Abstract
The global significance of Campylobacter jejuni and Campylobacter coli as gastrointestinal human pathogens has motivated numerous studies to characterize their population biology and evolution. These bacteria are a common component of the intestinal microbiota of numerous bird and mammal species and cause disease in humans, typically via consumption of contaminated meat products, especially poultry meat. Sequence-based molecular typing methods, such as multilocus sequence typing (MLST) and whole genome sequencing (WGS), have been instructive for understanding the epidemiology and evolution of these bacteria and how phenotypic variation relates to the high degree of genetic structuring in C. coli and C. jejuni populations. Here, we describe aspects of the relatively short history of coevolution between humans and pathogenic Campylobacter, by reviewing research investigating how mutation and lateral or horizontal gene transfer (LGT or HGT, respectively) interact to create the observed population structure. These genetic changes occur in a complex fitness landscape with divergent ecologies, including multiple host species, which can lead to rapid adaptation, for example, through frame-shift mutations that alter gene expression or the acquisition of novel genetic elements by HGT. Recombination is a particularly strong evolutionary force in Campylobacter, leading to the emergence of new lineages and even large-scale genome-wide interspecies introgression between C. jejuni and C. coli. The increasing availability of large genome datasets is enhancing understanding of Campylobacter evolution through the application of methods, such as genome-wide association studies, but MLST-derived clonal complex designations remain a useful method for describing population structure.
Pathogenic Campylobacters in meat products can cause gastroenteritis in humans. Features of their genomic sequences (e.g., mutations and gene transfer events) are shedding light on their epidemiology and evolution.
Campylobacter jejuni and Campylobacter coli remain among the most common causes of human bacterial gastroenteritis worldwide (Friedman et al. 2000). In high-income countries, Campylobacteriosis is much more common than gastroenteritis caused by Escherichia coli, Listeria, and Salmonella, and accounts for an estimated 2.5 million annual cases of gastrointestinal disease in the United States alone (Kessel et al. 2001). Infection with these bacteria is also a major cause of morbidity and mortality in low- and middle-income countries, although it is almost certainly underreported in these settings, especially as culture confirmation remains challenging. Poor understanding of the transmission of these food-borne pathogens to humans in all income settings has contributed to the failure of public health systems to adequately address this problem. As a consequence, over the past 20 years, much investment has been directed at understanding how these bacteria are transmitted from reservoir hosts to humans through the food chain.
Although the disease was first recognized by Theodor Escherich in 1886, who described the symptoms of intestinal Campylobacter infections in children as “cholera infantum” (Samie et al. 2007) or “summer complaint” (Condran and Murphy 2008), difficulties in the culture and characterization of these organisms precluded their recognition as major causes of disease until the 1970s. Campylobacteriosis is usually nonfatal and self-limiting; however, the symptoms of diarrhea, fever, abdominal pain, and nausea can be severe (Allos 2001), and sequelae, including Guillain–Barre syndrome and reactive arthritis, can have serious long-term consequences. Subsequently, recognition of the very high disease burden of human Campylobacter infection stimulated research on these bacteria and their relatives. Since the 1970s, C. coli and C. jejuni have been isolated from a wide range of wild and domesticated bird and mammal species, in which, typically, they are thought to cause few if any disease symptoms. Humans are usually infected by the consumption of contaminated food (especially poultry meat), water, milk, or contact with animals or animal feces (Niemann et al. 2003).
Most of what is known about these species comes from isolates obtained from humans with disease, the food chain, and the agricultural environment. It is, however, important to note that such isolates are by no means representative of natural Campylobacter populations, and it is becoming increasingly apparent that much of the diversity present among the Campylobacters is in strains that colonize wild animals. Increasing numbers of novel genotypes are being found as Campylobacter populations are analyzed in different animal species, especially wild birds (Carter et al. 2009; French et al. 2009); these populations undoubtedly contain many as-yet-undescribed lineages. Most human disease isolates from cases of gastroenteritis in countries, such as the United Kingdom and the United States, are C. jejuni, which typically accounts for 90% of cases in these settings, with the remaining ∼10% of cases mostly caused by C. coli. The majority of the genotypes isolated from human disease have also been isolated as commensal gastrointestinal inhabitants of domesticated and, especially, food animals. Furthermore, clinical isolates are a nonrandom subset of these strains. Asymptomatic carriage of C. jejuni and C. coli is thought to be rare in humans, especially among people in industrialized countries, suggesting that humans are not a primary host for these organisms in these settings and that people are sporadically, and frequently pathologically, infected via the food chain from animal reservoir hosts.
An understanding of the relatively short history of coevolution between humans and pathogenic Campylobacters can be obtained by examining their population structure and ecology. This approach has formed the basis of many recent investigations of the cryptic epidemiology of these organisms (Lang et al. 2010; Müllner et al. 2010; Thakur et al. 2010; Hastings et al. 2011; Jorgensen et al. 2011; Kittl et al. 2011; Magnússon et al. 2011; Sheppard et al. 2011a,b; Sproston et al. 2011; Read et al. 2013) and will be the focus of this review. Such studies have included molecular epidemiological and evolutionary analyses and, in the past 15 years or so, the application of high-throughput DNA sequencing technologies of increasing capacity has enhanced the integration of these two areas of investigation to their mutual benefit.
THE IMPACT OF SEQUENCE-BASED MOLECULAR TYPING
Many of the most important advances in understanding the epidemiology of Campylobacter infection were driven by the development of molecular typing methods, especially approaches that (1) used nucleotide sequence data, and (2) explicitly exploited evolutionary and population genetics paradigms. The first molecular methods to be used extensively, such as pulse field gel electrophoresis (PFGE) fingerprinting and serotyping, showed that Campylobacter in clinical samples were highly diverse comprising the two common species with numerous subtypes within them (Frost et al. 1998; Ribot et al. 2001). However, the most influential advances in understanding the evolutionary forces that shape this complex population arose from the development and application of multilocus sequence typing (MLST) (Maiden et al. 1998), which enabled evolutionary and population genetic analyses to be applied to isolate characterization data.
MLST is a genotyping method that indexes variation at several chromosomal loci-encoding housekeeping genes, that is, essential genes found in all isolates that are under stabilizing selection for the conservation of metabolic function of their products. In most MLST schemes, seven gene fragments of ∼400 bp are used, reflecting the technology available in the late 1990s when the technique was developed. Intriguingly, even this very limited amount of data (corresponding to ∼0.2% of the C. jejuni genome) has permitted extensive insights into the population biology and evolution of many bacteria (Maiden 2006). In MLST, each unique sequence at each locus is assigned an arbitrary allele number for cataloging and identification purposes and the unique combinations of these allelic variants found in isolates (allelic profiles or “sequence types,” [STs]) are also assigned unique arbitrary numbers. The system, thus, catalogs variation of ∼3300 bp with a single number from which the precise sequence of each MLST allele can be recovered by reference to a lookup table; for ease of use and portability, these are curated and accessible via a web-accessible database (Maiden et al. 1998).
Over the last decade, the “Sanger sequencing” chain-termination methods that were initially used for MLST have been increasingly replaced by highly parallel next-generation sequencing (NGS) techniques. This has provided numerous novel opportunities for studying Campylobacter (Farhat et al. 2014; Méric et al. 2014; Van Tonder et al. 2014). However, as correctly determined sequence data are absolute, the insights obtained from MLST-based analyses remain relevant and MLST data are “forward compatible” with NGS data (Jolley and Maiden 2014). For example, a major early finding was that related STs are grouped into “clonal complexes” that can be pragmatically defined as groups of isolates with STs that share identical alleles at four or more MLST loci with a definable “central genotype” (Dingle et al. 2008; Colles and Maiden 2012). The use of the clonal complex as a primary unit of analysis has been of great value in a wide range of studies of Campylobacter, and whole genome sequence (WGS) analyses have confirmed that the members of clonal complexes correspond to bacterial lineages, the members of which share a common ancestor and, therefore, frequently also share phenotypic properties.
FORCES OF GENETIC DIVERSIFICATION
As with many other bacterial pathogens, the clustering of Campylobacter isolates into “types,” however defined, reveals a complex population structure, which is a consequence of the evolutionary forces that have acted on the bacterial population over time. Bacterial evolution is dominated by the relative rates of (1) replication errors or damage, which generate point mutations, rearrangements, or deletions of various sizes, and (2) lateral or horizontal gene transfer (LGT or HGT, respectively), the acquisition of genetic variation from an external source, which is incorporated into the chromosome by recombination. Whereas the accumulation of mutations leads to the gradual, progressive divergence of clones from the ancestral genotype one polymorphism at a time, HGT can introduce large DNA stretches with multiple accumulated polymorphisms in a single genetic event. This can occur by replacement of homologous DNA with a sequence from another lineage, by the introduction of new genes into the chromosome by recombination in repetitive DNA regions, or by illegitimate recombination. The relative impact of these two processes is crucial in defining the population structure of a given bacterium, and is especially evident in the Campylobacter (Wilson et al. 2009).
A straightforward means of estimating of the relative importance of mutation and HGT in generating diversity from MLST data is the comparison of the number of different alleles at each locus for a given number of STs. For example, if all the new alleles were generated by the progressive accumulations of mutation, each new ST would contain a new allele at least one locus. Inspection of the total number of number of alleles (3225, with between 350 and 700 alleles per MLST locus) with the numbers of STs (6942) for the 25,869 Campylobacter isolates recorded in the PubMLST.org/campylobacter database on January 1, 2014 showed that there was an underrepresentation of alleles compared with the expectation under a clonal model of divergence. This straightforward observation showed that the large number of STs (genotypes) is mostly generated by the reassortment of existing alleles by continuous HGT and not by progressive mutation.
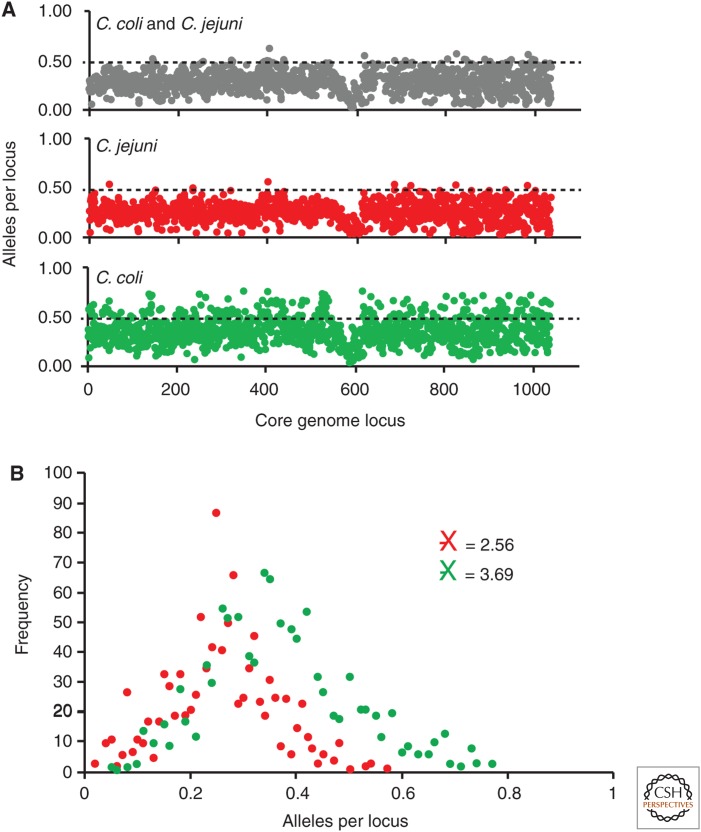
Although this provides strong evidence for the importance of HGT in shaping C. jejuni and C. coli populations, genetic exchange among lineages has not been sufficient to expunge all signals of common ancestry from Campylobacter. Therefore, in common with many bacteria, populations of these organisms contain a clonal frame that is disrupted to varying degrees by HGT. However, as the housekeeping gene fragments used for MLST are a very small fraction (0.2%) of the total genome, they are not necessarily representative of the entire genome; indeed, they were originally chosen as genes that were conserved across C. jejuni and C. coli to ensure that they were useful typing loci for the identification of lineages. In fact, genome sequence data shows that allelic diversity varies appreciably across the genome from loci that display little variation to those that are highly divergent (Fig. 1). Understanding how the effects of HGT and mutation interact in a complex fitness landscape to create the observed population genetic structure has proved instructive for both the evolutionary biology and pathogenicity of these bacteria.
Figure 1.
Allelic diversity in Campylobacter jejuni and Campylobacter coli genomes. (A) The average number of alleles per locus at 1035-core gene loci present 194 C. jejuni and C. coli isolates. (B) The distribution of allelic diversity showing that C. coli genes within the same isolate collection are, on average, more diverse, reflecting the deep branching 3-clade structure within this species.
RAPID ADAPTATION: PHASE VARIATION AND CONCERTED EVOLUTION
Adaptation of a bacterial pathogen or commensal to the host environment occurs, not only by the selection of certain genetic polymorphisms, but also through changes in gene expression. For example, studies in which C. jejuni isolates have been passaged through an animal host have shown increased rates of colonization and virulence in the passaged isolates (Coward et al. 2008). This rapid adaptation mechanism has been associated with enhanced host-interaction phenotypes, such as flagellar motility (Jerome and Mansfield 2014), and is linked to mechanisms that include large-scale genomic rearrangements and, in particular, mutations in homopolymeric tracts of DNA sequence. When it occurs within reading frames of genes, such instability in repetitive DNA sequence leads to frequent, reversible switches in expression of the associated genes. This process, phase variation, is observed in many bacteria and has been widely reported in C. jejuni in vitro and in vivo, providing these bacteria with rapid access to numerous phenotypes associated with host adaptation (Bayliss et al. 2012). Phase variable gene expression caused by on/off mutations in poly C/G tracts influences hundreds of genes in C. jejuni, but the details of the factors responsible for generating genetic diversity during host colonization and how this influences population structure remain incompletely understood (Bayliss et al. 2012).
POPULATION STRUCTURE IN C. jejuni AND C. coli
Although the epidemiology of C. jejuni and C. coli infections are superficially similar, the population genetic structure of these two species is conspicuously different. Although showing the evidence of high rates of HGT described above and, therefore, having a fundamentally nonclonal population structure, C. jejuni populations are nevertheless highly structured into clusters of related isolates, the MLST clonal complexes (Fig. 2A,B). From the data recoded in the PubMLST/campylobacter database on January 1, 2014, ∼80% of isolates belonged to one of the 44 clonal complexes that had been described for C. jejuni at that time. Extensive recombination within C. jejuni is shown by the presence of alleles with identical sequences in multiple, disparate (i.e., otherwise unrelated) lineages (Wimalarathna and Sheppard 2012). In contrast to this, C. coli have only two MLST-defined clonal complexes. The ST-828 clonal complex accounts for ∼70% of genotyped isolates submitted to PubMLST/campylobacter, with most of the other isolates sharing alleles with these and, therefore, being related. The second most-common clonal C. coli complex, ST-1150 complex, accounts for only 2% of isolates identified to date (Sheppard et al. 2010a); however, although these two clonal complexes dominate clinical and farm isolates, analysis of isolates from a broader ranges of sources show that they are both part of only one of three deep-branching clades within C. coli (Fig. 2A).
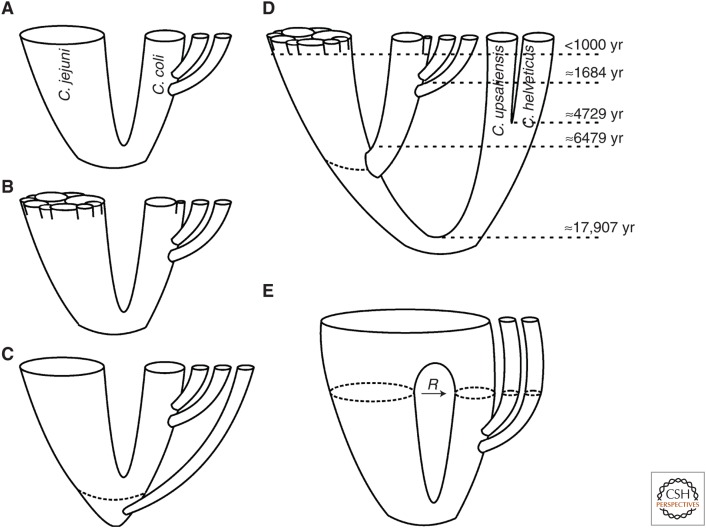
Figure 2.
Population structure and evolution of Campylobacter jejuni and Campylobacter coli. The tube diagrams illustrate (A) the two main human disease-associated species, (B) clonal complex and clade structure in C. jejuni and C. coli, (C) an example of New Zealand lineage branching from the common ancestor of C. jejuni and C. coli, (D) the timescale of evolution based on Sheppard et al. (2010a), and (E) a despeciation evolutionary scenario, resulting from high recombination (R) among the species. C. upsaliensis, Campylobacter upsaliensis; C. helveticus, Campylobacter helveticus.
The maintenance of the 3-clade C. coli population structure suggests that there are distinct gene pools with HGT occurring among members of the same clade, but being much rarer among members of different clades. Niche separation may provide an explanation for this limited gene flow with clade 1 organisms that dominate clinical and farm animal samples ecologically separated from clade 2 and 3 organisms, which are more abundant in water fowl and the riparian environment (Sheppard et al. 2011b). In addition to this potential ecological barrier, there are several genetic characteristics that distinguish clade 1 from the other two C. coli clades. First, there is relatively low synonymous sequence variation at MLST loci in C. coli clade 1, with a mean Ds of 0.006 per nucleotide compared with C. jejuni (0.016) and C. coli clades 2 (0.008) and 3 (0.013). This is consistent with a relatively recent genetic bottleneck in the evolution of clade 1. Second, the paucity of deep genetic structure within this clade, which predominantly comprises the ST-828 clonal complex, suggests relatively recent independent evolution. Third, the same allele is frequently found at individual MLST loci, suggesting recent clonal descent and high levels of genetic exchange within C. coli clade 1, estimated to be 5–10 times greater than in clades 2 and 3 (Sheppard et al. 2010a).
THE ECOLOGICAL BASIS OF POPULATION STRUCTURE
For both C. jejuni and C. coli, isolates from particular genotype clusters are often associated with given isolation sources, especially particular host animals. For example, although C. jejuni and C. coli are both found in chicken and cattle at a ratio of ∼9:1, this ratio is reversed in pigs, in which most of the isolates are C. coli (Miller et al. 2006). In the same way that species occupy different hosts, the genetic substructure within species can also be related to ecology. Within C. coli, the majority of isolates from clinical samples and agricultural animals belong to clade 1 with clades 2 and 3 C. coli more commonly associated with waterfowl and environmental sources that have, presumably, been contaminated by them (Sheppard et al. 2010a; Colles et al. 2011). There is also a strong host–genotype relationship at the MLST clonal complex, ST, and allele levels, particularly within C. jejuni (Miller et al. 2006; McCarthy et al. 2007; Sheppard et al. 2010b). This ecology-driven population structuring could be either a consequence or a cause of ancestral barriers to HGT among lineages. As particular clones become isolated in different hosts, they progressively diverge over time leading to the host-associated genetic clusters that are currently observed. This is particularly evident in wild birds, with different bird species hosting particular C. jejuni lineages (Colles et al. 2008a; Sheppard et al. 2011b; Griekspoor et al. 2013); indeed, the association is so strong that there is some congruence among phylogenies based on host species and Campylobacter lineage. This contrasts with an absence of strong phyogeographic structure in Campylobacter populations so that members of the same bird species from different continents, or even hemispheres, may have Campylobacter genotypes more similar than from different species isolated in the same locale.
The situation is quite different within agricultural animals. In this case, isolates from multiple clonal complexes are frequently sampled from the same animal species, and some of these clonal complexes are widely distributed among different farm animal hosts (Sheppard et al. 2014). A single chicken flock can contain Campylobacters belonging to more than 10 distinct clonal complexes (Colles et al. 2008b), and there is evidence for succession of clonal complexes within a single animal (Colles and Maiden 2014). The reasons for this are not fully understood; however, it is likely to reflect multiple historical colonization events of domestic animals from wild animals. Despite a strong focus on biosecurity as an agricultural control measure against Campylobacter infection, there is little evidence of direct transmission of Campylobacter between agricultural and wild animals. However, agriculture-associated genotypes do circulate within the farm environment, and it is possible that there are reservoir or vector hosts that are yet to be identified. We contend that agricultural animals represent a relatively new niche for Campylobacter in evolutionary terms, and it is possible that none of the lineages that have colonized has evolved a sufficient advantage to competitively exclude other strains. Just as in wild birds, the association of distinct lineages with ovine, bovine, and chicken hosts is stronger than spatial or temporal variation. For example, C. jejuni isolates from U.K. chickens are more similar to C. jejuni isolates from U.S. chickens than they are to isolates from U.K. cattle. This greater influence of host compared with geography in shaping C. jejuni populations has also been observed with isolates from cattle, pigs, and turkeys (Sheppard et al. 2010b), and almost certainly reflects transmission pathways associated with the global circulation of agricultural animals or adaptation to particular hosts.
The influence of human activity on the global Campylobacter population structure is particularly well illustrated by isolates from the New Zealand archipelago. This landmass has a well-described history of human colonization, first, by Polynesian populations, followed by the arrival of Europeans, and a more recent post-European influx. At least some of the native wild birds harbor Campylobacters that are closely related to C. coli and C. jejuni, but diverged before their common ancestor (Fig. 2C) (French et al. 2014), as well as lineages more closely related to European and U.S. isolates. It has been argued that this is a signature of geographical isolation combined with prehistoric and more recent human colonization events (French et al. 2014). This mixture of local and global genotypes is mirrored in the more recent epidemiology of Campylobacter in New Zealand in the early 2000s, with the emergence and expansion of an epidemic clone (ST-474) responsible for a large proportion of human infection in New Zealand, but rare elsewhere (Sears et al. 2011), simultaneously with the circulation of globally distributed lineages within the agricultural animals.
The presence of multiple lineages within a single host and the existence of lineages that are found in multiple hosts leads to questions about how lineages coexist without outcompeting each other. Wherever multiple clonal complexes are found in the same agricultural animals, they frequently share numerous alleles, indicating that they occupy the same physical space and can recombine; however, there are differences among lineages. For example, clonally related strains with very similar core genomes and identical MLST clonal complexes can differ at accessory genome loci if they are isolated from cattle or chicken (Sheppard et al. 2013a). Understanding hidden niche structure that exists within hosts is an important area of future research.
THE TIMESCALE OF Campylobacter EVOLUTION
In some cases, it is possible to position evolutionary events, such as lineage divergence, relative to one another. For example, it is clear that the emergence of clonal complexes in C. jejuni occurred after the ancestral split of C. jejuni from C. coli. This inference is robust, being supported by several lines of evidence, but lacks information as to when it occurred. Most trees of genetic relatedness among isolates are based on the number of nucleotide substitutions that separate two lineages, such that, for example, there may be one or two substitutions per gene separating isolates of the same C. jejuni clonal complex, but around 100 separating C. jejuni and C. coli. For a more complete understanding of the evolution of lineage divergence, it is desirable to derive an estimate of the timescale for the emergence of clonal complexes, clades, and species. By incorporating an estimate of mutation rate into a tree of genetic relatedness, the time taken for a certain number of substitutions to occur can be estimated. Then, by comparing estimated dates for each tree node with epidemiological or ecological data, it is possible to make inferences about the conditions that shaped the tree topology.
The fundamental challenge in determining the timescale of evolution is the accurate estimation of the mutation rate that is needed to calibrate the “molecular clock.” Difficulties in this process include (1) the sensitivity of the calibration to variation in the mutation rate of different genes, leading to different estimates depending on which genes are used, and (2) the overestimation of the mutation rate because of single recombination events that introduce numerous substitutions. A number of studies have addressed this issue and, by identifying genes that are under different selection pressures, it is possible to inform the choice of genes used for the mutation rate estimate. In practice, however, it may be simpler to use a genome-wide mutation rate average. Overestimation of mutation rate because of recombination can also be moderated by removal of recently recombined genes (Wilson et al. 2009; Cheng et al. 2013) or by constructing a tree that gives equal significance to individual point mutations and clusters of mutations that are likely to result from a single recombination event (Didelot and Falush 2007).
The ancestral diversification of Escherichia coli and Salmonella typhimurium, and a nucleotide substitution rate of 1% 16S rRNA divergence per 50 million years (Ochman and Wilson 1987) have been used to calibrate the molecular clock and date recent bacterial evolution (Achtman et al. 2004; Roumagnac et al. 2006). When such a calibration is applied, the ancestral diversification of C. jejuni and C. coli is estimated to have occurred ∼10 million years ago, and the divergence of the three C. coli clades ∼2.5 million years ago. However, this mutation rate estimate is quite low compared with, for example, some laboratory estimates for E. coli (Lenski et al. 1998) and Pseudomonas (Buckling et al. 2007). A number of studies have estimated lineage divergence using a more rapid rate of molecular evolution (Falush et al. 2001; Perez-Losada et al. 2007; Feng et al. 2008; Wilson et al. 2009; Sheppard et al. 2010a). One calibration of divergence in C. jejuni was based on the rate of nucleotide substitution in a longitudinal 3-year study of molecular variation in MLST data from a confined geographic location (Wilson et al. 2009). Using this estimate, derived from diversity in a natural population, the speciation of C. jejuni and C. coli was estimated to have occurred ∼6500 years ago. The divergence of the C. coli clades was estimated to have occurred 1000–1700 years ago, and clonal complex structure was even more recent (Fig. 2D) (Sheppard et al. 2010a).
Based on this estimate of diversification within C. jejuni and C. coli, with the observed species, clade, and clonal complex structure all <6500 years old, a correlation with changes in human ecology can be suggested. Agriculture originated in the Middle East (12,000 BC) and spread across Europe between 5000 and 3000 BC, accompanied by the establishment of the first major urban centers (Ammerman and Cavalii-Sforza 1984; McCorriston and Hole 1991; Zvelebil and Dolukhanov 1991). As with other important human diseases, such as plague (Achtman et al. 2004), the availability of a novel niche and enhanced opportunities for transmission between humans and animals can have a major effect on pathogen biology. In Campylobacter, agricultural animals represent a large and expanding multihost reservoir, providing opportunities for adaptation in different ways to that in the ancestral hosts. This is likely to have an impact on shaping the population structure that is observed today.
RECOMBINATION AND THE EMERGENCE OF NOVEL PHENOTYPES
C. jejuni and C. coli have the potential to evolve rapidly. First, because their large effective population sizes mean that even with a low mutation rate any given mutation is likely to occur regularly somewhere in the population (Wilson et al. 2009). Second, because HGT events can introduce large numbers of polymorphisms simultaneously, they generate novel phenotypes extremely rapidly. It has been estimated that HGT events occur at twice the rate of de novo mutation in these organisms. This evolvability has implications in the clinical context. Of particular concern is the increase in antimicrobial resistance among clinical isolates (Cody et al. 2010) and in isolates from other reservoir hosts, such as chickens (Wimalarathna et al. 2013). A study of the antimicrobial susceptibility of isolates from a U.K.-wide survey of C. jejuni and C. coli in retail poultry showed widespread resistance to tetracycline, quinolones (ciprofloxacin and naladixic acid), erythromycin, chloramphenicol, and aminoglycosides. Importantly, resistant isolates were widely distributed among lineages, indicating that acquisition was widespread and had occurred independently on multiple occasions (Wimalarathna et al. 2013). The clustering of resistance phenotypes also indicated that having acquired resistance, lineages expanded locally, presumably because they were at an advantage over their competitors.
This capacity for frequent HGT also impacts on another clinically important trait: the ability to colonize multiple hosts, and contaminate meat and poultry. In the first application of a formal genome-wide association mapping approach to bacteria, some of the genetic elements associated with adaptation to life in the cattle gut have been identified in C. jejuni genomes (Sheppard et al. 2013a). A seven-gene region associated with vitamin B5 biosynthesis was almost universally present in cattle isolates, but was frequently absent in isolates from chickens and wild birds. Laboratory experiments confirmed that isolates from cattle were better able to grow in a vitamin B5–depleted media and this is possibly advantageous in the gut of animals with a low vitamin B5 diet, such as ruminants (Sheppard et al. 2013a). More generally, the persistence of elements that are necessary for colonization of cattle in some isolates from chickens provides evidence for the mechanism by which these bacteria might achieve a “host generalist” lifestyle. It is not clear whether host generalism is a stable strategy or, as mentioned above, if it simply reflects the insufficient time for specialists to evolve in the novel agricultural niche. However, it is possible that the benefit of being able to continuously readapt to new hosts could offset the metabolic cost of maintaining genes that are not useful in the primary host.
INTERSPECIES HGT
Among the most intriguing characteristics of the biology of C. jejuni and C. coli is the observation of high levels of interspecies HGT among them (Sheppard et al. 2008). The identification of mosaic alleles (Sheppard et al. 2011b), mixed species multilocus STs (Sheppard et al. 2008), and genome-wide introgression (Sheppard et al. 2013b) provided evidence for progressive hybridization involving members of C. jejuni and C. coli. As these species are ∼12% divergent at the nucleotide sequence level—as far from each other at the nucleotide level as Salmonella enterica and E. coli or a marmoset and human—it is remarkable that some lineages appear to have exchanged almost a quarter of their genome. This finding is at odds with a number of assumptions concerning bacterial evolution and speciation, and has been challenged (Caro-Quintero et al. 2009; Lefébure et al. 2010); however, subsequent results from whole genome analyses are consistent with an evolutionary scenario, in which a single C. coli lineage (clade 1) has been progressively accumulating C. jejuni DNA. Introgression within this clade has led to the replacement of ∼10% and 23% of the C. coli core genome, as well as the import of novel DNA in the ST-828 and ST-1150 clonal complexes, respectively (Sheppard et al. 2013b).
If maintained over time across the genome, this level of interspecies HGT would lead to merging of the species within the agricultural niche in a process described as despeciation (Fig. 2E) (Sheppard et al. 2008). Whether this is the case or not, it is interesting to note that the cross-species exchange has not, so far, had a substantial impact on the gene pools of either C. jejuni or nonagricultural C. coli (clades 2 and 3). Furthermore, the existence of hybrids and the maintenance of alleles of C. jejuni origin within the C. coli gene pool shows that mechanistic barriers are not preventing interspecies gene flow and the hybrid lineages are not sufficiently maladapted to prevent their proliferation by presenting an “adaptive barrier” to the survival of these genotypes. Therefore, it is likely that ecological barriers to recombination have played a role in generating and maintaining the observed population structure in C. coli and C. jejuni and, remarkably, elements of the basic cellular machinery remain interchangeable even after a prolonged period of independent evolution.
CONCLUSIONS
Many advances have been made in understanding the biology of C. jejuni and C. coli by the application of population and evolutionary approaches. This has been greatly influenced by the increasing availability of genotype, and especially sequence-based, data from large numbers of isolates obtained from natural populations. As WGS data become increasingly available from comparable isolate collections with different phenotypes, it is becoming possible to identify the genes and gene networks that are involved in particular evolutionary processes and better understand how phenotypic factors, such as the animal host from which the isolate was sampled, influence the observed genetic structure in C. coli and C. jejuni populations.
The application of genome-wide association studies (GWAS) to bacteria (Sheppard et al. 2013a) has great potential for enhancing understanding of the genetic basis of phenotypic variation in Campylobacter and other organisms. As with similar approaches in human genomics, GWAS deviate from conventional, “bottom-up,” approaches that begin with the identification of a gene and then test its function—typically using knockout mutants—in favor of a “top-down” approach that groups isolates by phenotype, and then identifies genetic elements associated with a particular phenotype. Early applications of this approach have identified host-associated genes and alleles (Sheppard et al. 2013a), and there is considerable potential for unraveling other complex phenotypes, such as survival, through the food chain and virulence.
Much of the diversity within these two species of Campylobacter is found in wild-animal populations, especially birds, but an increasing body of evidence suggests that anthropogenic factors may have had a major influence on the evolution of this organism. The human population has doubled in the last 40 years, and this is accompanied with the intensification of agriculture and livestock farming on an industrial scale. To C. jejuni and C. coli, the dramatic increase in available hosts, particularly in respect of very large numbers of chickens present in broiler flocks, represents an enormous novel niche for exploitation. However, animal hosts in the modern human food chain are very different from the ancestral host species that Campylobacter species have colonized in many aspects, such as diet, life expectancy, and density. The impact of this novel niche structure on the colonizing Campylobacters are evident in various ways: (1) in the emergence of multihost C. jejuni lineages containing STs that are capable of colonizing both birds and mammals; (2) in the rapid expansion of a single C. coli lineage, which is common in agricultural animals and human disease; (3) in the widespread acquisition of antimicrobial resistance and the proliferation of resistant lineages; and (4), perhaps most strikingly, in the high levels of interspecies genetic exchange among agricultural C. jejuni and C. coli, which implies a breakdown in the barriers to gene flow that maintain these separate species. Understanding the effect of colonization of an agricultural niche from ancestral hosts not only enhances knowledge of key features of Campylobacter evolutionary biology but provides general insight into the emergence of zoonotic pathogens and the potential dangers of agricultural intensification.
Footnotes
Editor: Howard Ochman
Additional Perspectives on Microbial Evolution available at www.cshperspectives.org
REFERENCES
- Achtman M, Morelli G, Zhu P, Wirth T, Diehl I, Kusecek B, Vogler AJ, Wagner DM, Allender CJ, Easterday WR, et al. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc Natl Acad Sci 101: 17837–17842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allos B. 2001. Campylobacter jejuni infections: Update on emerging issues and trends. Clin Infect Dis 32: 1201–1206. [DOI] [PubMed] [Google Scholar]
- Ammerman AJ, Cavalii-Sforza LL. 1984. The neolithic transition and the genetics of populations in Europe. Princeton University Press, Princeton, NJ. [Google Scholar]
- Bayliss CD, Bidmos FA, Anjum A, Manchev VT, Richards RL, Grossier JP, Wooldridge KG, Ketley JM, Barrow PA, Jones MA, et al. 2012. Phase variable genes of Campylobacter jejuni exhibit high mutation rates and specific mutational patterns but mutability is not the major determinant of population structure during host colonization. Nucleic Acids Res 40: 5876–5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckling A, Brockhurst MA, Travisano M, Rainey PB. 2007. Experimental adaptation to high and low quality environments under different scales of temporal variation. J Evol Biol 20: 296–300. [DOI] [PubMed] [Google Scholar]
- Caro-Quintero A, Rodriguez-Castano GP, Konstantinidis KT. 2009. Genomic insights into the convergence and pathogenicity factors of Campylobacter jejuni and Campylobacter coli species. J Bacteriol 191: 5824–5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter PE, McTavish SM, Brooks HJ, Campbell D, Collins-Emerson JM, Midwinter AC, French NP. 2009. Novel clonal complexes with an unknown animal reservoir dominate Campylobacter jejuni isolates from river water in New Zealand. Appl Environ Microbiol 75: 6038–6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Connor TR, Siren J, Aanensen DM, Corander J. 2013. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol 30: 1224–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cody AJ, Clarke L, Bowler IC, Dingle KE. 2010. Ciprofloxacin-resistant campylobacteriosis in the UK. Lancet 376: 1987. [DOI] [PubMed] [Google Scholar]
- Colles FM, Maiden MC. 2012. Campylobacter sequence typing databases: Applications and future prospects. Microbiology 158: 2695–2709. [DOI] [PubMed] [Google Scholar]
- Colles FM, Maiden MC. 2014. Exploring the population dynamics of multiple Campylobacter strains in poultry. Campylobacter ecology and evolution (ed. Sheppard SK, Meric G). Caister Academic, Norfolk, England. [Google Scholar]
- Colles FM, Dingle KE, Cody AJ, Maiden MC. 2008a. Comparison of Campylobacter populations in wild geese with those in starlings and free-range poultry on the same farm. Appl Environ Microbiol 74: 3583–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colles FM, Jones TA, McCarthy ND, Sheppard SK, Cody AJ, Dingle KE, Dawkins MS, Maiden MC. 2008b. Campylobacter infection of broiler chickens in a free-range environment. Environ Microbiol 10: 2042–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colles FM, Ali JS, Sheppard SK, McCarthy ND, Maiden MC. 2011. Campylobacter populations in wild and domesticated Mallard ducks (Anas platyrhynchos). Environ Microbiol Rep 3: 574–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condran GA, Murphy J. 2008. Defining and managing infant mortality: A case study of Philadelphia, 1870–1920. Soc Sci Hist 32: 473–513. [Google Scholar]
- Coward C, van Diemen PM, Conlan AJ, Gog JR, Stevens MP, Jones MA, Maskell DJ. 2008. Competing isogenic Campylobacter strains exhibit variable population structures in vivo. Appl Environ Microbiol 74: 3857–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan CP, Lampen K, Corander J, Hanninen ML. 2013. Multilocus sequence types of environmental Campylobacter jejuni isolates and their similarities to those of human, poultry and bovine C. jejuni isolates. Zoonoses Public Health 60: 125–133. [DOI] [PubMed] [Google Scholar]
- Didelot X, Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175: 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle K, McCarthy N, Cody A, Peto T, Maiden M. 2008. Extended sequence typing of Campylobacter spp., United Kingdom. Emerg Infect Dis 14: 1620–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Kraft C, Taylor NS, Correa P, Fox JG, Achtman M, Suerbaum S. 2001. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: Estimates of clock rates, recombination size, and minimal age. Proc Natl Acad Sci 98: 15056–15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhat MR, Shapiro BJ, Sheppard SK, Colijn C, Murray M. 2014. A phylogeny-based sampling strategy and power calculator informs genome-wide associations study design for microbial pathogens. Genome Med 6: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Reeves PR, Lan R, Ren Y, Gao C, Zhou Z, Ren Y, Cheng J, Wang W, Wang J, et al. 2008. A recalibrated molecular clock and independent origins for the cholera pandemic clones. PLoS ONE 3: e4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French NP, Midwinter A, Holland B, Collins-Emerson J, Pattison R, Colles F, Carter P. 2009. Molecular epidemiology of Campylobacter jejuni isolates from wild-bird fecal material in children’s playgrounds. Appl Environ Microbiol 75: 779–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French N, Yu S, Biggs P, Holland B, Fearnheard P, Binney B, Fox A, Grove-White D, Leigh JW, Miller W, et al. 2014. Evolution of Campylobacter species in New Zealand Campylobacter ecology and evolution (ed. Sheppard SK). Caister Academic, Norfolk, England. [Google Scholar]
- Friedman CJ, Neiman J, Wegener HC, Tauxe RV. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialised nations. In Campylobacter (ed. Nachamkin I, Blaser MJ), pp. 121–138. ASM, Washington, DC. [Google Scholar]
- Frost JA, Oza AN, Thwaites RT, Rowe B. 1998. Serotyping scheme for Campylobacter jejuni and Campylobacter coli based on direct agglutination of heat-stable antigens. J Clin Microbiol 36: 335–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griekspoor P, Colles FM, McCarthy ND, Hansbro PM, Ashhurst-Smith C, Olsen B, Hasselquist D, Maiden MC, Waldenström J. 2013. Marked host specificity and lack of phylogeographic population structure of Campylobacter jejuni in wild birds. Mol Ecol 22: 1463–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings R, Colles FM, McCarthy ND, Maiden MC, Sheppard SK. 2011. Campylobacter genotypes from poultry transportation crates indicate a source of contamination and transmission. J Appl Microbiol 110: 266–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerome JP, Mansfield LS. 2014. Within-host evolution of Campylobacter jejuni. In Campylobacter ecology and evolution (ed. Sheppard SK). Caister Academic, Norfolk, England. [Google Scholar]
- Jolley KA, Maiden MC. 2014. Using MLST to study bacterial variation: Prospects in the genomic era. Future Microbiol 9: 623–630. [DOI] [PubMed] [Google Scholar]
- Jorgensen F, Ellis-Iversen J, Rushton S, Bull SA, Harris SA, Bryan SJ, Gonzalez A, Humphrey TJ. 2011. Influence of season and geography on Campylobacter jejuni and C. coli subtypes in housed broiler flocks reared in Great Britain. Appl Environ Microbiol 77: 3741–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessel AS, Gillespie IA, O’Brien SJ, Adak GK, Humphrey TJ, Ward LR. 2001. General outbreaks of infectious intestinal disease linked with poultry, England and Wales, 1992–1999. Commun Dis Public Health 4: 171–177. [PubMed] [Google Scholar]
- Kittl S, Kuhnert P, Hachler H, Korczak BM. 2011. Comparison of genotypes and antibiotic resistance of Campylobacter jejuni isolated from humans and slaughtered chickens in Switzerland. J Appl Microbiol 110: 513–520. [DOI] [PubMed] [Google Scholar]
- Lang P, Lefebure T, Wang W, Pavinski Bitar P, Meinersmann RJ, Kaya K, Stanhope MJ. 2010. Expanded multilocus sequence typing and comparative genomic hybridization of Campylobacter coli isolates from multiple hosts. Appl Environ Microbiol 76: 1913–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefébure T, Bitar PD, Suzuki H, Stanhope MJ. 2010. Evolutionary dynamics of complete Campylobacter pan-genomes and the bacterial species concept. Genome Biol Evol 2: 646–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski RE, Mongold JA, Sniegowski PD, Travisano M, Vasi F, Gerrish PJ, Schmidt TM. 1998. Evolution of competitive fitness in experimental populations of E. coli: What makes one genotype a better competitor than another? Antonie Van Leeuwenhoek 73: 35–47. [DOI] [PubMed] [Google Scholar]
- Magnússon SH, Guðmundsdóttir S, Reynisson E, Rúnarsson AR, Harðardóttir H, Gunnarson E, Georgsson F, Reiersen J, Marteinsson VT. 2011. Comparison of Campylobacter jejuni isolates from human, food, veterinary and environmental sources in Iceland using PFGE, MLST and fla-SVR sequencing. J Appl Microbiol 111: 971–981. [DOI] [PubMed] [Google Scholar]
- Maiden M. 2006. Multilocus sequence typing of bacteria. Annu Rev Microbiol 60: 561–588. [DOI] [PubMed] [Google Scholar]
- Maiden M, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, et al. 1998. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci 95: 3140–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy ND, Colles FM, Dingle KE, Bagnall MC, Manning G, Maiden MC, Falush D. 2007. Host-associated genetic import in Campylobacter jejuni. Emerg Infect Dis 13: 267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCorriston J, Hole F. 1991. The ecology of seasonal stress and the origin of agriculture in the Near East. Am Anthropol 93: 46–69. [Google Scholar]
- Méric G, Yahara K, Mageiros L, Pascoe B, Maiden MC, Jolley KA, Sheppard SK. 2014. A reference pan-genome approach to comparative bacterial genomics: Identification of novel epidemiological markers in pathogenic Campylobacter. PLoS ONE 9: e92798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WG, Englen MD, Kathariou S, Wesley IV, Wang G, Pittenger-Alley L, Siletz RM, Muraoka W, Fedorka-Cray PJ, Mandrell RE. 2006. Identification of host-associated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiology 152: 245–255. [DOI] [PubMed] [Google Scholar]
- Müllner P, Collins-Emerson JM, Midwinter AC, Carter P, Spencer SE, van der Logt P, Hathaway S, French NP. 2010. Molecular epidemiology of Campylobacter jejuni in a geographically isolated country with a uniquely structured poultry industry. Appl Environ Microbiol 76: 2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neimann J, Engberg J, Molbak K, Wegener HC. 2003. A case-control study of risk factors for sporadic Campylobacter infections in Denmark. Epidemiol Infect 130: 353–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Wilson AC. 1987. Evolution in bacteria: Evidence for a universal substitution rate in cellular genomes. J Mol Evol 26: 74–86. [DOI] [PubMed] [Google Scholar]
- Perez-Losada M, Crandall KA, Zenilman J, Viscidi RP. 2007. Temporal trends in gonococcal population genetics in a high prevalence urban community. Infect Genet Evol 7: 271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read DS, Woodcock DJ, Strachan NJ, Forbes KJ, Colles FM, Maiden MC, Clifton-Hadley F, Ridley A, Vidal A, Rodgers J, et al. 2013. Evidence for phenotypic plasticity among multihost Campylobacter jejuni and C. coli lineages, obtained using ribosomal multilocus sequence typing and Raman spectroscopy. Appl Environ Microbiol 79: 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot EM, Fitzgerald C, Kubota K, Swaminathan B, Barrett TJ. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J Clin Microbiol 39: 1889–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumagnac P, Weill FX, Dolecek C, Baker S, Brisse S, Chinh NT, Le TA, Acosta CJ, Farrar J, Dougan G, et al. 2006. Evolutionary history of Salmonella typhi. Science 314: 1301–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samie A, Obi CL, Barrett LJ, Powell SM, Guerrant RL. 2007. Prevalence of Campylobacter species, Helicobacter pylori and Arcobacter species in stool samples from the Venda region, Limpopo, South Africa: Studies using molecular diagnostic methods. J Infect 54: 558–566. [DOI] [PubMed] [Google Scholar]
- Sears A, Baker MG, Wilson N, Marshall J, Muellner P, Campbell DM, Lake RJ, French NP. 2011. Marked campylobacteriosis decline after interventions aimed at poultry, New Zealand. Emerg Infect Dis 17: 1007–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, McCarthy ND, Falush D, Maiden MC. 2008. Convergence of Campylobacter species: Implications for bacterial evolution. Science 320: 237–239. [DOI] [PubMed] [Google Scholar]
- Sheppard SK, Dallas JF, MacRae M, McCarthy ND, Sproston EL, Gormley FJ, Strachan NJ, Ogden ID, Maiden MC, Forbes KJ. 2009a. Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland 2005/6. Int J Food Microbiol 134: 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, Dallas JF, Strachan NJ, MacRae M, McCarthy ND, Wilson DJ, Gormley FJ, Falush D, Ogden ID, Maiden MC, et al. 2009b. Campylobacter genotyping to determine the source of human infection. Clin Infect Dis 48: 1072–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, Dallas JF, Wilson DJ, Strachan NJ, McCarthy ND, Jolley KA, Colles FM, Rotariu O, Ogden ID, Forbes KJ, et al. 2010a. Evolution of an agriculture-associated disease causing Campylobacter coli clade: Evidence from national surveillance data in Scotland. PLoS ONE 5: e15708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, Colles F, Richardson J, Cody AJ, Elson R, Lawson A, Brick G, Meldrum R, Little CL, Owen RJ, et al. 2010b. Host association of Campylobacter genotypes transcends geographic variation. Appl Environ Microbiol 76: 5269–5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, Colles FM, McCarthy ND, Strachan NJ, Ogden ID, Forbes KJ, Dallas JF, Maiden MC. 2011a. Niche segregation and genetic structure of Campylobacter jejuni populations from wild and agricultural host species. Mol Ecol 20: 3484–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, McCarthy ND, Jolley KA, Maiden MC. 2011b. Introgression in the genus Campylobacter: Generation and spread of mosaic alleles. Microbiology 157: 1066–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, Didelot X, Meric G, Torralbo A, Jolley KA, Kelly DJ, Bentley SD, Maiden MC, Parkhill J, Falush D. 2013a. Genome-wide association study identifies vitamin B5 biosynthesis as a host specificity factor in Campylobacter. Proc Natl Acad Sci 110: 11923–11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, Didelot X, Jolley KA, Darling AE, Pascoe B, Meric G, Kelly DJ, Cody A, Colles FM, Strachan NJ, et al. 2013b. Progressive genome-wide introgression in agricultural Campylobacter coli. Mol Ecol 22: 1051–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, Cheng L, Méric G, de Haan CP, Llarena AK, Marttinen P, Vidal A, Ridley A, Clifton-Hadley F, Connor TR, et al. 2014. Cryptic ecology among host generalist Campylobacter jejuni in domestic animals. Mol Ecol 23: 2442–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproston EL, Ogden ID, MacRae M, Dallas JF, Sheppard SK, Cody AJ, Colles FM, Wilson MJ, Forbes KJ, Strachan NJ. 2011. Temporal variation and host association in the Campylobacter population in a longitudinal ruminant farm study. Appl Environ Microbiol 77: 6579–6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur S, Zhao S, McDermott PF, Harbottle H, Abbott J, English L, Gebreyes WA, White DG. 2010. Antimicrobial resistance, virulence, and genotypic profile comparison of Campylobacter jejuni and Campylobacter coli isolated from humans and retail meats. Foodborne Pathog Dis 7: 835–844. [DOI] [PubMed] [Google Scholar]
- van Tonder AJ, Mistry S, Bray JE, Hill DM, Cody AJ, Farmer CL, Klugman KP, von Gottberg A, Bentley SD, Parkhill J, et al. 2014. Defining the estimated core genome of bacterial populations using a Bayesian decision model. PLoS Comput Biol 10: e1003788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DJ, Gabriel E, Leatherbarrow AJ, Cheesbrough J, Gee S, Bolton E, Fox A, Hart CA, Diggle PJ, Fearnhead P. 2009. Rapid evolution and the importance of recombination to the gastroenteric pathogen Campylobacter jejuni. Mol Biol Evol 26: 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimalarathna HML, Sheppard SK. 2012. Ecological factors that influence genetic structure in Campylobacter coli and Campylobacter jejuni. In Current topics in phylogenetics and phylogeography of terrestrial and aquatic systems (ed. Anamthawat-Jonsson K), pp. 1–12. InTech, Rijeka, Croatia. [Google Scholar]
- Wimalarathna HM, Richardson JF, Lawson AJ, Elson R, Meldrum R, Little CL, Maiden MC, McCarthy ND, Sheppard SK. 2013. Widespread acquisition of antimicrobial resistance among Campylobacter isolates from UK retail poultry and evidence for clonal expansion of resistant lineages. BMC Microbiol 13: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvelebil M, Dolukhanov P. 1991. The transition to farming in Eastern and Northern Europe. J World Prehist 5: 233–278. [Google Scholar]