Upon ribosomal stress, the central region of MDM2 is bound by ribosomal proteins, particularly ribosomal protein L11 (RPL11), leading to MDM2 inactivation and subsequent p53 activation. Zheng et al. solved the complex structure of human MDM2–RPL11 at 2.4 Å and show that formation of the MDM2–RPL11 complex induces substantial conformational changes in both proteins.
Keywords: cancer, RPL11—MDM2, p53 activation, molecular basis, complex structure
Abstract
The central region of MDM2 is critical for p53 activation and tumor suppression. Upon ribosomal stress, this region is bound by ribosomal proteins, particularly ribosomal protein L11 (RPL11), leading to MDM2 inactivation and subsequent p53 activation. Here, we solved the complex structure of human MDM2–RPL11 at 2.4 Å. MDM2 extensively interacts with RPL11 through an acidic domain and two zinc fingers. Formation of the MDM2–RPL11 complex induces substantial conformational changes in both proteins. RPL11, unable to bind MDM2 mutants, fails to induce the activation of p53 in cells. MDM2 mimics 28S rRNA binding to RPL11. The C4 zinc finger determines RPL11 binding to MDM2 but not its homolog, MDMX. Our results highlight the essential role of the RPL11–MDM2 interaction in p53 activation and tumor suppression and provide a structural basis for potential new anti-tumor drug development.
The tumor suppressor p53 plays a critical role in tumor suppression, and its inactivation has been linked to tumorigenesis. In >50% of human cancers, p53 loss-of-function mutations occur (Ozaki and Nakagawara 2011). For cancers in which p53 is not mutated, its functions are compromised because of defects that occur in other components of the pathway. p53 is a transcription factor that can up-regulate many downstream genes that are involved in cell cycle arrest, apoptosis, and senescence in response to a wide range of stress stimuli (Vousden and Prives 2009). Under normal physiological conditions, the anti-proliferative functions of p53 are strictly inhibited, mainly by the actions of an E3 ubiquitin ligase, MDM2, which mediates p53 ubiquitination and degradation. The MDM2 gene is also a p53 target, which forms a feedback loop that restrains p53 activity (Pant et al. 2013). The negative regulation of p53 by MDM2 has been genetically confirmed, as the early embryonic lethality caused by deletion of the MDM2 gene can be rescued by p53 deletion (Montes de Oca Luna et al. 1995).
MDM2 is a focal point for p53 activation in response to various stress signals, such as DNA damage, oncogenic activation, and ribosomal stresses (Zhou et al. 2012). Ribosomal stresses, which are caused by impaired ribosome biogenesis, can activate p53 through the inhibition of MDM2 by ribosomal proteins (RPs), notably RPL11 and RPL5 (Lohrum et al. 2003; Zhang et al. 2003; Fumagalli et al. 2012). Earlier studies have suggested that ribosomal stress can affect the integrity of the nucleolus, resulting in the passive release of RPs into the nucleoplasm, where they bind MDM2 and inhibit its activity (Zhang and Lu 2009; Miliani de Marval and Zhang 2011). Recently, it has been shown that RPL11 can actively accumulate in the nucleoplasm via the specific up-regulation of RPL11 mRNA translation induced by the inhibition of 40S ribosome biogenesis (Fumagalli et al. 2009; Bursac et al. 2014). Among the RPs that can bind to MDM2, RPL11 has been reported to play the most significant role in p53 activation and ribosomal stress sensing (Fumagalli et al. 2009; Zhou et al. 2012; Kim et al. 2014). The accumulation of RPL11 in the nucleoplasm appears to be the key mechanism for both MDM2 inactivation and p53 activation. Further studies have suggested that RPL11 in the nucleoplasm can be stabilized by forming a complex with RPL5 and 5S rRNA (Donati et al. 2013; Sloan et al. 2013).
MDM2 contains three domains: the N-terminal region, the C-terminal RING finger domain, and the central region that includes an acidic region and a C4 zinc finger domain. The N-terminal region interacts with p53 and down-regulates its activity (Kussie et al. 1996). The RING finger domain has ubiquitin ligase activity that can mediate MDM2 autoubiquitination and p53 ubiquitination and also provides binding sites for the MDM2 homolog MDMX (Honda and Yasuda 2000). Previous studies have suggested that the central region is involved in MDM2 targeting of p53 and also meditates interactions of MDM2 with ARF and RPs (Sherr 2006; Zhang and Lu 2009). The RPL11-binding region has been mapped to the central region (amino acid residues 63–125) (Zhang et al. 2003). However, how RPL11 precisely interacts with MDM2 remains to be elucidated.
MDMX is another key negative regulator of p53, which can bind to p53 and inhibit its transcriptional activity (Marine et al. 2007; Wade et al. 2013). Despite exhibiting high sequence homology with MDM2 in all three regions, including the C-terminal RING finger domain, MDMX does not possess any E3 ubiquitin ligase activity or bind to RPs. The central region of MDMX is similarly composed of an acidic region and a zinc finger domain. However, it remains unclear why RPs selectively bind MDM2 but not MDMX (Gilkes et al. 2006). Moreover, MDM2–RPL11 interaction not only prevents p53 degradation but also promotes MDMX ubiquitination and degradation in a MDM2-dependent manner (Li and Gu 2011), indicating that RPL11 uses a more complex mechanism to regulate p53 (Gilkes and Chen 2007).
The importance of the RPL11–MDM2–p53 pathway in tumor suppression has been demonstrated in mouse models and human tumors, and the pivotal role of RPL11 in p53 activation in response to ribosomal stresses has been well established. However, the structural basis and molecular mechanisms of the MDM2–RPL11 interaction remain poorly understood. Here, we solved the first structure of the human MDM2–RPL11 complex. In addition to the known C4 zinc finger domain, we identified another unanticipated zinc finger that forms intermolecularly within the complex. MDM2 fits tightly into the concave surface of RPL11 and forms a large number of interactions. Mutagenesis studies have shown that the two zinc fingers and some key interfacial residues are required to establish the RPL11–MDM2 interaction in vitro and in vivo. Interestingly, MDM2 binds to RPL11 in a manner mimicking 28S rRNA in the 60S ribosomal subunit. Moreover, we discovered the critical residues in MDMX that can block the MDMX–RPL11 interaction. Overall, our studies have yielded insights into the RPL11–MDM2–p53 surveillance pathway and may provide useful anti-cancer strategies.
Results
Crystal structure of the MDM2–RPL11 complex
As MDM2 could be easily degraded, we coexpressed a series of MDM2 and RPL11 fragments in the Escherichia coli cells and then copurified them through a series of procedures (see the Materials and Methods). After extensive trials, the MDM2290–437–RPL111–178 complex was found to be the most stable in vitro. A size exclusion chromatography assay (Supplemental Fig. 1A) showed that the complex eluted with two peaks. The first peak notably aggregated, but the second peak eluted at a molecular weight between 17 and 66 kDa, which was close to the calculated molecular weight (∼37 kDa). A further size exclusion chromatography assay and its corresponding SDS-PAGE results (Supplemental Fig. 1B) indicated that the products were highly purified and not degraded. After extensive trials and optimization, we eventually succeeded in obtaining crystals that diffracted to 2.4 Å resolution.
The MDM2–RPL11 complex structure was determined by molecular replacement and refined to a resolution of 2.4 Å in the space group of P3221, with Rwork = 19.2% and Rfree = 22.7% (Supplemental Table 1). The solved structure contained RPL11 and part of MDM2 (Fig. 1A,B; Supplemental Movie 1), most likely because of MDM2 degradation during the crystallization process. The final model included MDM2 residues 293–334, RPL11 residues 13–178 (residues 139–155 were missing), two zinc ions, one imidazole molecule, one β-mercaptoethanol molecule, and 84 water molecules. The number and position of these two zinc atoms were consistent with those obtained using the SHELX C/D program.
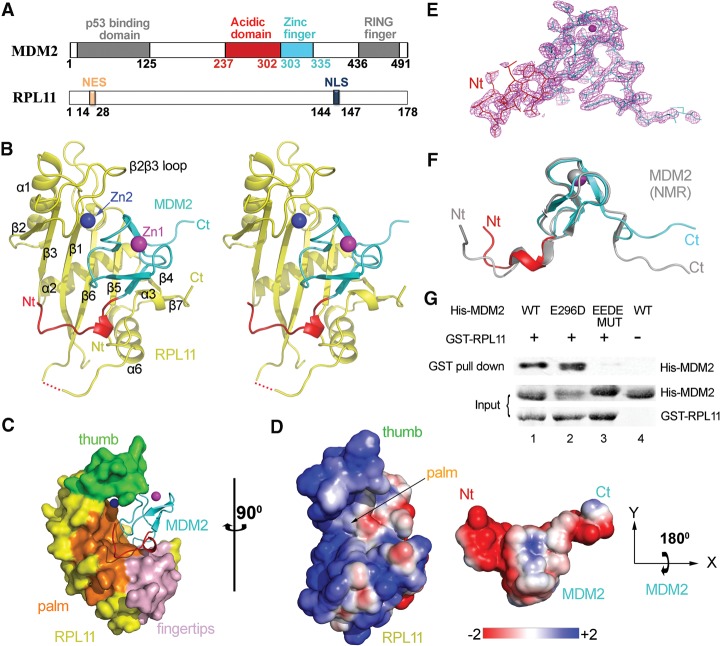
Figure 1.
Crystal structure of the human MDM2–RPL11 complex. (A) Domain organization of MDM2 and RPL11. Numbers indicate residues. (B) Overall structure of the MDM2–RPL11 complex. The domains of MDM2 are shown. (Yellow) RPL11. The two zinc ions are shown as spheres. (C) Surface representation of the MDM2–RPL11 complex. (Orange) β Sheets that formed the palm; (green) the β2β3 loop (thumb); (light pink) the fingertips. (D) The electrostatic potential of the binding interface. The surface potential is displayed as a color gradient ranging from red (negative) to blue (positive). Note that MDM2 is rotated 180° on its X-axis to show the interface. (E) The “omit” electron density map of MDM2, contoured at 1.5 σ. (F) Structural overlay of MDM2 in the MDM2–RPL11 structure and in a NMR structure (gray; Protein Data Bank [PDB] ID: 2C6A) (Yu et al. 2006). (G) The acidic domain of MDM2 is important for RPL11 binding. Purified GST-tagged RPL11 was used for GST pull-down of purified His-MDM2 wild type (WT) and its mutants, EEDE MUT (E292K/E293K/D294K/E296K).
The overall structure of RPL11 forms a hand-like shape (Fig. 1B). The fingertips consist of a parallel β sheet (β4 and β7), α3, and the loop following α3. The palm is composed of four anti-parallel β sheets, including β1, β3, β5, and β6. The four anti-parallel β sheets form a concave surface to which MDM2 is bound (Fig. 1C). Moreover, the β2β3 loop appears to be like a curving thumb that clamps MDM2 tightly by cooperating with the palm and fingertips (Fig. 1C). A sequence comparison of RPL11 in different species (Supplemental Fig. 2) also shows that the residues in the thumb are highly conserved. Moreover, analysis of the MDM2–RPL11 complex reveals that the interactions between them mainly include strong electrostatic interactions and hydrophobic interactions (Fig. 1D).
The MDM2 C4 zinc finger domain in the complex adopts a fold consisting of a 310 helix followed by two anti-parallel β strands, which shares significant structural similarity with the zinc ribbon protein family (Yu et al. 2006). A structural comparison between the MDM2 structure described in the present study and that observed in a NMR structure (Yu et al. 2006) shows that the core conformations of the C4 zinc finger domains are nearly identical, with an overall root mean square deviations (RMSD) of 0.8 Å. However, their overall conformations differed greatly, with an RMSD of 2.4 Å, and the differences occurred mostly at the N and C termini (Fig. 1F) because both termini of MDM2 in the aligned NMR structure would sterically crash the β6, β4, and β5 of RPL11 and hinder the binding of RPL11. Thus, to bind the C4 zinc finger of MDM2 tightly, the binding of RPL11 induces the N-terminal and C-terminal regions of MDM2 to undergo conformational changes and a sheet-to-coil transition (Fig. 1F). Overall, conformation changes of MDM2 are induced by the binding of RPL11.
The two zinc fingers in the MDM2–RPL11 complex are functionally significant
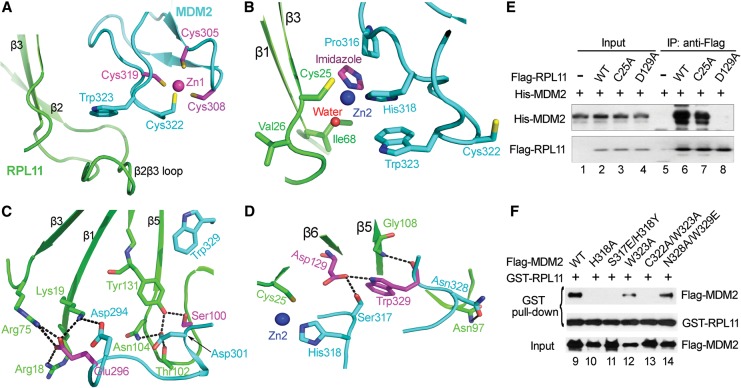
We unexpectedly observed two zinc ions in the MDM2–RPL11 complex (one intramolecular ion in the MDM2 C4 zinc finger and another intermolecular ion that was chelated by Cys25 in RPL11, by His318 in MDM2, by a water molecule, and by an imidazole molecule that was used in a buffer during crystallization) (Fig. 2A,B). The four cysteines that form the MDM2 C4 zinc finger were highly conserved, and any cysteine mutation (including C305F, C305S, C308W, C319R, and C322R) in the C4 zinc finger domain could lead to the collapse of the zinc finger fold, thus hindering the interaction between MDM2 and RPL11 (Macias et al. 2010; Zhang et al. 2011). For the intermolecular zinc finger, the C25A mutation also diminished this interaction in H1299 cells (Fig. 2E, lane 7), and the H318A mutation abolished this interaction (Fig. 2F, lane 10). Moreover, the π stacking between residues Trp323 and His318 was also very important for MDM2 stabilization and its interaction with RPL11 (Fig. 2B,F, lanes 12–13). In conclusion, these results emphasized the importance of maintaining the integrity of both zinc fingers.
Figure 2.
The two zinc fingers and extensive interactions between MDM2 and RPL11. (A,B) Close-up views of the two zinc fingers. Residues involved in zinc coordination are shown as sticks. A water molecule is shown as a red sphere. (C,D) Close-up views of MDM2–RPL11 interactions. Critical residues for the MDM2–RPL11 interaction are shown as sticks. Hydrogen bonds are indicated as dashed lines. Residues where cancer-associated mutations occur are colored magenta. (E) RPL11 mutants reduce the interaction of RPL11 with MDM2. H1299 cells transfected with Flag-tagged RPL11 wild-type (WT) and its mutants were immobilized on Flag beads and used to pull down purified His-MDM2. The resulting proteins were detected by Western blotting. (F) MDM2 mutants impair the interaction of MDM2 with RPL11. Mouse embryonic fibroblast cells from p53−/− MDM2−/− mice (double-knockout cells) were transfected with Flag-MDM2 wild type or its mutants. After 24 h, lysates were incubated with 100 μg of GST-RPL11 protein and glutathione sepharose beads.
An extensive binding interface establishes the MDM2–RPL11 interaction
Many studies have highlighted the importance of the interaction between MDM2 and RPL11 in the regulation of p53. Here, our results provide a structural basis for these observations. The total buried surface area between MDM2 and RPL11 reached 1173.4 Å2, as determined by analysis using the PISA server (Krissinel and Henrick 2007). There are many interactions at the interface, including hydrogen bonds and electrostatic, hydrophobic, and van der Waals interactions. Figure 1D shows that the acidic domain of MDM2 forms a negatively charged surface, which establishes an electrostatic interaction with the positively charged surface of the palm of RPL11. Notably, two key residues, Asp294 and Glu296, in the acidic domain of MDM2 form key salt bridges with two adjacent residues—Arg18 and Lys19—on the β1 sheet together with residue Arg75 on the loop that connects α2 and β3 in RPL11 (Fig. 2C). Consistently, mutation of the acidic residues to alkaline residues—MDM2 EEDE MUT (E292K/E293K/D294K/E296K)—abolishes RPL11-binding ability (Fig. 1G, lane 3). Moreover, Asp301 in the acidic domain of MDM2 forms an extensive array of hydrogen bonds with Thr102, Asn104, Tyr131, and Ser100 in RPL11 (Fig. 2C). Together, these findings demonstrate that the acidic domain of MDM2 is essential for RPL11 binding.
In addition to these interactions, the residues Ser317, Trp329, and Asn328 of MDM2 form hydrogen-bonding networks with Asp129, Gly108, and Asn97 in the center of the palm of RPL11 (Fig. 2D). These interactions are essential for the formation of a functional complex, which could be further confirmed by mutagenesis experiments (Fig. 2E,F, lanes 8,11,14). Disrupting the interactions described above greatly reduces the binding of RPL11 to MDM2. The large size of these interfacial surfaces and the extensive interactions suggest that strong binding exists between MDM2 and RPL11.
RPL11 loses the ability to rescue MDM2 mutation-induced p53 degradation in vivo
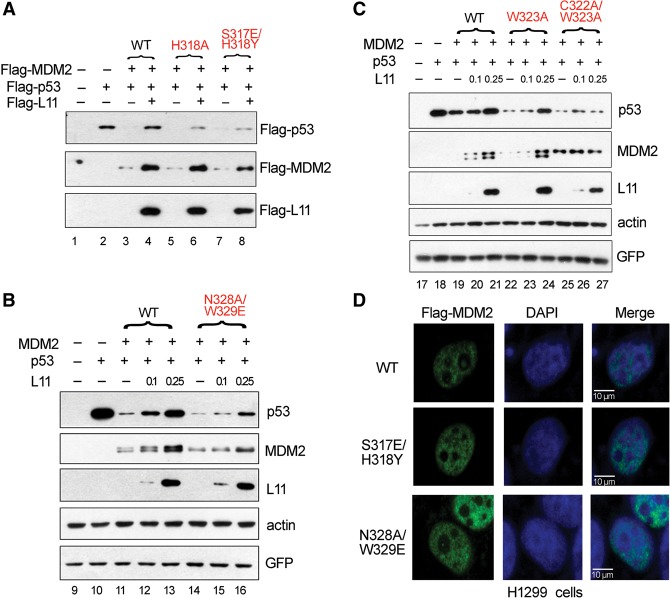
In a previous study (Zhang et al. 2011) and the mutagenesis experiments described above, key residues for the MDM2–RPL11 interaction were identified. To determine whether these mutants could escape RPL11-dependent inhibition by disrupting their interaction, we performed in vivo experiments. The reduction in p53 levels induced by MDM2 could be fully abolished by wild-type RPL11 (Fig. 3A, lanes 3–4). In contrast, the p53 degradation induced by the H318A, S317E/H318Y, and N328A/W329E mutants was less sensitive to RPL11-dependent inhibition in vivo (Fig. 3A [lanes 5–8], B [lanes 14–16]). Moreover, the W323A and C322A/W323A mutants also had a significant effect (Fig. 3C, lanes 22–27). These observations indicated that these mutations in MDM2 could impede p53 activation induced by RPL11 because of a reduced MDM2–RPL11 interaction. Therefore, these residues, including Ser317, His318, Asn328, and Trp329, were functionally important for the MDM2–RPL11 interaction and subsequent p53 activation. As shown in Figure 3D, there was no difference in localization between wild-type and mutant MDM2 proteins in H1299 cells. In the absence of endogenous p53 and MDM2, the MDM2 mutants (S317E/H318Y and N328A/W329E) and wild-type MDM2 were similarly able to promote self-ubiquitination (Supplemental Fig. 3). These two experiments suggested that the loss of the ability of RPL11 to rescue p53 from MDM2 mutant-induced degradation was not caused by the mislocalization or loss of the enzymatic activity of MDM2 but instead resulted from disruption of the MDM2–RPL11 interaction.
Figure 3.
Mutations of key residues in MDM2 impair the ability of RPL11 to up-regulate p53 through MDM2. (A–C) RPL11 cannot fully rescue MDM2 mutant-mediated degradation of p53. Double-knockout cells were transfected with plasmids encoding p53, an increasing amount of Flag-tagged RPL11, and MDM2 wild type or its mutants. Cells were harvested 24 h after transfection, and expression of the indicated proteins was examined by immunoblotting. (D) There was no difference in localization between wild-type MDM2 and its mutants. H1299 cells transfected with Flag-MDM2 or its mutants were immunostained with anti-Flag antibody. Nuclei were counterstained with DAPI. Images were acquired by fluorescence microscopy.
RPL11 undergoes conformational changes after binding MDM2
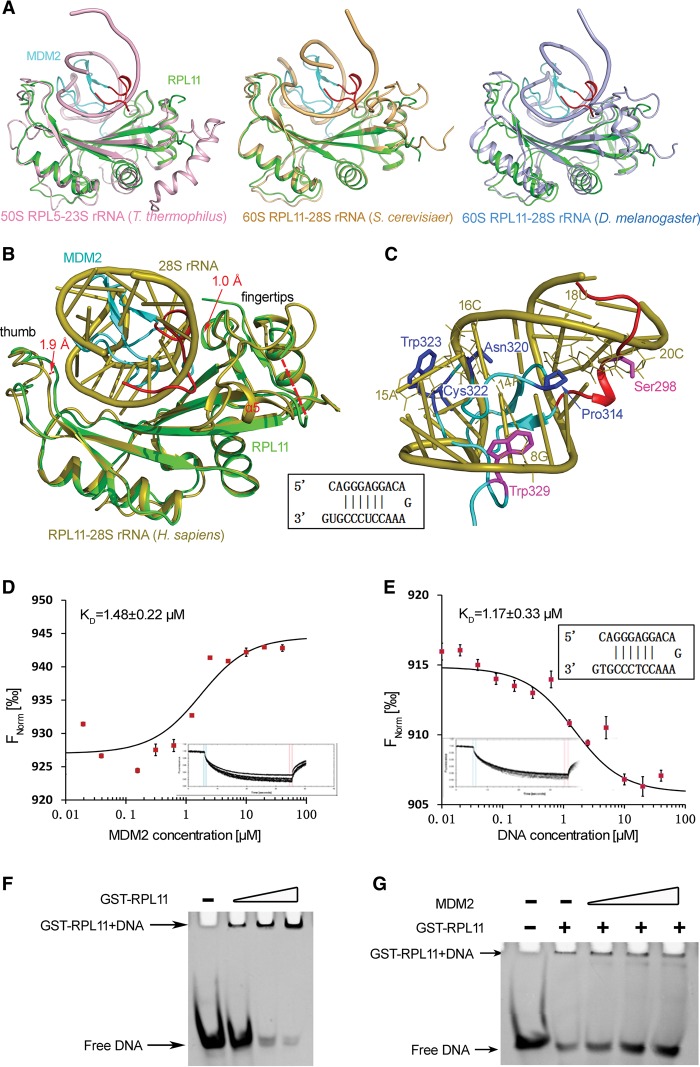
Under normal growth conditions, the p53-activating RPs are essential components of the ribosome. To date, many ribosomal structures from different species had been resolved (Ben-Shem et al. 2011; Anger et al. 2013; Gagnon et al. 2014). In this study, we compared the structures of RPL11 in the MDM2–RPL11 complex and its counterpart in the 60S ribosomal subunit from different species. Figure 4, A and B, show structural overlap between RPL11 in the MDM2–RPL11 complex and that in the 60S ribosomal subunit in Thermus thermophiles (Protein Data Bank [PDB] ID: 4W2E) (Gagnon et al. 2014), Saccharomyces cerevisiae (PDB ID: 4V88) (Ben-Shem et al. 2011), Drosophila melanogaster (PDB ID: 4V6W) (Anger et al. 2013), and Homo sapiens (PDB ID: 4V6X) (Anger et al. 2013). It was evident that the structures of RPL11 all have a hand-like conformation that was conserved from prokaryotes to eukaryotes. Moreover, a conserved self-double-stranded rRNA bound the palm region of RPL11 (Fig. 4A,B).
Figure 4.
MDM2 competes with 28S rRNA for binding to RPL11. (A) Structural comparison of RPL11 in the MDM2–RPL11 complex and its counterparts in the 60S ribosomal subunit from different species. RPL11 in the MDM2–RPL11 structure is shown in green. RPL11–rRNA in T. thermophiles (PDB ID: 4W2E) (Gagnon et al. 2014), S. cerevisiae (PDB ID: 4V88) (Ben-Shem et al. 2011), and D. melanogaster (PDB ID: 4V6W) (Anger et al. 2013) are shown in light pink, light orange, and light blue, respectively; only part of the rRNA is shown. (B) A structural comparison of RPL11 (cyan) in the MDM2–RPL11 complex and its counterpart (orange) in the 60S ribosomal subunit RPL11–28S rRNA in H. sapiens (PDB ID: 4V6X); only part of the 28S rRNA is shown (Anger et al. 2013). (C) A close-up view of the MDM2–RPL11 interaction shown in B. Interacting residues in RPL11 (green) and residues in MDM2 that mimic dsDNA riboses (magenta) or bases (blue). The G, A, C, and U of the 28S rRNA are annotated with their position numbers as 8G, 14A, 15A, 16C, 18U, and 20C, respectively. (D,E) Microscale thermophoresis (MST) measurements of the binding affinity of RPL11 for MDM2 and dsDNA. The resulting binding curve from plotting the FNorm (percentage) versus concentration was fit using a hyperbolic function to yield a KD of 1.48 μM ± 0.22 μM for MDM2 (D) and 1.17 μM ± 0.33 μM for dsDNA (E). (F,G) Electromobility shift assay results for dsDNA–RPL11 and dsDNA–RPL11–MDM2. dsDNA was used with an increasing amount of RPL11 proteins (F) or a constant amount of RPL11 mixed with an increasing amount of MDM2 proteins (G).
It was clear that the palm and fingertips of RPL11 moved ∼1.9 Å and ∼1.0 Å closer to MDM2 compared with those domains in the ribosome after MDM2 binding. These conformational changes allowed RPL11 to tightly bind with MDM2. Intriguingly, α5 became disordered after binding to MDM2, as judged by the complete lack of electron density that was observed. Based on the ribosomal structure, it was clear that α5 was key for binding to RPL5, 5S, and 28S rRNA. Thus, α5 was not necessary for MDM2 binding.
MDM2 binds to the RPL11 concave surface to mimic 28S rRNA in 60S ribosomes
Further analysis of the MDM2–RPL11 complex and 60S ribosomal subunit RPL11–28S rRNA in H. sapiens (PDB ID: 4V6X) showed that MDM2 bound to the concave surface of RPL11 through a large interface, which occupied the same structural space as the 28S rRNA in the structure of the RPL11–28S rRNA complex (Fig. 4B). A set of MDM2 residues structurally mimicked DNA bases and riboses on both strands of the 28S rRNA groove (Fig. 4C). Specifically, many residues were located at the palm and fingertip regions of RPL11 that interacted with the MDM2 residues, most of which mimicked base or ribose moieties of 28S rRNA (Fig. 4C). Among them, Ser298 and Trp329 mimicked the riboses of 20C and 8G, respectively. Additionally, the residues Pro314, Asn320, Cys322, and Trp323 mimicked the bases of 18U, 16C, 14A, and 15A, respectively (Fig. 4C). Consequently, RPL11 contacted MDM2 in a manner similar to that of 28S rRNA.
To validate our assumptions, we synthesized a self-dsDNA to mimic part of the 28S rRNA that bound to RPL11 in the human 60S ribosomal subunit and then performed microscale thermophoresis (MST) and electrophoretic mobility shift assay (EMSA) experiments. We found that MDM2 (KD = 1.48 μM ± 0.22 μM) and dsDNA (KD = 1.17 μM ± 0.33 μM) exhibited nearly equivalent binding affinities to RPL11 (Fig. 4D,E). These findings suggested that the binding affinities and capabilities between RPL11 and dsDNA or MDM2 were very similar. Accordingly, EMSA analysis showed that GST-RPL11 could bind to the dsDNA (Fig. 4F), and this interaction could be reduced by MDM2 in a dose-dependent manner (Fig. 4G), suggesting that MDM2 competed with dsDNA for GST-RPL11 binding. These observations strongly suggested that MDM2 bound to RPL11 in a mode similar to that of nucleic acid to RPL11.
The C4 zinc finger domain determines the specificity of RPL11 binding to MDM2 but not MDMX
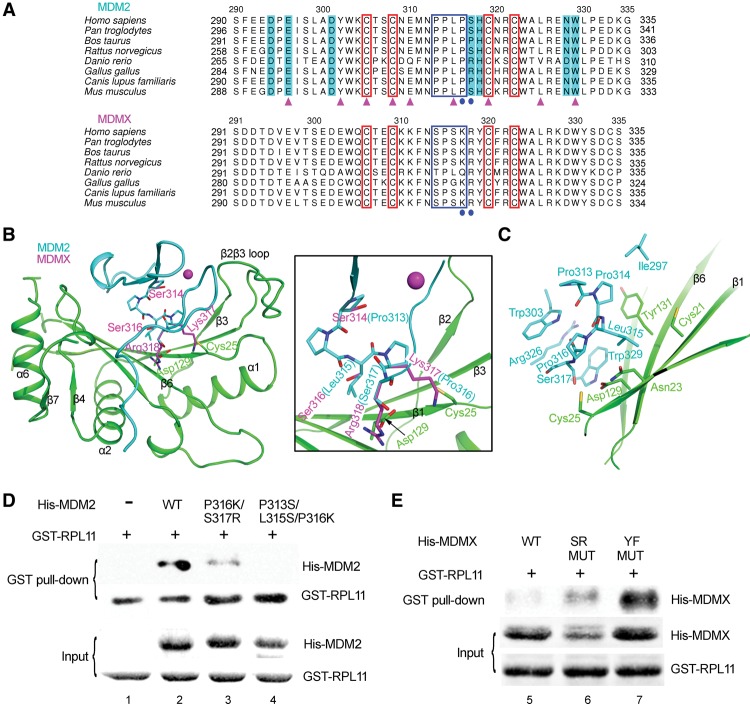
MDM2 and MDMX shared highly conserved functional domains, including the N-terminal p53-binding domain, the C-terminal RING finger domain, the central acidic domain, and the C4 zinc finger. However, RPL11 could selectively bind to MDM2 but not MDMX (Gilkes et al. 2006), implying that fundamental differences exist between these proteins in response to RPs. To uncover the mechanism behind this selectivity, we compared the sequences of the central domains of MDM2 and MDMX. The sequence alignment showed that, although both MDM2 and MDMX maintained a high level of conservation among many species, dramatic differences could be observed between the two proteins (Fig. 5A). In a further comparison of the MDM2 structure with that of MDMX (PDB ID: 2CR8), we observed that some residues in MDMX possess side chains significantly different from those in MDM2. For example, the RPL11 interfacial residues Pro316 and Ser317 in MDM2 were replaced by larger residues—Lys317 and Arg318—in MDMX (Fig. 5B). These larger side chains were predicted to induce strong steric hindrance and repulse RPL11. To test this hypothesis, we created the following two mutations in MDM2: Pro316 to Lys and Ser317 to Arg. As expected, we observed that the interaction between MDM2 and RPL11 was significantly reduced (Fig. 5D, lane 3). Furthermore, the conserved minimal motif PPLP (Yu et al. 2006) was located at the linker sequence between the two knuckles of the MDM2 zinc finger, creating hydrophobic binding networks with RPL11 (Fig. 5C). However, this motif was not conserved in MDMX (Fig. 5A). Replacement of the PPLP motif in MDM2 with the corresponding residues in MDMX (residues 313–316: PPLP → SPSK) resulted in the complete loss of the interaction between MDM2 and RPL11, demonstrating the importance of the hydrophobic interactions between MDM2 and RPL11 (Fig. 5D, lane 4). Therefore, it was the C4 zinc finger of MDMX that loses the ability to bind RPL11.
Figure 5.
The C4 zinc finger domain determines the specificity of RPL11 binding to MDM2 but not MDMX. (A) Sequence alignment of MDM2 with MDMX. The four conserved cysteine residues are marked by red boxes. The small motif PPLP is indicated by a blue box. The residues that differ between MDM2 and MDMX are labeled by blue circles. The residues interacting with RPL11 are shaded in cyan. The residues for which mutations have been found in cancers are indicated by magenta triangles. (B,C) Comparison of the residues located at the interface in MDM2 (cyan) and the corresponding residue in MDMX (magenta). (B) A close-up view of these residues is shown in the right panel. These marked differences in residues in MDMX would alter binding with RPL11. (C) The important hydrophobic core in MDM2 can interact with RPL11. Key residues in MDMX that can block the MDM2–RPL11 interaction are shown, and these residues in MDM2 were mutated into the corresponding residues in MDMX. (D,E) As shown by GST pull-down assays, the mutants in MDMX gain RPL11-binding ability. (SR MUT) 314–318 residues: SPSKR → PPLPS; (YF MUT) 314–321 residues: SPSKRYCF → PPLPSHCK.
To further verify our hypothesis, we reciprocally mutated the key residues in MDMX, which failed to support a MDMX–RPL11 interaction, into the corresponding residues from MDM2, including SR MUT (residues 314–318: SPSKR → PPLPS) and YF MUT (residues 314–321: SPSKRYCF → PPLPSHCK). As we expected, both mutants could bind RPL11 better than wild-type MDMX (Fig. 5E, cf. lanes 6,7,5). Thus, these binding assays resulting from both the structure-based gain-of-function and loss-of-function mutations confirmed that the C4 zinc finger domain was responsible for the exclusive binding preference of RPL11 for MDM2 but not MDMX.
Discussion
The only available structure of human RPL11 was solved in the 80S ribosome (Anger et al. 2013); however, its resolution was only 5.0 Å. Here, we presented the first high-resolution structure of a human MDM2–RPL11 complex. To form this complex, both MDM2 and RPL11 had to undergo substantial conformational changes compared with the apo structure or the structure in ribosome. The curving thumb and fingertips closed and caught MDM2 tightly in the palm (Fig. 4B). Moreover, a sheet-to-coil transition and RPL11-induced conformational changes could be observed for MDM2 (Fig. 1F). The binding interface of MDM2–RPL11 was extensive. RPL11, which could not interact with the MDM2 mutants, failed to induce p53 activation in vivo. Intriguingly, an intermolecular zinc finger could be identified in the complex, and this finding was further confirmed by the SAD method using the SHELEX C/D program (Sheldrick 2008). The zinc finger was critical for the RPL11–MDM2 interaction, as a disruptive mutation of MDM2, H318A, severely diminished this interaction (Fig. 2F, lane 10). Moreover, by generating structure-based gain-of-function and loss-of-function mutations, we found that the C4 zinc finger determined the recognition specificity of RPL11 for MDM2. This finding provided the answer to a long-standing question: “Why does RPL11 selectively bind to MDM2 but not MDMX?”
As a prominent negative regulator of p53, MDM2 is a tightly regulated protein. The central region of MDM2 is critical for sensing oncogenic and ribosomal stresses by binding to ARF and RPs, which then can transduce various signals to the p53 pathway. Our structural and functional results suggest that both the acidic domain and C4 zinc finger are essential for RPL11 binding. The importance of the C4 zinc finger of MDM2 in p53 regulation and tumor suppression has been reported previously (Lindstrom et al. 2007). Analysis of the MDM2–RPL11 structure has revealed that many cancer-associated MDM2 mutations occur in key residues that are essential for forming the C4 zinc finger, which raises the possibility that disruption of the C4 zinc finger structure could make MDM2 refractory to RPL11 inhibition, thereby contributing to tumorigenesis (Fig. 2A; Supplemental Table 2). In addition to the C4 zinc finger, our findings indicate that the acidic region and the key residues at the MDM2–RPL11 interface could also play an important role in their interaction and p53 activation, thereby contributing to tumor suppression. Indeed, MDM2 mutations at the RPL11-binding interface (W329L and W329G) and in the acidic region (E296D) can be observed in human cancers (Fig. 2C,D; Supplemental Table 2), underscoring the importance of these residues in tumor suppression. Consistently, these cancer-relevant mutations of MDM2 diminish the MDM2–RPL11 interaction as detected by MST (Supplemental Fig. 4).
Evidence that has accumulated over the previous decade has established the relevance of the RP–MDM2–p53 signaling pathway in human cancers (Teng et al. 2013; Bursac et al. 2014) and demonstrated that abnormalities in ribosome function can lead to tumorigenesis (Lai et al. 2009; Vlachos et al. 2012). RPs represent half of the ribosomes and have attracted intense research interest because of their extraribosomal roles in p53 regulation. Thus, imbalances in RPs lead to nucleolar stress and subsequent p53 activation. Furthermore, loss-of-function mutations in several RP genes have also been linked to cancer-prone genetic diseases (Fumagalli and Thomas 2011; Zhou et al. 2012; Teng et al. 2013). Notable examples include Diamond Blackfan anemia (DBA) (Badhai et al. 2009) and 5q syndrome (Ebert et al. 2008). In RPL11-associated cancers, we identified two mutations (S100L and D129V) at the MDM2–RPL11-binding surface (Fig. 2C,D; Supplemental Table 2). GST pull-down experiments have shown that these two mutations severely impaired the interaction of RPL11 with MDM2 (Supplemental Fig. 5). Our study suggests that the inability of RPL11 to bind MDM2 might be functionally important in RPL11-associated cancers.
MDM2 E3 ubiquitin ligase-mediated p53 degradation is generally accepted as the major mechanism for p53 regulation. Both MDM2 E3 ligase activity and MDM2:MDMX heterodimerization are essential for p53 regulation (Tollini et al. 2014). Several mechanisms have been proposed to explain how RP binding to the central domain of MDM2 inhibits the E3 ubiquitin ligase activity of its C-terminal RING finger (Zhang and Lu 2009). However, the exact connections between these two events remain nebulous. In this study, we found that α5 of RPL11 is missing in the MDM2–RPL11 structure (Fig. 4B; Supplemental Fig. 2). Interestingly, α5 is a key element for binding to RPL5 and 5S rRNA in the ribosomal structure. Superposition of MDM2–RPL11 onto the human ribosome (PDB ID: 4V6X) (Anger et al. 2013) has indicated that the orientation of MDM2 does not overlap with that of RPL5 and 5S rRNA. Recently, it has been shown that RPL11 in the nucleoplasm can be stabilized by forming a complex with RPL5 and 5S rRNA (Donati et al. 2013; Sloan et al. 2013). Because the association of RPL11 with MDM2 does not inhibit MDM2-mediated p53 ubiquitination in vitro (Zhang et al. 2003), it is possible that MDM2 could form a large complex with RPL11, RPL5, and 5S rRNA, resulting in an impaired ability to efficiently ubiquitinate p53 (Supplemental Fig. 6).
Currently, it is thought that rRNA is the most actively synthesized nucleic acid species within a cell and is highly abundant and essential for cell growth and proliferation. Equimolar concentrations of both rRNA and RPs are required for the biogenesis and homeostasis of ribosomes and to maintain inhibition of p53. In contrast, an imbalance between rRNA and RP synthesis leads to p53 activation in mammalian cells (Donati et al. 2011). However, rRNA is vulnerable to both endogenous and exogenous genotoxic agents. Previous studies have shown that inhibition of rRNA maturation can lead to nucleolar stress, which finally triggers RPL5/RPL11/5S rRNA complex binding to MDM2 and efficient activation of p53 (Fumagalli et al. 2009; Holzel et al. 2010; Donati et al. 2011, 2013; Sloan et al. 2013). This could be the method by which p53 can sense rRNA damage. Intriguingly, our structural analyses and biochemical data have both suggested that MDM2 mimics the structure of 28S rRNA when bound to RPL11. This region of 28S rRNA is conserved among different species and can be mimicked by a short DNA oligo that has been shown to bind to RPL11 in vitro (Fig. 4A,B). This finding is quite exciting and important because it suggests that nucleic acids could also be exploited for developing selective inhibitors of specific pathways for use in biological or therapeutic studies.
Most importantly, identification of RP–MDM2 interactions offers an opportunity for developing anti-cancer drugs that directly target MDM2. MDM2 inhibits not only p53 but also other tumor suppressors, such as Rb, through its E3 activity in MDM2-overexpressing tumors. The use of RPL11 to inhibit MDM2 not only might activate p53 but could also inhibit the other oncogenic properties of MDM2. Thus, this property of RPL11 could be exploited as part of a promising strategy for tumor treatment. It is conceivable that the design and development of chemical compounds that mimic the binding properties of RPL11 to MDM2 would yield promising drug candidates; our results provide a solid structural basis for this possibility.
Materials and methods
Cloning, expression, and purification of the MDM2–RPL11 complex
In this study, human MDM2 (residues 290–437) was cloned into pET28a expression vector, and human RPL11 (residues 1–178) was cloned into pET22b expression vector. The two correct plasmids were cotransfected into BL21 (DE3) cells. Cells were grown at 37°C until OD600 reached 0.6–0.8, and the proteins were expressed after induction with 0.2 mM IPTG. After 12 h, cells were harvested by centrifugation at 4000 rpm for 10 min, and bacterial paste was resuspended in buffer A containing 20 mM Tris (pH 8.0), 20 mM NaH2PO4, 300 mM NaCl, 5 mM imidazole, 0.1% Triton, 1 mM PMSF, 10 mM β-mercaptoethanol, 1 mM benzamidine, 30 μM leupeptin, and 100 μM pepstain A. After sonication, the cells were clarified at 18,300 rpm for 30 min. Cleared lysate was filtered with a 0.4-μm filter before being loaded onto a Profinity IMAC Ni-charged resin (Bio-Rad) column. The column was washed with three column volumes of buffer A and eluted in buffer A supplemented with 200 mM imidazole. The MDM2–RPL11 elution was further purified by Q Sepharose (GE Healthcare) to remove free RPL11. The wash fractions of the MDM2–RPL11 complex were concentrated and loaded onto a Superdex 200 10/30 column (GE Healthcare) in buffer B containing 20 mM Tris (pH 8.0), 150 mM NaCl, and 5 mM β-mercaptoethanol. The second peak fractions were collected and concentrated to 25 mg/mL for crystallography with buffer B supplemented with 2 mM tris (2-carboxyethyl) phosphine (TCEP).
Crystallization and data collection
Initial crystallization trials were performed at room temperature and 4°C by using the sitting drop vapor diffusion methods. After 10 d, a lot of small crystals could be observed in the well solution containing 10% PEG-8000 and 0.1 M imidazole (pH 8.0) at 4°C. After a series of optimizations of crystallization conditions—including precipitants, pH, and buffers—larger crystals with better diffraction were obtained. All of the crystals were transferred into the crystallization buffer supplemented with 15% ethylene glycol and flash-frozen in liquid nitrogen. Data were collected under cryocooled conditions (100 K) at the Photon Factory (KEK) on beamline NE3A with λ = 1.00 Å. Data were indexed, integrated, and scaled using HKL2000. Data collection and processing statistics are shown in Supplemental Table 1.
Structure determination and refinement
An initial molecular replacement solution was obtained from the BALBES (Long et al. 2008) software by using the structure of yeast RPL11 (PDB ID: 4V88) (Ben-Shem et al. 2011) as a model after extensive trials of ribosomal structures from different species. The solution had an MR score of 6.07 and an Rwork/Rfree value of 0.379/0.475. The primitive density map had a relatively clear outline, with several recognizable α helices and β-strand bundles. After removing the inappropriate main and side chains in the model using the program COOT (Emsley et al. 2010), we iteratively refined and built the model using the CCP4 suite (Winn et al. 2011). The best solution stopped at an Rwork/Rfree value of 0.371/0.428. At this point, the structure of MDM2 C4 zinc finger (PDB ID: 2C6A) (Yu et al. 2006) was fitted into electron density maps (Fig. 1E) using the graphics program COOT (Emsley et al. 2010). After 20 more cycles of manual rebuilding by COOT and refinement with REFMAC5 (Murshudov et al. 2011), the structure was refined to 2.4 Å with Rwork = 19.2% and Rfree = 22.7%. Ramachandran statistics indicated that 97% of residues lay in the favored regions, with 0% outliers. The crystal belonged to space group P3221, with one RPL11 molecule and one MDM2 molecule in the asymmetric unit (Supplemental Table 1).
As the data were collected at 1.0000 Å, however, the anomalous signal was strong, as analyzed by the SHELEX C program (Sheldrick 2008), revealing the existence of zinc atoms (Supplemental Fig. 7A). The number of zinc atoms and the position of the two zinc atoms in the solved structure were confirmed by the SHELEX D program (Supplemental Figs. 7B, 8). All structural figures were prepared by using the PyMOL program (https://www.pymol.org).
GST-mediated pull-down assay
Two-hundred micrograms of GST-RPL11 protein was immobilized on glutathione sepharose resin (GE Healthcare). Two-hundred micrograms of wild-type or mutant MDM2 proteins was incubated with the GST-RPL11-bound GST resin for 1 h at 4°C. The resin was extensively rinsed with buffer containing 20 mM Tris-HCl (pH 8.0), 300 mM NaCl, and 10 mM β–mercaptoethanol to wash unbound or nonspecific bound proteins. The beads were analyzed by Western blotting.
DNA annealing for EMSA and MST
The DNA oligonucleotide used for EMSA and MST was 5′-CAGGGAGGACAGAAACCTCCCGTG-3′. The oligonucleotide was annealed in 10 mM Tris–HCl (pH 8.0) and 50 mM NaCl. To get a self-dsDNA, annealing was performed by first heating the DNA oligonucleotide for 5 min at 95°C and then allowing it to cool rapidly to 4°C.
EMSA
Protein–dsDNA-binding interactions were assayed by EMSA. The 24-base-pair (bp) dsDNA (50 μM) was incubated with increasing amounts of GST-RPL11 proteins (0, 50, 150, and 300 μM) or a constant amount of RPL11 protein (300 μM) mixed with increasing amounts of MDM2 proteins (0, 100, and 200 μM) in buffer containing 40 mM Tris-HCl (pH 8.0), 300 mM NaCl, 5% glycerol, and 1 mM DTT for 30 min at room temperature. The DNA were separated by 10% native polyacrylamide gel in 0.5× TBS buffer (45 mM Tris, 45 mM boric acid). The results were visualized by staining with ethidium bromide.
MST
The binding affinity of dsDNA to RPL11 was calculated using MST. GST-RPL11 was labeled with fluorescent dye NT-647 (Cysteine Reactive) according to the manufacturer's protocol. A series of dsDNA and MDM2 solutions with different concentrations was prepared by consecutive twofold dilutions in buffer containing 20 mM HEPES (pH 8.0), 300 mM NaCl, and 0.05% Tween-20 from the highest concentration of 80 μM. The labeled 1 μM GST-RPL11 protein was mixed with dsDNA or MDM2 proteins prepared at a volume ratio of 1:1. The samples were loaded into silica capillaries after incubation at room temperature. After 30 min, the measurements were performed at 25°C by using 40% LED power and 20% laser power. Data analyses were performed by using the NTAnalysis software (NanoTemper Technologies).
Western blotting
Cells were lysed in 2× SDS sample buffer or lysis buffer (50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 1% Triton X-100, 1 mM DTT, 1× complete protease inhibitor cocktail, 10% glycerol). The protein samples were resolved by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane, which was blocked in 5% skim milk and probed with the indicated antibodies.
In vitro immunoprecipitation
H1299 cells transfected with wild-type or mutant Flag-RPL11 protein were lysed in lysis buffer as described above. One-hundred micrograms of His-MDM2 protein was incubated with Ni-NTA beads (GE Healthcare) for 30 min at 4°C, and, afterward, cell lysates were added. After another 30 min, the beads were analyzed by Western blotting.
Immunofluorescence microscopy
H1299 cells cultured on coverslips were transfected with the indicated plasmids. At 24 h after transfection, cells were sequentially fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.2% Triton X-100 for 15 min, blocked with 1% bovine serum albumin, and incubated with an anti-Flag antibody followed by a FITC-conjugated anti-rabbit IgG antibody. The cells were mounted with 4′,6-diamidino-2-phenylindole-containing medium (Beyotime Institute of Biotechnology), and the images were acquired with a fluorescence microscope.
Accession number
Coordinates and structure factors have been deposited in the PDB under accession code 4XXB.
Supplementary Material
Acknowledgments
We thank the staff at beamline NE3A at the Photon Factory (Tsukuba, Japan) and beamline 1W2B at the Beijing Synchrotron Radiation Facility for the excellent technical assistance during data collection. This work was supported financially by the National Basic Research Program of China (973 Program, 2011CB965304 to Zhongzhou Chen), the National Natural Science Foundation of China (31222032 and 31370720 to Zhongzhou Chen, and 31171331 to J.T.), and the State Key Laboratory of Agrobiotechnology (2013SKLAB1-6 to J.T. and Zhongzhou Chen).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.261792.115.
References
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R. 2013. Structures of the human and Drosophila 80S ribosome. Nature 497: 80–85. [DOI] [PubMed] [Google Scholar]
- Badhai J, Frojmark AS, Davey E, Schuster J, Dahl N. 2009. Ribosomal protein S19 and S24 insufficiency cause distinct cell cycle defects in Diamond-Blackfan anemia. Biochim Biophys Acta 1792: 1036–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. 2011. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334: 1524–1529. [DOI] [PubMed] [Google Scholar]
- Bursac S, Brdovcak MC, Donati G, Volarevic S. 2014. Activation of the tumor suppressor p53 upon impairment of ribosome biogenesis. Biochim Biophys Acta 1842: 817–830. [DOI] [PubMed] [Google Scholar]
- Donati G, Bertoni S, Brighenti E, Vici M, Trere D, Volarevic S, Montanaro L, Derenzini M. 2011. The balance between rRNA and ribosomal protein synthesis up- and downregulates the tumour suppressor p53 in mammalian cells. Oncogene 30: 3274–3288. [DOI] [PubMed] [Google Scholar]
- Donati G, Peddigari S, Mercer CA, Thomas G. 2013. 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2–p53 checkpoint. Cell Rep 4: 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, et al. 2008. Identification of RPS14 as a 5q− syndrome gene by RNA interference screen. Nature 451: 335–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S, Thomas G. 2011. The role of p53 in ribosomopathies. Semin Hematol 48: 97–105. [DOI] [PubMed] [Google Scholar]
- Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, Schwemberger S, Hall J, Babcock GF, Bernardi R, Pandolfi PP, Thomas G. 2009. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol 11: 501–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S, Ivanenkov VV, Teng T, Thomas G. 2012. Suprainduction of p53 by disruption of 40S and 60S ribosome biogenesis leads to the activation of a novel G2/M checkpoint. Genes Dev 26: 1028–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon MG, Lin J, Bulkley D, Steitz TA. 2014. Crystal structure of elongation factor 4 bound to a clockwise ratcheted ribosome. Science 345: 684–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes DM, Chen J. 2007. Distinct roles of MDMX in the regulation of p53 response to ribosomal stress. Cell Cycle 6: 151–155. [DOI] [PubMed] [Google Scholar]
- Gilkes DM, Chen L, Chen J. 2006. MDMX regulation of p53 response to ribosomal stress. EMBO J 25: 5614–5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzel M, Orban M, Hochstatter J, Rohrmoser M, Harasim T, Malamoussi A, Kremmer E, Langst G, Eick D. 2010. Defects in 18 S or 28 S rRNA processing activate the p53 pathway. J Biol Chem 285: 6364–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R, Yasuda H. 2000. Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene 19: 1473–1476. [DOI] [PubMed] [Google Scholar]
- Kim TH, Leslie P, Zhang Y. 2014. Ribosomal proteins as unrevealed caretakers for cellular stress and genomic instability. Oncotarget 5: 860–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797. [DOI] [PubMed] [Google Scholar]
- Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. 1996. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274: 948–953. [DOI] [PubMed] [Google Scholar]
- Lai K, Amsterdam A, Farrington S, Bronson RT, Hopkins N, Lees JA. 2009. Many ribosomal protein mutations are associated with growth impairment and tumor predisposition in zebrafish. Dev Dyn 238: 76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gu W. 2011. A critical role for noncoding 5S rRNA in regulating Mdmx stability. Mol cell 43: 1023–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom MS, Jin A, Deisenroth C, White Wolf G, Zhang Y. 2007. Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Mol Cell Biol 27: 1056–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. 2003. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3: 577–587. [DOI] [PubMed] [Google Scholar]
- Long F, Vagin AA, Young P, Murshudov GN. 2008. BALBES: a molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr 64: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias E, Jin A, Deisenroth C, Bhat K, Mao H, Lindstrom MS, Zhang Y. 2010. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 interaction. Cancer Cell 18: 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine JC, Dyer MA, Jochemsen AG. 2007. MDMX: from bench to bedside. J Cell Sci 120: 371–378. [DOI] [PubMed] [Google Scholar]
- Miliani de Marval PL, Zhang Y. 2011. The RP–Mdm2–p53 pathway and tumorigenesis. Oncotarget 2: 234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378: 203–206. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67: 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki T, Nakagawara A. 2011. Role of p53 in cell death and human cancers. Cancers (Basel) 3: 994–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant V, Xiong S, Jackson JG, Post SM, Abbas HA, Quintas-Cardama A, Hamir AN, Lozano G. 2013. The p53-Mdm2 feedback loop protects against DNA damage by inhibiting p53 activity but is dispensable for p53 stability, development, and longevity. Genes Dev 27: 1857–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. 2008. A short history of SHELX. Acta Crystallogr A 64: 112–122. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. 2006. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer 6: 663–673. [DOI] [PubMed] [Google Scholar]
- Sloan KE, Bohnsack MT, Watkins NJ. 2013. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep 5: 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng T, Thomas G, Mercer CA. 2013. Growth control and ribosomopathies. Curr Opin Genet Dev 23: 63–71. [DOI] [PubMed] [Google Scholar]
- Tollini LA, Jin A, Park J, Zhang Y. 2014. Regulation of p53 by Mdm2 E3 ligase function is dispensable in embryogenesis and development, but essential in response to DNA damage. Cancer Cell 26: 235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos A, Rosenberg PS, Atsidaftos E, Alter BP, Lipton JM. 2012. Incidence of neoplasia in Diamond Blackfan anemia: a report from the Diamond Blackfan Anemia Registry. Blood 119: 3815–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Prives C. 2009. Blinded by the light: the growing complexity of p53. Cell 137: 413–431. [DOI] [PubMed] [Google Scholar]
- Wade M, Li YC, Wahl GM. 2013. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 13: 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu GW, Allen MD, Andreeva A, Fersht AR, Bycroft M. 2006. Solution structure of the C4 zinc finger domain of HDM2. Protein Sci 15: 384–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lu H. 2009. Signaling to p53: ribosomal proteins find their way. Cancer Cell 16: 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. 2003. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol 23: 8902–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Xiao H, Chai SC, Hoang QQ, Lu H. 2011. Hydrophilic residues are crucial for ribosomal protein L11 (RPL11) interaction with zinc finger domain of MDM2 and p53 protein activation. J Biol Chem 286: 38264–38274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Liao JM, Liao WJ, Lu H. 2012. Scission of the p53-MDM2 loop by ribosomal proteins. Genes Cancer 3: 298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.