Reuter et al. show that Nab2, a poly(A)-binding protein important for correct poly(A) tail length and nuclear mRNA export, is present at all RNA polymerase III (RNAPIII) transcribed genes. Nab2 is required for the occupancy of RNAPIII and TFIIIB at target genes.
Keywords: RNA polymerase III, Nab2, tRNA, ncRNA, gene expression, TFIIIB
Abstract
RNA polymerase III (RNAPIII) synthesizes most small RNAs, the most prominent being tRNAs. Although the basic mechanism of RNAPIII transcription is well understood, recent evidence suggests that additional proteins play a role in RNAPIII transcription. Here, we discovered by a genome-wide approach that Nab2, a poly(A)-binding protein important for correct poly(A) tail length and nuclear mRNA export, is present at all RNAPIII transcribed genes. The occupancy of Nab2 at RNAPIII transcribed genes is dependent on transcription. Using a novel temperature-sensitive allele of NAB2, nab2-34, we show that Nab2 is required for the occupancy of RNAPIII and TFIIIB at target genes. Furthermore, Nab2 interacts with RNAPIII, TFIIIB, and RNAPIII transcripts. Importantly, impairment of Nab2 function causes an RNAPIII transcription defect in vivo and in vitro. Taken together, we establish Nab2, an important mRNA biogenesis factor, as a novel player required for RNAPIII transcription by stabilizing TFIIIB and RNAPIII at promoters.
RNA polymerase III (RNAPIII) is responsible for the production of most noncoding RNAs (ncRNAs) such as tRNAs, the 5S rRNA (encoded by RDN5), the RNA of the signal recognition particle (SCR1), the spliceosomal U6 snRNA (SNR6), and the RNA component of RNase P (RPR1). Transcription by RNAPIII is initiated at three classes of promoters, which are highly diverse, have a relatively small number of cis-acting elements, and require comparably few transcription factors (TFs) (White 2011; Orioli et al. 2012; Acker et al. 2013 and references therein). Type 1 promoters are exclusively present at 5S rRNA-encoding genes (RN5S in mammals, and RDN5 in yeast). Type 1 promoters have three internal elements: the A box, which is recognized by TFIIIC, and the intermediate element (IE) and the C box, which are recognized by TFIIIA. The most common RNAPIII promoters are of type 2 and drive transcription of tRNA genes as well as, e.g., the SCR1 and RPR1 genes. Type 2 promoters consist of A- and B-box elements, which together recruit TFIIIC. TFIIIC recruits TFIIIB to the transcription start site at type 1 and type 2 promoters. Type 3 promoters are relatively rare, and, in contrast to type 1 and 2 promoters, all of their elements lie 5′ of the transcription start site: the distal sequence element (DSE), the proximal sequence element (PSE), and the TATA box. The DSE recruits the TFs Oct1 and STAF, whereas the PSE recruits SNAPc, which in turn binds TFIIIB and recruits it together with the TATA box to the promoter (Schramm and Hernandez 2002 and references therein). Type 3 promoters are only present in vertebrates; e.g., at the U6 snRNA gene. The promoter of the Saccharomyces cerevisiae U6 snRNA gene SNR6 is of type 2 and contains an upstream TATA box in addition to the canonical A and B boxes (Brow and Guthrie 1990; Eschenlauer et al. 1993). At all three types of promoters, TFIIIB recruits RNAPIII. TFIIIB is also responsible for opening the dsDNA and thus for establishing a closed preinitiation complex and the transcription bubble (Acker et al. 2013 and references therein).
Recent evidence suggests that in addition to the main RNAPIII TFs described above, many other proteins might play a role in RNAPIII transcription. These are mainly proteins with known functions in RNAPII transcription: the tumor suppressors retinoblastoma protein and p53; Myc; various chromatin-modifying complexes like histone acetylases and deacetylases; the RNAPII coactivator PC4 and its S. cerevisiae ortholog, Sub1; and the RNAPII TFs Dst1/TFIIS, Yox1, Fkh1, Reb1, and Yap6 (White 2011; Acker et al. 2013; Gjidoda and Henry 2013 and references therein). However, the molecular function in RNAPIII transcription of most of these proteins has not been determined. Interestingly, the activity of an in vitro reconstituted yeast RNAPIII transcription system with purified RNAPIII, TFIIIC, and TFIIIB is very low, supporting the argument that additional proteins other than the established RNAPIII TFs are involved in RNAPIII transcription (Ducrot et al. 2006).
In a genome-wide study to determine the occupancy of nuclear mRNA-binding proteins on the S. cerevisiae genome (Meinel et al. 2013), we serendipitously observed that Nab2 localizes to all genes transcribed by RNAPIII. Nab2 is a nuclear polyadenylated RNA-binding protein required for 3′ end formation and mRNA export (Soucek et al. 2012 and references therein). It is present along the whole ORF of protein-coding genes as determined by chromatin immunoprecipitation (ChIP) experiments hybridized to high-density tiling arrays (ChIP–chip) (Meinel et al. 2013). As a member of the poly(A)-binding protein (PABP) family, Nab2 binds to the poly(A) tail of mRNAs during or shortly after their polyadenylation, regulating its length (Kelly et al. 2010; Soucek et al. 2012) and references therein). In addition to poly(A) RNA (Kelly et al. 2010), Nab2 also shows nonspecific RNA binding (Kelly et al. 2007), consistent with its binding to the body of mRNAs in vivo (Tuck and Tollervey 2013) and its at least partially RNA-dependent occupancy at an RNAPII reporter gene (Meinel et al. 2013). A second function of Nab2 in mRNP biogenesis is in mRNA export (Hector et al. 2002). After the mRNA is correctly processed and packaged by proteins in an mRNP, it is exported from the nucleus to the cytoplasm through the nuclear pore complex by the conserved S. cerevisiae mRNA exporter Mex67-Mtr2. Nab2, the TREX components Hpr1 and Sub2, and the SR protein Npl3 have been proposed to act as mRNA adaptors that recruit Mex67-Mtr2 to the mRNP (Strasser and Hurt 2001; Strasser et al. 2002; Gilbert and Guthrie 2004; Gwizdek et al. 2006; Iglesias et al. 2010).
Here, we show that Nab2 functions in RNAPIII transcription. Nab2 occupies RNAPIII genes genome-wide in a transcription-dependent manner. Importantly, impairment of Nab2 function leads to lower occupancy of RNAPIII at genes and a concomitant RNAPIII transcription defect. Nab2 interacts directly with RNAPIII, RNAPIII transcripts, and TFIIIB. Furthermore, the binding of TFIIIB with the target promoter DNA is stabilized by Nab2. Thus, we established Nab2, the first mRNA biogenesis factor, as a novel player essential for efficient RNAPIII transcription.
Results
Nab2 is recruited to RNAPIII genes
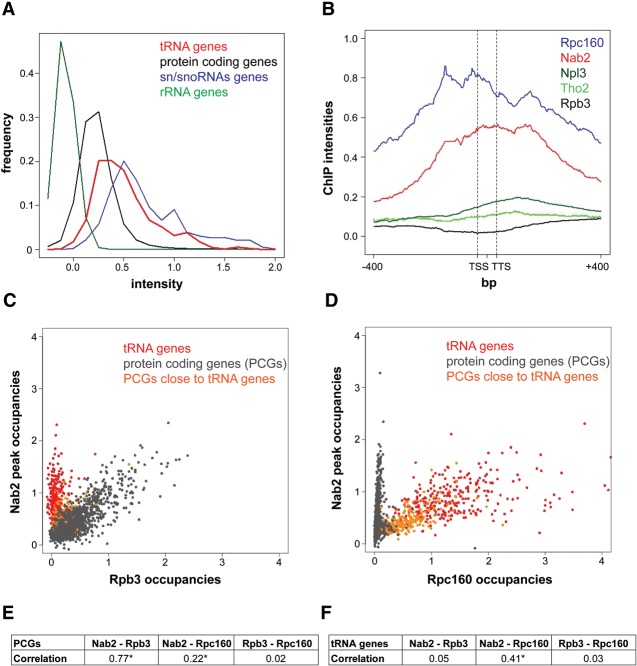
The nuclear PABP Nab2 is well known for its function in poly(A) tail length control and nuclear mRNA export. In a recent study, we determined the genome-wide occupancy of Nab2 and other nuclear mRNP components in S. cerevisiae by ChIP–chip (Meinel et al. 2013). As expected, Nab2 occupies protein-coding genes (Gonzalez-Aguilera et al. 2011; Meinel et al. 2013). However, we discovered that, in addition to RNAPII transcribed genes, Nab2 also localizes to genes transcribed by RNAPIII. The occupancies of Nab2 were calculated separately for genes encoding mRNAs, sn/snoRNAs, rRNAs, and tRNAs. Their frequencies were then plotted against the signal intensities for each gene class (Fig. 1A). The signal intensities for Nab2 are high for not only sn/snoRNA and protein-coding genes transcribed by RNAPII but also genes encoding tRNAs, which are transcribed by RNAPIII (Fig. 1A). In contrast, there is no signal for Nab2 at rRNA genes (Fig. 1A). Next, we calculated meta-tRNA gene occupancy profiles of Nab2 and Rpc160, the largest subunit of RNAPIII. The average nucleotide occupancy for Nab2 or Rpc160 was plotted for all intronless tRNA genes <76 base pairs (bp) after gene length normalization (Fig. 1B). The meta-tRNA gene occupancy profiles of Nab2 and Rpc160 are very similar (Fig. 1B), suggesting co-occupancy of Nab2 and RNAPIII at tRNA genes. Inspection of the individual traces revealed that the meta-tRNA gene occupancy profile of Nab2 reflects its occupancy at all tRNA genes (Supplemental Fig. S1). In contrast, meta-tRNA gene occupancy profiles of the RNAPII subunit Rpb3, the TREX component Tho2, and the SR protein Npl3 show that these proteins are absent from tRNA genes (Fig. 1B). Thus, Nab2 recruitment to tRNA genes is most likely independent of RNAPII and nuclear mRNP-binding proteins.
Figure 1.
Genome-wide localization of Nab2 to RNAPIII transcribed genes. (A) Density plots for Nab2 from ChIP–chip experiments. The signal intensities for probes mapping to tRNA, protein-coding, sn/snoRNA, and rRNA genes are plotted on the X-axis. The frequency for each intensity on the X-axis is plotted on the Y-axis. The green line (rRNAs) depicts the expected distribution for a genomic feature not occupied by Nab2. (B) Metaprofiles of Nab2-TAP, Rpc160-TAP (RNAPIII), Rpb3-TAP (RNAPII), TAP-Tho2, and TAP-Npl3 for all intronless tRNA genes <76 bp. Nab2 and Rpc160 are present on tRNA genes, whereas RNAPII, the TREX subunit Tho2, and the SR protein Npl3 are absent from tRNA genes. (TSS) Transcription start site; (TTS) transcription termination site. See also Supplemental Figure S1. (C) Nab2 occupancy correlates highly with RNAPII occupancy on protein-coding genes. Scatter plot of the peak occupancies of Nab2 (Y-axis) versus Rpb3 (RNAPII) (X-axis) at protein-coding genes. Protein-coding genes located in close proximity (<250 bp) to tRNA loci are labeled in orange in order to mark spillover effects; i.e., occupancies of RNAPIII at protein-coding genes due to high signals at a neighboring tRNA gene. (D) Nab2 occupancies correlate highly with RNAPIII occupancies on tRNA genes. Scatter plot of the peak occupancies of Nab2 (Y-axis) versus Rpc160 (RNAPIII) (X-axis) on tRNA genes. (E,F) Pearson's correlation coefficient of the occupancies of Nab2 and Rpb3, Nab2 and Rpc160, and Rpb3 and Rpc160 on protein-coding genes (PCGs) (E) and tRNA genes (F). Nab2 occupancy correlates with Rpb3 on protein-coding genes and with Rpc160 on tRNA genes. Asterisks indicate that, within 100,000 permutations, no similarly high Pearson's correlation coefficient was obtained, indicating that data sets are significantly positively correlated. The positive correlation coefficient at protein-coding genes for Nab2 and Rpc160 is most likely caused by spillover effects of tRNA genes (see C).
We then correlated the peak occupancies of Nab2, RNAPII (Rpb3), and RNAPIII (Rpc160). Scatter plots of the peak occupancies of Nab2 and the RNAPII subunit Rpb3 show that Nab2 and RNAPII occupancies are high at protein-coding genes (Fig. 1C, gray dots). In contrast, tRNA genes show occupancy of Nab2 but not RNAPII (Fig. 1C, red dots). Likewise, scatter plots of the peak occupancies of Nab2 and the RNAPIII subunit Rpc160 show that the Nab2 and RNAPIII peak occupancies are both high at tRNA genes (Fig. 1D, red dots), whereas protein-coding genes are occupied by Nab2 but not RNAPIII (Fig. 1C, gray dots). The occupancy of RNAPIII at some protein-coding genes is probably caused by spillover effects due to tRNA genes located nearby (Fig. 1D, orange dots). Consistently, Nab2 occupancies are strongly correlated with the RNAPII occupancies but not with the RNAPIII occupancies at protein-coding genes, whereas Nab2 occupancies correlate strongly with the RNAPIII occupancies but not with the RNAPII occupancies at tRNA genes (Fig. 1E,F). Thus, the occupancy of Nab2 at tRNA genes is specific and independent of RNAPII. Furthermore, Nab2 occupies not only tRNA genes but all genes transcribed by RNAPIII, such as RDN5, RPR1, SCR1, SNR6, and SNR52 (Supplemental Fig. S1). Taken together, Nab2 is present at all genes transcribed by RNAPIII, indicating a general role of Nab2 in RNAPIII transcription.
Recruitment of Nab2 to RNAPIII depends on active transcription
We wanted to determine whether the presence of Nab2 at RNAPIII genes depends on active transcription. To impair transcription by RNAPIII, we used the rpc25-S100P mutation, which impairs transcription initiation especially at the restrictive temperature (Zaros and Thuriaux 2005). We assessed the occupancy of Rpc160 (the largest subunit of RNAPIII) and Nab2 by ChIP at three exemplary RNAPIII genes: SNR6, tDNALys, and RPR1.
When RNAPIII transcription was impaired by shifting the rpc25-S100P mutant to the restrictive temperature (37°C), the occupancy of RNAPIII expectedly decreased significantly at the three RNAPIII genes, whereas the RNAPIII occupancy stayed unchanged in wild-type cells (Fig. 2A). Importantly, Nab2 occupancy also decreased significantly at the restrictive temperature in the rpc25-S100P mutant (Fig. 2B), suggesting that Nab2 recruitment depends on active transcription by RNAPIII.
Figure 2.
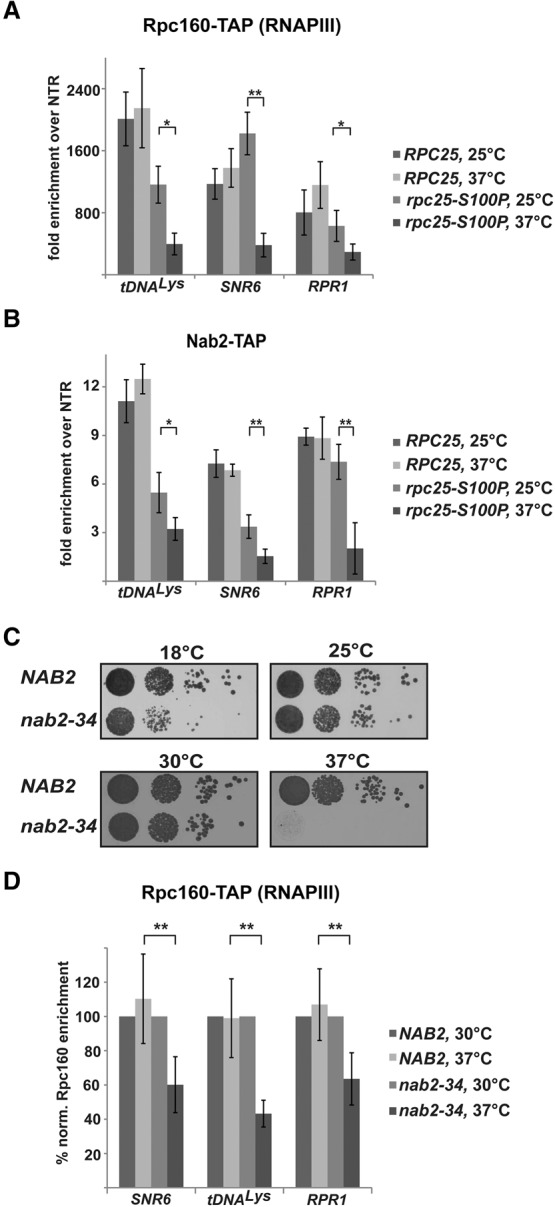
Nab2 functions in RNAPIII transcription. (A,B) Occupancy of Nab2 on RNAPIII transcribed genes is transcription-dependent. ChIP analysis of Rpc160 (A) and Nab2 (B) occupancies on selected RNAPIII genes in RPC25 and rpc25-S100P cells at the indicated temperatures. Data (n ≥ 3) represent the mean ± SD. (*) P < 0.05; (**) P < 0.01. (C) nab2-34 is a novel temperature-sensitive allele of NAB2. Growth of NAB2 (Δnab2 + pRS315-NAB2) and nab2-34 (Δnab2 + pRS315-nab2-34) cells at the indicated temperatures as assessed by dot spots on YPD plates. nab2-34 cells have a severe growth defect at 37°C (see also Supplemental Fig. S2). (D) Nab2 function is required for RNAPIII occupancy. ChIP analysis of Rpc160-TAP on selected RNAPIII transcribed genes in NAB2 and nab2-34 cells at 30°C and 37°C. The occupancies of Rpc160 in each strain were calculated relative to those at the permissive temperature. Data (n ≥ 3) represent the mean ± SD. (**) P < 0.01.
Nab2 is required for full occupancy of RNAPIII
The transcription-dependent, genome-wide presence of Nab2 at RNAPIII genes suggests a function for Nab2 in RNAPIII transcription. To study this potentially novel function of Nab2 in RNAPIII transcription, we first generated a new conditional allele of NAB2 as a tool. Previously generated alleles of NAB2 are not optimal: They cause a growth defect already at the permissive temperature (such as the nab2-1 and especially the nab2-1-GFP alleles) (Supplemental Fig. S2A), they do not cause pronounced temperature sensitivity (nab2-C437S), or they require a medium shift for depletion of Nab2 (nab2-td) (Marfatia et al. 2003; Gonzalez-Aguilera et al. 2011; Brockmann et al. 2012). After mutagenesis of NAB2, we screened for alleles that grow well at 30°C and are not viable at 37°C and identified one temperature-sensitive allele, nab2-34. On plates, nab2-34 cells grow as wild-type cells at the permissive temperature (30°C), are weakly impaired in growth at 25°C and 18°C, and are dead at the restrictive temperature of 37°C (Fig. 2C; Supplemental Fig. S2B). Growth curves in liquid culture revealed that the nab2-34 allele already confers a slight growth defect at 30°C. At 37°C, a growth retardation is first visible after 4 h, but nab2-34 cells continue to grow slowly for at least 9 h (Supplemental Fig. S2B).
We used this novel NAB2 allele to assess whether Nab2 is required for full occupancy of RNAPIII at the three exemplary genes. The occupancy of RNAPIII in NAB2 and nab2-34 cells at 30°C (permissive temperature) was normalized to 100%. RNAPIII occupancy does not change in NAB2 cells after shifting to 37°C (nonpermissive temperature) (Fig. 2D). In contrast, when Nab2 function is impaired in nab2-34 cells by shifting for 3 h to 37°C, the occupancy of RNAPIII decreases to ∼50% (Fig. 2B). The fact that a decrease in RNAPIII occupancy was observed before the nab2-34 cells displayed a growth defect suggests that the effect of Nab2 on RNAPIII occupancy is direct. Thus, Nab2 is required for full occupancy of RNAPIII, and Nab2 function is thus required for RNAPIII transcription.
Nab2 interacts directly with RNAPIII
Nab2 is likely to interact with RNAPIII if it functions in RNAPIII transcription. To assess a possible physical interaction of Nab2 with RNAPIII in vivo, we purified TAP-tagged Nab2 from S. cerevisiae by tandem affinity purification and tested for copurification of RNAPIII by Western blotting against HA-tagged Rpc160. Rpc160-HA and thus likely the whole RNAPIII complex copurify with Nab2 (Fig. 3A, lane 5). A strain that expresses HA-tagged Rpc160 but no TAP-tagged protein served as a negative control (Fig. 3A, lane 4). Conversely, we purified RNAPIII by tandem affinity purification from an Rpc160-TAP strain and assessed copurification of Nab2 by Western blotting. Nab2 indeed copurifies with RNAPIII (Fig. 3A, lane 6). Since the whole-cell protein extracts were treated with RNase A and DNase, the observed in vivo interaction between Nab2 and RNAPIII is most likely independent of RNA and DNA.
Figure 3.
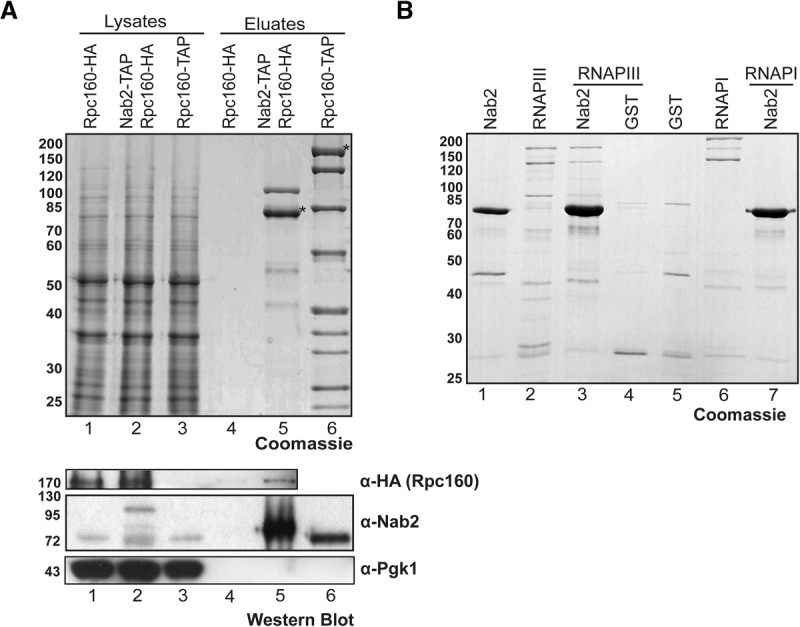
Nab2 interacts directly with RNAPIII. (A) Nab2 interacts with RNAPIII in vivo in an RNA- and DNA-independent manner. Tandem affinity purification of Nab2-TAP and Rpc160-TAP from lysates treated with DNase and RNase. To assess copurification of RNAPIII, Rpc160 was HA-tagged in the NAB2-TAP strain, and an RPC160-HA strain served as a negative control. (Top panel) Lysates and EGTA eluates were separated by SDS-PAGE and stained with Coomassie. TAP-tagged proteins are marked with an asterisk. (Lane 5) The additional band in the Nab2 purification corresponds to Kap104, the importin of Nab2. (Lane 6) The additional bands in the Rpc160 purification correspond to the core subunits of RNAPIII. Copurification of Nab2 with RNAPIII and vice versa was assessed by Western blotting with antibodies directed against HA (Rpc160) and Nab2. (Bottom panel) Pgk1 served as a negative control. (B) Nab2 interacts directly with RNAPIII. In vitro pull-down assay of recombinant GST-Nab2 with RNAPIII purified from S. cerevisiae. GST from Escherichia coli and RNAPI from S. cerevisiae served as negative controls.
In order to assess whether the in vivo interaction of Nab2 and RNAPIII is direct, we performed in vitro pull-down experiments. GST-Precission protease cleavage site–Nab2 was purified from Escherichia coli and bound to GST beads (Fig. 3B, lanes 1,3,7, Nab2). GST bound to beads served as a negative control (Fig. 3B, lanes 4,5). GST-Nab2 does not show high-molecular-weight bands without incubation with RNAPIII (Fig. 3B, lane 1). GST-Nab2 was then incubated with highly purified RNAPIII from S. cerevisiae, and bound proteins were eluted by cleavage with Precission protease. Importantly, purified RNAPIII (Fig. 3B, lane 2) bound to recombinant GST-Nab2 (Fig. 3B, lane 3). This interaction between Nab2 and RNAPIII is specific, as S. cerevisiae RNAPIII did not bind to GST alone (Fig. 3B, cf. lanes 4 and 5), and highly purified S. cerevisiae RNAPI did not bind to GST-Nab2 (Fig. 3B, cf. lanes 6 and 7). These experiments show that Nab2 interacts directly and specifically with RNAPIII, substantiating a function in RNAPIII transcription.
Nab2 binds RNAPIII transcripts in vivo
Since Nab2 binds poly(A) RNA and most likely functions in RNAPIII transcription, it could bind RNAPIII transcripts. Tuck and Tollervey (2013) recently determined all transcripts bound to 13 mRNA processing, export, and turnover proteins in S. cerevisiae in vivo—among them, Nab2. Analysis of these data revealed that Nab2 binds to RNAPIII transcripts (Fig. 4). A metaprofile of all intronless tRNA transcripts shows that Nab2 binds to the body of tRNAs (Fig. 4A). This metaprofile reflects binding to all tRNAs, as evident by the hit distribution for individual intronless and intron-containing tRNAs such as tRNAIle(AAU)E1, tRNALeu(UAG)L2, or tRNAIle(AAU)L1 (Fig. 4B,C). In addition to tRNAs, Nab2 also binds to all other known RNAPIII transcripts, such as the RNA of the signal recognition particle (Fig. 4B, SCR1), the RNA component of RNase P (Fig. 4D), and the 5S rRNA (Fig. 4E). Thus, Nab2 binds directly to all RNAPIII transcripts in vivo.
Figure 4.
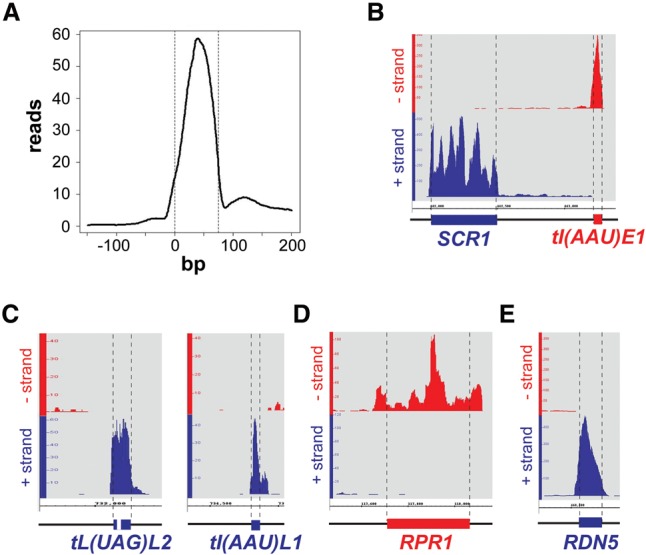
Nab2 binds to RNAPIII transcripts in vivo. (A) Metaprofile of Nab2 for nonspliced tRNAs. “0” indicates the transcription start site. (B–E) Hit distributions along selected RNAPIII transcripts relative to their strand orientation. (Blue) Plus strand; (red) minus strand. The 5′ and 3′ ends of the mature transcripts are indicated according to the current S. cerevisiae Genome Database (SGD) annotation by the dashed lines and colored bars. The precursor of tRNALeu(UAG)L2 contains an intron, and the precursor of tRNAIle(AAU)L1 contains a 3′ extension. RPR1 possesses an ∼84-bp 5′ extension and a 3′ extension. No precursors are known for the SCR1 RNA and the 5S rRNA. Note that scales differ due to different binding levels.
Interestingly, inspection of the traces of RNAPIII transcripts that are synthesized as precursors revealed that Nab2 already binds to the premature form of RNAPIII transcripts. For example, Nab2 binds to the intronic region of tRNALeu(UAG)L2 and the 3′ region of tRNAIle(AAU)L1 (Fig. 4C). The ∼84-bp 5′ extension of RPR1 (the RNA component of RNase P) that is cleaved off after transcription (Lee et al. 1991) is bound by Nab2 (Fig. 4D). In addition, Nab2 binds sequences downstream from the 3′ end of the mature RPR1, indicating that 3′ extended transcripts occur. Together with the findings that Nab2 binds directly to RNAPIII and is present at its target genes during transcription, this indicates that Nab2 probably binds to RNAPIII transcripts as they are being synthesized.
Nab2 is required for RNAPIII transcription in vivo
The data above suggest that Nab2 functions in RNAPIII transcription. We thus investigated whether Nab2 is needed to maintain RNAPIII transcript levels in vivo by assessing the levels of selected RNAPIII transcripts: the intron-containing tRNAIle(UAU)L, RPR1, and SNR6. The steady-state levels of these transcripts were determined by Northern blotting of total RNA extracted from wild-type and nab2-34 cells (Fig. 5A–C). The rpc25-S100P mutant was used as positive control (Fig. 5A–C). The amounts of intron-containing tRNAIle(UAU)L precursors (Fig. 5A), the RPR1 precursor (Fig. 5B), and the U6 snRNA (SNR6) (Fig. 5C) were measured and normalized to the amounts of the respective RNA in wild-type cells. After 6 h at the restrictive temperature, the levels of all three transcripts decreased significantly in nab2-34 cells (Fig. 5A–C). Since the levels of the pre-tRNAIle(UAU)L primary transcript and the RPR1 precursor decrease, it is likely that the de novo synthesis of these RNAs—i.e., RNAPIII transcription—is affected in nab2-34 cells. Thus, Nab2 function is most likely needed for full transcriptional activity of RNAPIII in vivo.
Figure 5.
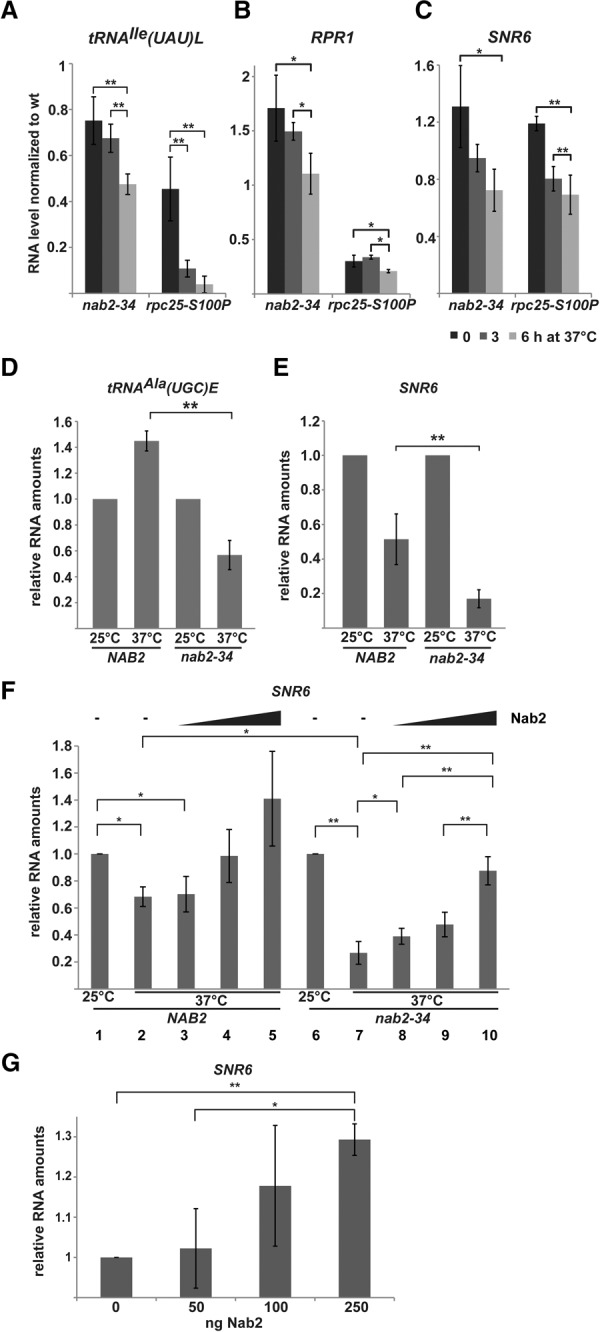
Nab2 is required for full RNAPIII transcriptional activity in vivo and in vitro. (A–C) Nab2 is required for RNAPIII transcription in vivo. Northern blot analysis of tRNAIle(UAU)L, RPR1, and the U6 snRNA (SNR6) using total RNA from nab2-34, rpc25-S100P, and the corresponding wild-type strains shifted to 37°C for the indicated times. Neosynthesis of tRNAIle, RPR1, and SNR6 is decreased in nab2-34 cells at 37°C. Quantification of the tRNAIle precursor (two highest bands), RPR1 precursor, and SNR6 levels in nab2-34 and rpc25-S100P normalized to the corresponding wild-type strains. Data (n ≥ 3) represent the mean ± SD. (*) P < 0.05. For Northern blots and loading control, see Supplemental Figure S3. (D,E) Nab2 is required for RNAPIII transcription in vitro. (D) In vitro transcription of tRNAAla(UGC)E in transcription-active extracts of NAB2 and nab2-34 cells at 25°C and 37°C. tRNAAla(UGC)E levels were normalized to the amount in the corresponding extract at 25°C, which was set to 1. Data represent mean ± SD. (**) P < 0.01. (E) Experiments as in D but with SNR6 as the reporter gene. Representative gels of the experiments quantified in D and E are shown in Supplemental Figure S4, A and B. (F) Recombinant Nab2 stimulates deficient transcription in nab2-34 extracts at 37°C. In vitro transcription assays were carried out with NAB2 (lanes 1–5) and nab2-34 (lanes 6–10) extracts at 25°C (lanes 1,6) or 37°C (lanes 2–5 and 7–10) in the absence (lanes 1,2,6,7) or presence (lanes 3–5,8–10) of increasing amounts of recombinant Nab2 (0, 100, 200, and 400 ng). The amount of SNR6 RNA was quantified, and the RNA amount at 25°C was set to 1. Data represent mean ± SD. (*) P < 0.05; (**) P < 0.01. A representative gel of the experiments quantified in F is shown in Supplemental Figure S4C. (G) Recombinant Nab2 stimulates RNAPIII transcription in a fully reconstituted in vitro transcription system. Increasing amounts of Nab2 (0, 50, 100, and 250 ng) were added to RNAPIII, TFIIIC, and TFIIIB preassembled on the SNR6 template DNA. The amount of SNR6 RNA was quantified, and the RNA amount without Nab2 was set to 1. Data (n = 3) represent mean ± SD. (*) P < 0.05; (**) P < 0.01. A representative gel of the experiments quantified in G is shown in Supplemental Figure S4D.
Nab2 functions directly in RNAPIII transcription
To assess whether Nab2 is directly involved in RNAPIII transcription, we performed in vitro transcription assays with two RNAPIII templates. The de novo synthesis of tRNAAla(UGC)E (Fig. 5D) and the U6 snRNA (SNR6) (Fig. 5E) in RNAPIII transcription-active extracts of wild-type and nab2-34 cells at 25°C and 37°C was determined. The synthesis of tRNAAla(UGC)E and SNR6 is significantly decreased in nab2-34 extracts compared with the transcription activity in wild-type extracts at 37°C (Fig. 5D,E). Thus, functional Nab2 is required for transcriptional activity in vitro, corroborating its function in RNAPIII transcription.
In order to assess whether the function of Nab2 in RNAPIII transcription is direct, we performed add-back experiments. In vitro transcription of the SNR6 gene was performed at 25°C and 37°C in NAB2 and nab2-34 transcription-active extracts. Increasing amounts of recombinant Nab2 were added to the transcription reactions at 37°C. Importantly, Nab2 stimulated the reduced transcriptional activity in nab2-34 extracts in a dose-dependent manner (Fig. 5F). To corroborate the direct function of Nab2 in RNAPIII transcription suggested by this add-back experiment, we used a fully reconstituted in vitro transcription assay consisting of RNAPIII, TFIIIB, TFIIIC, and template DNA. Also, in this system, addition of increasing amounts of Nab2 stimulated RNAPIII transcription (Fig. 5G). Taken together, the add-back experiment and the reconstituted transcription assay show that Nab2 has a direct function in RNAPIII transcription.
Nab2 is required for stable association of TFIIIB with the promoter DNA
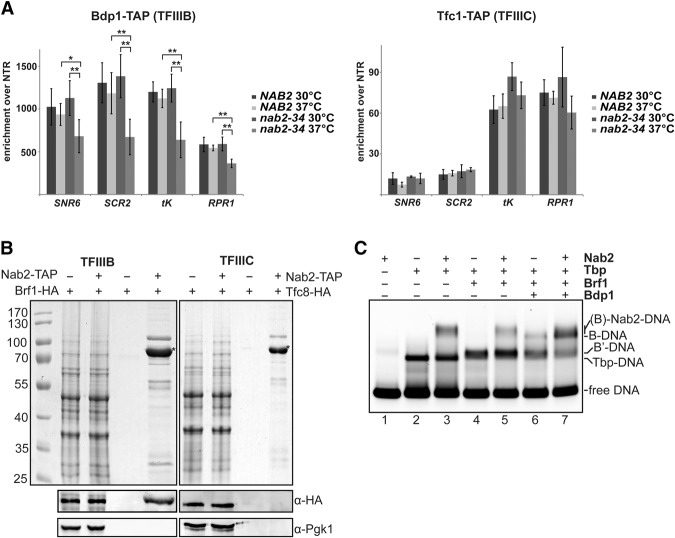
In order to get a first hint of Nab2's molecular function in RNAPIII transcription, we determined the occupancy profile of Nab2 over the longest RNAPIII transcribed gene, SCR1, and compared it with the profiles of RNAPIII, TFIIIB, and TFIIIC. RNAPIII, TFIIIB, and TFIIIIC show the expected profiles (Supplemental Fig. S5; Ghavi-Helm et al. 2008). Nab2 has its highest occupancies at the 5′ end of SCR1 (Supplemental Fig. S5E), compatible with a possible function of Nab2 in transcription initiation. Thus, we tested whether the occupancies of the RNAPIII transcription initiation factors TFIIIB and TFIIIC are affected when Nab2 function is compromised. As expected, RNAPIII occupancy decreases in the nab2-34 mutant at 37°C along the SCR1 gene (Supplemental Fig. S6A). Interestingly, the occupancy of TFIIIB is also reduced along the SCR1 gene and at selected RNAPIII transcribed genes in nab2-34 cells at the restrictive temperature (Fig. 6A, left panel; Supplemental Fig. S6B). This reduction of TFIIIB occupancy is specific, as the occupancy of TFIIIC does not decrease in nab2-34 cells at 37°C (Fig. 6A, right panel; Supplemental Fig. S6C). Thus, Nab2 is needed for full occupancy of TFIIIB in vivo. The reduced occupancy of TFIIIB at RNAPIII promoters in nab2-34 cells is most likely the reason for the decreased occupancy of RNAPIII and the decreased transcriptional activity in these cells (Figs. 2D, 5; Supplemental Fig. S6A). In order to recruit TFIIIB to the promoter or stabilize its binding, Nab2 should interact with TFIIIB in vivo. To test this, we purified Nab2 from S. cerevisiae by tandem affinity purification and performed Western blots to determine the presence of TFIIIB or TFIIIC subunits. Indeed, Nab2 specifically copurifies the TFIIIB subunit Brf1 and thus most likely the whole TFIIIB complex but does not interact with the TFIIIC subunit Tfc8 (Fig. 6B).
Figure 6.
Nab2 interacts with TFIIIB and stabilizes its binding to promoter DNA. (A) The occupancy of TFIIIB, but not TFIIIC, decreases when Nab2 function is impaired. ChIP analysis of the TAP-tagged TFIIIB subunit Bdp1 (left panel) and the TAP-tagged TFIIIC subunit Tfc1 (right panel) on selected RNAPIII transcribed genes in NAB2 and nab2-34 cells at 30°C and 37°C (see also Supplemental Figs. S5, S6). Data (n ≥ 3) represent the mean ± SD. (*) P < 0.05; (**) P < 0.01. (B) Nab2 interacts with TFIIIB but not TFIIIC in vivo. Tandem affinity purification of Nab2-TAP from a strain expressing C-terminally HA-tagged Brf1 (TFIIIB) or Tfc8 (TFIIIC). Strains without a TAP tag on Nab2 served as negative controls. (Left panel) Lysates and EGTA eluates were separated by SDS-PAGE and stained with Coomassie. Copurification of Brf1 (bottom panel) or Tfc8 (right panel) with Nab2 was assessed by Western blotting with antibodies directed against HA (Brf1/Tfc8). Pgk1 served as negative control. Tagged Nab2 is marked with an asterisk. (C) Nab2 increases the affinity of TFIIIB for DNA. EMSAs for Nab2-TFIIIB-DNA formation using TA-30-B6 DNA as template. Formed DNA–protein complexes are indicated at the right. Nab2 (500 ng) exclusively induces a supershift when Tbp is present on the promoter DNA and increases the affinity of Tbp/TFIIIB to target DNA (see also Supplemental Fig. S7).
These results indicate that Nab2 could increase the binding of TFIIIB to its cognate DNA. In order to test this directly, we performed gel shift assays (EMSAs). As a DNA substrate, we used TA-30-B6, a 76-bp-long dsDNA template derived from the SUP4 gene with the 6-bp TATA-box TATAAA and two 2-nucleotide (nt) mismatches at which Tbp binds the kinked DNA (Kassavetis et al. 1998). Since this DNA template requires specific interactions of the TFIIIB subunits (i.e., only a sequential binding of Tbp, Brf1, and Bdp1 is possible) in a TFIIIC-independent manner, we could test whether and, if so, at which step Nab2 increases the binding of TFIIIB subunits to the DNA. To first rule out an unspecific binding of Nab2 to DNA, we tested different amounts of Nab2 for binding to the cognate DNA template described above as well as four similar dsDNAs (dsDNA1+, dsDNA1−, dsDNA2+, and dsDNA2−), which are derived from TA-30-B6 with a scrambled nucleotide sequence with or without the 4-nt mismatches. Additionally, dsDNA1 contained no repeating nucleotides, whereas dsDNA2 contained double nucleotides (such as AA or TT). Nab2 bound to these DNAs only at a high concentration of Nab2, and the least to the TFIIIB DNA template TA-30-B6 (Supplemental Fig. S7). Using this probe, we then tested whether Nab2 increases the binding of TFIIIB subunits to the cognate DNA. When Tbp is already bound to the dsDNA, Nab2 can also bind to the DNA and consequently induces a supershift (Fig. 6C, cf. lanes 1–3). Similarly, Nab2 increases the amount of protein bound to the DNA and induces a supershift when either Tbp–Brf1 complexes or the whole TFIIIB complex are bound to the DNA (Fig. 6C, cf. lanes 4,5 and 6,7, respectively). Thus, Nab2 increases the amount of TFIIIB bound to DNA in vitro. Taken together, these results indicate that Nab2 increases TFIIIB binding to the promoter, thereby stimulating RNAPIII transcription.
Discussion
The PABP Nab2 has well-characterized roles in poly(A) tail length control and mRNA export. Here, we show that Nab2 also functions in RNAPIII transcription. Our genome-wide data on localization of several mRNA-binding proteins revealed that Nab2 occupies all RNAPIII transcribed genes (Fig. 1; Supplemental Fig. S1). This finding is consistent with the recent observation that Nab2 localizes to five genes transcribed by RNAPIII (Gonzalez-Aguilera et al. 2011). This work by Gonzalez-Aguilera et al. (2011) also implicated Nab2 in RNAPIII metabolism but excluded a role of Nab2 in RNAPIII transcription. However, nab2-1-GFP, the allele of NAB2 used in that study (encoded by plasmid pAC1038) (Marfatia et al. 2003), leads to extremely slow growth already at the permissive temperature (Supplemental Fig. S2A). We believe that the extremely slow growth of the nab2-1-GFP mutant most likely has secondary effects, causing the function of Nab2 in RNAPIII transcription to be missed.
Using our novel temperature-sensitive allele, nab2-34, we show here that Nab2 is important for RNAPIII transcription (Fig. 7). The function of Nab2 in RNAPIII transcription is most likely direct, since the in vitro RNAPIII transcription defect of nab2-34 extracts can be rescued by addition of recombinant Nab2 (Fig. 5F), and Nab2 stimulates transcription in a reconstituted in vitro transcription assay (Fig. 5G). Nab2 is essential for efficient RNAPIII transcription initiation, as it is required for full occupancy of TFIIIB (Fig. 6A), most likely mediated by its direct interaction with TFIIIB (Fig. 6B). Importantly, Nab2 increases the binding of Tbp and TFIIIB to promoter DNA in vitro (Fig. 6C). The reduced occupancy of TFIIIB is most likely the reason for the decreased occupancy of RNAPIII in nab2-34 cells at the restrictive temperature (Fig. 2; Supplemental Fig. S6A). Thus, Nab2 functions to recruit TFIIIB and thus RNAPIII to the gene or increase their stability at the gene during transcription initiation (Fig. 7).
Figure 7.
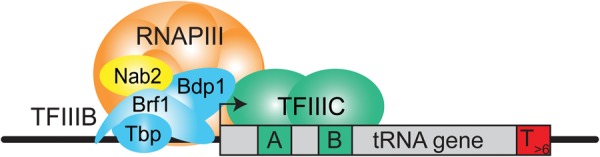
Model of Nab2 function in RNAPIII transcription. See the text for details.
Since Nab2 interacts directly with precursors of RNAPIII transcripts (Fig. 4), it probably binds to RNAPIII transcripts already during transcription. This cotranscriptional binding of Nab2 to RNAPIII transcripts might be necessary for efficient transcription elongation, similar to the function of mRNA-binding proteins in RNAPII transcription. mRNA-binding protein complexes such as the TREX and THSC complexes prevent formation of so-called R loops (RNA:DNA hybrids plus a displaced DNA strand) during transcription elongation by RNAPII, probably by binding to the mRNA (Aguilera and Garcia-Muse 2012 and references therein). Likewise, Nab2 could bind to the nascent RNA during transcription elongation by RNAPIIII, thus reducing the ability of the RNA to rehybridize with the transiently opened DNA double strand. In contrast, Nab2 is most likely not necessary for the processing of RNAPIII transcripts, since precursors of pre-tRNAs or other RNAPIII transcripts do not accumulate in nab2-34 cells at the restrictive temperature (Supplemental Fig. S3). As tRNAs are spliced in the cytoplasm (Hopper and Huang 2015), it is also unlikely that Nab2 is required for nuclear tRNA export. Taken together, Nab2 might have two functions in RNAPIII transcription: One in initiation, as shown here, and a second one in elongation based on its interaction with precursors of RNPIII transcripts. It will be interesting to determine the exact functions of Nab2 in RNAPIII transcription.
In recent years, several proteins involved in RNAPII transcription have been implicated in RNAPIII transcription (see above). In higher cells, RNAPII and RNAPII TFs localize to RNAPIII transcribed genes. However, this is not the case in S. cerevisiae (Barski et al. 2010; Raha et al. 2010; Venters et al. 2011), consistent with our finding that RNAPII, the TREX component Tho2, and the SR protein Npl3 are not present at RNAPIII genes (Fig. 1; Supplemental Fig. S1). Thus, the presence of Nab2 at RNAPIII genes is independent of its association with RNAPII. Importantly, Nab2 is the first mRNA biogenesis factor with a function in RNAPIII transcription. Interestingly, proteins involved in both RNAPII and RNAPIII transcription or metabolism, such as Nab2 and Dst1 in yeast (also see above), could coordinate these two important cellular pathways. Whether this is the case remains to be determined.
Nab2 is highly conserved throughout evolution and is named ZC3H14 in Homo sapiens, Msut-2 in Mus musculus, ZC3H14 in Rattus norvegicus, SUT-2 in Caenorhabditis elegans, dNab2 in Drosophila melanogaster, and nab2 in Schizosaccharomyces pombe. The functions of Nab2 homologs in these diverse organisms also seem to be conserved: Poly(A) binding has been shown in H. sapiens, M. musculus, R. norvegicus, and D. melanogaster (Pak et al. 2011; Kelly et al. 2014), and its requirement for correct poly(A) tail length has been shown in D. melanogaster and probably H. sapiens (Pak et al. 2011). Human ZC3H14 is ubiquitously expressed and exists in four different splice variants: Isoforms 1, 2, 3, and 3short contain a predicted classical nuclear localization signal (cNLS) and localize to the nucleus, whereas isoform 4 contains an alternative first exon lacking the NLS and consequently localizes to the cytoplasm (Leung et al. 2009). Interestingly, mutations in ZC3H14 cause a form of autosomal recessive intellectual disability in humans, and Nab2 is needed for normal neuronal function in Drosophila (Pak et al. 2011). Thus, Nab2 is important for proper cellular functions, especially in neuronal cells (Kelly et al. 2015). Since Nab2 is already known to have important and conserved cellular functions in 3′ end processing and mRNA export, it will be of great interest to assess whether its function in RNAPIII transcription is also conserved.
In conclusion, we show here that Nab2, an important protein in mRNP biogenesis but with no previous implication in transcription, has a second, pivotal role in RNAPIII transcription. Here, one function of Nab2 is to efficiently assemble the RNAPIII initiation complex by increasing the binding of TFIIIB to the promoter DNA.
Materials and methods
Yeast strains and plasmids
Yeast strains, plasmids, and DNA sequences used in this study are listed in Supplemental Tables S1–S3.
Generation of a NAB2 temperature-sensitive allele
The temperature-sensitive allele of NAB2, nab2-34, was generated essentially according to Chanarat et al. (2011). Briefly, the ORF of NAB2 was randomly mutagenized by error-prone PCR, and the resulting PCR fragments were cotransformed with plasmid pRS315-NAB2 cut with NotI and XhoI, which removes the ORF of NAB2, into the NAB2 shuffle strain (Δnab2 + pRS316-NAB2). Transformants were selected on SDC(−leu) plates, replica-plated on plates containing 5-fluoroorotic acid to shuffle out pRS316-NAB2, and tested for thermosensitivity at 37°C. One allele was identified and named nab2-34.
ChIP experiments and analysis of the ChIP–chip and CRAC data
The ChIP experiments were performed as in Rother et al. (2010), and the ChIP–chip experiments were performed as in Meinel et al. (2013). RPC25 and rpc25-S100P cells were grown for 10 h at 25°C or shifted to 37°C. NAB2 and nab2-34 cells were grown for 3 h at 30°C or shifted to 37°C. ChIP–chip data sets of Nab2-TAP, TAP-Npl3, TAP-Tho2, and Rpb3-TAP cells were used from Meinel et al. (2013); the Rpc160-TAP ChIP–chip data are available at ArrayExpress (http://www.ebi.ac.uk/arrayexpress), accession number E-MTAB-3700.
The ChIP–chip data were normalized according to Meinel et al. (2013). Data normalization and analysis were carried out using R (CRAN). The metaprofiles for tRNA genes were calculated by averaging the occupancies of all intronless tRNA. To analyze the significance of the Pearson's correlation coefficient for the peak occupancies at protein-coding or tRNA genes for Nab2 and Rpb3 or Nab2 and Rpc160, the correlation coefficients of 100,000 random permutations of the data sets were calculated and compared with the correlation coefficients of the nonpermutated data set.
The Nab2 CRAC data from Tuck and Tollervey (2013) were inspected for Nab2 binding to the different RNAPIII transcripts using Integrated Genome Browser (Nicol et al. 2009) and R (CRAN). For the CRAC tRNA metaprofile, the reads for all intronless tRNAs <76 bp were averaged and plotted versus the position relative to the first nucleotide of the tRNA gene.
Protein purification
RNAPI and RNAPIII were purified from S. cerevisiae by tandem affinity purification of Rpa190-TAP and Rpc160-TAP, respectively, according to Rother et al. (2006) with the following modifications: All whole-cell protein extracts were treated with RNase and DNase prior to the purification. For the in vivo interaction assay of Nab2 with RNAPIII, the proteins/protein complexes were purified to the EGTA eluate. For in vitro interaction assays, proteins were purified with buffer containing 800 mM NaCl until the TEV eluate. GST- or His6-tagged recombinant Nab2 was produced in E. coli Bl21 DE3 transformed with pGex6-P1-NAB2 or pET21a-NAB2 at 37°C by addition of 0.3 mM isopropyl-1-thio-β-D-galactopyranoside at an OD600 of 0.6 for 3 h. Cells were harvested, washed, resuspended in NETN buffer (1 M NaCl, 1 mM EDTA, 50 mM Tris-HCl at pH 7.5, 1% NP-40, 10 µM ZnCl2, 2 mM DTT, protease inhibitors) and lysed by sonication. Lysates were incubated with glutathione sepharose 4B (GE Healthcare) or Ni-NTA agarose (Qiagen) and purified according to the manufacturers’ protocols. His-tagged Nab2 was further purified by ion exchange chromatography using a MonoS 5/50 GL column (GE Healthcare) with a step elution from 0 M NaCl to 0.6 M NaCl, 50 mM MES (pH 6.5), 10 µM ZnCl2, and 2 mM DTT. To exchange buffers, a PD-10 desalting column (GE Healthcare), equilibrated with the respective assay buffer, was used. For the fully reconstituted in vitro transcription assay, proteins were purified as follows: Bdp1, Brf1, and Tbp were purified essentially as described in Kassavetis et al. (1998) and references therein, Nab2 was purified from E. coli, and RNAPIII (Rpc160-TAP) and TFIIIC (Tfc1-TAP) were purified from from S. cerevisiae as described above for RNAPIII under stringent conditions until the TEV eluate.
In vitro interaction assay
Purified GST or GST-Nab2 bound to beads and treated with 25 µg/mL DNase I and 100 µg/mL RNase A (Thermo Scientific) was incubated with equal amounts of TAP-purified RNAPI or RNAPIII in buffer containing 20 mM HEPES (pH 7.6), 1 mM DTT, 75 mM NaCl, and 2 mM MgCl2 for 1 h at 16°C. After washing, proteins were eluted by incubation with 5 µL of Precission protease for 2 h at 16°C in 150 µL of buffer and analyzed by SDS-PAGE.
RNA extraction
Total RNA was extracted from S. cerevisiae grown to 0.6–0.8 OD600 in YPD at 30°C or shifted to 37°C for the indicated times with 300 µL of buffer (10 mM Tris-HCl at pH 8.0, 1 mM EDTA, 100 mM NaCl, 1% SDS, 2% Triton X-100), 300 µL of glass beads, and 300 µL of phenol by vortexing. After centrifugation at 13,200 rpm for 5 min at room temperature, the RNA was washed once with chloroform and precipitated using standard conditions. The RNA was dissolved in DEPC water, and the DNA was digested with 10 U/100 µL DNase I (Thermo Scientific) in the presence of 80 U/100 µL RiboLock RNase inhibitor (Thermo Scientific) for 15 min at 37°C. RNA concentration and integrity were tested on 2% agarose denaturing formaldehyde gels.
Northern blotting
Five micrograms of total RNA was separated by electrophoresis on 6% or 9% polyacrylamide gels containing 6 M urea. After blotting onto a nylon membrane, radiolabeled oligonucleotides were hybridized in Church buffer. After washing three times with 2× SSC buffer and 0.1% SDS, the blots were exposed overnight to a storage phosphor screen and analyzed by a Typhoon 9400 PhosphorImager (Amersham Bioscience). Signals were quantified with ImageQuant 5.2.
In vitro transcription assay
RNAPIII transcription-active extracts were generated, and in vitro transcription experiments were performed essentially as described in Schultz et al. (1991). In brief, 30 µg of whole-cell extract was mixed with 4 µL of 5× transcription buffer (100 mM HEPES at pH 7.9, 400 mM KCl, 25 mM MgCl2, 5 mM EDTA, 10% [v/v] glycerol); 1 mM DTT; 200 ng of α-Amanitin; 20 U of RiboLock RNase inhibitor (Thermo Scientific); 500 µM ATP, CTP, and UTP; 50 µM GTP; 100 ng of plasmid template, and 5 µCi [α-32P]GTP (3000 Ci/mmol; Hartmann Analytic). Reactions were incubated for 30 min at 25°C or 37°C. The fully in vitro reconstituted transcription experiments were carried out as described in Huet et al. (1996). Reaction mixtures were set up with 20 mM HEPES (pH 7.9); 90 mM KCl; 5 mM MgCl2; 0.1 mM EDTA; 10% (v/v) glycerol; 1 mM DTT; 10 U of RiboLock RNase inhibitor; 0.6 mM ATP, CTP, and GTP; 0.03 mM UTP; 100 ng of plasmid template; and 10 µCi [α-32P] UTP (3000 Ci/mmol; Hartmann Analytic). Proteins were added in the described order and contained varying amounts of Nab2. Transcription mixes were incubated for 30 min at 25°C. After phenol/chloroform extraction and ethanol precipitation, the dried RNA pellets were resuspended in formamide loading buffer and separated on a 9% polyacrylamide gel with 7 M urea in TAE buffer. Dried gels were exposed overnight to a storage phosphor screen or autoradiographed and analyzed with a Typhoon FLA 9400 or FLA 9500 PhosphorImager (GE Healthcare). Signals were quantified with ImageQuant 5.2 or TL.
EMSAs
EMSAs were done as previously described in Kassavetis et al. (1998). Briefly, protein–DNA complexes were formed in 40 mM Tris-HCl (pH 8.0), 7 mM MgCl2, 3 mM DTT, 5% (v/v) glycerol, 100 µg/mL BSA, 50 mM NaCl, and 100 ng of ssDNA, and 5 pmol of 6-FAM 5′-labeled dsDNA was used instead of poly(dG–dC)–poly(dG–dC). Proteins were added as indicated. Reaction mixtures were incubated for 60 min at 25°C and separated on a 2% (w/v) agarose gel in 1× TAE. Gels were analyzed with a Typhoon FLA 9500 and ImageQuant TL software.
Supplementary Material
Acknowledgments
We thank Cornelia Burkert-Kautzsch for the Rpc160 ChIP–chip experiments, Susanne Röther for the strain RPA190-TAP, and Sittinan Chanarat, Larissa Knüppel, and Florence Gauye for support of this project. Plasmid pBS-SNR6 was a gift of Dr. Hung-Ta Chen. We are grateful to Yuh Min Chook (University of Texas Southwestern Medical Center) and Dietmar Martin (Gene Center, Ludwig-Maximilians-University) for critical reading of the manuscript. L.M.R. and D.M.M. were supported by the International Max Planck Research School for Molecular and Cellular Life Sciences, K.S. was supported by the Deutsche Forschungsgemeinschaft, an ERC Starting Grant, the Ludwig-Maximilians-Universität LMUMentoring program, and the Center for Integrated Protein Science Munich.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.266205.115.
References
- Acker J, Conesa C, Lefebvre O. 2013. Yeast RNA polymerase III transcription factors and effectors. Biochim Biophys Acta 1829: 283–295. [DOI] [PubMed] [Google Scholar]
- Aguilera A, Garcia-Muse T. 2012. R loops: from transcription byproducts to threats to genome stability. Mol Cell 46: 115–124. [DOI] [PubMed] [Google Scholar]
- Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, Cui K, White RJ, Zhao K. 2010. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol 17: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann C, Soucek S, Kuhlmann SI, Mills-Lujan K, Kelly SM, Yang JC, Iglesias N, Stutz F, Corbett AH, Neuhaus D, et al. 2012. Structural basis for polyadenosine-RNA binding by Nab2 Zn fingers and its function in mRNA nuclear export. Structure 20: 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow DA, Guthrie C. 1990. Transcription of a yeast U6 snRNA gene requires a polymerase III promoter element in a novel position. Genes Dev 4: 1345–1356. [DOI] [PubMed] [Google Scholar]
- Chanarat S, Seizl M, Strasser K. 2011. The Prp19 complex is a novel transcription elongation factor required for TREX occupancy at transcribed genes. Genes Dev 25: 1147–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducrot C, Lefebvre O, Landrieux E, Guirouilh-Barbat J, Sentenac A, Acker J. 2006. Reconstitution of the yeast RNA polymerase III transcription system with all recombinant factors. J Biol Chem 281: 11685–11692. [DOI] [PubMed] [Google Scholar]
- Eschenlauer JB, Kaiser MW, Gerlach VL, Brow DA. 1993. Architecture of a yeast U6 RNA gene promoter. Mol Cell Biol 13: 3015–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Michaut M, Acker J, Aude JC, Thuriaux P, Werner M, Soutourina J. 2008. Genome-wide location analysis reveals a role of TFIIS in RNA polymerase III transcription. Genes Dev 22: 1934–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W, Guthrie C. 2004. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol Cell 13: 201–212. [DOI] [PubMed] [Google Scholar]
- Gjidoda A, Henry RW. 2013. RNA polymerase III repression by the retinoblastoma tumor suppressor protein. Biochim Biophys Acta 1829: 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Aguilera C, Tous C, Babiano R, de la Cruz J, Luna R, Aguilera A. 2011. Nab2 functions in the metabolism of RNA driven by polymerases II and III. Mol Biol Cell 22: 2729–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwizdek C, Iglesias N, Rodriguez MS, Ossareh-Nazari B, Hobeika M, Divita G, Stutz F, Dargemont C. 2006. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc Natl Acad Sci 103: 16376–16381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector RE, Nykamp KR, Dheur S, Anderson JT, Non PJ, Urbinati CR, Wilson SM, Minvielle-Sebastia L, Swanson MS. 2002. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J 21: 1800–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK, Huang HY. 2015. Quality control pathways for nucleus-encoded eukaryotic tRNA biosynthesis and subcellular trafficking. Mol Cell Biol 35: 2052–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J, Manaud N, Dieci G, Peyroche G, Conesa C, Lefebvre O, Ruet A, Riva M, Sentenac A. 1996. RNA polymerase III and class III transcription factors from Saccharomyces cerevisiae. Methods Enzymol 273: 249–267. [DOI] [PubMed] [Google Scholar]
- Iglesias N, Tutucci E, Gwizdek C, Vinciguerra P, Von Dach E, Corbett AH, Dargemont C, Stutz F. 2010. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev 24: 1927–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis GA, Kumar A, Ramirez E, Geiduschek EP. 1998. Functional and structural organization of Brf, the TFIIB-related component of the RNA polymerase III transcription initiation complex. Mol Cell Biol 18: 5587–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SM, Pabit SA, Kitchen CM, Guo P, Marfatia KA, Murphy TJ, Corbett AH, Berland KM. 2007. Recognition of polyadenosine RNA by zinc finger proteins. PNAS 104: 12306–12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SM, Leung SW, Apponi LH, Bramley AM, Tran EJ, Chekanova JA, Wente SR, Corbett AH. 2010. Recognition of polyadenosine RNA by the zinc finger domain of nuclear poly(A) RNA-binding protein 2 (Nab2) is required for correct mRNA 3′-end formation. J Biol Chem 285: 26022–26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SM, Leung SW, Pak C, Banerjee A, Moberg KH, Corbett AH. 2014. A conserved role for the zinc finger polyadenosine RNA binding protein, ZC3H14, in control of poly(A) tail length. RNA 20: 681–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SM, Bienkowski R, Banerjee A, Melicharek DJ, Brewer ZA, Marenda DR, Corbett AH, Moberg KH. 2015. The Drosophila ortholog of the Zc3h14 RNA binding protein acts within neurons to pattern axon projection in the developing brain. Dev Neurobiol 10.1002/dneu.22301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Rohlman CE, Molony LA, Engelke DR. 1991. Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol Cell Biol 11: 721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung SW, Apponi LH, Cornejo OE, Kitchen CM, Valentini SR, Pavlath GK, Dunham CM, Corbett AH. 2009. Splice variants of the human ZC3H14 gene generate multiple isoforms of a zinc finger polyadenosine RNA binding protein. Gene 439: 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfatia KA, Crafton EB, Green DM, Corbett AH. 2003. Domain analysis of the Saccharomyces cerevisiae heterogeneous nuclear ribonucleoprotein, Nab2p. Dissecting the requirements for Nab2p-facilitated poly(A) RNA export. J Biol Chem 278: 6731–6740. [DOI] [PubMed] [Google Scholar]
- Meinel DM, Burkert-Kautzsch C, Kieser A, O'Duibhir E, Siebert M, Mayer A, Cramer P, Soding J, Holstege FC, Strasser K. 2013. Recruitment of TREX to the transcription machinery by its direct binding to the phospho-CTD of RNA polymerase II. PLoS Genet 9: e1003914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SG Jr, Raja A, Loraine AE. 2009. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25: 2730–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orioli A, Pascali C, Pagano A, Teichmann M, Dieci G. 2012. RNA polymerase III transcription control elements: themes and variations. Gene 493: 185–194. [DOI] [PubMed] [Google Scholar]
- Pak C, Garshasbi M, Kahrizi K, Gross C, Apponi LH, Noto JJ, Kelly SM, Leung SW, Tzschach A, Behjati F, et al. 2011. Mutation of the conserved polyadenosine RNA binding protein, ZC3H14/dNab2, impairs neural function in Drosophila and humans. Proc Natl Acad Sci 108: 12390–12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, Struhl K, Snyder M. 2010. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci 107: 3639–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rother S, Clausing E, Kieser A, Strasser K. 2006. Swt1, a novel yeast protein, functions in transcription. J Biol Chem 281: 36518–36525. [DOI] [PubMed] [Google Scholar]
- Rother S, Burkert C, Brunger KM, Mayer A, Kieser A, Strasser K. 2010. Nucleocytoplasmic shuttling of the La motif-containing protein Sro9 might link its nuclear and cytoplasmic functions. RNA 16: 1393–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm L, Hernandez N. 2002. Recruitment of RNA polymerase III to its target promoters. Genes Dev 16: 2593–2620. [DOI] [PubMed] [Google Scholar]
- Schultz MC, Choe SY, Reeder RH. 1991. Specific initiation by RNA polymerase I in a whole-cell extract from yeast. Proc Natl Acad Sci 88: 1004–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek S, Corbett AH, Fasken MB. 2012. The long and the short of it: the role of the zinc finger polyadenosine RNA binding protein, Nab2, in control of poly(A) tail length. Biochim Biophys Acta 1819: 546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Hurt E. 2001. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413: 648–652. [DOI] [PubMed] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308. [DOI] [PubMed] [Google Scholar]
- Tuck AC, Tollervey D. 2013. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell 154: 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck-Walsh C, et al. 2011. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol Cell 41: 480–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ. 2011. Transcription by RNA polymerase III: more complex than we thought. Nat Rev Genetics 12: 459–463. [DOI] [PubMed] [Google Scholar]
- Zaros C, Thuriaux P. 2005. Rpc25, a conserved RNA polymerase III subunit, is critical for transcription initiation. Mol Microbiol 55: 104–114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.