the basis for the further accumulation of driver mutations. The notion that TP53 is found mutated in apparently normal bronchial epithelium accompanying SCLC further suggests that these mutations could serve as an initiating event in SCLC development (Wistuba et al. 2000b). Strikingly, a recent study in which 110 SCLC samples were sequenced discovered previously unidentified genomic rearrangements in another TP53 family member, TP73, in a substantial fraction of cases (George et al. 2015). Specifically, these genomic alterations comprised the deletion of exons 2 and 3 of TP73, resulting in a known oncogenic transcript that exerts a dominant-negative function toward wild-type TP53 family members (Tannapfel et al. 2008; George et al. 2015). These findings point to an even broader involvement of the p53 family members in the tumorigenesis of SCLC.
The second tumor suppressor that is inactivated in nearly all SCLC is the retinoblastoma susceptibility gene (RB1) (Harbour et al. 1988; George et al. 2015). RB1 was first identified as a tumor suppressor in retinoblastoma and was also found deleted in prostatic NE carcinoma (Friend et al. 1988; Tan et al. 2014). Interestingly, these cancers arise from neuronal progenitors, linking loss of RB1 more specifically to tumors of neuronal lineage.
The retinoblastoma protein is a member of a family of pocket proteins that includes P107 (RBL1) and P130 (RBL2). While RB1 loss is a signature mutation in SCLC, mutations in the other family members are rarely observed in SCLC (Helin et al. 1997; Modi et al. 2000). With regard to its function, the RB1 protein has a central role in cell cycle regulation, where it suppresses the transition of cells from G1 to S phase (Weinberg 1995). In addition, RB1 also plays a role in the regulation of differentiation, as mutant forms of this protein that fail to inhibit cell cycle progression still retain their ability to promote cellular differentiation (Sellers et al. 1998). Recently, RB1 was shown to globally repress pluripotency networks in somatic cells through direct binding to known pluripotency genes, such as Oct4 and Sox2; the latter is amplified in 27% of SCLC (Peifer et al. 2012). Consequently, Rb1 loss leads to derepression of these factors and a gain in pluripotency, making cells more amenable to reprogramming (Kareta et al. 2015). Loss of RB1 in SCLC is also strongly associated with augmented expression of enhancer of zeste 2 (EZH2) (Coe et al. 2013; Hubaux et al. 2013). Interestingly, EZH2 was shown to be expressed at high levels in proliferating neural stem cells and has been implicated in neuronal progenitor maintenance and lineage specification (Sher et al. 2008; Pereira et al. 2010). In addition, EZH2 was shown to regulate the phenotypic switch between basal and secretory cells in the lung (Snitow et al. 2015). Accumulating evidence thus suggests that RB1 loss is associated with an increase in cell plasticity.
Amplification or transcriptional up-regulation of one of the MYC proto-oncogenes—MYC, MYCN, or MYCL—has been identified in 20%–30% of SCLC cases (Kiefer et al. 1987; Krystal et al. 1988). The MYC family proteins are transcriptional activators able to drive the expression of a wide variety of genes that contribute to cell cycle progression and developmental regulation (Grandori and Eisenman 1997). Amplification of the three MYC genes is mutually exclusive, suggesting that the capacity to drive SCLC development is shared between the family members even though activation of the individual genes seems to result in quite distinct patterns of expression (Kim et al. 2006). The exact mechanism of MYC-mediated transformation in SCLC cells is not completely understood. MYC has been implicated in the control of pluripotency, self-renewal, and epithelial-to-mesenchymal transition, processes that are strongly implicated in cellular transformation (Chappell and Dalton 2013). Analysis of mouse models of SCLC with targeted MYC overexpression can help to dissect its function further.
In addition to activation of MYC signaling, the activation of the oncogenic phosphatidylinositol 3 kinase (PI3K) pathway has been also observed at high frequencies in SCLC. It was discovered that phosphatase and tensin homolog (PTEN), an inhibitor of this pathway, is lost in a substantial fraction of SCLC cases. Initial allelotyping studies showed that homozygous deletions of PTEN were present in ∼10% of SCLC cell lines (Forgacs et al. 1998; Yokomizo et al. 1998). Using next-generation sequencing, mutations and amplifications in other members of this pathway have been identified in 20%–40% of all studied SCLC tumors. Importantly, all of these alterations occurred in a mutually exclusive fashion (Peifer et al. 2012; Ross et al. 2014; Umemura et al. 2014). Activation of this pathway was shown to facilitate aberrant regulation of proliferation, survival, and migration, giving the tumor cell a selective advantage (Wojtalla et al. 2013).
A recent study identified the presence of alterations in NOTCH family members in a quarter of analyzed SCLC samples (George et al. 2015). Since NOTCH signaling is critical in regulation of the NE compartment size in lung development, the observed inactivating mutations could contribute to tumorigenesis by allowing expansion of NE tumor cell mass.
The high frequencies of alterations found in TP53, RB1, and the PI3K pathway as well as in MYC and NOTCH family members suggested that their respective activation or inactivation is an important driver for SCLC. The formal proof for this has been provided by the generation of specific mouse models with these exact lesions in the lung, which is discussed in a later section.
Several studies on recurrent alterations have indicated that up-regulation of anti-apoptotic signaling is another common event in SCLC. In particular, expression of the anti-apoptotic protein BCL2 could be detected in 75%–90% of primary SCLC cases, whereas it was only present in 10%–30% of all other lung cancers (Pezzella et al. 1993; Jiang et al. 1995). As activation of MYC, which is observed frequently in SCLC, is associated with an increase in apoptosis, the up-regulation of BCL2 may permit escape from this side effect (Fanidi et al. 1992). Interestingly, BCL2 expression is transcriptionally regulated by p53, and the almost uniform loss of this tumor suppressor could therefore be a prime reason for the up-regulation of BCL2 in SCLC (Miyashita et al. 1994). Thus, BCL2 up-regulation might be a consequence rather than a cause for tumorigenesis in SCLC.
Another striking observation in the studies on recurrent alterations in SCLC is the high number of aberrations found in epigenetic regulators. Mutations in the histone acetyltransferases CREBBP and EP300 and the histone methyltransferases MLL, MLL2, and EZH2 were found in independent studies at frequencies of 4%–6% for each gene (Peifer et al. 2012; Rudin et al. 2012; Ross et al. 2014; Umemura et al. 2014). Mutations in these genes could be a major source of genome-wide alterations in epigenetic regulation; however, how they affect SCLC tumorigenesis remains an open question.
Currently, there is no consensus on the importance of other drivers for SCLC. This is mostly due to the fact that studies on the genetic alterations in SCLC are being performed on a limited number of samples per study and that validations of additional driver mutations are time-consuming. In addition, the exceptionally high mutational load in SCLC complicates the identification of driver mutations in the vast background of passenger mutations. The most interesting other changes that have been found in SCLC are alterations in a number of SOX family members as well as mutations in FGFR1, EGFR, and the Hedgehog signaling pathway (Peifer et al. 2012; Rudin et al. 2012; Ross et al. 2014; Umemura et al. 2014).
Models of SCLC
Both in vitro and in vivo model systems are indispensable to study the role of recurrent genetic and epigenetic aberrations in SCLC. Cell lines have been established from a fair number of SCLCs and therefore are an easily accessible source of material for research (Gazdar and Minna 1996; Gazdar et al. 2010). However, these cell lines have been adapted to tissue culture conditions and therefore have lost some of the features of the primary tumor. Patient-derived xenograft (PDX) models partly overcome this drawback, as transplanted pieces of the tumor retain at least some of the heterogeneity and structural features of the primary tumor (Gu et al. 1988; Daniel et al. 2009). Both cell lines and PDX models have been shown to harbor genetic alterations almost identical to those observed in the original tumor; however, they do differ in their expression profiles (Daniel et al. 2009; Gazdar et al. 2010; Leong et al. 2014).
Genetically engineered mouse models (GEMMs) for SCLC have been successfully introduced as a system for validation of known driver mutations. The GEMMs reproduce the NE nature of the tumor and share many of the histopathological features observed in the human disease (Kwon and Berns 2013). Importantly, whereas tumors that develop in the mouse models harbor mostly genomic rearrangements, the human tumors additionally contain a high load of point mutations caused by cigarette smoke exposure. Most of the SCLC models are based on site-specific Cre-Lox-mediated conditional deletion of the two key tumor suppressors, Trp53 and Rb1, combined with either deletion of an additional suppressor (P130 or Pten) or activation of an oncogene (Mycl) (Table 1). In the majority of the models, cells in the lung were targeted using intranasal or intratracheal adenoviral delivery of Cre recombinase.
Table 1.
Mouse models of SCLC

In the first mouse model generated in our laboratory in 2003, conditional deletion of Rb1 and Trp53 was accomplished by intratracheal adenoviral delivery of Cre using the ubiquitous CMV promoter, thus leading to a sporadic inactivation of both genes in a variety of respiratory epithelial cell types. This approach resulted in tumors that closely resembled human SCLC in its histopathology, molecular characteristics, and metastatic behavior (Meuwissen et al. 2003). Despite the nonspecific nature of the viral promoter, SCLC was the predominant tumor type, suggesting that the cells from which SCLC originates are particularly sensitive to the loss of Rb1. Strikingly, Rb1 loss in Scgb1a1(CC10)-expressing cells in the developing lung did not affect club and ciliated cell lineage specification but resulted in an increase in the number of CGRP-positive cells. Similarly, when Rb1 was inactivated in adult lungs, multifocal hypercellular CGRP-positive epithelial lesions located at airway branch points and bronchio–alveolar duct junctions were observed. Thus, NE cell lineage specification was uniquely augmented upon Rb1 loss, with RB1 promoting non-NE specification while suppressing NE cell fate (Wikenheiser-Brokamp 2004). In line with this, germline RB1 mutations confer an increased risk of SCLC development (Kleinerman et al. 2000).
To test the relevance of additional candidate tumor suppressor genes, these genes were inactivated together with Rb1 and Trp53. Inactivation of P130, a member of the RB family, is associated with higher grade and poorer survival in a subset of SCLC patients (Schaffer et al. 2010). Deletion of P130 in addition to Rb1 and Trp53 in mice resulted in accelerated tumor development while maintaining the overall histopathological and metastatic features of SCLC (Schaffer et al. 2010).
PTEN mutations are found in a significant number of human SCLC cases. Interestingly, in the original Trp53/Rb1 mouse model, chromosome 19, which harbors the Pten gene, was frequently lost (McFadden et al. 2014). It was hypothesized that inactivation of Pten may be the driving force behind chromosome 19 loss, establishing Pten as a potent tumor suppressor in SCLC. To test the role of Pten loss in SCLC initiation and progression, three groups generated triple Trp53/Rb1/Pten conditional knockout mouse models. All three models resulted in a dramatic acceleration of tumor progression, thus proving the key role of the PI3K pathway in SCLC. Cui et al. (2014) used the ubiquitous CMV promoter, while McFadden et al. (2014) used the NE-specific CGRP promoter. Using the CMV promoter, homozygous deletion of all three genes shifted the tumor phenotype toward adenocarcinoma with NE differentiation (Cui et al. 2014), while tumors with heterozygous loss of PTEN in most cases maintained a full NE marker profile. It is likely that, given the nonspecific nature of CMV, cells other than NE cells gave rise to the non-NE tumor component. Indeed, when the NE-specific CGRP promoter was used, the vast majority of tumors displayed NE features (McFadden et al. 2014). Interestingly, a significant proportion of adenocarcinomas, termed NSCLC-NE, express markers of NE differentiation, although the etiology of these neoplasms remains unknown (Bhattacharjee et al. 2001; Gazdar et al. 2015).
Another group targeted CreER into the endogenous CGRP locus and used a lineage tracing approach to characterize the early events following inactivation of the Trp53, Rb1, and Pten in the NE compartment (Song et al. 2012). A potential drawback of this model is that all CGRP-positive cells within the mouse tissues were targeted; on the other hand, with this approach, a much larger proportion of PNECs is switched. Proliferation of PNECs as early as 1 wk after induction was observed, which developed into early hyperplastic lesions, the likely precursors of SCLC tumors. In addition, early tumors displayed an invasive phenotype, with tumor cells found in and around vessels (Song et al. 2012).
Tumors of SCLC patients show frequent amplifications of one of the MYC family of oncogenes. To test the oncogenic activity of MYC in SCLC, Mycl under the control of a ubiquitous promoter was introduced together with the targeted deletion of Rb1 and Trp53 (Huijbers et al. 2014). Adenoviral delivery of Cre under the control of a CMV promoter significantly accelerated tumor formation, showing the potency of Mycl as a driver. The resulting tumors displayed the typical NE features; however, metastases were rare.
To address the role of the Hedgehog pathway, members of which were found overexpressed in SCLC tumor samples, the pathway was activated in the respiratory epithelium using the Ad-Cre system. Constitutive activation of the Hedgehog signaling molecule Smoothened (SMO) in the background of Trp53 and Rb1 loss promoted the initiation and progression of mouse SCLC in vivo (Park et al. 2011b).
In a recent publication, the consequence of Notch pathway activation was tested in the Trp53/Rb1/Rbl2 mouse model. Conditional overexpression of an activated form of NOTCH2 or NOTCH1 resulted in a significant reduction in the number of tumors as well as increased survival, establishing Notch as a tumor suppressor in SCLC (George et al. 2015). Interestingly, elevated NOTCH signaling abrogated expression of NE markers, consistent with its key role in regulating NE differentiation.
A thorough histopathological evaluation of mouse models of SCLC was published this year (Gazdar et al. 2015). Although the original Trp53/Rb1 double-conditional mouse model was found to best resemble human SCLC, the combination of the models described above appears to represent the entire spectrum of high-grade NE carcinomas of the lung, thus providing a platform for the dissection of molecular mechanisms of initiation, multistage progression, heterogeneity, and metastatic properties of these tumor subtypes.
Existing and future mouse models should allow for efficient screening of newly discovered candidate drivers as well as comprehensive evaluation of novel therapies. In this regard, the Jacks laboratory (Sánchez-Rivera et al. 2014) has described an elegant and rapid system to assess cooperating mutations in the model of adenocarcinoma. They tested CRISPR/Cas9-induced inactivation of putative tumor suppressor genes in relevant genetic backgrounds by intratracheal administration of lentiviral vectors carrying both the CRISPR/Cas9 system and Cre recombinase (Sánchez-Rivera et al. 2014). The same approach can be taken in models of SCLC described above, with the aim to test the contribution of newly identified candidate tumor suppressors such as Ep300 or Crebbp. Alternatively, a similar system in which CRISPR/Cas9 is linked to a transactivation domain can be used to test the effects of overexpression of proto-oncogenes such as Fgfr1, Sox2, and Pik3ca (Cheng et al. 2013; Hsu et al. 2014). In support of the applicability of current genetically engineered SCLC mouse models for drug validation, the Trp53/Rb1/P130 model was used to show the proapoptotic effect of tricyclic anti-depressants and related molecules on both untreated and cisplatin-treated SCLC tumors (Jahchan et al. 2013). In addition, a newly discovered sensitivity of SCLC to transcription targeting drugs, in particular to inhibition of cyclin-dependent kinase 7, was successfully tested in the original double-knockout model (Christensen et al. 2014). The same model has also been used to test the efficacy of anti-apoptotic inhibitors both as single agents and in a combination regimen (Faber et al. 2015).
Dissection of the cell of origin of SCLC
Several laboratories have used the Trp53/Rb1 SCLC mouse model with the aim to identify the cell of origin of this tumor type. Sutherland et al. (2011) used three different viruses, each targeting a distinct respiratory epithelial population. CGRP virus, which targets NE cells of the lung, was most effective in generating SCLC, with 27 out of 30 animals developing SCLC (Sutherland et al. 2011). Interestingly, the Sftpc (SPC) virus, which targeted AT2 cells as well as BASCs, also resulted in SCLC, although with lower frequency and longer latency. The CC10 virus, which targets club cells and BASCs, resulted mainly in bronchiolar and alveolar hyperplasia, while hardly any tumors were formed (Sutherland et al. 2011).
In a similar approach, the Kim and Sage laboratories (Park et al. 2011a) used CMV, CC10, and SPC viruses to delete Trp53 and Rb1; however, in their case, neither the CC10 virus nor the SPC virus was effective in generating SCLC. A possible explanation for this discrepancy could be in the difference between the Cre systems, with the Berns laboratory (Sutherland et al. 2011) using constitutive Cre as compared with an inducible CreER system used by the Kim group (Park et al. 2011a). The constitutive Cre might have allowed for some background switching in NE cells after prolonged times, or, alternatively, since the NE tumors that were found by Sutherland et al. (2011) occurred far over a year, they might have escaped detection in the other study.
Using lineage tracing, CGRP-CreER mice were crossed to ROSA26-mTmG and Trp53/Rb1 conditional animals. In this system, CGRP-expressing cells were permanently labeled with eGFP after administration of tamoxifen, allowing the examination of early events following tumor initiation (Song et al. 2012). This analysis showed that almost all PNECs within early hyperplastic lesions were lineage-labeled and likely were the cells that led to tumor development. Thus, based on the Rb1/p53 model, all three groups show convincing evidence for NE cells serving as the predominant cell of origin for SCLC.
However, these results do not exclude the existence of a yet-unidentified NE cell progenitor that can act as an alternative cell of origin or the appearance of distinct tumor-initiating cells under conditions of injury or inflammation or in the presence of additional driver mutations. The remarkable plasticity of various putative stem cell compartments in the lung supports this claim. It is interesting to point out that, in the presence of CC10-hASH1, which is a strong NE lineage differentiation factor, in combination with CC10-driven SV40 large T antigen, club cells gave rise to adenocarcinoma with clear NE differentiation (Linnoila et al. 2000). Large T antigen is known to interfere with the RB1, TP53, and CREBBP/EP300 pathways, all of which are mutated in human SCLC (Peifer et al. 2012; Rudin et al. 2012). Thus, the right combination of oncogenic drivers with lineage differentiation factors could create a favorable environment for possible transdifferentiation and broaden the source of cancer-initiating cell populations.
Therapeutic targets
The clinical management of SCLC has been difficult due to the aggressive nature of this tumor, as it is generally fatal within 2–4 mo after diagnosis when left untreated (Kato et al. 1969). Whereas patients with LD have been shown to benefit from surgery or thoracic radiotherapy in combination with chemotherapy, patients with ED rely solely on the systemically delivered chemotherapy to target the disseminated tumor cells. Different treatment modalities for SCLC have therefore been evaluated to increase both the survival rate and the quality of life of these patients. This has led to a standard first-line treatment that combines the two DNA-damaging compounds cisplatin and etoposide (Johnson 1999). As SCLC frequently metastasizes to the brain, an additional treatment consisting of prophylactic cranial irradiation (PCI) is administered to patients that show a response to chemotherapy at the primary site. Despite a high initial response rate of 45%–80%, patients quickly relapse after treatment cessation (Chute et al. 1999). The treatment therefore has limited effects on overall survival of SCLC patients, and the 5-yr survival rate has remained <7% for decades (Shepherd et al. 2007).
The development of resistance has become a major obstacle in the management of SCLC. Several mechanisms of resistance have been brought forward in which the up-regulation of multidrug transporters, the alteration of expression levels of topoisomerases, increased cellular adhesion, the maintenance of cancer-initiating cells, and the plasticity of SCLC have all been implicated (Stewart 2010). An example of the potential plasticity of lung cancer cells has been observed in the case of transdifferentiation of primary NSCLC to SCLC following treatment with EGFR inhibitors (Sequist et al. 2011; Norkowski et al. 2013; Popat et al. 2013; Kuiper et al. 2014). Importantly, whereas the primary tumors were chemoresistant, the recurring tumors showed many of the characteristics of SCLC, including the loss of RB1 and an increased sensitivity to chemotherapy (Niederst et al. 2015). In the case of primary SCLC, a degree of plasticity was observed following the introduction of active RAS, where cells with the NE phenotype acquired a mesenchymal marker profile. This phenotypic transition may also represent a mechanism of resistance exploited by SCLC cells (Calbo et al. 2011).
A recent study reporting on a novel model of human SCLC opens up new possibilities in the ongoing search for the mechanisms behind the acquired resistance to chemotherapeutics. Hodgkinson et al. (2014) showed that circulating tumor cells from SCLC can be isolated from the blood of the patient and subsequently grown as xenografts. These circulating tumor cell-derived xenografts (CTXs) reflected the donor's response to platinum and etoposide (Hodgkinson et al. 2014). The isolation of tumor cells from the blood of a patient can be performed without the need for invasive biopsies, but, more importantly, the tumor cells can also be isolated longitudinally, allowing the comparison of cells isolated before the start of the treatment and at the moment of tumor progression.
As cancer cells rely on the continuous oncogenic signals induced by genetic aberrations, these lesions become a potential Achilles' heel of these cells and could thus form a solid basis for therapeutic intervention. Most clinical trials that used novel targeted therapeutics for SCLC have failed to show a significant clinical benefit over the existing treatment options (Sgambato et al. 2013). With the accumulation of more data on the drivers of SCLC, however, newly targeted therapies have been tested in clinical trials (Table 2). Below, we discuss some of the more promising agents.
Table 2.
Targeted therapies for SCLC in clinical trialsa

As alterations in TP53 and RB1 have been found in nearly all SCLC patients, restoration of their function is an attractive route to treat these tumors. In the case of SCLC where mutant forms of p53 are expressed, the restoration of the wild-type function would be an attractive strategy. In line with this notion, introduction of a wild-type TP53 allele in vitro induces apoptosis in TP53 mutated tumor cells (Adachi et al. 1996). The advent of molecular compounds that can revert the properties of mutant p53 back to its wild-type functions therefore brings new possibilities for the treatment of SCLC (Zandi et al. 2011). At present, one of these compounds, PRIMA-1MET, entered early phase clinic trials for a subset of solid tumors, and the results of those are eagerly awaited (Duffy et al. 2014).
Restoration of RB1 function using small molecules is not possible at present. However, the disruption of RB1 function does induce targetable changes in tumor cells. One such change is an increase in the expression of EZH2 (Byers et al. 2012; Hubaux et al. 2013). Inhibition of EZH2 in SCLC cell lines reduced cell growth and induced apoptosis, suggesting that this transcriptional regulator is worth examining for the treatment of RB1-deficient SCLC (Coe et al. 2013; Poirier et al. 2015).
The amplification of MYC family members has been reported in up to 30% of SCLC samples. MYC amplification is often observed in previously treated SCLC patients, suggesting that MYC up-regulation can lead to chemoresistance (Brennan et al. 1991; Johnson et al. 1996). MYC is therefore an attractive therapeutic target, and several indirect methods have been devised to target MYC-dependent cells. It has been shown that JQ1, a bromodomain inhibitor, is able to down-regulate MYC-driven transcription, leading to cell cycle arrest and senescence (Delmore et al. 2011; Zuber et al. 2011). Disrupting the structure of Aurora kinase A (AURKA) by specific AURKA inhibitors is another means to interfere with MYC function (Gustafson et al. 2014; Silva et al. 2014). Indeed, it could be shown that AURKA inhibition was especially effective in SCLC cell lines bearing MYC amplifications (Sos et al. 2012). Interestingly, alisertib, a highly selective AURKA inhibitor, produced high response rates as a single agent in relapsed SCLC (Byers and Rudin 2014; Melichar et al. 2015). It is therefore tempting to speculate that relapsed SCLC, which generally shows high MYC expression, is responsive to AURKA inhibition.
As the involvement of the PI3K pathway in SCLC development has been clearly established, this pathway has also been the subject of therapeutic intervention. The inhibition of individual components of the pathway leads to a potent induction of apoptosis and reversion of chemoresistance in preclinical studies (Razzini et al. 2000; Wu et al. 2005). In clinical trials, however, single agent administration of mTOR inhibitors exerted only limited anti-tumor activity (Tarhini et al. 2010). Combination therapies using more potent dual PI3K/mTOR inhibitors that circumvent the reactivation of the PI3K pathway through a negative feedback loop may possibly be more effective (Sun et al. 2005).
A variety of targetable receptor tyrosine kinases, such as KIT, MET, FGFR, IGF1R, and EGFR, are found amplified, overexpressed, or mutated in SCLC (Rygaard et al. 1993; Warshamana-Greene et al. 2005; Norkowski et al. 2013). Multiple compounds targeting these tyrosine kinases have already progressed into the clinic for other tumor types. Unfortunately, inhibition of these receptors in SCLC has not yet led to beneficial responses. It has to be noted that most of these studies were performed in unselected patient groups, and careful stratification of patients using suitable biomarkers might improve the response rates.
The overexpression of the anti-apoptotic protein BCL2 has been demonstrated in almost all SCLC cell lines and tumor samples (Jiang et al. 1995). Even though inhibition of this protein showed promising effects in cell lines and xenografts, the clinical benefit of inhibition of BCL2 was limited. Antisense oligonucleotides and several compounds inhibiting BCL2 did not improve the outcome in SCLC patients (Rudin et al. 2008; Baggstrom et al. 2011; Langer et al. 2014). However, a recent study brought new hope for the application of BCL2 inhibitors, as the combination of an mTOR inhibitor with a BCL2 inhibitor induced marked apoptosis in cell lines, xenografts, and GEMMs of SCLC (Faber et al. 2015).
The observation that PARP1 is highly expressed in SCLC also offers perspectives for the treatment of this cancer (Byers et al. 2012). In preclinical work, the administration of different PARP inhibitors already exhibited good activity when administered as a single agent, an effect that was also observed in a subset of SCLC patients in an early phase clinical trial (Cardnell et al. 2013; Wainberg et al. 2014). Since SCLC is initially extremely sensitive to chemotherapeutics, the outcomes of currently ongoing trials that combine PARP inhibitors with chemotherapy are eagerly anticipated.
In addition to targeting of specific molecular aberrations in SCLC, the NE character of this tumor type has been explored as a putative target for treatment. The SCLC cells retain NE features, such as high expression of ASCL1 and high secretion of the mitogenic factor bombesin (Westerman et al. 2002). Possibly the most promising NE-specific target for therapy is NCAM1 (CD56), which is expressed in SCLC cells. Preclinical work on lorvotuzumab, a toxin-conjugated anti-NCAM1 antibody, showed impressive response rates in SCLC xenograft models. Here the addition of lorvotuzumab to standard chemotherapeutics showed superior responses over the chemotherapy treatment alone (Whiteman et al. 2014). Disappointingly, clinical trials using this agent were stopped prematurely due to a lack of additional benefits.
Interestingly, a number of pathways involved in normal lung development, such as the Fgfr, Wnt, and Notch pathways, are aberrantly regulated in SCLC. These pathways are therefore also actively pursued as potential targets for therapeutic intervention (Table 2). The Hedgehog pathway plays a central role in development and stem cell renewal and is an essential pathway in lung regeneration following injury (Varjosalo and Taipale 2008). A key transcription factor in this pathway, the glioma-associated oncogene homolog 1 (GLI1), is frequently found overexpressed in SCLC tumor samples but not in cell lines (Vestergaard et al. 2006). Targeting the Hedgehog pathway in patients with SCLC, however, provided no significant benefit when added to cisplatin and etoposide.
SCLC is characterized by high vascularization and is associated with increased serum levels of vascular endothelial growth factor (VEGF) (Salven et al. 1998; Lucchi et al. 2002). Consequently, trials testing a number of anti-angiogenic compounds in SCLC patients were initiated. Bevacizumab, a humanized monoclonal antibody targeting VEGF, showed the most promising results in clinical trials (Lu et al. 2013). Currently, however, conflicting results on the clinical benefit of this agent for SCLC patients suggest that careful patient selection, possibly on the basis of VEGF expression levels, might be needed to better assess the potential benefits of this treatment (Jalal et al. 2010; Spigel et al. 2013).
Immunotherapy is showing promising activity in lung cancer (Villaruz et al. 2014). The most recent approaches include the checkpoint blockade, in which T-cell modulatory activities are inhibited by blocking signaling through PD1, PDL1, and CTLA-4. The effectiveness of these approaches likely depends on the presence of tumor-specific neoantigens as well as the ability of tumor cells to properly present these antigens. Tumors with a high load of somatic mutations would generate a higher diversity of neoantigens and therefore would more likely respond to checkpoint blockade (Brown et al. 2014; Snyder et al. 2014). Lung cancers, including SCLC, are characterized by a high degree of genomic instability as well as a clear smoking-associated mutational profile and therefore are likely to generate a variety of tumor-specific antigens (Alexandrov et al. 2013). On the other hand, SCLC is reported to have a severe reduction in the levels of both MHC class I and class II (Doyle et al. 1985; Yazawa et al. 1999), a means of escaping anti-cancer immunity. Combining checkpoint inhibition with radiotherapy and chemotherapy, which may boost MHC levels and better expose antigen repertoire, may be promising (Reits 2006; Bracci et al. 2013). Currently, a number of phase I and phase II clinical trials in patients with extensive and limited stage SCLC are testing the efficacy and tolerability of the standard treatment in combination with checkpoint inhibitors (Villaruz et al. 2014).
Future perspective
SCLC is an aggressive disease characterized by early metastatic spread and a rapid development of resistance. In the last decades, utilization of a variety of in vitro and in vivo model systems of SCLC helped to uncover key driver mutations and identify NE cells as the most likely cells of origin for this cancer type. In spite of these insights, no significant therapeutic improvements were achieved over 30 years. It is evident that a more detailed understanding of SCLC is required to develop more effective therapeutic approaches. Unique features dictated by both the cell of origin and the genetic lesion shape the resulting tumors and their capacity to quickly develop resistance to therapy. Therefore, detailed insight into signaling in NE cells and molecular changes that lead to their transformation may help to identify better ways to inhibit SCLC progression.
An in-depth analysis of genetic and epigenetic alterations that take place within the tumor cells in the course of acquisition of resistance is of utmost importance. Thus, expansion of a currently limited tumor tissue collection is urgently needed, with samples taken longitudinally, representing different stages of disease progression. In this regard, CTX models hold great promise and can facilitate the identification of the diversity of resistance mechanisms in individual patients.
Our understanding of the mechanisms underlying resistance and metastatic progression can come from detailed analysis of tumor heterogeneity, including further delineation of distinct cancer-initiating populations, as well as characterization of the intrinsic or acquired plasticity of SCLC, such as NE-to-mesenchymal transition. Advanced GEMMs that reflect complex genetic and cellular heterogeneity of SCLC and model both primary and resistant diseases can represent an important step forward. Development of mouse models that will allow tracing of NE cells and help to assess their potential plasticity that may arise following transformation will be of interest. For efficient utilization, these models should have a relatively short latency and allow for easy estimation of tumor burden, thus providing a tool to monitor the effectiveness of therapeutic interventions. Furthermore, GEMMs that combine key driver alterations with carcinogen-induced mutations might be an ideal model for preclinical testing of immunotherapeutic interventions.
With the availability of an increasing number of drugs for targeted therapy, new possibilities for the treatment of SCLC arise. At present, the results from early phase clinical trials using these compounds are generally disappointing, urging a further quest for suitable biomarkers for patient stratification and the identification of synergistic combination therapies.
Considering continuous improvements in molecular and computational profiling, easier access to tumor material, and rapid development of novel complex in vivo model systems, we should be able to offer better treatments to SCLC patients in the years to come.
Acknowledgments
We thank Brigid Hogan and Ji-Ying Song for sharing figures and providing images. This work was supported by a grant from the Dutch Cancer Society and a Synergy European Research Council grant to A.B.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.263145.115.
References
- Adachi J, Ookawa K, Shiseki M, Okazaki T, Tsuchida S, Morishita K, Yokota J. 1996. Induction of apoptosis but not G1 arrest by expression of the wild-type p53 gene in small cell lung carcinoma. Cell Growth Differ 7: 879–886. [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale A-L, et al. 2013. Signatures of mutational processes in human cancer. Nature 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali FR, Cheng K, Kirwan P, Metcalfe S, Livesey FJ, Barker RA, Philpott A. 2014. The phosphorylation status of Ascl1 is a key determinant of neuronal differentiation and maturation in vivo and in vitro. Development 141: 2216–2224. [DOI] [PubMed] [Google Scholar]
- Azzopardi JG. 1959. Oat-cell carcinoma of the bronchus. J Pathol Bacteriol 78: 513–519. [DOI] [PubMed] [Google Scholar]
- Baggstrom MQ, Qi Y, Koczywas M, Argiris A, Johnson EA, Millward MJ, Murphy SC, Erlichman C, Rudin CM, Govindan R. 2011. A phase II study of AT-101 (Gossypol) in chemotherapy-sensitive recurrent extensive-stage small cell lung cancer. J Thorac Oncol 6: 1757–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard WG. 1926. The nature of the ‘oat-celled sarcoma’ of the mediastinum. J Pathol Bacteriol 29: 241–244. [Google Scholar]
- Bensch KG, Gordon GB, Miller LR. 1965. Electron microscopic and biochemical studies on the bronchial carcinoid tumor. Cancer 18: 592–602. [DOI] [PubMed] [Google Scholar]
- Bensch KG, Corrin B, Pariente R, Spencer H. 1968. Oat-cell carcinoma of the lung. Its origin and relationship to bronchial carcinoid. Cancer 22: 1163–1172. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. 2001. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci 98: 13790–13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges M, Linnoila RI, van de Velde HJ, Chen H, Nelkin BD, Mabry M, Baylin SB, Ball DW. 1997. An achaete-scute homologue essential for neuroendocrine differentiation in the lung. Nature 386: 852–855. [DOI] [PubMed] [Google Scholar]
- Bracci L, Schiavoni G, Sistigu A, Belardelli F. 2013. Immune-based mechanisms of cytotoxic chemotherapy: implications for the design of novel and rationale-based combined treatments against cancer. Cell Death Differ 21: 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan J, O'Connor T, Makuch RW, Simmons AM, Russell E, Linnoila RI, Phelps RM, Gazdar AF, Ihde DC, Johnson BE. 1991. myc family DNA amplification in 107 tumors and tumor cell lines from patients with small cell lung cancer treated with different combination chemotherapy regimens. Cancer Res 51: 1708–1712. [PubMed] [Google Scholar]
- Brown SD, Warren RL, Gibb EA, Martin SD, Spinelli JJ, Nelson BH, Holt RA. 2014. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res 24: 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers LA, Rudin CM. 2014. Small cell lung cancer: where do we go from here? Cancer 121: 664–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers LA, Wang J, Nilsson MB, Fujimoto J, Saintigny P, Yordy J, Giri U, Peyton M, Fan YH, Diao L, et al. 2012. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov 2: 798–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calbo J, van Montfort E, Proost N, van Drunen E, Beverloo HB, Meuwissen R, Berns A. 2011. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell 19: 244–256. [DOI] [PubMed] [Google Scholar]
- Cardnell RJ, Feng Y, Diao L, Fan Y-H, Masrorpour F, Wang J, Shen Y, Mills GB, Minna JD, Heymach JV, et al. 2013. Proteomic markers of DNA repair and PI3K pathway activation predict response to the PARP inhibitor BMN 673 in small cell lung cancer. Clin Cancer Res 19: 6322–6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso WV. 2006. Regulation of early lung morphogenesis: questions, facts and controversies. Development 133: 1611–1624. [DOI] [PubMed] [Google Scholar]
- Carvajal LA, Manfredi JJ. 2013. Another fork in the road—life or death decisions by the tumour suppressor p53. EMBO Rep 14: 414–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda S, Ang CE, Davila J, Pak C, Mall M, Lee QY, Ahlenius H, Jung SW, Südhof TC, Wernig M. 2014. Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Rep 3: 282–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J, Dalton S. 2013. Roles for MYC in the establishment and maintenance of pluripotency. Cold Spring Harb Perspect Med 3: a014381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R. 2013. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res 23: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CL, Kwiatkowski N, Abraham BJ, Carretero J, Al-Shahrour F, Zhang T, Chipumuro E, Herter-Sprie GS, Akbay EA, Altabef A, et al. 2014. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell 26: 909–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chute JP, Chen T, Feigal E, Simon R, Johnson BE. 1999. Twenty years of phase III trials for patients with extensive-stage small-cell lung cancer: perceptible progress. J Clin Oncol 17: 1794–1801. [DOI] [PubMed] [Google Scholar]
- Coe BP, Thu KL, Aviel-Ronen S, Vucic EA, Gazdar AF, Lam S, Tsao M-S, Lam WL. 2013. Genomic deregulation of the E2F/Rb pathway leads to activation of the oncogene EZH2 in small cell lung cancer. PLoS One 8: e71670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Augert A, Rongione M, Conkrite K, Parazzoli S, Nikitin AY, Ingolia N, MacPherson D. 2014. PTEN is a potent suppressor of small cell lung cancer. Mol Cancer Res 12: 654–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel VC, Marchionni L, Hierman JS, Rhodes JT, Devereux WL, Rudin CM, Yung R, Parmigiani G, Dorsch M, Peacock CD, et al. 2009. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res 69: 3364–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146: 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle A, Martin WJ, Funa K, Gazdar A, Carney D, Martin SE, Linnoila I, Cuttitta F, Mulshine J, Bunn P. 1985. Markedly decreased expression of class I histocompatibility antigens, protein, and mRNA in human small-cell lung cancer. J Exp Med 161: 1135–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy MJ, Synnott NC, McGowan PM, Crown J, O'Connor D, Gallagher WM. 2014. p53 as a target for the treatment of cancer. Cancer Treat Rev 40: 1153–1160. [DOI] [PubMed] [Google Scholar]
- Faber AC, Farago AF, Costa C, Dastur A, Gomez-Caraballo M, Robbins R, Wagner BL, Rideout WM, Jakubik CT, Ham J, et al. 2015. Assessment of ABT-263 activity across a cancer cell line collection leads to a potent combination therapy for small-cell lung cancer. Proc Natl Acad Sci 112: E1288–E1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanidi A, Harrington EA, Evan GI. 1992. Cooperative interaction between c-myc and bcl-2 proto-oncogenes. Nature 359: 554–556. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Shin H-R, Bray F, Forman D, Mathers C, Parkin DM. 2010. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127: 2893–2917. [DOI] [PubMed] [Google Scholar]
- Forgacs E, Biesterveld EJ, Sekido Y, Fong K, Muneer S, Wistuba II, Milchgrub S, Brezinschek R, Virmani A, Gazdar AF, et al. 1998. Mutation analysis of the PTEN/MMAC1 gene in lung cancer. Oncogene 17: 1557–1565. [DOI] [PubMed] [Google Scholar]
- Friend SH, Dryja TP, Weinberg RA. 1988. Oncogenes and tumor-suppressing genes. N Engl J Med 318: 618–622. [DOI] [PubMed] [Google Scholar]
- Gazdar AF, Minna JD. 1996. NCI series of cell lines: an historical perspective. J Cell Biochem Suppl 24: 1–11. [DOI] [PubMed] [Google Scholar]
- Gazdar AF, Gao B, Minna JD. 2010. Lung cancer cell lines: useless artifacts or invaluable tools for medical science? Lung Cancer 68: 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF, Savage TK, Johnson JE, Berns A, Sage J, Linnoila RI, MacPherson D, McFadden DG, Farago A, Jacks T, et al. 2015. The comparative pathology of genetically engineered mouse models for neuroendocrine carcinomas of the lung. J Thorac Oncol 10: 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, Leenders F, Lu X, Fernández-Cuesta L, Bosco G, et al. 2015. Comprehensive genomic profiles of small cell lung cancer. Nature 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan R. 2006. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol 24: 4539–4544. [DOI] [PubMed] [Google Scholar]
- Grandori C, Eisenman RN. 1997. Myc target genes. Trends Biochem Sci 22: 177–181. [DOI] [PubMed] [Google Scholar]
- Gu J, Linnoila RI, Seibel NL, Gazdar AF, Minna JD, Brooks BJ, Hollis GF, Kirsch IR. 1988. A study of myc-related gene expression in small cell lung cancer by in situ hybridization. Am J Pathol 132: 13–17. [PMC free article] [PubMed] [Google Scholar]
- Gustafson WC, Meyerowitz JG, Nekritz EA, Chen J, Benes C, Charron E, Simonds EF, Seeger R, Matthay KK, Hertz NT, et al. 2014. Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer Cell 26: 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour JW, Lai SL, Whang-Peng J, Gazdar AF, Minna JD, Kaye FJ. 1988. Abnormalities in structure and expression of the human retinoblastoma gene in SCLC. Science 241: 353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori S, Matsuda M, Tateishi R, Nishihara H, Horai T. 1972. Oat-cell carcinoma of the lung. Clinical and morphological studies in relation to its histogenesis. Cancer 30: 1014–1024. [DOI] [PubMed] [Google Scholar]
- Helin K, Holm K, Niebuhr A, Eiberg H, Tommerup N, Hougaard S, Poulsen HS, Spang-Thomsen M, Norgaard P. 1997. Loss of the retinoblastoma protein-related p130 protein in small cell lung carcinoma. Proc Natl Acad Sci 94: 6933–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F, Polanski R, Burt DJ, Simpson KL, Morris K, et al. 2014. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med 20: 897–903. [DOI] [PubMed] [Google Scholar]
- Hogan BLM, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CCW, Niklason L, Calle E, Le A, Randell SH, et al. 2014. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Stem Cell 15: 123–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong KU, Reynolds SD, Giangreco A, Hurley CM, Stripp BR. 2001. Clara cell secretory protein-expressing cells of the airway neuroepithelial body microenvironment include a label-retaining subset and are critical for epithelial renewal after progenitor cell depletion. Am J Respir Cell Mol Biol 24: 671–681. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR–Cas9 for genome engineering. Cell 157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubaux R, Thu KL, Coe BP, MacAulay C, Lam S, Lam WL. 2013. EZH2 promotes E2F-driven SCLC tumorigenesis through modulation of apoptosis and cell-cycle regulation. J Thorac Oncol 8: 1102–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbers IJ, Bin Ali R, Pritchard C, Cozijnsen M, Kwon M-C, Proost N, Song J-Y, de Vries H, Badhai J, Sutherland K, et al. 2014. Rapid target gene validation in complex cancer mouse models using re-derived embryonic stem cells. EMBO Molecular Medicine 6: 212–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Udaka N, Yazawa T, Okudela K, Hayashi H, Sudo T, Guillemot F, Kageyama R, Kitamura H. 2000. Basic helix–loop–helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development 127: 3913–3921. [DOI] [PubMed] [Google Scholar]
- Jackman DM, Johnson BE. 2005. Small-cell lung cancer. Lancet 366: 1385–1396. [DOI] [PubMed] [Google Scholar]
- Jahchan NS, Dudley JT, Mazur PK, Flores N, Yang D, Palmerton A, Zmoos A-F, Vaka D, Tran KQT, Zhou M, et al. 2013. A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors. Cancer Discov 3: 1364–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalal S, Bedano P, Einhorn L, Bhatia S, Ansari R, Bechar N, Koneru K, Govindan R, Wu J, Yu M, et al. 2010. Paclitaxel plus bevacizumab in patients with chemosensitive relapsed small cell lung cancer: a safety, feasibility, and efficacy study from the Hoosier Oncology Group. J Thorac Oncol 5: 2008–2011. [DOI] [PubMed] [Google Scholar]
- Jiang SX, Sato Y, Kuwao S, Kameya T. 1995. Expression of bcl-2 oncogene protein is prevalent in small cell lung carcinomas. J Pathol 177: 135–138. [DOI] [PubMed] [Google Scholar]
- Johnson DH. 1999. Management of small cell lung cancer: current state of the art. Chest 116: 525S–530S. [DOI] [PubMed] [Google Scholar]
- Johnson BE, Russell E, Simmons AM, Phelps R, Steinberg SM, Ihde DC, Gazdar AF. 1996. MYC family DNA amplification in 126 tumor cell lines from patients with small cell lung cancer. J Cell Biochem Suppl 24: 210–217. [DOI] [PubMed] [Google Scholar]
- Kalemkerian GP, Akerley W, Bogner P, Borghaei H, Chow LQ, Downey RJ, Gandhi L, Ganti AKP, Govindan R, Grecula JC, et al. 2013. Small cell lung cancer. J Natl Compr Canc Netw 11: 78–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamangar F, Dores GM, Anderson WF. 2006. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol 24: 2137–2150. [DOI] [PubMed] [Google Scholar]
- Kareta MS, Gorges LL, Hafeez S, Benayoun BA, Marro S, Zmoos A-F, Cecchini MJ, Spacek D, Batista LFZ, O'Brien M, et al. 2015. Inhibition of pluripotency networks by the Rb tumor suppressor restricts reprogramming and tumorigenesis. Stem Cell 16: 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Ferguson TB, Bennett DE, Burford TH. 1969. Oat cell carcinoma of the lung. A review of 138 cases. Cancer 23: 517–524. [DOI] [PubMed] [Google Scholar]
- Kiefer PE, Bepler G, Kubasch M, Havemann K. 1987. Amplification and expression of protooncogenes in human small cell lung cancer cell lines. Cancer Res 47: 6236–6242. [PubMed] [Google Scholar]
- Kim YH, Girard L, Giacomini CP, Wang P, Hernandez-Boussard T, Tibshirani R, Minna JD, Pollack JR. 2006. Combined microarray analysis of small cell lung cancer reveals altered apoptotic balance and distinct expression signatures of MYC family gene amplification. Oncogene 25: 130–138. [DOI] [PubMed] [Google Scholar]
- Kleinerman RA, Tarone RE, Abramson DH, Seddon JM, Li FP, Tucker MA. 2000. Hereditary retinoblastoma and risk of lung cancer. J Natl Cancer Inst 92: 2037–2039. [DOI] [PubMed] [Google Scholar]
- Kotton DN, Morrisey EE. 2014. Lung regeneration: mechanisms, applications and emerging stem cell populations. Nat Med 20: 822–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal G, Birrer M, Way J, Nau M, Sausville E, Thompson C, Minna J, Battey J. 1988. Multiple mechanisms for transcriptional regulation of the myc gene family in small-cell lung cancer. Mol Cell Biol 8: 3373–3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper JL, Heideman DAM, Thunnissen E, Paul MA, van Wijk AW, Postmus PE, Smit EF. 2014. Incidence of T790M mutation in (sequential) rebiopsies in EGFR-mutated NSCLC-patients. Lung Cancer 85: 19–24. [DOI] [PubMed] [Google Scholar]
- Kwon M-c, Berns A. 2013. Mouse models for lung cancer. Mol Oncol 7: 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer CJ, Albert I, Ross HJ, Kovacs P, Blakely LJ, Pajkos G, Somfay A, Zatloukal P, Kazarnowicz A, Moezi MM, et al. 2014. Randomized phase II study of carboplatin and etoposide with or without obatoclax mesylate in extensive-stage small cell lung cancer. Lung Cancer 85: 420–428. [DOI] [PubMed] [Google Scholar]
- Leong TL, Marini KD, Rossello FJ, Jayasekara SN, Russell PA, Prodanovic Z, Kumar B, Ganju V, Alamgeer M, Irving LB, et al. 2014. Genomic characterisation of small cell lung cancer patient-derived xenografts generated from endobronchial ultrasound-guided transbronchial needle aspiration specimens. PLoS One 9: e106862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnoila RI. 2006. Functional facets of the pulmonary neuroendocrine system. Lab Invest 86: 425–444. [DOI] [PubMed] [Google Scholar]
- Linnoila RI, Sahu A, Miki M, Ball DW, DeMayo FJ. 2000. Morphometric analysis of CC10-hASH1 transgenic mouse lung: a model for bronchiolization of alveoli and neuroendocrine carcinoma. Exp Lung Res 26: 595–615. [DOI] [PubMed] [Google Scholar]
- Lu H-Y, Wang X-J, Mao W-M. 2013. Targeted therapies in small cell lung cancer. Oncol Lett 5: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchi M, Mussi A, Fontanini G, Faviana P, Ribechini A, Angeletti CA. 2002. Small cell lung carcinoma (SCLC): the angiogenic phenomenon. Eur J Cardiothorac Surg 21: 1105–1110. [DOI] [PubMed] [Google Scholar]
- McFadden DG, Papagiannakopoulos T, Taylor-Weiner A, Stewart C, Carter SL, Cibulskis K, Bhutkar A, McKenna A, Dooley A, Vernon A, et al. 2014. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell 156: 1298–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melichar B, Adenis A, Lockhart AC, Bennouna J, Dees EC, Kayaleh O, Obermannova R, DeMichele A, Zatloukal P, Zhang B, et al. 2015. Safety and activity of alisertib, an investigational aurora kinase A inhibitor, in patients with breast cancer, small-cell lung cancer, non-small-cell lung cancer, head and neck squamous-cell carcinoma, and gastro-oesophageal adenocarcinoma: a five-arm phase 2 study. Lancet Oncol 16: 395–405. [DOI] [PubMed] [Google Scholar]
- Metzger RJ, Klein OD, Martin GR, Krasnow MA. 2008. The branching programme of mouse lung development. Nature 453: 745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. 2003. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell 4: 181–189. [DOI] [PubMed] [Google Scholar]
- Micke P, Faldum A, Metz T, Beeh K-M, Bittinger F, Hengstler J-G, Buhl R. 2002. Staging small cell lung cancer: Veterans Administration Lung Study Group versus International Association for the Study of Lung Cancer—what limits limited disease? Lung Cancer 37: 271–276. [DOI] [PubMed] [Google Scholar]
- Miyashita T, Krajewski S, Krajewska M, Wang HG, Lin HK, Liebermann DA, Hoffman B, Reed JC. 1994. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene 9: 1799–1805. [PubMed] [Google Scholar]
- Modi S, Kubo A, Oie H, Coxon AB, Rehmatulla A, Kaye FJ. 2000. Protein expression of the RB-related gene family and SV40 large T antigen in mesothelioma and lung cancer. Oncogene 19: 4632–4639. [DOI] [PubMed] [Google Scholar]
- Morimoto M, Nishinakamura R, Saga Y, Kopan R. 2012. Different assemblies of Notch receptors coordinate the distribution of the major bronchial Clara, ciliated and neuroendocrine cells. Development 139: 4365–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederst MJ, Sequist LV, Poirier JT, Mermel CH, Lockerman EL, Garcia AR, Katayama R, Costa C, Ross KN, Moran T, et al. 2015. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun 6: 6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norkowski E, Ghigna M-R, Lacroix L, Le Chevalier T, Fadel É, Dartevelle P, Dorfmuller P, Thomas de Montpréville V. 2013. Small-cell carcinoma in the setting of pulmonary adenocarcinoma: new insights in the era of molecular pathology. J Thorac Oncol 8: 1265–1271. [DOI] [PubMed] [Google Scholar]
- Otterson G, Lin A, Kay F. 1992. Genetic etiology of lung cancer. Oncology 6: 97–104. [PubMed] [Google Scholar]
- Park K-S, Liang M-C, Raiser DM, Zamponi R, Roach RR, Curtis SJ, Walton Z, Schaffer BE, Roake CM, Zmoos A-F, et al. 2011a. Characterization of the cell of origin for small cell lung cancer. Cell Cycle 10: 2806–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K-S, Martelotto LG, Peifer M, Sos ML, Karnezis AN, Mahjoub MR, Bernard K, Conklin JF, Szczepny A, Yuan J, et al. 2011b. A crucial requirement for Hedgehog signaling in small cell lung cancer. Nat Med 17: 150–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Fernández-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, Plenker D, Leenders F, Sun R, Zander T, et al. 2012. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 44: 1104–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JD, Sansom SN, Smith J, Dobenecker MW, Tarakhovsky A, Livesey FJ. 2010. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci 107: 15957–15962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzella F, Turley H, Kuzu I, Tungekar MF, Dunnill MS, Pierce CB, Harris A, Gatter KC, Mason DY. 1993. bcl-2 protein in non-small-cell lung carcinoma. N Engl J Med 329: 690–694. [DOI] [PubMed] [Google Scholar]
- Poirier JT, Gardner EE, Connis N, Moreira AL, de Stanchina E, Hann CL, Rudin CM. 2015. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene 10.1038/onc.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popat S, Wotherspoon A, Nutting CM, Gonzalez D, Nicholson AG, O'Brien M. 2013. Transformation to ‘high grade’ neuroendocrine carcinoma as an acquired drug resistance mechanism in EGFR-mutant lung adenocarcinoma. Lung Cancer 80: 1–4. [DOI] [PubMed] [Google Scholar]
- Post LC, Ternet M, Hogan BL. 2000. Notch/Delta expression in the developing mouse lung. Mech Dev 98: 95–98. [DOI] [PubMed] [Google Scholar]
- Rawlins EL, Clark CP, Xue Y, Hogan BLM. 2009. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development 136: 3741–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzini G, Berrie CP, Vignati S, Broggini M, Mascetta G, Brancaccio A, Falasca M. 2000. Novel functional PI 3-kinase antagonists inhibit cell growth and tumorigenicity in human cancer cell lines. FASEB J 14: 1179–1187. [DOI] [PubMed] [Google Scholar]
- Reits EA. 2006. Radiation modulates the peptide repertoire, enhances MHC class I expression, and induces successful antitumor immunotherapy. J Exp Med 203: 1259–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw AA, Haja J, Lozano RL, Wilbur DC. 2005. Distinguishing carcinoid tumor from small cell carcinoma of the lung: correlating cytologic features and performance in the College of American Pathologists Non-Gynecologic Cytology Program. Arch Pathol Lab Med 129: 614–618. [DOI] [PubMed] [Google Scholar]
- Reynolds SD, Giangreco A, Power JH, Stripp BR. 2000. Neuroepithelial bodies of pulmonary airways serve as a reservoir of progenitor cells capable of epithelial regeneration. Am J Pathol 156: 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried T, Petersen I, Holtgreve-Grez H, Speicher MR, Schröck E, du Manoir S, Cremer T. 1994. Mapping of multiple DNA gains and losses in primary small cell lung carcinomas by comparative genomic hybridization. Cancer Res 54: 1801–1806. [PubMed] [Google Scholar]
- Rock JR, Hogan BLM. 2011. Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu Rev Cell Dev Biol 27: 493–512. [DOI] [PubMed] [Google Scholar]
- Rock JR, Randell SH, Hogan BLM. 2010. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis Model Mech 3: 545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JS, Wang K, Elkadi OR, Tarasen A, Foulke L, Sheehan CE, Otto GA, Palmer G, Yelensky R, Lipson D, et al. 2014. Next-generation sequencing reveals frequent consistent genomic alterations in small cell undifferentiated lung cancer. J Clin Pathol 67: 772–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM, Salgia R, Wang X, Hodgson LD, Masters GA, Green M, Vokes EE. 2008. Randomized phase II Study of carboplatin and etoposide with or without the bcl-2 antisense oligonucleotide oblimersen for extensive-stage small-cell lung cancer: CALGB 30103. J Clin Oncol 26: 870–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, Bergbower EA, Guan Y, Shin J, Guillory J, et al. 2012. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 44: 1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rygaard K, Nakamura T, Spang-Thomsen M. 1993. Expression of the proto-oncogenes c-met and c-kit and their ligands, hepatocyte growth factor/scatter factor and stem cell factor, in SCLC cell lines and xenografts. Br J Cancer 67: 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgia R, Skarin AT. 1998. Molecular abnormalities in lung cancer. J Clin Oncol 16: 1207–1217. [DOI] [PubMed] [Google Scholar]
- Salven P, Ruotsalainen T, Mattson K, Joensuu H. 1998. High pre-treatment serum level of vascular endothelial growth factor (VEGF) is associated with poor outcome in small-cell lung cancer. Int J Cancer 79: 144–146. [DOI] [PubMed] [Google Scholar]
- Sánchez-Rivera FJ, Papagiannakopoulos T, Romero R, Tammela T, Bauer MR, Bhutkar A, Joshi NS, Subbaraj L, Bronson RT, Xue W, et al. 2014. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 516: 428–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer BE, Park KS, Yiu G, Conklin JF, Lin C, Burkhart DL, Karnezis AN, Sweet-Cordero EA, Sage J. 2010. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res 70: 3877–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers WR, Novitch BG, Miyake S, Heith A, Otterson GA, Kaye FJ, Lassar AB, Kaelin WG. 1998. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev 12: 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, et al. 2011. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 3: 75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgambato A, Casaluce F, Maione P, Rossi A, Sacco PC, Panzone F, Ciardiello F, Gridelli C. 2013. Medical treatment of small cell lung cancer: state of the art and new development. Expert Opin Pharmacother 14: 2019–2031. [DOI] [PubMed] [Google Scholar]
- Shepherd FA, Crowley J, Van Houtte P, Postmus PE, Carney D, Chansky K, Shaikh Z, Goldstraw P. 2007. The International Association for the Study of Lung Cancer lung cancer staging project: proposals regarding the clinical staging of small cell lung cancer in the forthcoming (seventh) edition of the tumor, node, metastasis classification for lung cancer. J Thorac Oncol 2: 1067–1077. [DOI] [PubMed] [Google Scholar]
- Sher F, Rossler R, Brouwer N, Balasubramaniyan V, Boddeke E, Copray S. 2008. Differentiation of neural stem cells into oligodendrocytes: involvement of the polycomb group protein Ezh2. Stem Cells 26: 2875–2883. [DOI] [PubMed] [Google Scholar]
- Shivapurkar N, Virmani AK, Wistuba II, Milchgrub S, Mackay B, Minna JD, Gazdar AF. 1999. Deletions of chromosome 4 at multiple sites are frequent in malignant mesothelioma and small cell lung carcinoma. Clin Cancer Res 5: 17–23. [PubMed] [Google Scholar]
- Silva A, Wang J, Lomahan S, Tran TA, Grenlin L, Suganami A, Tamura Y, Ikegaki N. 2014. Aurora kinase A is a possible target of OSU03012 to destabilize MYC family proteins. Oncol Rep 32: 901–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snitow ME, Li S, Morley MP, Rathi K, Lu MM, Kadzik RS, Stewart KM, Morrisey EE. 2015. Ezh2 represses the basal cell lineage during lung endoderm development. Development 142: 108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, et al. 2014. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 371: 2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Yao E, Lin C, Gacayan R, Chen M-H, Chuang P-T. 2012. Functional characterization of pulmonary neuroendocrine cells in lung development, injury, and tumorigenesis. Proc Natl Acad Sci 109: 17531–17536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sos ML, Dietlein F, Peifer M, Schöttle J, Balke-Want H, Müller C, Koker M, Richters A, Heynck S, Malchers F, et al. 2012. A framework for identification of actionable cancer genome dependencies in small cell lung cancer. Proc Natl Acad Sci 109: 17034–17039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spigel DR, Waterhouse DM, Lane S, Legenne P, Bhatt K. 2013. Efficacy and safety of oral topotecan and bevacizumab combination as second-line treatment for relapsed small-cell lung cancer: an open-label multicenter single-arm phase II study. Clin Lung Cancer 14: 356–363. [DOI] [PubMed] [Google Scholar]
- Stevens TP, McBride JT, Peake JL, Pinkerton KE, Stripp BR. 1997. Cell proliferation contributes to PNEC hyperplasia after acute airway injury. Am J Physiol 272: L486–493. [DOI] [PubMed] [Google Scholar]
- Stewart DJ. 2010. Tumor and host factors that may limit efficacy of chemotherapy in non-small cell and small cell lung cancer. Crit Rev Oncol Hematol 75: 173–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S-Y, Rosenberg LM, Wang X, Zhou Z, Yue P, Fu H, Khuri FR. 2005. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res 65: 7052–7058. [DOI] [PubMed] [Google Scholar]
- Sutherland KD, Proost N, Brouns I, Adriaensen D, Song J-Y, Berns A. 2011. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell 19: 754–764. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Nau MM, Chiba I, Birrer MJ, Rosenberg RK, Vinocour M, Levitt M, Pass H, Gazdar AF, Minna JD. 1989. p53: a frequent target for genetic abnormalities in lung cancer. Science 246: 491–494. [DOI] [PubMed] [Google Scholar]
- Tan HL, Sood A, Rahimi HA, Wang W, Gupta N, Hicks J, Mosier S, Gocke CD, Epstein JI, Netto GJ, et al. 2014. Rb loss is characteristic of prostatic small cell neuroendocrine carcinoma. Clin Cancer Res 20: 890–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannapfel A, John K, Mise N, Schmidt A, Buhlmann S, Ibrahim SM, Putzer BM. 2008. Autonomous growth and hepatocarcinogenesis in transgenic mice expressing the p53 family inhibitor DNp73. Carcinogenesis 29: 211–218. [DOI] [PubMed] [Google Scholar]
- Tarhini A, Kotsakis A, Gooding W, Shuai Y, Petro D, Friedland D, Belani CP, Dacic S, Argiris A. 2010. Phase II study of everolimus (RAD001) in previously treated small cell lung cancer. Clin Cancer Res 16: 5900–5907. [DOI] [PubMed] [Google Scholar]
- Toyooka S, Tsuda T, Gazdar AF. 2003. The TP53 gene, tobacco exposure, and lung cancer. Hum Mutat 21: 229–239. [DOI] [PubMed] [Google Scholar]
- Umemura S, Mimaki S, Makinoshima H, Tada S, Ishii G, Ohmatsu H, Niho S, Yoh K, Matsumoto S, Takahashi A, et al. 2014. Therapeutic priority of the PI3K/AKT/mTOR pathway in small cell lung cancers as revealed by a comprehensive genomic analysis. J Thorac Oncol 9: 1324–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lommel A. 2001. Pulmonary neuroendocrine cells (PNEC) and neuroepithelial bodies (NEB): chemoreceptors and regulators of lung development. Paediatr Respir Rev 2: 171–176. [DOI] [PubMed] [Google Scholar]
- Van Lommel A, Bollé T, Fannes W, Lauweryns JM. 1999. The pulmonary neuroendocrine system: the past decade. Arch Histol Cytol 62: 1–16. [DOI] [PubMed] [Google Scholar]
- van Meerbeeck JP, Fennell DA, De Ruysscher DKM. 2011. Small-cell lung cancer. Lancet 378: 1741–1755. [DOI] [PubMed] [Google Scholar]
- Varjosalo M, Taipale J. 2008. Hedgehog: functions and mechanisms. Genes Dev 22: 2454–2472. [DOI] [PubMed] [Google Scholar]
- Vaughan AE, Brumwell AN, Xi Y, Gotts JE, Brownfield DG, Treutlein B, Tan K, Tan V, Liu FC, Looney MR, et al. 2015. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature 517: 621–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergaard J, Pedersen MW, Pedersen N, Ensinger C, Tümer Z, Tommerup N, Poulsen HS, Larsen LA. 2006. Hedgehog signaling in small-cell lung cancer: frequent in vivo but a rare event in vitro. Lung Cancer 52: 281–290. [DOI] [PubMed] [Google Scholar]
- Villaruz LC, Kalyan A, Zarour H, Socinski MA. 2014. Immunotherapy in lung cancer. Transl Lung Cancer Res 3: 2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virmani AK, Fong KM, Kodagoda D, McIntire D, Hung J, Tonk V, Minna JD, Gazdar AF. 1998. Allelotyping demonstrates common and distinct patterns of chromosomal loss in human lung cancer types. Genes Chromosomes Cancer 21: 308–319. [DOI] [PubMed] [Google Scholar]
- Wainberg ZA, Rafii S, Ramanathan RK, Mina L, Byers LA, Chugh R, Goldman JW, Sachdev JC, Matei DE, Wheler JJ, et al. 2014. Safety and antitumor activity of the PARP inhibitor BMN673 in a phase 1 trial recruiting metastatic small-cell lung cancer (SCLC) and germline BRCA-mutation carrier cancer patients. J Clin Oncol 32: 5s. [Google Scholar]
- Warshamana-Greene GS, Litz J, Buchdunger E, García-Echeverría C, Hofmann F, Krystal GW. 2005. The insulin-like growth factor-I receptor kinase inhibitor, NVP-ADW742, sensitizes small cell lung cancer cell lines to the effects of chemotherapy. Clin Cancer Res 11: 1563–1571. [DOI] [PubMed] [Google Scholar]
- Watson WL, Berg JW. 1962. Oat cell lung cancer. Cancer 15: 759–768. [DOI] [PubMed] [Google Scholar]
- Weinberg RA. 1995. The retinoblastoma protein and cell cycle control. Cell 81: 323–330. [DOI] [PubMed] [Google Scholar]
- Westerman BA, Neijenhuis S, Poutsma A, Steenbergen RDM, Breuer RHJ, Egging M, van Wijk IJ, Oudejans CBM. 2002. Quantitative reverse transcription-polymerase chain reaction measurement of HASH1 (ASCL1), a marker for small cell lung carcinomas with neuroendocrine features. Clin Cancer Res 8: 1082–1086. [PubMed] [Google Scholar]
- Whiteman KR, Johnson HA, Mayo MF, Audette CA, Carrigan CN, LaBelle A, Zukerberg L, Lambert JM, Lutz RJ. 2014. Lorvotuzumab mertansine, a CD56-targeting antibody-drug conjugate with potent antitumor activity against small cell lung cancer in human xenograft models. MAbs 6: 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikenheiser-Brokamp KA. 2004. Rb family proteins differentially regulate distinct cell lineages during epithelial development. Development 131: 4299–4310. [DOI] [PubMed] [Google Scholar]
- Wistuba II, Behrens C, Virmani AK, Mele G, Milchgrub S, Girard L, Fondon JW, Garner HR, McKay B, Latif F, et al. 2000a. High resolution chromosome 3p allelotyping of human lung cancer and preneoplastic/preinvasive bronchial epithelium reveals multiple, discontinuous sites of 3p allele loss and three regions of frequent breakpoints. Cancer Res 60: 1949–1960. [PubMed] [Google Scholar]
- Wistuba II, Berry J, Behrens C, Maitra A, Shivapurkar N, Milchgrub S, Mackay B, Minna JD, Gazdar AF. 2000b. Molecular changes in the bronchial epithelium of patients with small cell lung cancer. Clin Cancer Res 6: 2604–2610. [PMC free article] [PubMed] [Google Scholar]
- Witschi H. 2001. A short history of lung cancer. Toxicol Sci 64: 4–6. [DOI] [PubMed] [Google Scholar]
- Wojtalla A, Fischer B, Kotelevets N, Mauri FA, Sobek J, Rehrauer H, Wotzkow C, Tschan MP, Seckl MJ, Zangemeister-Wittke U, et al. 2013. Targeting the phosphoinositide 3-kinase p110-α isoform impairs cell proliferation, survival, and tumor growth in small cell lung cancer. Clin Cancer Res 19: 96–105. [DOI] [PubMed] [Google Scholar]
- Wu C, Wangpaichitr M, Feun L, Kuo MT, Robles C, Lampidis T, Savaraj N. 2005. Overcoming cisplatin resistance by mTOR inhibitor in lung cancer. Mol Cancer 4: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazawa T, Kamma H, Fujiwara M, Matsui M, Horiguchi H, Satoh H, Fujimoto M, Yokoyama K, Ogata T. 1999. Lack of class II transactivator causes severe deficiency of HLA-DR expression in small cell lung cancer. J Pathol 187: 191–199. [DOI] [PubMed] [Google Scholar]
- Yokomizo A, Tindall DJ, Drabkin H, Gemmill R, Franklin W, Yang P, Sugio K, Smith DI, Liu W. 1998. PTEN/MMAC1 mutations identified in small cell, but not in non-small cell lung cancers. Oncogene 17: 475–479. [DOI] [PubMed] [Google Scholar]
- Zandi R, Selivanova G, Christensen CL, Gerds TA, Willumsen BM, Poulsen HS. 2011. PRIMA-1Met/APR-246 induces apoptosis and tumor growth delay in small cell lung cancer expressing mutant p53. Clin Cancer Res 17: 2830–2841. [DOI] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478: 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo W, Zhang T, Wu DZ, Guan SP, Liew A-A, Yamamoto Y, Wang X, Lim SJ, Vincent M, Lessard M, et al. 2015. p63+Krt5+ distal airway stem cells are essential for lung regeneration. Nature 517: 616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]