Abstract
Cell growth is a highly regulated, plastic process. Its control involves balancing positive regulation of anabolic processes with negative regulation of catabolic processes. Although target of rapamycin (TOR) is a major promoter of growth in response to nutrients and growth factors, AMP-activated protein kinase (AMPK) suppresses anabolic processes in response to energy stress. Both TOR and AMPK are conserved throughout eukaryotic evolution. Here, we review the fundamentally important roles of these two kinases in the regulation of cell growth with particular emphasis on their mutually antagonistic signaling.
TOR is a major promoter of cell growth in response to nutrients and growth factors. In contrast, AMPK promotes a catabolic response when nutrients and energy are low.
An efficient homeostatic response to maintain cellular energy despite a noncontinuous supply of nutrients is crucial for the survival of organisms. Cells have, therefore, evolved a host of molecular pathways to sense both intra- and extracellular nutrients and thereby quickly adapt their metabolism to changing conditions. The target of rapamycin (TOR) and AMP-activated protein kinase (AMPK) signaling pathways control growth and metabolism in a complementary manner with TOR promoting anabolic processes under nutrient- and energy-rich conditions, whereas AMPK promotes a catabolic response when cells are low on nutrients and energy. Both pathways are highly conserved from yeast to human. This review summarizes the cross talk between TOR and AMPK in different organisms.
TOR SIGNALING IN MAMMALS
TOR is a conserved Ser/Thr protein kinase that belongs to the phosphoinositide-3-kinase (PI3K)-related kinase (PIKK) family (Wullschleger et al. 2006; Laplante and Sabatini 2012). TOR was originally identified in the budding yeast Saccharomyces cerevisiae (Heitman et al. 1991; Kunz et al. 1993), and in mammalian cells shortly thereafter (Brown et al. 1994; Chiu et al. 1994; Sabatini et al. 1994; Sabers et al. 1995). TOR exists in two conserved and structurally and functionally distinct multiprotein complexes, rapamycin-sensitive TOR complex 1 (TORC1), and rapamycin-insensitive TOR complex 2 (TORC2) (see Table 1) (Loewith et al. 2002; Reinke et al. 2004). Mammalian TOR complex (mTORC)1 consists of three core components: the catalytic subunit mammalian TOR (mTOR), regulatory-associated protein of target of rapamycin (RAPTOR), and mammalian lethal with SEC13 protein 8 (mLST8). mTORC2 is comprised of four different core proteins: mTOR, rapamycin-insensitive companion of target of rapamycin (RICTOR), mammalian stress-activated protein kinase interacting protein (mSIN1), and mLST8. mTORC1, whose localization is well characterized, is mainly on the lysosome when active (Bar-Peled and Sabatini 2012). mTORC2 is at mitochondria-associated endoplasmic reticulum (ER) membranes (MAM) (Betz et al. 2013). For a detailed review of mTOR localization, the reader is referred to Betz and Hall (2013).
Table 1.
TORC1, TORC2, TSC1/2, RHEB, and AMPK homologs across different species
| Sc | Sp | Dd | Ce | Dm | At | Hs | |
|---|---|---|---|---|---|---|---|
| TORC1 | TOR1 TOR2 |
Tor1 Tor2 |
Tor | TOR | TOR | TOR | mTOR |
| Kog1 | Mip1 | Raptor | DAF-15 | RAPTOR | RAPTOR1A RAPTOR1B |
RAPTOR | |
| Lst8 | Pop3 | Lst8 | C10H11.8 | LST8 | LST8-1 LST8-2 | LST8 | |
| Tco89 | Tco89 | - | - | - | - | - | |
| - | Toc1 | - | - | - | - | - | |
| - | - | - | - | - | - | PRAS40 | |
| - | - | - | - | - | - | DEPTOR | |
| TORC2 | TOR2 | Tor1 | Tor | TOR | TOR | TOR | mTOR |
| Avo1 | Sin1 | RipA | - | SIN1 | - | SIN1 | |
| Avo2 | - | - | - | - | - | - | |
| Avo3 | Ste20 | PiaA | RICTOR | RICTOR | - | RICTOR | |
| Lst8 | Pop3 | Lst8 | C10H11.8 | LST8 | LST8-1 LST8-2 | LST8 | |
| Bit61 | Bit61 | - | - | - | - | PRR5 | |
| - | - | - | - | - | DEPTOR | ||
| TSC1/2 | - | Tsc1 | - | - | TSC1 | - | TSC1 |
| - | Tsc2 | Tsc2 | - | TSC2 | - | TSC2 | |
| RHEB | Rhb1 | Rhb1 | Rheb | RHEB-1 | RHEB | - | RHEB |
| AMPK | Snf1 | Ssp2 Ppk9 | SnfA | AAK-1 AAK-2 |
SNF1A | KIN10 KIN11 | AMPKα1 AMPKα2 |
| Gal83 Sip1 Sip2 |
Amk2 | PrkaB | AAKB-1 AAKB-2 |
AMPKB1 | KINβ1 KINβ2 KINβ3 |
AMPKβ1 AMPKβ2 |
|
| Snf4 | Cbs2 | PrkaG | AAKG-1 AAKG-2 AAKG-3 AAKG-4 AAKG-5 |
SNF4Aγ | KINγ KINβγ |
AMPKγ1 AMPKγ2 AMPKγ3 |
At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Dd, Dictyostelium discoideum; Dm, Drosophila melanogaster; Hs, Homo sapiens; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe. AMPK, AMP-activated protein kinase; DEPTOR, DEP domain-containing mTOR-interacting protein; mTOR, mammalian target of rapamycin; RAPTOR, regulatory-associated protein of target of rapamycin. RHEB, RAS homolog enriched in brain; RICTOR, rapamycin-insensitive companion of target of rapamycin; TORC1, target of rapamycin complex 1; TORC2, target of rapamycin complex 2; TSC1/2, tuberous sclerosis complex 1/2. (-) indicates that there are no shown/obvious homologs of the indicated proteins in the corresponding organisms.
Upstream Regulators of mTOR Signaling
mTORC1 is activated by a variety of upstream signals, most importantly, nutrients, growth factors, and cellular energy. Branched chain amino acids, in particular, leucine, are the most effective nutrient activators of mTORC1. Amino-acid sensing involves the RAG family of small GTPases (Kim et al. 2008; Sancak et al. 2008; Duran and Hall 2012; Jewell et al. 2013). The Rags form heterodimers of RagA or RagB with RagC or RagD. Amino-acid sufficiency promotes the formation of the active form of the Rag heterodimer (RagA/BGTP-RagC/DGDP), which binds directly to RAPTOR, and thereby recruits mTORC1 to the lysosome. Once on the lysosome, mTORC1 encounters the small GTPase RHEB (RAS homolog enriched in brain), and the association of mTORC1 with RHEB leads to mTORC1 activation. Under amino acid starvation, the Rag heterodimers assume an inactive configuration (RagA/BGDP-RagC/DGTP), which is unable to recruit mTORC1 to the lysosomal surface. Thus, mTORC1 remains cytosolic and inactive. Multiple mechanisms have been proposed to explain how amino acids are sensed to activate mTORC1. For example, amino acids could be sensed in the lumen of the lysosome through an inside-out mechanism that requires the v-ATPase (vacuolar-ATPase) and RAGULATOR, a pentameric complex that acts as a guanine nucleotide exchange factor (GEF) and activates the RAG complex at the lysosomal surface (Sancak et al. 2010; Zoncu et al. 2011; Bar-Peled et al. 2012). Others have proposed that at least leucine and glutamine are sensed through glutaminolysis, which stimulates mTORC1 via the production of α-ketoglutarate (Duran et al. 2012, 2013). These two models are not mutually exclusive as both may act on the Rags and RAGULATOR (Fig. 1). Recently, new regulators of the Rags have been characterized. Two independent studies identified a multiprotein complex, termed GATOR, that interacts with the Rags. GATOR is composed of two subcomplexes: GATOR1 and GATOR2. GATOR1 has GTPase activating protein (GAP) activity for RagA/B and negatively regulates mTORC1 signaling by promoting the inactive RagA/BGDP conformation (Bar-Peled et al. 2013; Panchaud et al. 2013). GATOR2 is a negative regulator of GATOR1 (Bar-Peled et al. 2013). More recently, FOLLICULIN (FLCN) and its binding partners folliculin-interacting protein 1/2 (FNIP1/2) have been shown to act as a positive regulator of Rag-mTORC1 signaling by acting as a GAP for RagC/D to promote the active RagC/DGDP conformation (Petit et al. 2013; Tsun et al. 2013). It is unclear whether GATOR and FLCN play a role in amino-acid sensing.
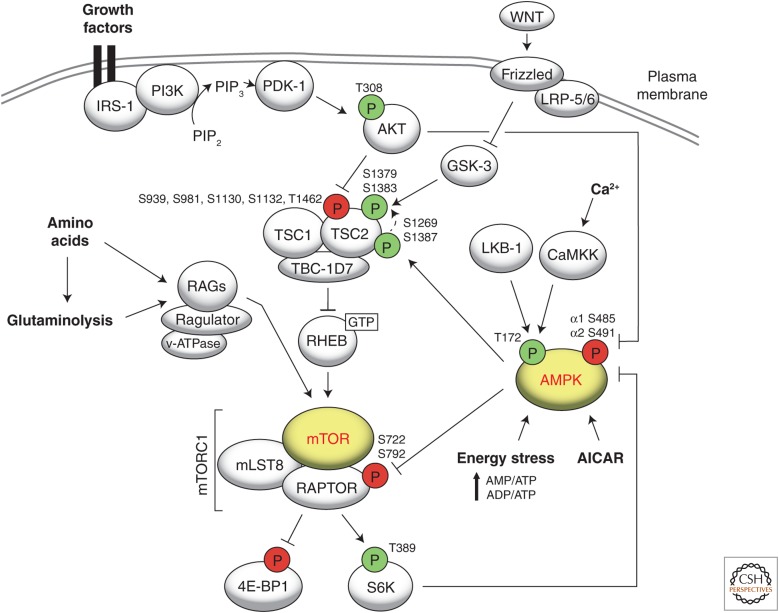
Figure 1.
The model shows the main components in cross talk between mammalian target of rapamycin (mTOR) and AMP-activated protein kinase (AMPK). See the main text for details. Phosphorylation depicted in green indicates an activation signal. Phosphorylation depicted in red indicates an inhibitory signal.
Growth factors stimulate mTORC1 via the PI3K-phosphoinositide-dependent kinase-1-AKT (PI3K-PDK-1-AKT) pathway. AKT controls mTORC1 signaling by phosphorylating and inactivating tuberous sclerosis complex 2 (TSC2), TUBERIN (Fig. 1) (Dan et al. 2002; Inoki et al. 2002; Manning et al. 2002). AKT phosphorylates TSC2 on Ser939, Ser981, Ser1130, Ser1132, and Thr1462. TSC2 associates with tuberous sclerosis complex 1 (TSC1), HAMARTIN and TBC-1D7 to form the tumor suppressor tuberous sclerosis complex (TSC), which functions as a GAP for the small GTPase RHEB (Garami et al. 2003; Inoki et al. 2003a; Tee et al. 2003; Zhang et al. 2003; Dibble et al. 2012). RHEB is anchored to the surface of the lysosome (Saito et al. 2005) and in its active GTP-loaded form provides an essential stimulatory signal to mTORC1 (Long et al. 2005; Sancak et al. 2007). Two recent reports suggest that translocation of the TSC complex to the lysosome is a key determinant of mTORC1 inactivation (Demetriades et al. 2014; Menon et al. 2014). Growth factor signaling, although necessary, cannot efficiently activate mTOR when amino acids are limiting (Hara et al. 1998; Kim et al. 2008; Sancak et al. 2008). Nutrients are a key activator of TORC1 already in unicellular organisms, whereas growth factor signaling was grafted onto the TOR pathway only later with the advent of multicellularity. This reflects the primordial role of nutrients as a growth-regulating and TORC1-activating input. The energy status of the cell (AMP:ATP ratio) is signaled to mTORC1 through AMPK, a master sensor of intracellular energy status (Inoki et al. 2003b; Gwinn et al. 2008). The mechanism by which energy status is signaled to mTORC1 is discussed in detail below. Apart from the above three inputs, mTOR is also regulated by various other signaling pathways, such as Hippo, Wnt, and Notch, as reviewed in Shimobayashi and Hall (2014).
mTORC2 is involved in various cellular and developmental processes, but its mechanism of activation remains poorly understood. mTORC2 is activated in a PI3K-dependent manner by association with the ribosome (Zinzalla et al. 2011). Thus, mTORC2 activation is responsive to growth factors, but independent of nutrients and energy.
Downstream Functions of mTOR Signaling
Downstream functions of mTORC1 include activation of protein synthesis, ribosome biogenesis, lipogenesis, nucleotide synthesis, and inhibition of autophagy. Protein synthesis and ribosome biogenesis are particularly important readouts because they determine the growth capacity of the cell and are extremely energy-demanding processes. mTORC1 promotes protein synthesis by phosphorylating the eukaryotic initiation factor 4E (eIF4E)-binding protein 1 (4E-BP1) and ribosomal S6 kinase (S6K) (Fig. 1) (reviewed in Ma and Blenis 2009; Huang and Fingar 2014). The phosphorylation of 4E-BP1 prevents its binding to eIF4E, thereby enabling eIF4E to promote cap-dependent translation. The stimulation of S6K activity by mTORC1 leads to increased translation initiation and elongation. mTOR controls ribosome biogenesis by increasing translation of ribosomal proteins (reviewed in Mayer and Grummt 2006) and transcription of ribosomal RNAs (rRNAs) by Pol III (5S rRNA) or Pol I (the other three rRNAs) (Goodfellow and White 2007; Kantidakis et al. 2010). In addition, mTORC1 promotes processing of pre-rRNA (Iadevaia et al. 2012). Furthermore, mTORC1 promotes lipogenic gene expression by activating the sterol-regulatory element-binding protein (SREBP) transcription factors (for review, see Ricoult and Manning 2013). mTORC1 stimulates nucleotide synthesis by phosphorylating carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase (CAD) through S6K. CAD phosphorylation promotes its activation and thereby the de novo synthesis of pyrimidines (Ben-Sahra et al. 2013; Robitaille et al. 2013). mTORC1 inhibits autophagy by phosphorylating the proautophagic kinase Unc-51-like kinase 1 (ULK1) (Fig. 2) (Kim et al. 2011). On activation by growth factors, mTORC2 phosphorylates the AGC kinases AKT, serum- and glucocorticoid-regulated kinase (SGK), and protein kinase C (PKC) to control a variety of processes (for review, see Cybulski and Hall 2009; Oh and Jacinto 2011).
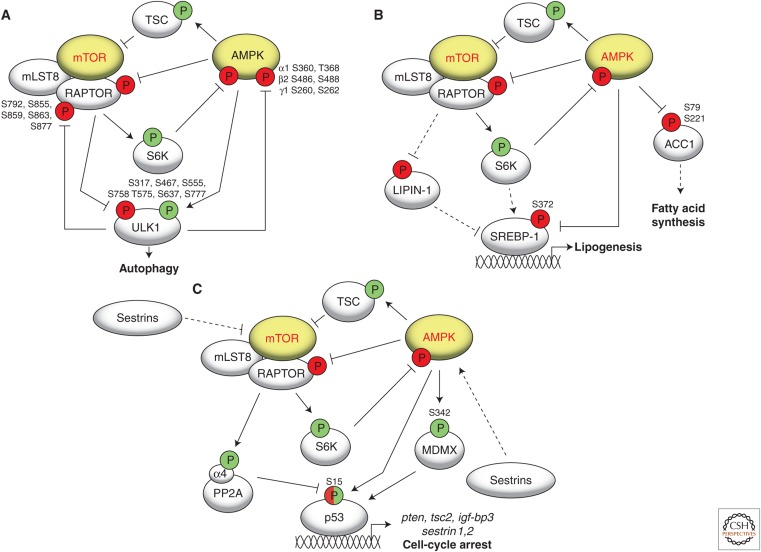
Figure 2.
Cross talk between mTOR and AMPK signaling in regulating (A) autophagy, (B) lipid synthesis, and (C) p53 in regulating the cell cycle.
AMPK SIGNALING IN MAMMALS
In eukaryotes, strict maintenance of cellular energy, as reflected in relative concentrations of ATP, ADP, and AMP (adenosine tri-, di-, and monophosphate), is of paramount importance for the control of all energy requiring metabolic processes. AMPK, activated on energy stress (high intracellular AMP and ADP), plays a crucial role in maintaining this energy balance (Yeh et al. 1980; Oakhill et al. 2011; Gowans et al. 2013; Hardie 2014, 2015). Like TOR, AMPK is a multiprotein complex comprising three subunits: (1) the α catalytic subunit, (2) the β subunit containing a glycogen-binding domain (GBD) and binding sites for both the α and γ subunits, and (3) the γ subunit, which contains nucleotide-binding sites (Bateman domains) (Bateman 1997). Table 1 summarizes the different subunits of AMPK (for recent findings on the structure of AMPK, the reader is referred to Xiao et al. 2013; Calabrese et al. 2014).
Upstream Regulation of AMPK
Activation of AMPK requires phosphorylation on Thr172 in the activation loop of the α subunit (Hawley et al. 1996). Liver kinase B1 (LKB-1) (Hawley et al. 1996; Sakamoto et al. 2006), calmodulin-dependent protein kinase kinase β (CaMKKβ) (Hawley et al. 2005; Hurley et al. 2005; Woods et al. 2005), and transforming growth factor β–activated kinase (TAK1) (Momcilovic et al. 2006) are known upstream kinases of AMPK (Fig. 1). Although LKB-1 activates AMPK when AMP or ADP levels are high (energy stress), CaMKKβ activates AMPK in response to an increase in cytoplasmic Ca2+ levels on, for example, ER stress (Davies et al. 1995; Hawley et al. 1996, 2005; Hurley et al. 2005; Woods et al. 2005; Sakamoto et al. 2006). Whether TAK1 activates AMPK directly or via LKB-1 remains to be determined. A recent report suggests that AXIN, originally discovered as an inhibitor of WNT signaling (Zeng et al. 1997), is required for AMP-triggered AMPK activation by LKB-1 at the lysosomal surface (Zhang et al. 2013). It was also recently shown that the small molecule A-769662 (and AMP) can directly activate AMPK allosterically and independently of upstream kinase signaling, that is, in the absence of AMPK Thr172 phosphorylation (Scott et al. 2014).
Downstream Functions of AMPK
AMPK acts as a metabolic checkpoint by activating catabolic processes and inhibiting anabolic processes, in part, by negatively regulating mTORC1 signaling. Thus, mTORC1 and AMPK work in opposing ways in the regulation of cell growth and metabolism. The cross talk between AMPK and mTOR signaling is discussed in detail below. By virtue of its inhibitory effect on mTORC1 signaling, AMPK inhibits protein synthesis and promotes autophagy (Gwinn et al. 2008; Inoki et al. 2012). AMPK promotes autophagy by directly phosphorylating and activating ULK1 (Fig. 2A) (Egan et al. 2011; Kim et al. 2011). In addition, AMPK modulates carbohydrate metabolism by increasing intracellular glucose levels (Holmes et al. 1999; Kurth-Kraczek et al. 1999; Barnes et al. 2002), and reduces lipid synthesis by inhibiting and acetyl CoA carboxylase 1 (ACC1) and acetyl CoA carboxylase 2 (ACC2) (Ha et al. 1994), fatty acid synthase (FAS) (An et al. 2007), glycerol-3-phosphate acyltransferase (GPAT) (Muoio et al. 1999), and 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) (Beg and Brewer 1982).
EVOLUTION OF TOR–AMPK SIGNALING FROM YEAST TO MAMMALS
TOR Signaling
Although almost all eukaryotes have a single TOR gene, lower eukaryotes, such as S. cerevisiae or Schizosaccharomyces pombe, have two TOR genes. In budding yeast, TORC1 contains either TOR1 or TOR2, but TORC2 is assembled from only TOR2 (see Table 1) (Loewith et al. 2002; Reinke et al. 2004). Rapamycin inhibits TORC1 and growth in most eukaryotes, with worms (Caenorhabditis elegans) and plants (Arabidopsis thaliana) being exceptions (Long et al. 2002; Mahfouz et al. 2006). In S. pombe, rapamycin treatment is not sufficient to cause a growth defect (Takahara and Maeda 2012). S. cerevisiae lacks TSC homologs, but possesses an RHEB homolog (Rhb1). However, Rhb1 in S. cerevisiae does not seem to function upstream of TORC1 (Urano et al. 2000). In contrast, in S. pombe, Tsc1 and Tsc2 are present and regulate TORC1 via the RHEB ortholog Rhb1 (Aspuria et al. 2007). The slime mold Dictyostelium discoideum has orthologs of TSC2 and RHEB, in addition to all the core components of mTORC1 (Lee et al. 2005). C. elegans contains RHEB-1 and the TORC1 components, but lacks TSC (Long et al. 2002). In Drosophila melanogaster, like in mammals, TORC1 signaling is regulated by TSC1, TSC2, and RHEB (Oldham et al. 2000; Zhang et al. 2000, 2003; Gao and Pan 2001; Gao et al. 2002; Saucedo et al. 2003; Stocker et al. 2003). A. thaliana contains TORC1, but is devoid of RHEB and the TSC complex (Vernoud et al. 2003; Diaz-Troya et al. 2008). Rag homologs are found in all the above model organisms except A. thaliana. Thus, TORC1 is highly conserved yet flexible in the composition of its upstream regulators (see Table 1).
AMPK Signaling
Like mammalian AMPK, S. cerevisiae AMPK is heterotrimeric (nomenclature of mammalian AMPK subunits and its homologs in other organisms are summarized in Table 1). The AMPKα ortholog Snf1 is required primarily for the adaptation to glucose limitation, but is also involved in responses to other environmental stresses (reviewed in Hedbacker and Carlson 2008). Snf1 is activated on glucose or nitrogen starvation and on sodium or alkaline stress (Orlova et al. 2006; Hong and Carlson 2007). The activation of Snf1 requires the phosphorylation of Thr210 within the conserved activation loop (Thr210 in Snf1 corresponds to Thr172 in mammalian AMPKα) (Estruch et al. 1992). S. pombe has two homologs of mammalian AMPKα: Ppk9 and Ssp2 (see Table 1). Ssp2 is required for the response to nitrogen starvation (Valbuena and Moreno 2012). The AMPKα homologs in C. elegans (AAK1 and AAK2) and D. melanogaster (SNF1A) are activated by AMP (Pan and Hardie 2002; Apfeld et al. 2004). In D. melanogaster, AMPK is important for cellular homeostasis and survival on energy deprivation (Tschape et al. 2002; Lee et al. 2007; Spasic et al. 2008). In A. thaliana, the Snf1 and AMPKα homologs are the sucrose nonfermenting-1-related protein kinase 1 (SnRK1) subfamily members, KIN10 and KIN11. Plants also express two other large plant-specific subfamilies, namely, SnRK2 and SnRK3 (reviewed in Polge and Thomas 2007; Ghillebert et al. 2011). However, unlike KIN10 and KIN11 (Bhalerao et al. 1999), several SnRK2 and SnRK3 family members do not complement yeast snf1 mutations (Hrabak et al. 2003) and are, thus, not further considered in this review. It is predicted that KIN10 and KIN11 require phosphorylation of Thr175 and Thr176, respectively, for activation. These residues are equivalent to Thr172 in mammalian AMPKα (Bhalerao et al. 1999; Sugden et al. 1999). However, KIN10 is not allosterically activated by AMP (Mackintosh et al. 1992). KIN10 and KIN11 sense decreasing energy levels caused by nutrient deprivation, environmental stress, or alternate light–dark cycles (Polge and Thomas 2007; Baena-Gonzalez and Sheen 2008). Thus, like TOR, AMPK is conserved from yeast to human.
EVOLUTION OF CROSS TALK BETWEEN TOR AND AMPK SIGNALING
TORC1 and AMPK are both important nutrient sensors that have broadly opposing effects on metabolism. The cross talk between TORC1 and AMPK signaling can be grouped into two categories. We refer to the situations in which AMPK and TORC1 regulate each other directly as “direct cross talk,” and if they converge to regulate downstream functions as “indirect cross talk.”
Direct Cross Talk
AMPK Regulation of TORC1
mTORC1 was shown early on to be inhibited by the AMPK activator AICAR (5-amino-1-β-d-ribofuranosyl-imidazole-4-carboxamide) (Bolster et al. 2002; Kimura et al. 2003). However, the molecular mechanism of mTORC1 inhibition by AICAR was not well understood. Interesting mechanistic insights came from observations in nutrient-starved mouse embryonic fibroblasts (MEFs), in which AMPK was shown to phosphorylate and activate the TSC complex, and thereby to inactivate RHEB and mTORC1 signaling (Fig. 1) (Inoki et al. 2003b). AMPK phosphorylates TSC2 on Thr1227 and Ser1345, residues corresponding to Thr1269 and Ser1387 in human TSC2, respectively. AMPK-mediated phosphorylation of TSC2 primes TSC2 for subsequent phosphorylation by glycogen synthase kinase 3β (GSK-3β) on Ser1337 and Ser1341 (Ser1379 and Ser 1383 in humans, respectively). This phosphorylation by GSK-3β, although not absolutely essential, further stabilizes the TSC complex to enhance the inhibitory effect of AMPK on mTORC1 signaling (Fig. 1) (Inoki et al. 2006). Although the AMPK sites in TSC2 are conserved in vertebrates, they are not present in D. discoideum or S. pombe (Huang and Manning 2008; Serfontein et al. 2011; van Dam et al. 2011). Moreover, the TSC complex is absent in S. cerevisiae, C. elegans, and A. thaliana. Thus, not all eukaryotes use an AMPK–TSC axis to regulate TORC1 signaling.
AMPK activation (treatment with 2-deoxyglucose, AICAR, or phenformin) can still cause partial inhibition of mTORC1 in TSC2-deleted MEFs (Hahn-Windgassen et al. 2005; Gwinn et al. 2008), indicating that AMPK can also inhibit the mTORC1 pathway in a TSC-independent manner. A breakthrough was the identification of RAPTOR as an AMPK substrate (Gwinn et al. 2008). AMPK phosphorylates RAPTOR on Ser722 and Ser792, thus inhibiting mTORC1 directly (Fig. 1). Although Ser722 and its flanking residues in RAPTOR are conserved only in vertebrates, Ser792, along with the critical flanking residues, are conserved from yeast to mammals (Gwinn et al. 2008), suggesting that AMPK-mediated phosphorylation of RAPTOR could be a highly conserved cross talk mechanism between AMPK and TORC1 (Fig. 1). However, it is not clear whether the AMPK site is present in Kog1 (the S. cerevisiae RAPTOR ortholog) because of a low degree of sequence conservation of the AMPK target site (Hardie 2011). The significance of these in-silico predictions remains to be experimentally confirmed. A recent report suggests that Snf1 is required for TORC1 inactivation under glucose starvation conditions (Hughes Hallett et al. 2014), but it remains unclear whether this is a direct or indirect effect of Snf1 on TORC1.
TORC1 Regulation of AMPK
Insulin signaling inhibits AMPK in a variety of tissues, including cardiomyocytes, adipocytes, and hepatocytes (Witters and Kemp 1992; Gamble and Lopaschuk 1997; Kovacic et al. 2003; Clark et al. 2004; Horman et al. 2006). This decrease in AMPK activity was attributed to inhibitory phosphorylation of AMPKα1-Ser485 or AMPKα2-Ser491 by AKT (Kovacic et al. 2003; Horman et al. 2006; Berggreen et al. 2009; Ning et al. 2011; Valentine et al. 2014). However, a study in mouse hypothalamus showed that leptin signals through the mTORC1 effector S6K to phosphorylate these sites (Dagon et al. 2012), suggesting that these sites are phosphorylated by different kinases in different tissues (Fig. 1). As mentioned earlier, phosphorylation of Thr172 is required for AMPK activity. Two studies showed that inhibitory phosphorylation of AMPKα1-Ser485 or AMPKα2-Ser491 does not lead to loss of Thr172 phosphorylation, suggesting that the inhibitory phosphorylation of AMPK serves as an off switch that inhibits AMPK even when AMPKα is phosphorylated at Thr172 (Dagon et al. 2012; Valentine et al. 2014). However, in some other studies, Ser485/491 phosphorylation is associated with a decrease in Thr172 phosphorylation (Kovacic et al. 2003; Horman et al. 2006; Berggreen et al. 2009). A recent report supports the notion that AKT-mediated phosphorylation of AMPKα1 at Ser485 reduces Thr172 phosphorylation. Interestingly, the equivalent site on AMPKα2 (Ser491) is not an AKT target, but is phosphorylated by AMPK α2 itself (Hawley et al. 2014).
In mouse liver, rapamycin promotes phosphorylation of AMPKα on Thr172, which, in turn, leads to an increase in AMPK activity (Chaveroux et al. 2013). A similar effect of rapamycin was observed in S. cerevisiae, in which rapamycin induced phosphorylation of Snf1 on Thr210 (Orlova et al. 2006). Nomura et al. (2010), however, did not observe an increase of Snf1 phosphorylation on Thr210 after rapamycin treatment. This difference could be caused by Snf1 overexpression in the former study. These observations suggest that, at least in budding yeast and mammals, TORC1 may also negatively regulate AMPK by decreasing phosphorylation of Thr172 (mammals) or Thr210 (S. cerevisiae) in an indirect fashion.
Cross Talk between mTORC1 and AMPK at the Lysosomal Surface
A recent report suggests that, on glucose starvation, the v-ATPase-RAGULATOR complex binds AXIN/LKB-1 to promote AMPK phosphorylation and activation at the lysosomal surface. Concurrently, AXIN inhibits GEF activity of RAGULATOR toward Rags, causing mTORC1 inactivation (Zhang et al. 2014). This regulation seems to occur only under glucose starvation conditions because AXIN is not required for mTORC1 inactivation on amino-acid starvation (Zhang et al. 2014). Thus, it appears that v-ATPase-RAGULATOR, known to be involved in mTORC1 activation by amino, acids, is also required for AMPK activation under glucose starvation at the lysosomal surface. Although this intriguing model awaits further confirmation, these observations suggest that lysosomal surfaces may represent a key platform, in which nutrients are sensed in a reciprocal manner by both mTORC1 and AMPK. Thus, the presence of AMPK, TSC, and mTORC1 on the surface of the lysosome could facilitate AMPK-mTORC1 cross talk via AMPK-TSC and AMPK-RAPTOR interactions.
Indirect Cross talk
Opposing Roles of TORC1 and AMPK in Controlling Autophagy
Autophagy is a process in which cytoplasmic components, including macromolecules and organelles, are degraded in the lysosome during periods of low nutrient availability. Although rapamycin was long known to induce autophagy in mammals (Blommaart et al. 1995), the underlying molecular mechanism remained elusive. The identification of ULK1 as a direct target of both mTORC1 and AMPK was an important step toward understanding how nutrient sensors regulate autophagy. On nutrient stimulation, mTORC1 prevents autophagy by phosphorylating ULK1 on Ser758, thereby inhibiting its interaction with AMPK (Hosokawa et al. 2009; Kim et al. 2011). On nutrient stress, AMPK, on the one hand, inhibits mTORC1 to prevent Ser758 phosphorylation on ULK1, leading to ULK1-AMPK interaction and, on the other hand, phosphorylates ULK1 on multiple sites (Ser317, Ser467, Ser555, Thr575, Ser637, and Ser777) to activate ULK1 and autophagy (Fig. 2A) (Egan et al. 2011; Kim et al. 2011). As another level of regulation, ULK1 exerts a negative feedback signal on both mTORC1 and AMPK. ULK1 phosphorylates RAPTOR on residues Ser855, Ser859, Ser863, Ser877, and Ser792. This phosphorylation inhibits mTORC1 activity by weakening the interaction of mTORC1 with its substrates (Dunlop et al. 2011; Jung et al. 2011). ULK1 also phosphorylates all three AMPK subunits, AMPKα1 (S360/T368), AMPKβ2 (S486/T488), and AMPKγ1 (S260/T262) (residues refer to rat AMPK), to dampen AMPK activation (Fig. 2A) (Loffler et al. 2011). Thus, ULK1 not only induces, but also down-regulates autophagy. In S. cerevisiae, inhibition of TORC1 results in dephosphorylation of Atg13, which leads to phosphorylation of the proautophagic kinase Atg1 (ULK1 ortholog) and activation of autophagy (Kamada et al. 2010; Kijanska et al. 2010; Yeh et al. 2010; Kraft et al. 2012). Whether TORC1 also directly phosphorylates Atg1 remains to be shown. ATG1 and ATG13 were identified as multicopy suppressors of the glycogen-deficient phenotype of cells lacking Snf1. Autophagy is impaired in snf1 mutant cells starved of nitrogen or entering stationary phase. Thus, it was proposed that Snf1 is a positive regulator of autophagy, probably, via Atg1 (Wang et al. 2001). However, further investigation is required to elucidate the mechanisms by which Snf1 controls autophagy.
TORC1 inhibition promotes autophagy in S. pombe (Takahara and Maeda 2012), D. discoideum (Otto et al. 2003), C. elegans, D. melanogaster, and A. thaliana (Scott et al. 2004; Hansen et al. 2008; Liu and Bassham 2010), and the respective ULK1 ortholog in each organism has been shown to be required for induction of autophagy (Tekinay et al. 2006; Kohda et al. 2007; Scott et al. 2007; Egan et al. 2011; Suttangkakul et al. 2011). Whereas the role of AMPK in autophagy needs to be shown in S. pombe and D. discoideum, AMPK activation has been shown to trigger autophagy in C. elegans, D. melanogaster, and A. thaliana (Diaz-Troya et al. 2008; Lippai et al. 2008; Egan et al. 2011). In these cases, it is likely that AMPK mediates its effect on autophagy via phosphorylation of ULK1 orthologs. Interestingly, in C. elegans, UNC-51 (ULK1 ortholog) has two conserved AMPK sites (Ser555 and Ser574), indicating that UNC-51 could be a conserved AMPK target (Egan et al. 2011). However, difficulties in aligning the poorly conserved Ser/Thr-rich domain in ULK1 and its homologs makes it difficult to assess whether the residues are conserved in other eukaryotes.
Opposing Roles of AMPK and TORC1 in Controlling Lipid Metabolism
AMPK and mTORC1 show opposing effects on lipid metabolism. Two basic building blocks of membranes, fatty acids and sterols, are under control of the SREBP transcription factors (Foretz et al. 1999; Shimano et al. 1999; Shimomura et al. 1999). mTORC1 activates SREBPs and lipid synthesis via two mechanisms. First, in an S6K-dependent fashion, TORC1 regulates the maturation of SREBPs to promote de novo lipid synthesis (Porstmann et al. 2008; Duvel et al. 2010). Second, in an S6K-independent fashion, mTORC1 directly phosphorylates the phosphatidic acid phosphatase LIPIN-1 (a negative regulator of SREBP-1), preventing its translocation into the nucleus and, thus, promoting SREBP transcriptional activity (Fig. 2B) (Peterson et al. 2011). Conversely, AMPK activation by AICAR or 2-deoxyglucose inhibits nuclear accumulation of SREBP-1 (Porstmann et al. 2008). AMPK phosphorylates SREBP-1 on Ser372 to prevent its nuclear translocation, leading to inhibition of lipogenesis and lipid accumulation (Li et al. 2011). Yet another way by which AMPK inhibits lipogenesis is by phosphorylating and inactivating the acetyl CoA carboxylases ACC1 and ACC2 on Ser79 and Ser221, respectively (Fig. 2B) (Munday et al. 1988). Rapamycin promotes phosphorylation of ACC1 on Ser79 in mouse liver (Chaveroux et al. 2013). It would be of interest to determine whether this increase in ACC1-Ser79 phosphorylation is mediated by AMPK.
In S. cerevisiae, Snf1 phosphorylates and inactivates Acc1 (Mitchelhill et al. 1994; Woods et al. 1994). However, the phosphorylation sites in Acc1 remain to be determined. snf1 mutant cells have a higher amount of fatty acids under glucose-limiting conditions (Usaite et al. 2009; Zhang et al. 2011). Thus, the role of AMPK in the regulation of lipogenesis seems to be evolutionarily conserved. Furthermore, in cells lacking the PP2A-like phosphatase Sit4, lipid droplet content is reduced possibly because of the hyperactivation of Snf1 (Bozaquel-Morais et al. 2010). Two independent groups showed that deletion of SIT4 leads to a constitutive increase in phosphorylation of Snf1 on Thr210 (Bozaquel-Morais et al. 2010; Ruiz et al. 2011). Because Sit4 is negatively controlled by TORC1 (Di Como and Arndt 1996; Jacinto et al. 2001; Rohde et al. 2004), these results suggest a connection between TORC1 and AMPK in regulating lipid droplet biogenesis (Bozaquel-Morais et al. 2010). Further studies are required to elucidate the cross talk between TORC1 and Snf1 in lipid droplet formation.
Opposing Roles of TORC1 and AMPK in Controlling Aging
Aging is defined as an accumulation of cellular damage over time, resulting in disease and death of the organism. Genetic and chemical inhibition of TORC1 have been shown to increase the life span of mice (Harrison et al. 2009; Miller et al. 2011), C. elegans (Vellai et al. 2003; Jia et al. 2004; Pan et al. 2007; Korta et al. 2012), D. melanogaster (Kapahi et al. 2004; Bjedov et al. 2010), and S. cerevisiae (Kaeberlein et al. 2005). Dietary restriction (DR) also extends life span and appears to act largely through inhibition of TORC1. DR is unable to further increase life span when TORC1 is inactive in D. melanogaster (Kapahi et al. 2004; Bjedov et al. 2010) and S. cerevisiae (Kaeberlein et al. 2005). The mutual antagonism of TOR and AMPK is also present in controlling aging. For example, in rats, it was observed that AMPK activity declines with age (Ljubicic and Hood 2009), and activation of AMPK leads to life span extension (Anisimov 2010). Furthermore, metformin increases life span and activates AMPK in mice (Martin-Montalvo et al. 2013). Activation of AMPK also extends life span in C. elegans (Apfeld et al. 2004; Curtis et al. 2006; Schulz et al. 2007), D. melanogaster (Lee et al. 2010), and A. thaliana (Thelander et al. 2004; Baena-Gonzalez and Sheen 2008). Taken together, these observations indicate important and opposing roles of AMPK and TOR signaling in controlling aging. However, in S. cerevisiae, the role of AMPK and TOR in controlling aging seems to differ from what is known in other organisms. In S. cerevisiae, cells lacking Sip2 (one of the β subunits of AMPK) have a shortened life span and this phenotype has been attributed to an increase in Snf1 activity (Ashrafi et al. 2000). Lu et al. (2011) recently proposed that as yeast cells age, Sip2 becomes deacetylated, thus reducing the inhibition of Snf1 and inducing aging via phosphorylation of the TORC1 substrate Sch9. Snf1 would directly activate Sch9 by phosphorylating residues different from those targeted by TORC1. Further experiments are required to determine how Snf1 is involved in aging in budding yeast.
Aging is accompanied by an inhibition of autophagy (Cuervo 2008; Mizushima et al. 2008). DR and rapamycin could extend life span by activating autophagy because an autophagy deficiency leads to a decrease in turnover of cellular components and accumulation of defective organelles within the cell. Furthermore, reduced autophagy affects various essential processes in mice, such as immune regulation (Levine et al. 2011), lymphocyte survival (Miller et al. 2008), maintenance of fetal hematopoietic stem cells (HSCs) (Mortensen et al. 2011), and various liver functions (Zhang and Cuervo 2008), ultimately decreasing life span. A brain-specific knockout of the autophagy gene Atg5 or Atg7 in mice causes accelerated neuronal degeneration and shorter life span (Hara et al. 2006; Komatsu et al. 2006). Furthermore, inhibition of autophagy by B-cell lymphoma-2 (BCL-2, an antiapoptotic protein) binding to BECLIN1 (Bcl-2-interacting protein, autophagy related protein) allows tumor formation, leading to a shorter life span (Pattingre et al. 2005; reviewed in Pattingre and Levine 2006). The above findings suggest an important role for TOR- and AMPK-regulated autophagy in determining life span.
Opposing Roles of TORC1 and AMPK in the Control of Transcription Factors
p53, SREBP-1 (discussed above), HIF1α, Gln3, and Msn2 are some of the important transcription factors regulated by both AMPK and TORC1. The tumor suppressor p53 is a sensor of genotoxic stress that protects cells from DNA damage by inducing cell-cycle arrest. AMPK and mTORC1 regulate p53 in opposite ways. Although AMPK phosphorylates p53 on Ser15 to stabilize it (Jones et al. 2005), mTORC1 inhibits this phosphorylation (Fig. 2C). mTORC1 does this by directly phosphorylating the α4 subunit of the PP2A (protein phosphatase 2A) phosphatase, thereby activating it to dephosphorylate phospho-Ser15 and destabilize p53 (Kong et al. 2004; Feng 2010). In addition, AMPK directly phosphorylates MDMX on Ser342, thereby protecting p53 from ubiquitylation (He et al. 2014). Conversely, p53 activates AMPK and inhibits mTORC1 signaling by increasing expression of SESTRINS (SESTRIN1 and SESTRIN2). SESTRINS are highly conserved proteins encoded by genes whose expression is up-regulated in cells exposed to a variety of stresses, such as DNA damage, oxidative stress, and hypoxia. SESTRINS activate AMPK via an unknown mechanism. Active AMPK leads to activation of TSC2 and a decrease in mTORC1 activity (Budanov and Karin 2008). Recent reports suggest that SESTRINS also inhibit mTORC1 in an AMPK-independent manner through the GATOR complex (Chantranupong et al. 2014; Parmigiani et al. 2014) or via RagA/B (Peng et al. 2014), thus preventing mTORC1 lysosomal localization in response to amino acids. Furthermore, p53 inhibits mTORC1 by increasing expression of genes that negatively regulate mTORC1, such as igf-bp3 (insulin-like growth-factor-binding protein 3), pten, and tsc2 (Fig. 2C) (reviewed in Budanov 2011). Thus, the interplay between AMPK, mTORC1, and p53 balances the growth-inhibiting response to cellular stress versus a commitment to cell growth.
Under hypoxic conditions, many organisms show a conserved transcriptional response mediated by the heterodimeric transcription factor hypoxia inducible factor 1 (HIF1) (Wang and Semenza 1993). HIF1, composed of α and β subunits, regulates a gene- expression program required for adaptation to decreasing concentrations of oxygen (hypoxia). HIF1α subunits have a very short half-life under normoxic conditions. However, under hypoxic conditions, the degradation of HIF1α is delayed (reviewed in Weidemann and Johnson 2008). Although mTORC1 up-regulates HIF1α protein synthesis (Hudson et al. 2002), depletion of AMPKα leads to elevated HIF1α levels in MEFs (Shackelford et al. 2009; Faubert et al. 2013). In addition, inhibition of mTORC1 signaling reduces HIF1α levels in AMPKα-null cells (Faubert et al. 2013). Besides the positive signals from mTORC1 to HIF1α, HIF1α shows a negative feedback loop on mTORC1 signaling by stabilizing and activating the TSC complex via the transcriptional activation of REDD1 (regulated in development and DNA damage responses 1) (Brugarolas et al. 2004; DeYoung et al. 2008). These findings show the intricate network between energy-sensing signals (AMPK) and growth-inducing signals (mTORC1) in regulating HIF1α in opposing ways.
In S. cerevisiae, TORC1 and Snf1 converge in the regulation of the two transcription factors Gln3 and Msn2. Gln3 mediates activation of nitrogen catabolite-repressible (NCR) genes. TORC1 and Snf1 control the phosphorylation status of Gln3 in response to nitrogen and glucose, respectively (Beck and Hall 1999; Bertram et al. 2002). Although TORC1 phosphorylation keeps Gln3 in the cytoplasm, Snf1-dependent phosphorylation correlates with increased accumulation of Gln3 in the nucleus. The opposing effects of the two different phosphorylation events suggest that TORC1- and Snf1-dependent phosphorylation occur at distinct sites (Bertram et al. 2002). However, these sites remain to be identified. The TORC1 and Snf1 pathways also regulate the subcellular localization of Msn2, a transcriptional activator of stress-response-element (STRE)-regulated genes. Active Snf1 kinase prevents nuclear localization of Msn2 in response to TORC1 inhibition by rapamycin. Thus, Snf1 and TORC1 may act at similar steps in the regulation of Msn2 (Mayordomo et al. 2002). Finally, TORC1 and Snf1 phosphorylate Hcm1 to inhibit and stimulate, respectively, nuclear translocation of this transcription factor (Rodriguez-Colman et al. 2013). The nuclear translocation of Hcm1 leads to a shift in metabolism from fermentation to respiration. Based on these findings in budding yeast, AMPK and TORC1 may also participate in the regulation of the subcellular localization of nutrient-related transcription factors in higher eukaryotes.
FUTURE DIRECTIONS
AMPK and mTOR are energy sensors and metabolic regulators that integrate multiple inputs to mediate cellular homeostasis. Cellular energy and nutrient status dictate whether mTORC1 signaling drives anabolic processes or, conversely, whether AMPK is activated to redirect cell metabolism toward catabolic processes, such as autophagy. An increasing number of metabolites and signaling intermediates are being shown to regulate AMPK and TOR signaling. Further understanding of the relationship between TOR and AMPK in coordinating amino-acid and energy-sensing pathways in mammals and other model organisms will provide new insights into the regulation of whole body energy homeostasis. The precise spatiotemporal coordination of TOR and AMPK in the cell remains unclear. The lysosomal surface appears to serve as a platform in which both mTORC1 and AMPK could interact in response to nutrient availability. Finally, the complex interplay of AMPK and mTOR in regulating cellular energy balance is rapidly gaining clinical relevance in diabetes and cancer. Novel therapies for metabolic disease may emerge from pharmacological or nutritional manipulation of this interplay.
ACKNOWLEDGMENTS
A.G. is the recipient of a Federation of European Biochemical Societies (FEBS) long-term Fellowship. We acknowledge support from the Louis Jeantet Foundation, the Swiss National Science Foundation, and the Canton of Basel.
Footnotes
Editors: Rebecca Heald, Iswar K. Hariharan, and David B. Wake
Additional Perspectives on Size Control in Biology: From Organelles to Organisms available at www.cshperspectives.org
REFERENCES
- An Z, Wang H, Song P, Zhang M, Geng X, Zou MH. 2007. Nicotine-induced activation of AMP-activated protein kinase inhibits fatty acid synthase in 3T3L1 adipocytes: A role for oxidant stress. J Biol Chem 282: 26793–26801. [DOI] [PubMed] [Google Scholar]
- Anisimov VN. 2010. Metformin for aging and cancer prevention. Aging 2: 760–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld J, O’Connor G, McDonagh T, DiStefano PS, Curtis R. 2004. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev 18: 3004–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K, Lin SS, Manchester JK, Gordon JI. 2000. Sip2p and its partner snf1p kinase affect aging in S. cerevisiae. Genes Dev 14: 1872–1885. [PMC free article] [PubMed] [Google Scholar]
- Aspuria PJ, Sato T, Tamanoi F. 2007. The TSC/Rheb/TOR signaling pathway in fission yeast and mammalian cells: Temperature sensitive and constitutive active mutants of TOR. Cell Cycle 6: 1692–1695. [DOI] [PubMed] [Google Scholar]
- Baena-Gonzalez E, Sheen J. 2008. Convergent energy and stress signaling. Trends Plant Sci 13: 474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes K, Ingram JC, Porras OH, Barros LF, Hudson ER, Fryer LG, Foufelle F, Carling D, Hardie DG, Baldwin SA. 2002. Activation of GLUT1 by metabolic and osmotic stress: Potential involvement of AMP-activated protein kinase (AMPK). J Cell Sci 115: 2433–2442. [DOI] [PubMed] [Google Scholar]
- Bar-Peled L, Sabatini DM. 2012. SnapShot: mTORC1 signaling at the lysosomal surface. Cell 151: 1390–1390 e1391. [DOI] [PubMed] [Google Scholar]
- Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. 2012. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 150: 1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. 2013. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340: 1100–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A. 1997. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem Sci 22: 12–13. [DOI] [PubMed] [Google Scholar]
- Beck T, Hall MN. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692. [DOI] [PubMed] [Google Scholar]
- Beg ZH, Brewer HB Jr., 1982. Modulation of rat liver 3-hydroxy-3-methylglutaryl-CoA reductase activity by reversible phosphorylation. Fed Proc 41: 2634–2638. [PubMed] [Google Scholar]
- Ben-Sahra I, Howell JJ, Asara JM, Manning BD. 2013. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 339: 1323–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berggreen C, Gormand A, Omar B, Degerman E, Goransson O. 2009. Protein kinase B activity is required for the effects of insulin on lipid metabolism in adipocytes. Am J Physiol Endocrinol Metab 296: E635–E646. [DOI] [PubMed] [Google Scholar]
- Bertram PG, Choi JH, Carvalho J, Chan TF, Ai W, Zheng XF. 2002. Convergence of TOR-nitrogen and Snf1-glucose signaling pathways onto Gln3. Mol Cell Biol 22: 1246–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz C, Hall MN. 2013. Where is mTOR and what is it doing there? J Cell Biol 203: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz C, Stracka D, Prescianotto-Baschong C, Frieden M, Demaurex N, Hall MN. 2013. Feature article: mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc Natl Acad Sci 110: 12526–12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao RP, Salchert K, Bako L, Okresz L, Szabados L, Muranaka T, Machida Y, Schell J, Koncz C. 1999. Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc Natl Acad Sci 96: 5322–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L. 2010. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab 11: 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. 1995. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem 270: 2320–2326. [DOI] [PubMed] [Google Scholar]
- Bolster DR, Crozier SJ, Kimball SR, Jefferson LS. 2002. AMP-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mTOR) signaling. J Biol Chem 277: 23977–23980. [DOI] [PubMed] [Google Scholar]
- Bozaquel-Morais BL, Madeira JB, Maya-Monteiro CM, Masuda CA, Montero-Lomeli M. 2010. A new fluorescence-based method identifies protein phosphatases regulating lipid droplet metabolism. PLoS ONE 5: e13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. 1994. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369: 756–758. [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG Jr. 2004. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev 18: 2893–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budanov AV. 2011. Stress-responsive sestrins link p53 with redox regulation and mammalian target of rapamycin signaling. Antioxid Redox Signal 15: 1679–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budanov AV, Karin M. 2008. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 134: 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese MF, Rajamohan F, Harris MS, Caspers NL, Magyar R, Withka JM, Wang H, Borzilleri KA, Sahasrabudhe PV, Hoth LR, et al. 2014. Structural basis for AMPK activation: Natural and synthetic ligands regulate kinase activity from opposite poles by different molecular mechanisms. Structure 22: 1161–1172. [DOI] [PubMed] [Google Scholar]
- Chantranupong L, Wolfson RL, Orozco JM, Saxton RA, Scaria SM, Bar-Peled L, Spooner E, Isasa M, Gygi SP, Sabatini DM. 2014. The sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep 9: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaveroux C, Eichner LJ, Dufour CR, Shatnawi A, Khoutorsky A, Bourque G, Sonenberg N, Giguere V. 2013. Molecular and genetic crosstalks between mTOR and ERRα are key determinants of rapamycin-induced nonalcoholic fatty liver. Cell Metab 17: 586–598. [DOI] [PubMed] [Google Scholar]
- Chiu MI, Katz H, Berlin V. 1994. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc Natl Acad Sci 91: 12574–12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark H, Carling D, Saggerson D. 2004. Covalent activation of heart AMP-activated protein kinase in response to physiological concentrations of long-chain fatty acids. Eur J Biochem 271: 2215–2224. [DOI] [PubMed] [Google Scholar]
- Cuervo AM. 2008. Autophagy and aging: Keeping that old broom working. Trends Genet 24: 604–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis R, O’Connor G, DiStefano PS. 2006. Aging networks in Caenorhabditis elegans: AMP-activated protein kinase (aak-2) links multiple aging and metabolism pathways. Aging Cell 5: 119–126. [DOI] [PubMed] [Google Scholar]
- Cybulski N, Hall MN. 2009. TOR complex 2: A signaling pathway of its own. Trends Biochem Sci 34: 620–627. [DOI] [PubMed] [Google Scholar]
- Dagon Y, Hur E, Zheng B, Wellenstein K, Cantley LC, Kahn BB. 2012. p70S6 kinase phosphorylates AMPK on serine 491 to mediate leptin’s effect on food intake. Cell Metab 16: 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan HC, Sun M, Yang L, Feldman RI, Sui XM, Ou CC, Nellist M, Yeung RS, Halley DJ, Nicosia SV, et al. 2002. Phosphatidylinositol 3-kinase/Akt pathway regulates tuberous sclerosis tumor suppressor complex by phosphorylation of tuberin. J Biol Chem 277: 35364–35370. [DOI] [PubMed] [Google Scholar]
- Davies SP, Helps NR, Cohen PT, Hardie DG. 1995. 5′-AMP inhibits dephosphorylation, as well as promoting phosphorylation, of the AMP-activated protein kinase. Studies using bacterially expressed human protein phosphatase-2C α and native bovine protein phosphatase-2AC. FEBS Lett 377: 421–425. [DOI] [PubMed] [Google Scholar]
- Demetriades C, Doumpas N, Teleman AA. 2014. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 156: 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. 2008. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev 22: 239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Troya S, Perez-Perez ME, Florencio FJ, Crespo JL. 2008. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy 4: 851–865. [DOI] [PubMed] [Google Scholar]
- Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, Finan PM, Kwiatkowski DJ, Murphy LO, Manning BD. 2012. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell 47: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como CJ, Arndt KT. 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev 10: 1904–1916. [DOI] [PubMed] [Google Scholar]
- Dunlop EA, Hunt DK, Acosta-Jaquez HA, Fingar DC, Tee AR. 2011. ULK1 inhibits mTORC1 signaling, promotes multisite Raptor phosphorylation and hinders substrate binding. Autophagy 7: 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran RV, Hall MN. 2012. Regulation of TOR by small GTPases. EMBO Rep 13: 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran RV, Oppliger W, Robitaille AM, Heiserich L, Skendaj R, Gottlieb E, Hall MN. 2012. Glutaminolysis activates Rag-mTORC1 signaling. Mol Cell 47: 349–358. [DOI] [PubMed] [Google Scholar]
- Duran RV, MacKenzie ED, Boulahbel H, Frezza C, Heiserich L, Tardito S, Bussolati O, Rocha S, Hall MN, Gottlieb E. 2013. HIF-independent role of prolyl hydroxylases in the cellular response to amino acids. Oncogene 32: 4549–4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. 2010. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 39: 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. 2011. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331: 456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch F, Treitel MA, Yang X, Carlson M. 1992. N-terminal mutations modulate yeast SNF1 protein kinase function. Genetics 132: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubert B, Boily G, Izreig S, Griss T, Samborska B, Dong Z, Dupuy F, Chambers C, Fuerth BJ, Viollet B, et al. 2013. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab 17: 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z. 2010. p53 regulation of the IGF-1/AKT/mTOR pathways and the endosomal compartment. Cold Spring Harb Perspect Biol 2: a001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foretz M, Guichard C, Ferre P, Foufelle F. 1999. Sterol regulatory element binding protein-1c is a major mediator of insulin action on the hepatic expression of glucokinase and lipogenesis-related genes. Proc Natl Acad Sci 96: 12737–12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble J, Lopaschuk GD. 1997. Insulin inhibition of 5′ adenosine monophosphate-activated protein kinase in the heart results in activation of acetyl coenzyme A carboxylase and inhibition of fatty acid oxidation. Metabolism 46: 1270–1274. [DOI] [PubMed] [Google Scholar]
- Gao X, Pan D. 2001. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev 15: 1383–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Zhang Y, Arrazola P, Hino O, Kobayashi T, Yeung RS, Ru B, Pan D. 2002. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat Cell Biol 4: 699–704. [DOI] [PubMed] [Google Scholar]
- Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G. 2003. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell 11: 1457–1466. [DOI] [PubMed] [Google Scholar]
- Ghillebert R, Swinnen E, Wen J, Vandesteene L, Ramon M, Norga K, Rolland F, Winderickx J. 2011. The AMPK/SNF1/SnRK1 fuel gauge and energy regulator: Structure, function and regulation. FEBS J 278: 3978–3990. [DOI] [PubMed] [Google Scholar]
- Goodfellow SJ, White RJ. 2007. Regulation of RNA polymerase III transcription during mammalian cell growth. Cell Cycle 6: 2323–2326. [DOI] [PubMed] [Google Scholar]
- Gowans GJ, Hawley SA, Ross FA, Hardie DG. 2013. AMP is a true physiological regulator of AMP-activated protein kinase by both allosteric activation and enhancing net phosphorylation. Cell Metab 18: 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30: 214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha J, Daniel S, Broyles SS, Kim KH. 1994. Critical phosphorylation sites for acetyl-CoA carboxylase activity. J Biol Chem 269: 22162–22168. [PubMed] [Google Scholar]
- Hahn-Windgassen A, Nogueira V, Chen CC, Skeen JE, Sonenberg N, Hay N. 2005. Akt activates the mammalian target of rapamycin by regulating cellular ATP level and AMPK activity. J Biol Chem 280: 32081–32089. [DOI] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. 2008. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet 4: e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. 1998. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem 273: 14484–14494. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441: 885–889. [DOI] [PubMed] [Google Scholar]
- Hardie DG. 2011. Cell biology. Why starving cells eat themselves. Science 331: 410–411. [DOI] [PubMed] [Google Scholar]
- Hardie DG. 2014. AMPK—Sensing energy while talking to other signaling pathways. Cell Metab 20: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. 2015. AMPK: Positive and negative regulation, and its role in whole-body energy homeostasis. Curr Opin Cell Biol 33: 1–7. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. 2009. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460: 392–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Davison M, Woods A, Davies SP, Beri RK, Carling D, Hardie DG. 1996. Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J Biol Chem 271: 27879–27887. [DOI] [PubMed] [Google Scholar]
- Hawley SA, Pan DA, Mustard KJ, Ross L, Bain J, Edelman AM, Frenguelli BG, Hardie DG. 2005. Calmodulin-dependent protein kinase kinase-β is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab 2: 9–19. [DOI] [PubMed] [Google Scholar]
- Hawley SA, Ross FA, Gowans GJ, Tibarewal P, Leslie NR, Hardie DG. 2014. Phosphorylation by Akt within the ST loop of AMPK-α1 down-regulates its activation in tumour cells. Biochem J 459: 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Zhang YW, Lee JH, Zeng SX, Wang YV, Luo Z, Dong XC, Viollet B, Wahl GM, Lu H. 2014. AMP-activated protein kinase Induces p53 by phosphorylating MDMX and inhibiting its activity. Mol Cell Biol 34: 148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedbacker K, Carlson M. 2008. SNF1/AMPK pathways in yeast. Front Biosci 13: 2408–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN. 1991. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253: 905–909. [DOI] [PubMed] [Google Scholar]
- Holmes BF, Kurth-Kraczek EJ, Winder WW. 1999. Chronic activation of 5′-AMP-activated protein kinase increases GLUT-4, hexokinase, and glycogen in muscle. J Appl Physiol 87: 1990–1995. [DOI] [PubMed] [Google Scholar]
- Hong SP, Carlson M. 2007. Regulation of snf1 protein kinase in response to environmental stress. J Biol Chem 282: 16838–16845. [DOI] [PubMed] [Google Scholar]
- Horman S, Vertommen D, Heath R, Neumann D, Mouton V, Woods A, Schlattner U, Wallimann T, Carling D, Hue L, et al. 2006. Insulin antagonizes ischemia-induced Thr172 phosphorylation of AMP-activated protein kinase α-subunits in heart via hierarchical phosphorylation of Ser485/491. J Biol Chem 281: 5335–5340. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. 2009. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 20: 1981–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, et al. 2003. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132: 666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Fingar DC. 2014. Growing knowledge of the mTOR signaling network. Semin Cell Dev Biol 36: 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Manning BD. 2008. The TSC1-TSC2 complex: A molecular switchboard controlling cell growth. Biochem J 412: 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson CC, Liu M, Chiang GG, Otterness DM, Loomis DC, Kaper F, Giaccia AJ, Abraham RT. 2002. Regulation of hypoxia-inducible factor 1α expression and function by the mammalian target of rapamycin. Mol Cell Biol 22: 7004–7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes Hallett JE, Luo X, Capaldi AP. 2014. State transitions in the TORC1 signaling pathway and information processing in Saccharomyces cerevisiae. Genetics 198: 773–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley RL, Anderson KA, Franzone JM, Kemp BE, Means AR, Witters LA. 2005. The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J Biol Chem 280: 29060–29066. [DOI] [PubMed] [Google Scholar]
- Iadevaia V, Zhang Z, Jan E, Proud CG. 2012. mTOR signaling regulates the processing of pre-rRNA in human cells. Nucleic Acids Res 40: 2527–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL. 2002. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 4: 648–657. [DOI] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL. 2003a. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 17: 1829–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL. 2003b. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115: 577–590. [DOI] [PubMed] [Google Scholar]
- Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, et al. 2006. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 126: 955–968. [DOI] [PubMed] [Google Scholar]
- Inoki K, Kim J, Guan KL. 2012. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu Rev Pharmacol Toxicol 52: 381–400. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Guo B, Arndt KT, Schmelzle T, Hall MN. 2001. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol Cell 8: 1017–1026. [DOI] [PubMed] [Google Scholar]
- Jewell JL, Russell RC, Guan KL. 2013. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol 14: 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K, Chen D, Riddle DL. 2004. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development 131: 3897–3906. [DOI] [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. 2005. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell 18: 283–293. [DOI] [PubMed] [Google Scholar]
- Jung CH, Seo M, Otto NM, Kim DH. 2011. ULK1 inhibits the kinase activity of mTORC1 and cell proliferation. Autophagy 7: 1212–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. 2005. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310: 1193–1196. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. 2010. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol 30: 1049–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantidakis T, Ramsbottom BA, Birch JL, Dowding SN, White RJ. 2010. mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc Natl Acad Sci 107: 11823–11828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. 2004. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol 14: 885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijanska M, Dohnal I, Reiter W, Kaspar S, Stoffel I, Ammerer G, Kraft C, Peter M. 2010. Activation of Atg1 kinase in autophagy by regulated phosphorylation. Autophagy 6: 1168–1178. [DOI] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. 2008. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 10: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura N, Tokunaga C, Dalal S, Richardson C, Yoshino K, Hara K, Kemp BE, Witters LA, Mimura O, Yonezawa K. 2003. A possible linkage between AMP-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) signalling pathway. Genes Cells 8: 65–79. [DOI] [PubMed] [Google Scholar]
- Kohda TA, Tanaka K, Konomi M, Sato M, Osumi M, Yamamoto M. 2007. Fission yeast autophagy induced by nitrogen starvation generates a nitrogen source that drives adaptation processes. Genes Cells 12: 155–170. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441: 880–884. [DOI] [PubMed] [Google Scholar]
- Kong M, Fox CJ, Mu J, Solt L, Xu A, Cinalli RM, Birnbaum MJ, Lindsten T, Thompson CB. 2004. The PP2A-associated protein α4 is an essential inhibitor of apoptosis. Science 306: 695–698. [DOI] [PubMed] [Google Scholar]
- Korta DZ, Tuck S, Hubbard EJ. 2012. S6K links cell fate, cell cycle and nutrient response in C. elegans germline stem/progenitor cells. Development 139: 859–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacic S, Soltys CL, Barr AJ, Shiojima I, Walsh K, Dyck JR. 2003. Akt activity negatively regulates phosphorylation of AMP-activated protein kinase in the heart. J Biol Chem 278: 39422–39427. [DOI] [PubMed] [Google Scholar]
- Kraft C, Kijanska M, Kalie E, Siergiejuk E, Lee SS, Semplicio G, Stoffel I, Brezovich A, Verma M, Hansmann I, et al. 2012. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J 31: 3691–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN. 1993. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73: 585–596. [DOI] [PubMed] [Google Scholar]
- Kurth-Kraczek EJ, Hirshman MF, Goodyear LJ, Winder WW. 1999. 5′ AMP-activated protein kinase activation causes GLUT4 translocation in skeletal muscle. Diabetes 48: 1667–1671. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. 2012. mTOR signaling in growth control and disease. Cell 149: 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Comer FI, Sasaki A, McLeod IX, Duong Y, Okumura K, Yates JR III, Parent CA, Firtel RA. 2005. TOR complex 2 integrates cell movement during chemotaxis and signal relay in Dictyostelium. Mol Biol Cell 16: 4572–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Koh H, Kim M, Kim Y, Lee SY, Karess RE, Lee SH, Shong M, Kim JM, Kim J, et al. 2007. Energy-dependent regulation of cell structure by AMP-activated protein kinase. Nature 447: 1017–1020. [DOI] [PubMed] [Google Scholar]
- Lee JH, Budanov AV, Park EJ, Birse R, Kim TE, Perkins GA, Ocorr K, Ellisman MH, Bodmer R, Bier E, et al. 2010. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science 327: 1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. 2011. Autophagy in immunity and inflammation. Nature 469: 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xu S, Mihaylova MM, Zheng B, Hou X, Jiang B, Park O, Luo Z, Lefai E, Shyy JY, et al. 2011. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab 13: 376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippai M, Csikos G, Maroy P, Lukacsovich T, Juhasz G, Sass M. 2008. SNF4Aγ, the Drosophila AMPK γ subunit is required for regulation of developmental and stress-induced autophagy. Autophagy 4: 476–486. [DOI] [PubMed] [Google Scholar]
- Liu Y, Bassham DC. 2010. TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS ONE 5: e11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljubicic V, Hood DA. 2009. Diminished contraction-induced intracellular signaling towards mitochondrial biogenesis in aged skeletal muscle. Aging Cell 8: 394–404. [DOI] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 10: 457–468. [DOI] [PubMed] [Google Scholar]
- Loffler AS, Alers S, Dieterle AM, Keppeler H, Franz-Wachtel M, Kundu M, Campbell DG, Wesselborg S, Alessi DR, Stork B. 2011. Ulk1-mediated phosphorylation of AMPK constitutes a negative regulatory feedback loop. Autophagy 7: 696–706. [DOI] [PubMed] [Google Scholar]
- Long X, Spycher C, Han ZS, Rose AM, Muller F, Avruch J. 2002. TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Curr Biol 12: 1448–1461. [DOI] [PubMed] [Google Scholar]
- Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. 2005. Rheb binds and regulates the mTOR kinase. Curr Biol 15: 702–713. [DOI] [PubMed] [Google Scholar]
- Lu JY, Lin YY, Sheu JC, Wu JT, Lee FJ, Chen Y, Lin MI, Chiang FT, Tai TY, Berger SL, et al. 2011. Acetylation of yeast AMPK controls intrinsic aging independently of caloric restriction. Cell 146: 969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J. 2009. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10: 307–318. [DOI] [PubMed] [Google Scholar]
- Mackintosh RW, Davies SP, Clarke PR, Weekes J, Gillespie JG, Gibb BJ, Hardie DG. 1992. Evidence for a protein kinase cascade in higher plants. 3-Hydroxy-3-methylglutaryl-CoA reductase kinase. Eur J Biochem 209: 923–931. [DOI] [PubMed] [Google Scholar]
- Mahfouz MM, Kim S, Delauney AJ, Verma DP. 2006. Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 18: 477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. 2002. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell 10: 151–162. [DOI] [PubMed] [Google Scholar]
- Martin-Montalvo A, Mercken EM, Mitchell SJ, Palacios HH, Mote PL, Scheibye-Knudsen M, Gomes AP, Ward TM, Minor RK, Blouin MJ, et al. 2013. Metformin improves healthspan and lifespan in mice. Nat Commun 4: 2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Grummt I. 2006. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 25: 6384–6391. [DOI] [PubMed] [Google Scholar]
- Mayordomo I, Estruch F, Sanz P. 2002. Convergence of the target of rapamycin and the Snf1 protein kinase pathways in the regulation of the subcellular localization of Msn2, a transcriptional activator of STRE (stress response element)-regulated genes. J Biol Chem 277: 35650–35656. [DOI] [PubMed] [Google Scholar]
- Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD. 2014. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156: 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK, Mizushima NN, Iwasaki A, He YW, Swat W, et al. 2008. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy 4: 309–314. [DOI] [PubMed] [Google Scholar]
- Miller RA, Harrison DE, Astle CM, Baur JA, Boyd AR, de Cabo R, Fernandez E, Flurkey K, Javors MA, Nelson JF, et al. 2011. Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci 66: 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchelhill KI, Stapleton D, Gao G, House C, Michell B, Katsis F, Witters LA, Kemp BE. 1994. Mammalian AMP-activated protein kinase shares structural and functional homology with the catalytic domain of yeast Snf1 protein kinase. J Biol Chem 269: 2361–2364. [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. 2008. Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momcilovic M, Hong SP, Carlson M. 2006. Mammalian TAK1 activates Snf1 protein kinase in yeast and phosphorylates AMP-activated protein kinase in vitro. J Biol Chem 281: 25336–25343. [DOI] [PubMed] [Google Scholar]
- Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, Sadighi-Akha E, Stranks AJ, Glanville J, Knight S, Jacobsen SE, et al. 2011. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med 208: 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munday MR, Campbell DG, Carling D, Hardie DG. 1988. Identification by amino acid sequencing of three major regulatory phosphorylation sites on rat acetyl-CoA carboxylase. Eur J Biochem 175: 331–338. [DOI] [PubMed] [Google Scholar]
- Muoio DM, Seefeld K, Witters LA, Coleman RA. 1999. AMP-activated kinase reciprocally regulates triacylglycerol synthesis and fatty acid oxidation in liver and muscle: Evidence that sn-glycerol-3-phosphate acyltransferase is a novel target. Biochem J 338: 783–791. [PMC free article] [PubMed] [Google Scholar]
- Ning J, Xi G, Clemmons DR. 2011. Suppression of AMPK activation via S485 phosphorylation by IGF-I during hyperglycemia is mediated by AKT activation in vascular smooth muscle cells. Endocrinology 152: 3143–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura W, Maeta K, Kita K, Izawa S, Inoue Y. 2010. Methylglyoxal activates Gcn2 to phosphorylate eIF2α independently of the TOR pathway in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 86: 1887–1894. [DOI] [PubMed] [Google Scholar]
- Oakhill JS, Steel R, Chen ZP, Scott JW, Ling N, Tam S, Kemp BE. 2011. AMPK is a direct adenylate charge-regulated protein kinase. Science 332: 1433–1435. [DOI] [PubMed] [Google Scholar]
- Oh WJ, Jacinto E. 2011. mTOR complex 2 signaling and functions. Cell Cycle 10: 2305–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S, Montagne J, Radimerski T, Thomas G, Hafen E. 2000. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev 14: 2689–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova M, Kanter E, Krakovich D, Kuchin S. 2006. Nitrogen availability and TOR regulate the Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot Cell 5: 1831–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. 2003. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J Biol Chem 278: 17636–17645. [DOI] [PubMed] [Google Scholar]
- Pan DA, Hardie DG. 2002. A homologue of AMP-activated protein kinase in Drosophila melanogaster is sensitive to AMP and is activated by ATP depletion. Biochem J 367: 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, Kapahi P. 2007. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell 6: 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchaud N, Peli-Gulli MP, De Virgilio C. 2013. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci Signal 6: ra42. [DOI] [PubMed] [Google Scholar]
- Parmigiani A, Nourbakhsh A, Ding B, Wang W, Kim YC, Akopiants K, Guan K-L, Karin M, Budanov AV. 2014. Sestrins inhibit mTORC1 kinase activation through the GATOR complex. Cell Rep 9: 1281–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Levine B. 2006. Bcl-2 inhibition of autophagy: A new route to cancer? Cancer Res 66: 2885–2888. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. 2005. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122: 927–939. [DOI] [PubMed] [Google Scholar]
- Peng M, Yin N, Li MO. 2014. Sestrins function as guanine nucleotide dissociation inhibitors for Rag GTPases to control mTORC1 signaling. Cell 159: 122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, et al. 2011. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146: 408–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit CS, Roczniak-Ferguson A, Ferguson SM. 2013. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol 202: 1107–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polge C, Thomas M. 2007. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci 12: 20–28. [DOI] [PubMed] [Google Scholar]
- Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. 2008. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab 8: 224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke A, Anderson S, McCaffery JM, Yates J 3rd, Aronova S, Chu S, Fairclough S, Iverson C, Wedaman KP, Powers T. 2004. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J Biol Chem 279: 14752–14762. [DOI] [PubMed] [Google Scholar]
- Ricoult SJ, Manning BD. 2013. The multifaceted role of mTORC1 in the control of lipid metabolism. EMBO Rep 14: 242–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN. 2013. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science 339: 1320–1323. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Colman MJ, Sorolla MA, Vall-Llaura N, Tamarit J, Ros J, Cabiscol E. 2013. The FOX transcription factor Hcm1 regulates oxidative metabolism in response to early nutrient limitation in yeast. Role of Snf1 and Tor1/Sch9 kinases. Biochim Biophys Acta 1833: 2004–2015. [DOI] [PubMed] [Google Scholar]
- Rohde JR, Campbell S, Zurita-Martinez SA, Cutler NS, Ashe M, Cardenas ME. 2004. TOR controls transcriptional and translational programs via Sap-Sit4 protein phosphatase signaling effectors. Mol Cell Biol 24: 8332–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Xu X, Carlson M. 2011. Roles of two protein phosphatases, Reg1-Glc7 and Sit4, and glycogen synthesis in regulation of SNF1 protein kinase. Proc Natl Acad Sci 108: 6349–6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. 1994. RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78: 35–43. [DOI] [PubMed] [Google Scholar]
- Sabers CJ, Martin MM, Brunn GJ, Williams JM, Dumont FJ, Wiederrecht G, Abraham RT. 1995. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem 270: 815–822. [DOI] [PubMed] [Google Scholar]
- Saito K, Araki Y, Kontani K, Nishina H, Katada T. 2005. Novel role of the small GTPase Rheb: Its implication in endocytic pathway independent of the activation of mammalian target of rapamycin. J Biochem 137: 423–430. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Zarrinpashneh E, Budas GR, Pouleur AC, Dutta A, Prescott AR, Vanoverschelde JL, Ashworth A, Jovanovic A, Alessi DR, et al. 2006. Deficiency of LKB1 in heart prevents ischemia-mediated activation of AMPKα2 but not AMPKα1. Am J Physiol Endocrinol Metab 290: E780–E788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. 2007. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell 25: 903–915. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. 2003. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol 5: 566–571. [DOI] [PubMed] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. 2007. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab 6: 280–293. [DOI] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP. 2004. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 7: 167–178. [DOI] [PubMed] [Google Scholar]
- Scott RC, Juhasz G, Neufeld TP. 2007. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol 17: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JW, Ling N, Issa SM, Dite TA, O’Brien MT, Chen ZP, Galic S, Langendorf CG, Steinberg GR, Kemp BE, et al. 2014. Small molecule drug A-769662 and AMP synergistically activate naïve AMPK independent of upstream kinase signaling. Chem Biol 21: 619–627. [DOI] [PubMed] [Google Scholar]
- Serfontein J, Nisbet RE, Howe CJ, de Vries PJ. 2011. Conservation of structural and functional elements of TSC1 and TSC2: A bioinformatic comparison across animal models. Behav Genet 41: 349–356. [DOI] [PubMed] [Google Scholar]
- Shackelford DB, Vasquez DS, Corbeil J, Wu S, Leblanc M, Wu CL, Vera DR, Shaw RJ. 2009. mTOR and HIF-1α-mediated tumor metabolism in an LKB1 mouse model of Peutz–Jeghers syndrome. Proc Natl Acad Sci 106: 11137–11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Yahagi N, Amemiya-Kudo M, Hasty AH, Osuga J, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, Harada K, et al. 1999. Sterol regulatory element-binding protein-1 as a key transcription factor for nutritional induction of lipogenic enzyme genes. J Biol Chem 274: 35832–35839. [DOI] [PubMed] [Google Scholar]
- Shimobayashi M, Hall MN. 2014. Making new contacts: The mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol 15: 155–162. [DOI] [PubMed] [Google Scholar]
- Shimomura I, Bashmakov Y, Ikemoto S, Horton JD, Brown MS, Goldstein JL. 1999. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc Natl Acad Sci 96: 13656–13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spasic MR, Callaerts P, Norga KK. 2008. Drosophila alicorn is a neuronal maintenance factor protecting against activity-induced retinal degeneration. J Neurosci 28: 6419–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, Breuer S, Thomas G, Hafen E. 2003. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol 5: 559–565. [DOI] [PubMed] [Google Scholar]
- Sugden C, Crawford RM, Halford NG, Hardie DG. 1999. Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5′-AMP. Plant J 19: 433–439. [DOI] [PubMed] [Google Scholar]
- Suttangkakul A, Li F, Chung T, Vierstra RD. 2011. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara T, Maeda T. 2012. TORC1 of fission yeast is rapamycin-sensitive. Genes Cells 17: 698–708. [DOI] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. 2003. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol 13: 1259–1268. [DOI] [PubMed] [Google Scholar]
- Tekinay T, Wu MY, Otto GP, Anderson OR, Kessin RH. 2006. Function of the Dictyostelium discoideum Atg1 kinase during autophagy and development. Eukaryot Cell 5: 1797–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelander M, Olsson T, Ronne H. 2004. Snf1-related protein kinase 1 is needed for growth in a normal day–night light cycle. EMBO J 23: 1900–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschape JA, Hammerschmied C, Muhlig-Versen M, Athenstaedt K, Daum G, Kretzschmar D. 2002. The neurodegeneration mutant lochrig interferes with cholesterol homeostasis and Appl processing. EMBO J 21: 6367–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. 2013. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell 52: 495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano J, Tabancay AP, Yang W, Tamanoi F. 2000. The Saccharomyces cerevisiae Rheb G-protein is involved in regulating canavanine resistance and arginine uptake. J Biol Chem 275: 11198–11206. [DOI] [PubMed] [Google Scholar]
- Usaite R, Jewett MC, Oliveira AP, Yates JR 3rd, Olsson L, Nielsen J. 2009. Reconstruction of the yeast Snf1 kinase regulatory network reveals its role as a global energy regulator. Mol Syst Biol 5: 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valbuena N, Moreno S. 2012. AMPK phosphorylation by Ssp1 is required for proper sexual differentiation in fission yeast. J Cell Sci 125: 2655–2664. [DOI] [PubMed] [Google Scholar]
- Valentine RJ, Coughlan KA, Ruderman NB, Saha AK. 2014. Insulin inhibits AMPK activity and phosphorylates AMPK Ser(485/491) through Akt in hepatocytes, myotubes and incubated rat skeletal muscle. Arch Biochem Biophys 562: 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam TJ, Zwartkruis FJ, Bos JL, Snel B. 2011. Evolution of the TOR pathway. J Mol Evol 73: 209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. 2003. Genetics: Influence of TOR kinase on lifespan in C. elegans. Nature 426: 620. [DOI] [PubMed] [Google Scholar]
- Vernoud V, Horton AC, Yang Z, Nielsen E. 2003. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol 131: 1191–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Semenza GL. 1993. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci 90: 4304–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wilson WA, Fujino MA, Roach PJ. 2001. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol Cell Biol 21: 5742–5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidemann A, Johnson RS. 2008. Biology of HIF-1α. Cell Death Differ 15: 621–627. [DOI] [PubMed] [Google Scholar]
- Witters LA, Kemp BE. 1992. Insulin activation of acetyl-CoA carboxylase accompanied by inhibition of the 5′-AMP-activated protein kinase. J Biol Chem 267: 2864–2867. [PubMed] [Google Scholar]
- Woods A, Munday MR, Scott J, Yang X, Carlson M, Carling D. 1994. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J Biol Chem 269: 19509–19515. [PubMed] [Google Scholar]
- Woods A, Dickerson K, Heath R, Hong SP, Momcilovic M, Johnstone SR, Carlson M, Carling D. 2005. Ca2+/calmodulin-dependent protein kinase kinase-β acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab 2: 21–33. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. 2006. TOR signaling in growth and metabolism. Cell 124: 471–484. [DOI] [PubMed] [Google Scholar]
- Xiao B, Sanders MJ, Carmena D, Bright NJ, Haire LF, Underwood E, Patel BR, Heath RB, Walker PA, Hallen S, et al. 2013. Structural basis of AMPK regulation by small molecule activators. Nat Commun 4: 3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh LA, Lee KH, Kim KH. 1980. Regulation of rat liver acetyl-CoA carboxylase. Regulation of phosphorylation and inactivation of acetyl-CoA carboxylase by the adenylate energy charge. J Biol Chem 255: 2308–2314. [PubMed] [Google Scholar]
- Yeh YY, Wrasman K, Herman PK. 2010. Autophosphorylation within the Atg1 activation loop is required for both kinase activity and the induction of autophagy in Saccharomyces cerevisiae. Genetics 185: 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Fagotto F, Zhang T, Hsu W, Vasicek TJ, Perry WL 3rd, Lee JJ, Tilghman SM, Gumbiner BM, Costantini F. 1997. The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell 90: 181–192. [DOI] [PubMed] [Google Scholar]
- Zhang C, Cuervo AM. 2008. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med 14: 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP. 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev 14: 2712–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. 2003. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol 5: 578–581. [DOI] [PubMed] [Google Scholar]
- Zhang J, Vaga S, Chumnanpuen P, Kumar R, Vemuri GN, Aebersold R, Nielsen J. 2011. Mapping the interaction of Snf1 with TORC1 in Saccharomyces cerevisiae. Mol Syst Biol 7: 545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YL, Guo H, Zhang CS, Lin SY, Yin Z, Peng Y, Luo H, Shi Y, Lian G, Zhang C, et al. 2013. AMP as a low-energy charge signal autonomously initiates assembly of AXIN-AMPK-LKB1 complex for AMPK activation. Cell Metab 18: 546–555. [DOI] [PubMed] [Google Scholar]
- Zhang CS, Jiang B, Li M, Zhu M, Peng Y, Zhang YL, Wu YQ, Li TY, Liang Y, Lu Z, et al. 2014. The lysosomal v-ATPase-ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab 20: 526–540. [DOI] [PubMed] [Google Scholar]
- Zinzalla V, Stracka D, Oppliger W, Hall MN. 2011. Activation of mTORC2 by association with the ribosome. Cell 144: 757–768. [DOI] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. 2011. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science 334: 678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]