Abstract
Tubulointerstitial hypoxia plays a critical role in the pathogenesis of kidney injury, and hypoxia-inducible factor (HIF)-1 is a master regulator of cellular adaptation to hypoxia. Aside from oxygen molecules, factors that modify HIF-1 expression and functional operation remain obscure. Therefore, we sought to identify novel HIF-1-regulating genes in kidney. A short-hairpin RNA library consisting of 150 hypoxia-inducible genes was derived from a microarray analysis of the rat renal artery stenosis model screened for the effect on HIF-1 response. We report that CCAAT/enhancer-binding protein δ (CEBPD), a transcription factor and inflammatory response gene, is a novel HIF-1 regulator in kidney. CEBPD was induced in the nuclei of tubular epithelial cells in both acute and chronic hypoxic kidneys. In turn, CEBPD induction augmented HIF-1α expression and its transcriptional activity. Mechanistically, CEBPD directly bound to the HIF-1α promoter and enhanced its transcription. Notably, CEBPD was rapidly induced by inflammatory cytokines, such as IL-1β in a nuclear factor-κB-dependent manner, which not only increased HIF-1α expression during hypoxia, but was also indispensable for the non-hypoxic induction of HIF-1α. Thus our study provides novel insight into HIF-1 regulation in tubular epithelial cells and offers a potential hypoxia and inflammation link relevant in both acute and chronic kidney diseases.
Keywords: cell signaling, hypoxia, inflammation, proximal tubule
Oxygen homeostasis underlies many developmental and physiological processes, and, its disturbances have key roles in the pathogenesis of many human diseases, including kidney disease.1 The kidney requires a complicated vasculature to maintain its structure and function and is thus highly susceptible to ischemic insults. Indeed, tubulointerstitial fibrosis is an end-stage feature in chronic kidney disease (CKD), which is frequently associated with the loss of peritubular capillary networks reminiscent of hypoxia.2 To date, there is an increasing body of evidence showing that tubulointerstitial hypoxia is not only a key player in ischemic acute kidney injury (AKI) but is also a critical mediator toward end-stage renal disease in CKD settings.3, 4
Hypoxia-inducible factors (HIFs) regulate adaptive responses to hypoxia at the molecular, cellular, and tissue levels. HIF is a heterodimeric transcription factor, consisting of an oxygen-labile α subunit (HIF-1α, HIF-2α, and HIF-3α) and a constitutively expressed β subunit.5, 6 Under normoxia, HIF-α is hydroxylated by prolyl hydroxylase domain-containing protein (PHDs) and undergoes proteasomal degradation assisted by the von Hippel-Lindau (VHL) tumor-suppressor protein. In hypoxia, the lack of oxygen inhibits PHD activity and disallows hydroxylation of HIF-α. Stabilized HIF-α translocates to the nucleus, dimerizes with HIF-1β, and transactivates its target genes.
In response to hypoxia in kidney, HIF-1α is expressed in tubular epithelial cells, including proximal and distal tubules, connecting tubules, and collecting ducts.7 Functional studies both in vivo and in vitro show that HIF-1 appears to have protective roles in various acute and chronic kidney injuries.8, 9, 10, 11, 12, 13, 14, 15
In addition to its expression control by protein stabilization, HIF-1α undergoes transcriptional and translational regulation, particularly in tumor and inflammatory cells via the phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin, nuclear factor (NF)-κB, and extracellular signal–regulated kinase mitogen-activated protein kinase pathways.16, 17, 18 However, it remains unknown whether the transcriptional and translational control of HIF-1α is operative in cells of noncancerous tissues, such as renal tubular cells, and which, if any, genes are responsible for the HIF-1α upstream signaling.
After taking into account the accumulating knowledge of HIF-1 on the pathogenesis of both AKI and CKD, we designed this study to identify novel HIF-1 regulators through RNAi library screening.
RESULTS
Identification of HIF-1 regulating genes through short hairpin RNA (shRNA) library screening
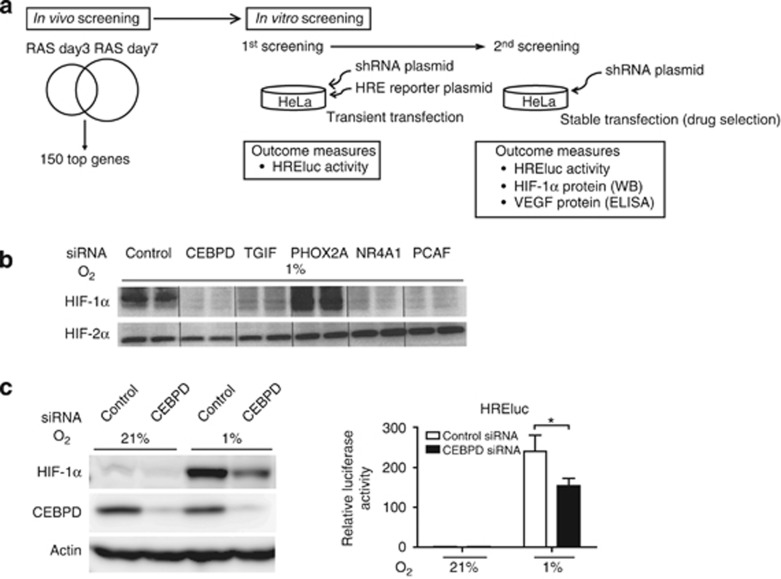
To identify which genes are relevant in HIF-1 regulation, we created a shRNA library against the top 150 hypoxia-inducible genes screened from a microarray analysis of the rat renal artery stenosis (RAS) model, a representative chronic hypoxic kidney injury model (Supplementary Table S1 online).19 For the initial screening, the impact of candidate genes on HIF-1 was evaluated in vitro through a hypoxia-responsive element (HRE)-driven luciferase (HREluc) reporter assay (Figure 1a); shRNA transfection against a bona fide HIF-1-regulating gene would affect HREluc activity. The hypoxic induction (defined as (luciferase activity in 1% O2)/(luciferase activity in 21% O2)) was also calculated to exclude the nonspecific effect. The shRNA-transfected clones showing hypoxic changes within 50 percentile compared with the positive control (shRNA anti-HIF-1α) were extracted as hit clones. Eighteen hit genes proceeded to the second screening.
Figure 1.
The identification of hypoxia-inducible factor (HIF)-1-regulating genes. (a) Scheme of short hairpin RNA (shRNA) library screening. (b) HeLa cells transfected with small interfering RNA (siRNA) against the five candidate genes, CCAAT/enhancer-binding protein δ (CEBPD), transforming growth factor-beta-induced factor (TGIF), nuclear receptor superfamily 4A member 1 (NR4A1), paired mesoderm homeobox protein 2A (PHOX2A), or P300/CBP-associated factor (PCAF), were treated under hypoxia for 16 h to confirm the shRNA study results. The cells were processed for immunoblot analysis of HIF-1α and HIF-2α. Four genes except for PHOX2A showed a reduction in HIF-1α protein expression under hypoxia. The data are representative of three independent experiments, shown in duplicate. (c) Immunoblot analyses of HIF-1α (left panel) and HREluc activity (right panel) are shown representatively for siRNA-mediated CEBPD knockdown in HeLa cells. Knockdown of CEBPD decreased HIF-1α and hypoxic induction of HREluc activity in HeLa cells. (right panel) The ratio of luciferase reporter activity (firefly/TK-Renilla) to that of control siRNA under normoxia is indicated. Bar graph (mean±s.e.m. from three independent experiments) statistics performed using two-way analysis of variance (ANOVA) with Bonferroni post-hoc tests. *P<0.05. RAS, renal artery stenosis.
Stable HeLa cells transfected with shRNA against candidate genes were further assessed for the hypoxic induction of HREluc, HIF-1α protein, and supernatant vascular endothelial growth factor (VEGF, an endogenous HIF-1 target) expression (Supplementary Figure S1 online). At this stage, five genes were selected as novel, potential HIF-1-regulating genes: CCAAT/enhancer-binding protein δ (CEBPD); transforming growth factor-beta-induced factor (TGIF); paired mesoderm homeobox protein 2A (PHOX2A); nuclear receptor superfamily 4A member 1 (NR4A1); and P300/CBP-associated factor (PCAF). Intercellular adhesion molecule 1 (ICAM1); leucine-rich alpha-2-glycoprotein 1 (LRG1); cytochrome P-450, family 4, subfamily B, polypeptide 1 (CYP4B1); and renin (RENIN) were extracted as hit genes by HREluc activity criteria but did not have positive effects on HIF-1α or VEGF and were therefore not selected as candidates (data not shown).
Finally, small interfering RNAs (siRNAs) against these five candidates were designed and transfected into HeLa cells, and the above experiments were repeated. PHOX2A was excluded because of a failure to reduce HIF-1α expression by siRNA, whereas the remaining four matched the criteria. As a result, we identified four novel genes––TGIF, NR4A1, PCAF, and CEBPD––that functionally regulated HIF-1α protein expression and its transcriptional activity (Figure 1b). None of them had an effect on HIF-2α (Figure 1b). Of these genes, CEBPD had the most pronounced effect on HIF-1α expression, which we investigated further (Figure 1c).
Enhanced CEBPD expression in tubular epithelial cells of acute and chronic hypoxic injury
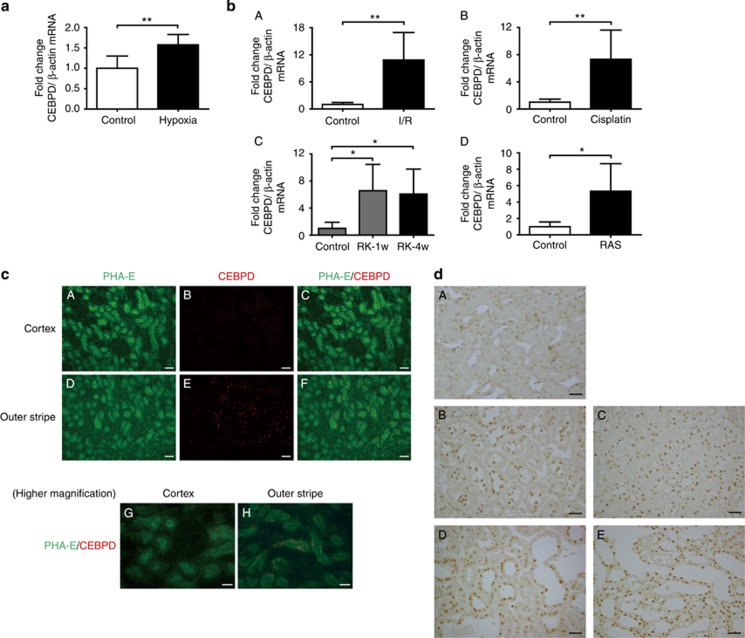
CEBPD is a member of the highly conserved C/EBP family of basic region leucine zipper transcription factors. Although its basic expression level is low in most normal tissues and cells, CEBPD is rapidly upregulated by a variety of stimuli, including inflammatory signals.20, 21 We investigated whether CEBPD expression in vivo correlates with hypoxia in the kidney. Rodent kidneys were subjected to a series of hypoxic stimuli and tested for CEPBD expression. CEBPD was induced in kidneys exposed to systemic hypoxia (8% O2, 6 h), as well as in models of kidney injuries (ischemia–reperfusion (I/R) injury, cisplatin nephrotoxicity, RAS, and 5/6 nephrectomy (remnant kidney (RK)); Figure 2a and b). Although the pathogenesis of these models is multifactorial, they serve as models of acute and chronic ischemic kidney injury. Cisplatin nephrotoxicity is hypoxic in its outer medulla because of the reduced blood flow caused by the toxin,10 and activation of the renin–angiotensin system results in decreased peritubular capillary flow with subsequent hypoxia in RK.22
Figure 2.
CCAAT/enhancer-binding protein δ (CEBPD) is expressed in hypoxic kidney tubular cells in vivo. (a) Mice were exposed to 8% O2 for 6 h using a hypoxia chamber (hypoxia group, n=6). The mice at ambient oxygen were used as the control (control group, n=5). The whole kidney was examined for CEBPD expression by real-time quantitative (q)RT-PCR. (b) The CEBPD expression levels in the renal cortex of acute and chronic rat hypoxic injuries were evaluated by qRT–PCR. (A) The ischemia–reperfusion (I/R) group (control group, n=6; I/R group, n=6). (B) The cisplatin nephrotoxicity group (control group, n=6; cisplatin group, n=7). (C) The remnant kidney (RK) group (control group, n=5; RK-1w group, n=5; RK-4w group, n=5). (D) The RAS group (control group, n=6; RAS group, n=6). (c) Fluorescent images of control kidney tissues for CEBPD and Phaseolus vulgaris Erythroagglutinin (PHA-E), a marker for proximal tubules. (A–C) Cortex and (D–F) outer stripe (OS) of the outer medulla, × 200. Bar=50 μm. CEBPD is stained in the S3 segment of proximal tubules in the OS. (G) Cortex and (H) OS of the outer medulla for further higher magnification, × 400. Bar=30 μm. (d) (A) Control kidney and four hypoxic kidney models ((B) I/R group, (C) cisplatin group, (D) RK group, and (E) RAS group) were stained with an anti-CEBPD antibody. CEBPD is stained in the nuclei of tubular cells in all four models compared with the control kidney. Original magnifications, × 400. Bar=30 μm. Bar graph (combined results from two independent experiments, shown as mean±s.d.) statistics performed using Student's t-test or one-way analysis of variance (ANOVA) with Dunnett's post-hoc tests. *P<0.05 and **P<0.01.
Immunofluorescence studies of kidney sections using anti-CEBPD antibody and Phaseolus vulgaris Erythroagglutinin (PHA-E), a cell-specific marker for proximal tubular cells, demonstrated that in the control kidney CEBPD was most evidently expressed in the S3 segment of proximal tubules in the outer stripe of the outer medulla, physiologically hypoxic and the most susceptible portion of kidney to hypoxia (Figure 2c).7 Upon hypoxic stimulation, CEBPD was robustly induced predominantly in the nuclei of proximal tubular cells (Figure 2d). CEBPD co-localized with PHA-E in the hypoxic models and did not co-localize with Tamm–Horsfall protein (THP, a marker for thick ascending limbs) or with calcium-binding protein (CaBP, a marker for ascending limb or distal tubules; data not shown). These data demonstrate that CEBPD is induced by hypoxia predominantly in proximal tubular cells.
Hypoxia increases CEBPD expression via HIF-1-independent pathway and regulates HIF-1α expression in human proximal tubular cells
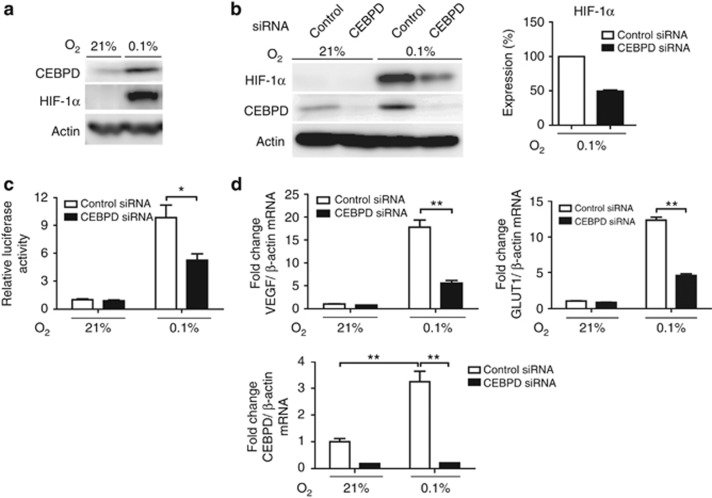
CEBPD induction in proximal tubules following a series of hypoxic injury conditions prompted us to investigate whether these events occur in a cell-autonomous manner. In HK-2 cells, hypoxia increased CEBPD expression (Figure 3a). CEBPD mRNA expression was increased by threefold, which was not blunted by HIF-1α siRNA (Supplementary Figure S2A online). This suggested that the hypoxic induction of CEBPD was mediated independently of HIF-1. Conversely, CEBPD knockdown by siRNA resulted in reduced HIF-1α protein expression under hypoxia (Figure 3b), which was accompanied by a subsequent reduction in HIF transcriptional activity (Figure 3c) and reduced expression of the endogenous HIF-1 target genes glucose transporter 1 (GLUT1) and VEGF (Figure 3d). We further investigated whether other members of the C/EBP family might similarly regulate HIF-1. CCAAT/enhancer-binding protein β (CEBPB), a known homologue to CEBPD, was not induced by hypoxia nor did it regulate HIF-1α expression (Supplementary Figure S2B online). In contrast, CEBPD overexpression significantly increased HIF-1α protein levels (Supplementary Figure S2C online). Interestingly, HIF-1α protein upregulation by CEBPD overexpression was more pronounced in 1% hypoxia but was evident at near anoxic conditions (0.1% hypoxia). These data collectively demonstrate that CEBPD regulates endogenous HIF-1 in tubular cells, which is consistent with previous report in cancer cell line,23 and this effect is strictly retained within the context of the CEBPD–HIF-1 axis.
Figure 3.
Hypoxia-inducible factor (HIF)-1 regulation by CCAAT/enhancer-binding protein δ (CEBPD) in tubular epithelial cells. (a) CEBPD protein is upregulated by hypoxia (0.1% O2) in HK-2 cells. (b) Knockdown of CEBPD decreased HIF-1α protein expression. Right panel shows densitometrical quantification of HIF-1α protein under hypoxic condition of three independent experiments. (c) Knockdown of CEBPD significantly decreased HREluc activity under hypoxia. The ratio of luciferase reporter activity (firefly/CMV-Renilla) to that of control small interfering RNA (siRNA) under normoxia is indicated. (d) Real-time qRT-PCR for HIF-1 target genes, glucose transporter 1 (GLUT1) and vascular endothelial growth factor (VEGF), also demonstrated that their induction under hypoxia is dependent on CEBPD. Bar graph (mean±s.e.m. or representative of three independent experiments) statistics performed using two-way analysis of variance (ANOVA) with Bonferroni post-hoc tests. *P<0.05 and **P<0.01.
Cross-talk of inflammation and hypoxia in kidney
CEBPD is an inflammatory response gene, which contributes to both the stimulation and the resolution of inflammation.20, 24 CEBPD is induced by bacterial lipopolysaccharide (LPS), interferon-γ, interleukin (IL)-1β, tumor necrosis factor (TNF)-α, and IL-6 in myeloid cells.24, 25, 26 On the other hand, HIF-1α is induced under normoxic condition by LPS, growth factors such as epidermal growth factor or insulin, and inflammatory cytokines such as IL-1β or TNF-α,27, 28, 29 depending on the cell type, as well as the pathophysiologial contexts.
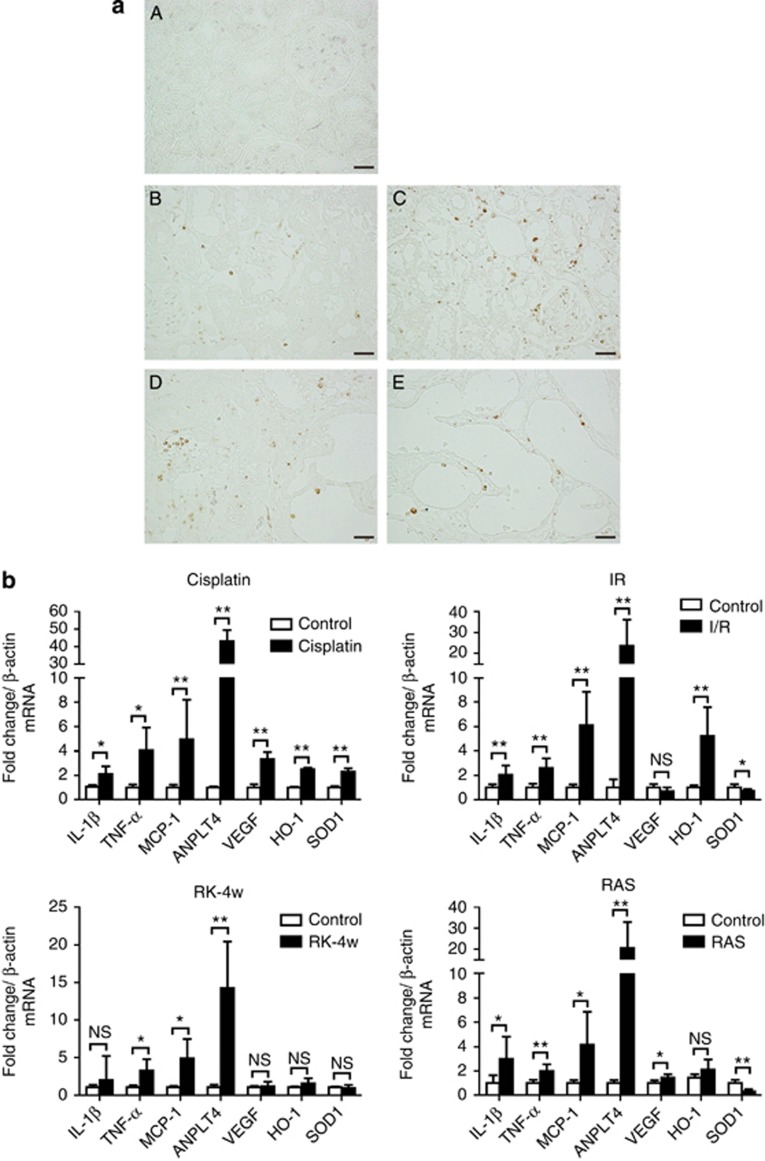
These facts led us to hypothesize that tissue inflammation might fine-tune HIF-1 through CEBPD expression. In the kidney, inflammation is mediated by resident tubular cells and inflammatory cells, most commonly by macrophages.30, 31 Macrophages are present in the tubulointerstitium of all of the above-indicated hypoxic kidney models (Figure 4a). Macrophages and tubular cells under inflammatory conditions produce various cytokines and chemokines, including IL-1β, TNF-α, and monocyte chemotactic protein-1. These were confirmed in all the above hypoxic kidney models by real-time quantitative (q)RT-PCR of the diseased renal cortex (Figure 4b). Endogenous HIF-1 target genes, including ANPLT4 (angiopoietin-like 4), VEGF, SOD1 (superoxidase dismutase 1), and HO-1 (hemeoxygenase-1), were overall upregulated except for the RK model, in which the insufficient expression of HIF-target genes is reported (Figure 4b).32 This confirmed that inflammation and hypoxia coexist and may have interactions in kidney diseases.
Figure 4.
Hypoxia and inflammation coexist in kidney. (a) Macrophage infiltration was visualized by endothelin-1 staining for (A) control, (B) I/R injury, (C) cisplatin nephrotoxicity, (D) RK-4w, and (E) RAS day 7 kidneys. Original magnification, × 400. Bar=30 μm. (b) Real-time qRT-PCR analysis of the renal cortex for inflammatory cytokines (interleukin (IL)-1β, tumor necrosis factor (TNF)-α, and monocyte chemotactic protein-1 (MCP-1)), and HIF-1 target genes (vascular endothelial growth factor (VEGF), angiopoietin-like 4 (ANPLT4), hemeoxygenase-1 (HO-1), and superoxidase dismutase 1 (SOD1)) were performed in cisplatin nephrotoxicity (control group, n=5; cisplatin group, n=5), I/R injury (control group, n=6; I/R group, n=6), RK-4w (control group, n=5; RK-4w group, n=5), and RAS d7 (control group, n=6; RAS group, n=6). Bar graph (combined results from two independent experiments, shown as mean±s.d.) statistics performed using Student's t-test. *P<0.05 and **P<0.01.
CEBPD regulates HIF-1 in human tubular cells under inflammatory stimuli
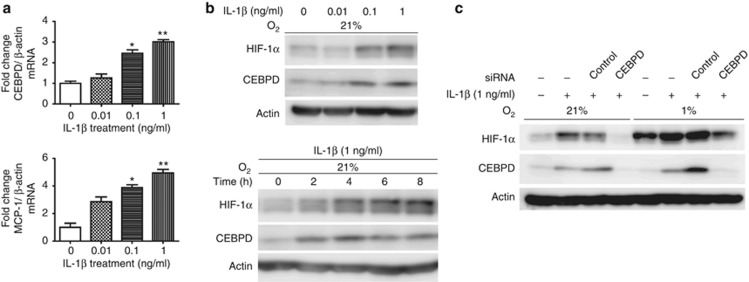
HK-2 cells were accordingly treated with various inflammatory stimuli, including LPS, TNF-α, and IL-1β (Supplementary Figure S3A and B online, Figure 5a and b). Whereas neither LPS nor TNF-α influenced the expression of CEBPD or HIF-1α (Supplementary Figures 3A and B online), IL-1β induced CEBPD and HIF-1α protein expression in a dose- and time-dependent manner under normoxia (Figure 5a and b). The parallel measurement of IL-6 or monocyte chemotactic protein-1 confirmed the efficacy of each treatment (Supplementary Figure S3A online, Figure 5a). A time-lapse study revealed that the induction of CEBPD preceded that of HIF-1α (Figure 5b), suggesting the existence of a CEBPD/HIF-1 pathway; indeed, HIF-1α induction by IL-1β was shown to be CEBPD dependent through siRNA knockdown and overexpression experiments (Figure 5c and Supplementary Figure S3C online). This regulation was also observed in A549 cells, a human lung carcinoma cell line (data not shown). These results indicate that CEBPD regulates the expression of HIF-1α protein not only under hypoxia but also under inflammatory conditions.
Figure 5.
CCAAT/enhancer-binding protein δ/hypoxia-inducible factor 1 (CEBPD/HIF-1) pathway activation under normoxia by interleukin (IL)-1β. (a) HK-2 cells were treated with IL-1β (0–1 ng/ml) under 21% O2 for 6 h and assessed for CEBPD and monocyte chemotactic protein-1 (MCP-1) mRNA by real-time qRT-PCR. CEBPD and MCP-1 mRNA levels were increased in a dose-dependent manner. (b) Immunoblot analysis of CEBPD and HIF-1α expression levels in HK-2 cells under IL-1β treatment. CEBPD and HIF-1α protein levels increased in a dose- and time-dependent manner. (Upper panel) HK-2 cells were treated with IL-1β (0–1 ng/ml) under 21% O2 or 1% O2 for 6 h. (Lower panel) HK-2 cells were treated with IL-1β (1 ng/ml) for 0–8 h under normoxia. (c) HK-2 cells transfected with small interfering RNA (siRNA) against CEBPD were treated with IL-1β for 6 h under 21% O2 or 1% O2 and assessed for HIF-1α expression through immunoblot analysis. Knockdown of CEBPD reduced the HIF-1α induction by IL-1β both under normoxia and hypoxia. Bar graph (mean±s.e.m. of three independent experiments) statistics performed using one-way analysis of variance (ANOVA) with Dunnett's post-hoc tests. *P<0.05 and **P<0.01.
CEBPD regulates HIF-1α at the transcriptional level
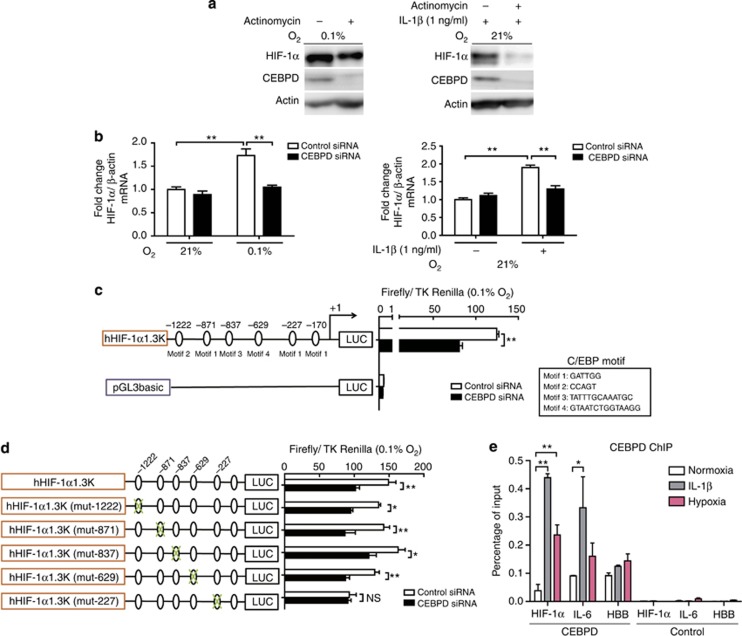
HIF-α expression is tightly regulated at multiple steps. While the posttranslational ubiquitination subsequent to oxygen-dependent hydroxylation by PHD is the dominant step,16 the quantity of HIF-α is also influenced through changes in the transcriptional and translational rates, particularly in tumor and inflammatory cells.18 Because CEBPD regulated HIF-1α expression under various O2 concentrations, including those under which the PHD activity is already severely impaired, we hypothesized that the latter mechanisms may be involved in the control of HIF-1α by CEBPD. In HK-2 cells, treatment with actinomycin, a transcription inhibitor, decreased HIF-1α expression under hypoxia or IL-1β treatment (Figure 6a). HIF-1α mRNA showed a CEBPD-dependent increase under both conditions (Figure 6b). Cycloheximide, a protein synthesis inhibitor, did not show differences in HIF-1α protein half-lives between control and CEBPD siRNA groups, whereas MG132, a proteasome inhibitor, clearly demonstrated a decrease in the HIF-1α synthesis in the CEBPD siRNA group (Supplementary Figure S4A and B online), which indicated that CEBPD regulates HIF-1α at the transcription or the translation level. This was also confirmed by an experiment using a 2-oxoglutarate analog, DMOG, which disallows PHD enzymatic activity and thus prevents HIF-α degradation (Supplementary Figure S4C online). Furthermore, PI3K/AKT or mammalian target of rapamycin signaling has been implicated in the transcriptional or translational regulation of HIF-1α in several tumor cells. However, pretreatment with specific inhibitors, PI3K inhibitor (LY294002) and rapamycin, had no effect on the expression of HIF-1α, suggesting that these pathways were not involved in our study (Supplementary Figure S4D online).27, 29, 33
Figure 6.
CCAAT/enhancer-binding protein δ (CEBPD) mechanistically regulates hypoxia-inducible factor (HIF)-1α at the transcription level. (a) Transcription inhibition reduces the induction of HIF-1α protein both under hypoxic and interleukin (IL)-1β-treated conditions. HK-2 cells pretreated with actinomycin for 20 min were either exposed to hypoxia (0.1% O2; left panel) or treated with IL-1β (1 ng/ml; right panel) for 6 h and assessed for HIF-1α and CEBPD protein expression levels through immunoblot analysis. (b) Small interfering RNA (siRNA)-mediated CEBPD knockdown cancelled the increase in HIF-1α mRNA under hypoxia (left panel) or IL-1β (right panel). Data are the mean±s.e.m. of three independent experiments. *P<0.05 and **P<0.01 (two-way analysis of variance (ANOVA) with Bonferroni's post-hoc tests). (c) Position and nucleotide sequence for the binding of the C/EBP transcription factors are shown as motifs 1–4. The hHIF-1α1.3K promoter activity transfected either with siRNA against control or CEBPD together with TK-Renilla vector was measured under hypoxia. The activity of the pGL3 basic control vector was not influenced by the treatment. (d) Site-directed mutagenesis against the hHIF-1α1.3K promoter was performed to identify the responsible site. Out of the five candidate sites, a mutation at −227/−222 bp site decreased HIF-1α promoter activity. This result signified that this site −227/−222 was functionally responsible for the transcriptional activation of HIF-1α. (e) Chromatin immunoprecipitation–PCR revealed CEBPD–HIF-1α interaction following hypoxia or IL-1β treatment. CEBPD on HIF-1α promoter (−227/−222 bp site) was significantly enriched under hypoxia or IL-1β compared with normoxia. Cross-linked DNA–protein complexes in the normoxia, hypoxia, and IL-1β groups were captured by anti-CEBPD antibody, and enriched nucleotide fragments were quantitatively amplified by PCR using promoters specific to HIF-1α, IL-6 (positive control under IL-1β treatment), and HBB (negative control). This result indicates that CEBPD is recruited to the HIF-1α promoter under both conditions. (c–e) The data are the mean±s.d. of a representative of three independent experiments. *P<0.05 and **P<0.01 (two-way ANOVA with Bonferroni's post-hoc tests).
These results altogether indicated that HIF-1α expression levels under these conditions were regulated at the transcriptional and/or the translational level. Consistently, the activity of 1.3k human HIF-1α promoter was dependent on CEBPD (Figure 6c). siRNA knockdown of CEBPD significantly decreased HIF-1α promoter activity.
The HIF-1α promoter contains six potential binding motifs for C/EBP (motif 1–4) within its 1.3-kb region (Figure 6c);21, 34 therefore, we tested for potential binding of CEBPD to the promoter. Because a pilot transfection of the minimal promoter in close proximity to the HIF-1α transcriptional start site did not reveal significant changes following CEBPD stimulation (data not shown), we speculated that the potential CEBPD binding sites might be one (or more) of the five C/EBP motifs indicated in Figure 6c. Mutations were introduced against each of the candidate C/EBP motifs, and the mutant promoters were tested for the CEBPD response. A mutation at the −227/−222 bp site cancelled the CEBPD-dependent increase in promoter activity (Figure 6d). Chromatin immunoprecipitation (ChIP) –PCR analysis confirmed CEBPD recruitment to this HIF-1α promoter (−227/−222 bp) under hypoxia or IL-1β treatment (Figure 6e). Collectively, these results indicate that CEBPD binds to the HIF-1α promoter and transcriptionally upregulates its expression in response to hypoxia or inflammatory stimulation.
The NF-κB pathway is involved in CEBPD activation under hypoxic or inflammatory stimuli
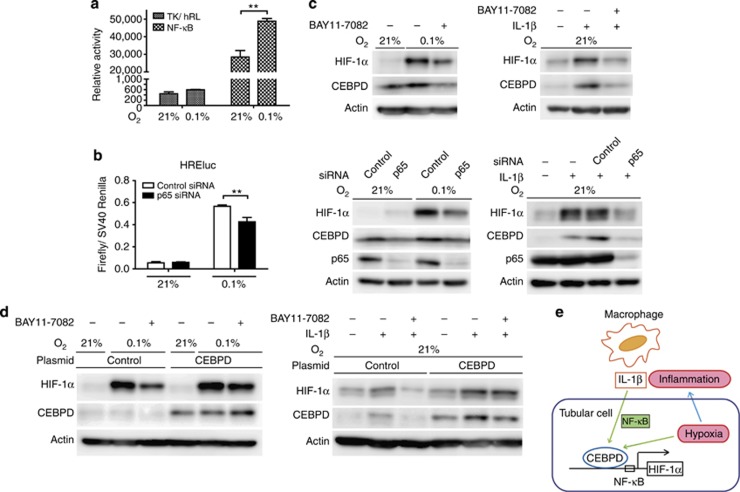
The expression levels of both CEBPD and HIF-1α are regulated by NF-κB signaling in certain cell types, although the detailed pathways of the latter remain largely unknown.19, 35 In HK-2 cells, NF-κB activity was increased by hypoxia (Figure 7a); conversely, siRNA knockdown of p65, a component of NF-κB, led to a decrease in HREluc, indicating that the HIF activity was partially mediated by NF-κB (Figure 7b). On the basis of our previous observation that CEBPD augments HIF-1α expression in hypoxia and allows its normoxic, IL-1β-dependent accumulation, we hypothesized that NF-κB signaling might underlie hypoxic- or IL-1β-stimulated CEBPD expression, which is responsible for the induction of HIF-1α. Upon NF-κB inhibition by the IκB-α phosphorylation and degradation inhibitor (BAY11-7082) or p65 siRNA, the expression levels of both CEBPD and HIF-1α significantly decreased (Figure 7c). Conversely, stable CEBPD overexpression partially rescued the reduced HIF-1α expression caused by NF-κB inhibition (Figure 7d). In summary, these results signify a novel NF-κB/CEBPD/HIF-1 pathway in which CEBPD was primarily responsible for the NF-κB-mediated fine-tuning of HIF-1α (Figure 7e).
Figure 7.
The nuclear factor (NF)-κB pathway regulates the CCAAT/enhancer-binding protein δ/hypoxia-inducible factor 1 (CEBPD/HIF-1) pathway under hypoxia or interleukin (IL)-1β treatment. (a) NF-κB reporter activity was measured under normoxia or hypoxia. The pRL-TK vector served as a control. Hypoxia (0.1% O2, 6 h) increased NF-κB activity in HK-2 cells. (b) Inhibition of the NF-κB pathway by p65 knockdown significantly decreased HREluc activity under hypoxia in HK-2 cells. (c) (Upper panel) HK-2 cells were pretreated with BAY11-7082 (IKK inhibitor, 10 μM) or dimethyl sulfoxide (for control) for 30 min and then treated with hypoxia (upper left panel) or IL-1β (1 ng/ml; upper right panel) for 6 h. CEBPD and HIF-1α protein induction under both conditions were inhibited. The same phenomena were observed using small interfering RNA (siRNA) against p65-mediated NF-κB inhibition (lower left panel for hypoxia and lower right panel for IL-1β). (d) Human CEBPD stable overexpression HK-2 clones were generated using a retroviral system. Upon NF-κB inhibition by BAY11-7082 (10 μM), HIF-1α protein induction under hypoxia (left panel) or IL-1β (1 ng/ml; right panel) was reduced in control HK-2 cells. In stable CEBPD-expressed HK-2 cells, the HIF-1α protein induction was restored under both conditions, indicating the necessity for HIF-1α protein regulation by CEBPD. The endogenous CEBPD protein is hardly detected, because exposure time was optimized for the overexpressed protein. Bar graph (mean±s.e.m. or representative of at least three independent experiments) statistics performed using two-way analysis of variance with Bonferroni's post-hoc tests. **P<0.01. (e) A scheme of a pathway proposed in this study. CEBPD is upregulated in tubular epithelial cells, either by hypoxia or IL-1β via the NF-κB-dependent pathway and regulates HIF-1 expression and its transcriptional activity.
DISCUSSION
In this study, we identified novel regulators of HIF-1α expression in kidney using shRNA library screening. Out of the four identified genes, CEBPD had the most striking effect, which was induced by hypoxia or inflammation in tubular epithelial cells via HIF-1-independent, NF-κB-dependent pathways, and CEBPD regulated HIF-1α expression through transcriptional upregulation.
CEBPD is a transcription factor originally identified as an inflammatory response gene.24, 25 Although a small amount of mRNA is observed in the lung, adipose tissue, and intestine, its expression is generally low in the majority of normal tissues, including the kidney.36, 37 CEBPD is involved in a wide range of cell type–specific processes, such as differentiation and proliferation, thus contributing to various physiological and pathological conditions. CEBPD deficiency causes genomic instability in mouse embryo fibroblasts,38 and CEBPD is a tumor-suppressor gene in breast, cervical, and liver cancers.33 With regard to the kidney, CEBPD is upregulated after LPS stimulation.24 However, its functional role in renal pathophysiology is largely unknown. Here, we found that CEBPD was expressed in hypoxic proximal tubular cells and augmented HIF-1α expression.
The role of CEBPD on HIF-1 regulation was not restricted to hypoxic settings. We demonstrated that CEBPD also regulated HIF-1α under normoxic conditions by inflammatory stimuli. As C/EBP family members have important roles in inflammation by regulating the functions of both cytokine-producing inflammatory cells and parenchymal cells, CEBPD-mediated, O2-independent HIF-1 induction under inflammation is expected to have an important role in kidney disease. This regulatory pathway appeared to be common to other types of parenchymal cells to some extent because similar results were obtained in other cell lines, such as A549 lung cancer cells.
There is a growing body of evidence that hypoxia and inflammation coexist and have interactive roles in various tissues.39, 40 Hypoxia promotes inflammation by increasing vascular permeability and facilitating production of inflammatory mediators by recruited leukocytes or resident parenchymal cells. Conversely, inflamed tissues often become severely hypoxic because of an increase in the metabolic demands of cells and a reduction in metabolic substrates. Thus it is not surprising that HIF-1α activation is also achieved by growth factors and cytokines. The functional role of HIF-1 in an inflammatory milieu is best exemplified in the protection of mucosal barrier in a hapten-based murine colitis model.41 Furthermore, in myeloid cells, HIF-1 is essential in differentiation and functions even under normoxia. HIF-1α depletion in these cells results in the impairment of cell aggregation, motility, invasiveness, and bacterial killing.42
Clearly, the importance of HIF within the context of inflammation also applies to kidney diseases. In experimental CKD, HIF activation by cobalt chloride reduced macrophage infiltration in the RK and Thy-1 nephritis models.10, 11 Conditional knockout of HIF-1α/2α in myeloid cells showed more severe inflammation in the unilateral ureteral obstruction model in mice, and conversely HIF activation by myeloid-specific Vhl-knockout suppressed inflammation and fibrosis.43 These interactions of structural cells (e.g., epithelial cells, endothelial cells, and fibroblasts), inflammatory cells (e.g., macrophages, dendritic cells, and leukocytes), and soluble mediators (e.g., growth factors, cytokines, and chemokines) coordinate in the progression of and resolution of kidney injury. However, the signaling pathways that regulate this cross-talk warrant further investigation. On the basis of our results and the close associations between macrophages and tubular epithelial cells, we propose that CEBPD may provide a mechanistic link between inflammation and hypoxia signaling through HIF-1, which possibly underlies a substantially broader spectrum of pathophysiological contexts, including renal pathophysiology, than previously thought. In this regard, whether CEBPD activation provides an additional mode of protection in ischemic kidney diseases requires further in vivo investigation. In previous studies, systemic activations of HIFs by cobalt chloride, PHD inhibitors, inhibiting Factor Inhibiting HIF, or gene transfer of constitutively active HIF (HIF/VP16) protected kidney against its injury,8, 9, 10, 11, 12, 13, 14, 15 which signified that HIF-1 activation was not optimal in diseased kidney. In the present study, lack of methods to modulate CEBPD activity in vivo hindered us from validating the hypothesis that further manipulation to increase the CEBPD activity would ameliorate CKD progression through HIF-1. Alternatively, it is also possible that factors other than CEBPD insufficiency––for example, indoxyl sulfate effect on HIF-1 transcriptional activity in RK model32––may contribute to the incommensurate HIF-1 activity in diseased models. It is also reported that adipose-derived stem cells derived from end-stage kidney disease patients show impaired HIF and angiogenic response, because of weakened expression of PCAF.44 Further studies are needed to fully elucidate the regulating mechanism of HIF-1 expression and its activity, which are determined by multiple factors that vary among different kidney diseases. In our study, the CEBPD/HIF-1 pathway is activated in various time point from several hours to several weeks, by either hypoxic or inflammatory stimuli. In this regard, it remains an open question how long this pathway persists, and whether there is ever diminishing signal for this pathway. CEBPD promoter is known to undergo methylation and becomes silenced in cancer tissues.45, 46 These epigenetic modifications may also have roles in the regulation of CEBPD in kidney.
We further found that CEBPD regulated HIF-1α at the transcription level by mechanistically binding to its promoter. In inflammation, LPS or reactive oxygen species have also been reported to increase HIF-1α mRNA expression through activating a NF-κB-binding site in the HIF-1α gene promoter.35, 47, 48 In agreement with these reports in macrophages, we demonstrated that the induction of CEBPD by hypoxia or IL-1β, and subsequent expression of HIF-1α, required NF-κB activity. In addition, CEBPD overexpression partially rescued HIF-1α protein synthesis under NF-κB inhibition, indicating that CEBPD was the key regulator of HIF-1α downstream of NF-κB. Interestingly, the NF-κB binding site (−197/−188 bp)48 is adjacent to the responsible CEBPD binding site (−227/−222 bp) in the HIF-1α promoter. In this regard, it remains an open question whether these two elements synergistically function to increase HIF-1α transcription. Indeed, C/EBP members have been reported to physically and functionally interact with transcription factors other than the bZIP proteins.49, 50, 51 The interaction of CEBPD with NF-κB has also been reported,52, 53 and this may account for the mechanism of stimulus-specific transcriptional responses, as observed in IL-1β stimulation.
In conclusion, we have discovered the CEBPD-HIF-1 pathway as a new HIF-1-regulating fundamental mechanism in tubular epithelial cells for the cellular protection against kidney injury. The data also point to an important role for NF-κB in this process and provide a potential link between hypoxia and inflammation through the cross-talk of macrophages and tubular cells, which in particular provide insights into how the tubular environment in the kidney is maintained under pathophysiological conditions. The results of our present study advance our understanding of the HIF-1 response in the kidney and may open up possibilities to control various aspects of the hypoxic response by experimentally manipulating CEBPD/HIF-1 signaling.
METHODS
Cell culture
HK-2, a human proximal tubular cell line, was cultured in Dulbecco's modified Eagle's medium/F-12 (Nissui, Tokyo, Japan) supplemented with 10% fetal calf serum. HeLa cervical cancer cells and A549 lung cancer cells were maintained in Dulbecco's modified Eagle's medium (Nissui) containing 10% fetal calf serum. 0.1% O2 stimulation was induced with an anaerobic bag (Anaerocult A sachets, Merck Chemicals, Darmstadt, Germany). 1% O2 stimulation was accomplished by exposing cells to 1% O2/5% CO2, with the balance as nitrogen, in a multigas incubator, APM-30D (ASTEC, Fukuoka, Japan).
Reagents
LPS (E Coli 0111:B4, L3012), actinomycin, MG132, cycloheximide, and BAY11-7082 were purchased from Sigma (Sigma-Aldrich, St. Louis, MO). LY294002, human angiotensin II, and rapamycin were purchased from Calbiochem (Calbiochem, Gibbstown, NJ). Recombinant human IL-1β (Roche, Basel, Switzerland), DMOG (Cayman Chemical, Ann Arbor, MI), and recombinant human TNF-α (R&D Systems, Minneapolis, MN) were used.
Luciferase reporter assay
The HIF-1 transcriptional activity was measured using dual-luciferase reporter assays (Promega, Madison, WI) in 24-well culture dishes. 500 ng pGL3 (Promega) vector driven by 7 × hypoxia-responsive elements (pHREluc)9 and 25 ng Renilla luciferase vector (pRL-CMV, pRL-TK, or pRL-SV40) were co-transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA); 24 h later, the cells were exposed to hypoxia (1% or 0.1%) for 16 h. The cells were processed for the dual-luciferase assay in fixed protein aliquots, and HREluc activity was measured using a Lumat 9507 luminometer (EG and Berthold, Bad Wildbad, Germany). The relative light unit value of the firefly luciferase was divided by that of the Renilla luciferase to correct for the transfection efficiency. To measure the HIF-1α promoter activity, a similar method as above was used. The human 1.3-kb HIF-1α promoter (hHIF-1α1.3K promoter) fragment was PCR amplified from human genomic DNA using PrimeStar HS DNA polymerase (Takara Bio, Shiga, Japan) and primers with SacI or NheI overhangs. Digested and purified fragments were inserted into the pGL3basic reporter vector (Promega). Mutations of the potential C/EBP-binding sites (screened by TRANSFAC, indicated in Figure 6) of the hHIF-1α1.3k promoter were introduced through PCR. The PCR primers are listed in Table 1. NF-κB reporter activity was measured using pNF-kappaB RE-TK hRluc (F) (RIKEN BIOResource Center, Tsukuba, Japan), and pRL-TK was used as control. The transfection efficacy was corrected to the pGL3-basic vector.
Table 1. A list of primers used in this study.
| Gene | Use | Species | F/R | Sequence (5′→3′) |
|---|---|---|---|---|
| CEBPD | RT–PCR | Human | Forward | GAAGAGCGCCGGCAAGAGGG |
| Reverse | CGGCGATGTTGTTGCGCTCG | |||
| CEBPD | RT–PCR | Rat | Forward | GAATTGCTACAGTTTCTTGG |
| Reverse | ATGCGCAGTCTCTTCCTC | |||
| CEBPD | RT–PCR | Mouse | Forward | TCGACTTCAGCGCCTACATTGACT |
| Reverse | CCGCTTTGTGGTTGCTGTTGAAGA | |||
| HIF-1α | RT–PCR | Human | Forward | CCATTAGAAAGCAGTTCCGC |
| Reverse | TGGGTAGGAGATGGAGATGC | |||
| GLUT1 | RT–PCR | Human | Forward | CTTCACTGTCGTGTCGCTGT |
| Reverse | CCAGGACCCACTTCAAAGAA | |||
| VEGF | RT–PCR | Human | Forward | TCTGAGCAAGGCCCACAGGGA |
| Reverse | CCCTGATGAGATCGAGTACATCTT | |||
| IL-6 | RT–PCR | Human | Forward | AGTTCCTGCAGAAAAAGGCA |
| Reverse | AAAGCTGCGCAGAATGAGAT | |||
| MCP-1 | RT–PCR | human | Forward | CCCCAGTCACCTGCTGTTAT |
| Reverse | AGATCTCCTTGGCCACAATG | |||
| IL-1β | RT–PCR | Rat | Forward | CACCTCTCAAGCAGAGCACAG |
| Reverse | GGGTTCCATGGTGAAGTCAAC | |||
| HIF-1α | ChIP–PCR | Human | Forward | TCTCCTTTCTCTTTCCTCCG |
| Reverse | GGTTCCTCGAGATCCAATG | |||
| IL-6 | ChIP–PCR | Human | Forward | TTTCCCCCTAGTTGTGTCTT |
| Reverse | AATCTTTGTTGGAGGGTGAG | |||
| HBB | ChIP–PCR | Human | Forward | GGGCTGAGGGTTTGAAGTCC |
| Reverse | CATGGTGTCTGTTTGAGGTTGC | |||
| CEBPD | Vector | Human | Forward | CAGCTCGAGGTGACAGCCTCGCTTGGA |
| Reverse | CGTACGTATGGGTCGTTGCTGAGTCTCT | |||
| CEBPB | Vector | Human | Forward | CAGCTCGAGAAACTTTAGCGAGTCAGAG |
| Reverse | CGTACGTAAAATTACCGACGGGCTCCC | |||
| hHIF-1α1.3K | Reporter assay | Human | Forward | GCGAGCTCACACCTCTGAGTAGCTGGA |
| Reverse | GCGCTAGCTACCGAGCTGCAGCTCATT | |||
| hHIF-1α1.3K (mut-1222) | Reporter assay | Human | Forward | GTAGGTCTACTGAACGTTTAAAAAGAGGTG |
| Reverse | CACCTCTTTTTAAACGTTCAGTAGACCTAC | |||
| hHIF-1α1.3K (mut-871) | Reporter assay | human | Forward | CTCTTTCCCTGAAAACGTTTTATATGCTTA |
| Reverse | TAAGCATATAAAACGTTTTCAGGGAAAGAG | |||
| hHIF-1α1.3K (mut-837) | Reporter assay | Human | Forward | TTAACATAGTTTCAGATTTTAAGAT |
| Reverse | GGTATAAATGACTTTAATAAGCATA | |||
| hHIF-1α1.3K (mut-629) | Reporter assay | Human | Forward | AAAGACCCCGTTGTAACACATCTG |
| Reverse | ACCTAACCATGGGTCAATGTCACC | |||
| hHIF-1α1.3K (mut-227) | Reporter assay | Human | Forward | GCTGACCGCCTCCTGGTACCCTGAGAGCGG |
| Reverse | CCGCTCTCAGGGTACCAGGAGGCGGTCAGC | |||
| HIF-1α | siRNA | Human | Forward | AUAUGAUUGUGUCUCCAGCGGCUGG |
| Reverse | CCAGCCGCUGGAGACACAAUCAUAU | |||
| Negative control | siRNA | Human | Forward | GUACCGCACGUCAUUCGUAUC |
| Reverse | UACGAAUGACGUGCGGUACG |
Abbreviations: CEBPD, CCAAT/enhancer-binding protein δ ChIP–PCR, chromatin immunoprecipitation–PCR; GLUT1, glucose transporter 1; HIF-1α, hypoxia-inducible factor 1α IL, interleukin; MCP-1, monocyte chemotactic protein-1; RT–PCR, reverse transcriptase–PCR; siRNA, small interfering RNA; VEGF, vascular endothelial growth factor.
Plasmids and transient or stable overexpression
For transient overexpression studies, cDNA encoding human CEBPB or CEBPD was amplified from HK-2 cells using the primers listed in Table 1 and subcloned into the pMD20-T vector (Takara Bio). Each fragment was then subcloned into pcDNA3.1 (−) (Promega). For CEBPB/CEBPD double transfections, each vector amount was reduced to half the dose of single vector transfection (5 μg each, making total 10 μg vector/dish). HK-2 clones, which stably overexpress CEBPD, were generated through retrovirus transduction (Platinum Retrovirus Expression System, Pantropic, Cell Biolabs, San Diego, CA). The above CEBPD in the pMD20-T vector was further subcloned into the retroviral vector, pMXs-IRES-Puro, using the XhoI-SnaBI site. The plasmids were transfected to the packaging cell line, and, the culture supernatants were collected 24 and 48 h later, passed through 0.22-μm filters, and added to the cells. Drug selection by 4 μg/ml puromycin was initiated 72 h after infection.
shRNA library screening
The shRNA library screening was performed using essentially the same method as in our previous work.54 The shRNA expression plasmid library was constructed against the top 150 continuously upregulated genes at days 3 and 7 in the RAS model, based on the pcPUR hU6 vector (Tsukuba, Ibaraki, Japan), which contained a human U6 promoter, a puromycin resistance gene, and BspMI cloning sites. Two shRNA vectors were used against each gene. For the initial screening, the shRNA plasmid, the pHRE-luc vector, and the pRL-CMV vector were transfected into HeLa cells using Lipofectamine 2000 according to the manufacturer's instructions. Forty-eight hours after transfection, the cells were cultured in 1% hypoxic conditions for 16 h and evaluated for HRE reporter activity. The pcPUR hU6 vector containing a targeted sequence against the HIF-1α gene and 7 × tandem thymidine repeats (T7) served as positive control and negative control, respectively. A clone fulfilling the following two criteria in two independent experiments was considered a hit clone: (1) its HREluc activity change was within the range of 50% compared with positive control; and (2) its HREluc activity ratio (1% O2/21% O2) was within the range of 50% compared with positive control. The hit clones in the first screening proceeded to the second screening. HeLa cells stably expressing candidate shRNA plasmids were used. The shRNA plasmids were transfected into HeLa cells using Lipofectamine 2000. Drug selection with 1 μg/ml puromycin was initiated after 48 h. The selected cells in bulk were then cultured in 1% O2 condition for 16 h and assessed for HIF-1α protein levels through immunoblotting, and supernatant VEGF levels were assessed using enzyme-linked immunosorbent assay (ELISA) (R&D Systems). For HREluc measurements, the pHRE-luc vector and the pRL-CMV vector were further transfected into these HeLa cells and treated as described above. These three measurements were then scored together for the degree of HIF-1 regulation.
siRNA transfection
Control siRNA (Sigma-Aldrich), p65 siRNA (SASI_Hs01_00171091, Sigma-Aldrich), HIF-1α siRNA (Invitrogen), CEBPD siRNA (L-010453-00, Thermo Fischer Scientific, Waltham, MA), and CEBPB siRNA (L-006423-00, Thermo Fischer Scientific) were used for the siRNA experiments. The target siRNA sequences are listed in Table 1. HeLa or HK-2 cells were transfected with 5 nM siRNA using Lipofectamine 2000 according to the manufacturer's instruction. After 24 or 48 h, the siRNA-treated cells were processed for further experiments.
Real-time qRT–PCR
Real-time qRT–PCR assays were performed as previously described.32, 55 The data were calibrated to the β-actin value. The corresponding primers are described in the literature and Table 1.
Immunoblotting analysis
The cells were collected for immunoblotting analysis as previously described.32, 55 The primary antibodies were as follows: anti-HIF1α (Novus Biologicals, Littleton, CO); anti-CEBPD (Santa Cruz Biotechnology, Santa Cruz, CA); anti-CEBPB (Santa Cruz Biotechnology); anti-p65 (Santa Cruz Biotechnology); anti-phospho-Akt (Ser473) (Cell Signaling Technology, Danvers, MA); anti-phospho-p70 S6 kinase (Thr389) (Cell Signaling Technology); and anti-actin (Sigma-Aldrich). Horseradish peroxidase–conjugated anti-rabbit immunoglobulin G (IgG; Bio-Rad Laboratories, Hercules, CA) or anti-mouse IgG (Bio-Rad Laboratories) antibodies were used as secondary antibodies. The signals were detected with the ECL Plus reagent (Thermo Scientific) using the chemiluminescence protocol.
Chromatin immunoprecipitation
The binding of CEBPD to the HIF-1α (ChIP) promoter was assessed using the chromatin immunoprecipitation assay. HK-2 cells were cross-linked for 10 min using 1% paraformaldehyde and sonicated into fragments. The fragmented DNA forming a complex with CEBPD was immunoprecipitated with an anti-CEBPD rabbit polyclonal antibody (Santa Cruz Biotechnology) and protein Sepharose beads (GE Healthcare, Buckinghamshire, UK). The precipitated DNA was used as a template for PCR reactions with the primers listed in Table 1. The IL-6 promoter region has known IL-1β-induced CEBPD-binding site, and this was used as a positive control.56 HBB is the negative-control primer, which was designed for the promoter regions of chromosome 2.
In vivo experiments
Male C57BL/6NJc1 mice aged 7 weeks (Nippon Seibutsu Zairyo Center, Tokyo, Japan) were exposed to 8% hypoxia for 6 h using an oxygen controller, ProOx110 (BioSpherix, Lacona, NY). The mice exposed to ambient air were used as controls. The mice were euthanized, and the kidneys were harvested and stored at −80 °C for qRT–PCR analysis. Other animal experiments were performed using male Wistar rats aged 6–8 weeks (Nippon Seibutsu Zairyo Center) under pentobarbital anesthesia. Sham-control rats received only laparotomy, except for the cisplatin nephrotoxicity model, where they received intraperitoneal saline injection. At euthanasia, the kidneys were harvested, fixed in formalin for immunohistochemical analysis, snap-frozen in OCT (Optimal Cutting Temperature), or stored at −80 °C for qRT–PCR analysis. For acute hypoxic kidney models, I/R injury and cisplatin nephrotoxicity models were adopted. Two distinct models of renal disease, the RAS model and the RK (5/6 nephrectomy) model, were used for chronic hypoxic kidney models. The surgical procedures were performed as previously described.8, 9, 10, 11, 19 Briefly, for I/R injury, after a midline abdominal incision and right kidney removal, left renal arteries and veins were occluded for 45 min with microaneurysm clamps. The body temperature was kept constant at 37 °C during the operation. Twenty-four hours following clamp release, the rats were euthanized. Cisplatin nephrotoxicity was created by injecting 6 mg/kg cisplatin intraperitoneally, and the rats were euthanized 3 days later. RAS was induced by placing a U-shaped silver clip (0.23 mm internal diameter) around the left renal artery through a midline abdominal incision. The RK model was constructed in two steps. One week prior to disease induction, the right kidney was removed. At day 0, infarction of approximately two-thirds of the left kidney was accomplished by ligation of the posterior and two anterior branches of the main renal artery. The rats were euthanized either at 1 or 4 weeks. All mice and rats were housed in the animal care facility of the University of Tokyo under standardized pathogen-free conditions (25 °C, 50% humidity, 12 h light/dark cycle) with food and water available ad libitum. All experiments were performed in accordance with the guidelines of the Committee on Ethical Animal Care and Use at the University of Tokyo Graduate School of Medicine (M-P10-077).
Histopathology and immunohistochemistry
The kidney tissues were prepared and stained as previously described.32, 55 Briefly, the kidneys were either immersion fixed in neutralized formalin solutions and embedded in paraffin or snap-frozen in OCT compound. Periodic acid–Schiff staining was used for routine histological examination. An indirect immunoperoxidase method was used to identify the following antigens: CEBPD with anti-CEBPD rabbit polyclonal antibody (Santa Cruz Biotechnology); and monocytes/macrophages with murine monoclonal IgG1 antibody endothelin-1 (Chemicon, Temecula, CA). Briefly, sections (3 μm) were deparaffinized and brought to water through graded ethanols. Antigen retrieval was performed by boiling sections in a microwave in 1 mmol/l EDTA, pH 6.0. Endogenous peroxidase activity was quenched using 3% hydrogen peroxide; nonspecific protein binding was blocked using 2% normal horse serum (Sigma-Aldrich), and endogenous biotin activity was quenched using the Avidin/Biotin Blocking Kit (Vector Laboratories, Burlingame, CA). After blocking, the tissue sections were incubated with the indicated primary antibody overnight at 4 °C. The corresponding biotinylated secondary antibodies anti-rabbit IgG (Vector Laboratories) or anti-mouse IgG (Vector Laboratories) were applied followed by an R.T.U. Vectastain Kit (Vector Laboratories). The immunoreactive antigens sites were detected with hydrogen peroxide and diaminobenzidine (Wako, Osaka, Japan). For immunofluorescent staining, frozen tissue sections (4 μm) were briefly air dried and fixed in 100% methanol. The sections were then blocked with 2% normal horse serum and incubated with primary antibodies: anti-CEBPD rabbit polyclonal antibody (Santa Cruz Biotechnology), biotinylated PHA-E (Vector Laboratories), THP (Cappel, West Chester, PA), or CaBP28k (Sigma-Aldrich) overnight at 4 °C. The sections were incubated at room temperature for 40 min with secondary antibodies: Texas Red-conjugated anti-rabbit IgG (Molecular Probes, Eugene, OR), Alexa488 (Molecular Probes), fluorescein isothiocyanate–conjugated (FITC) anti-goat IgG (DAKO, Tokyo, Japan), and FITC anti-mouse IgG (Southern Biotechnology, Birmingham, AL). The images were captured on a BZ-9000 fluorescence microscope (Keyence, Osaka, Japan) with the BZ-analyzer software (Keyence).
Statistical analysis
All data are reported as the means±s.e.m. or the means±s.d. The data for two groups were analyzed using unpaired Student's t-test. The differences among more than two groups were analyzed using a one-way analysis of variance (ANOVA) with post-Dunnett's test or a two-way ANOVA with post-Bonferroni's test. Differences with a P-value<0.05 were considered significant. GraphPad Prism version 5.04 for Windows (GraphPad Software, San Diego, CA) was used for data analysis.
Acknowledgments
This work was supported by Grant-in-Aids for Scientific Research 24390213 (to MN) and 22790781 (to TT) from the Japan Society for the Promotion of Science, the Naito Kinen Foundation (to JY) and Kyowa Hakko Kirin, Tokyo, Japan. We thank Dr Makiko Koike (Kyowa Hakko Kirin) for the helpful discussions.
All the authors declared no competing interests.
Footnotes
SUPPLEMENTARY MATERIAL
Figure S1. Second shRNA library screening for HIF-1 regulating genes.
Figure S2. CEBPD/HIF-1 pathway induction by hypoxia, Related to Figure 3.
Figure S3. CEBPD/HIF-1 pathway induction by inflammatory stimuli, Related to Figure 5.
Figure S4. Mechanism of HIF-1 regulation by CEBPD, Related to Figure 6.
Table S1. A list of top 150 genes in the renal artery stenosis microarray.
Supplementary material is linked to the online version of the paper at http://www.nature.com/ki
Supplementary Material
References
- Semenza GL. Oxygen sensing, homeostasis, and disease. N Engl J Med. 2011;365:537–547. doi: 10.1056/NEJMra1011165. [DOI] [PubMed] [Google Scholar]
- Bohle A, Mackensen-Haen S, Wehrmann M. Significance of postglomerular capillaries in the pathogenesis of chronic renal failure. Kidney Blood Press Res. 1996;19:191–195. doi: 10.1159/000174072. [DOI] [PubMed] [Google Scholar]
- Fine LG, Bandyopadhay D, Norman JT. Is there a common mechanism for the progression of different types of renal diseases other than proteinuria? Towards the unifying theme of chronic hypoxia. Kidney Int. 2000;57 (Suppl 75:S22–S26. [PubMed] [Google Scholar]
- Nangaku M. Chronic hypoxia and tubulointerstitial injury: a final common pathway to end-stage renal failure. J Am Soc Nephrol. 2006;17:17–25. doi: 10.1681/ASN.2005070757. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Wiesener M, Bernhardt W, et al. The human HIF (hypoxia-inducible factor)-3alpha gene is a HIF-1 target gene and may modulate hypoxic gene induction. Biochem J. 2009;424:143–151. doi: 10.1042/BJ20090120. [DOI] [PubMed] [Google Scholar]
- Rosenberger C, Mandriota S, Jürgensen JS, et al. Expression of hypoxia-inducible factor-1α and -2α in hypoxic and ischemic rat kidneys. J Am Soc Nephrol. 2002;13:1721–1732. doi: 10.1097/01.asn.0000017223.49823.2a. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Makino Y, Tanaka T, et al. Induction of renoprotective gene expression by cobalt ameliorates ischemic injury of the kidney in rats. J Am Soc Nephrol. 2003;14:1825–1832. doi: 10.1097/01.asn.0000074239.22357.06. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Miyata T, Inagi R, et al. Hypoxia in renal disease with proteinuria and/or glomerular hypertension. Am J Pathol. 2004;165:1979–1992. doi: 10.1016/S0002-9440(10)63249-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Kojima I, Ohse T, et al. Hypoxia-inducible factor modulates tubular cell survival in cisplatin nephrotoxicity. Am J Physiol Renal Physiol. 2005;289:F1123–F1133. doi: 10.1152/ajprenal.00081.2005. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Kojima I, Ohse T, et al. Cobalt promotes angiogenesis via hypoxia-inducible factor and protects tubulointerstitium in the remnant kidney model. Lab Invest. 2005;85:1292–1307. doi: 10.1038/labinvest.3700328. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Matsumoto M, Inagi R, et al. Induction of protective genes by cobalt ameliorates tubulointerstitial injury in the progressive Thy1 nephritis. Kidney Int. 2005;68:2714–2725. doi: 10.1111/j.1523-1755.2005.00742.x. [DOI] [PubMed] [Google Scholar]
- Bernhardt WM, Câmpean V, Kany S, et al. Preconditional activation of hypoxia-inducible factors ameliorates ischemic acute renal failure. J Am Soc Nephrol. 2006;17:1970–1978. doi: 10.1681/ASN.2005121302. [DOI] [PubMed] [Google Scholar]
- Bernhardt WM, Gottmann U, Doyon F, et al. Donor treatment with a PHD-inhibitor activating HIFs prevents graft injury and prolongs survival in an allogenic kidney transplant model. Proc Natl Acad Sci USA. 2009;106:21276–21281. doi: 10.1073/pnas.0903978106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill P, Shukla D, Tran MG, et al. Inhibition of hypoxia inducible factor hydroxylases protects against renal ischemia-reperfusion injury. J Am Soc Nephrol. 2008;19:39–46. doi: 10.1681/ASN.2006090998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- Blouin CC, Pagé EL, Soucy GM, et al. Hypoxic gene activation by lipopolysaccharide in macrophages: implication of hypoxia-inducible factor 1alpha. Blood. 2004;103:1124–1130. doi: 10.1182/blood-2003-07-2427. [DOI] [PubMed] [Google Scholar]
- Rius J, Guma M, Schachtrup C, et al. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature. 2008;453:807–811. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima I, Tanaka T, Inagi R, et al. Metallothionein is upregulated by hypoxia and stabilizes hypoxia-inducible factor in the kidney. Kidney Int. 2009;75:268–277. doi: 10.1038/ki.2008.488. [DOI] [PubMed] [Google Scholar]
- Takiguchi M. The C/EBP family of transcription factors in the liver and other organs. Int J Exp Pathol. 1998;79:369–391. doi: 10.1046/j.1365-2613.1998.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manotham K, Tanaka T, Matsumoto M, et al. Evidence of tubular hypoxia in the early phase in the remnant kidney model. J Am Soc Nephrol. 2004;15:1277–1288. doi: 10.1097/01.asn.0000125614.35046.10. [DOI] [PubMed] [Google Scholar]
- Min Y, Ghose S, Boelte K, et al. C/EBP-δ regulates VEGF-C autocrine signaling in lymphangiogenesis and metastasis of lung cancer through HIF-1α. Oncogene. 2011;30:4901–4909. doi: 10.1038/onc.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam T, An MR, Papaconstantinou J. Differential expression of three C/EBP isoforms in multiple tissues during the acute phase response. J Biol Chem. 1992;267:5021–5024. [PubMed] [Google Scholar]
- Juan TS, Wilson DR, Wilde MD, et al. Participation of the transcription factor C/EBP delta in the acute-phase regulation of the human gene for complement component C3. Proc Natl Acad Sci USA. 1993;90:2584–2588. doi: 10.1073/pnas.90.7.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamurugan K, Sharan S, Klarmann KD, et al. FBXW7α attenuates inflammatory signalling by downregulating C/EBPδ and its target gene Tlr4. Nat Commun. 2013;4:1662. doi: 10.1038/ncomms2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treins C, Giorgetti-Peraldi S, Murdaca J, et al. Insulin stimulates hypoxia-inducible factor 1 through a phosphatidylinositol 3-kinase/target of rapamycin-dependent signaling pathway. J Biol Chem. 2002;277:27975–27981. doi: 10.1074/jbc.M204152200. [DOI] [PubMed] [Google Scholar]
- Zhong H, Chiles K, Feldser D, et al. Modulation of hypoxia-inducible factor 1alpha expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Res. 2000;60:1541–1545. [PubMed] [Google Scholar]
- Stiehl DP, Jelkmann W, Wenger RH, et al. Normoxic induction of the hypoxia-inducible factor 1alpha by insulin and interleukin-1beta involves the phosphatidylinositol 3-kinase pathway. FEBS Lett. 2002;512:157–162. doi: 10.1016/s0014-5793(02)02247-0. [DOI] [PubMed] [Google Scholar]
- Ricardo SD, van Goor H, Eddy AA. Macrophage diversity in renal injury and repair. J Clin Invest. 2008;118:3522–3530. doi: 10.1172/JCI36150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung AC, Lan HY. Chemokines in renal injury. J Am Soc Nephrol. 2011;22:802–809. doi: 10.1681/ASN.2010050510. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Yamaguchi J, Higashijima Y, et al. Indoxyl sulfate signals for rapid mRNA stabilization of Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 2 (CITED2) and suppresses the expression of hypoxia-inducible genes in experimental CKD and uremia. FASEB J. 2013;27:4059–4075. doi: 10.1096/fj.13-231837. [DOI] [PubMed] [Google Scholar]
- Balamurugan K, Wang JM, Tsai HH, et al. The tumour suppressor C/EBPδ inhibits FBXW7 expression and promotes mammary tumour metastasis. EMBO J. 2010;29:4106–4117. doi: 10.1038/emboj.2010.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada S, Yamamoto H, Nishihara T, et al. DNA binding specificity of the CCAAT/enhancer-binding protein transcription factor family. J Biol Chem. 1996;271:3891–3896. doi: 10.1074/jbc.271.7.3891. [DOI] [PubMed] [Google Scholar]
- Litvak V, Ramsey SA, Rust AG, et al. Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat Immunol. 2009;10:437–443. doi: 10.1038/ni.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Umek RM, McKnight SL. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- Kinoshita S, Akira S, Kishimoto T. A member of the C/EBP family, NF-IL6 beta, forms a heterodimer and transcriptionally synergizes with NF-IL6. Proc Natl Acad Sci USA. 1992;89:1473–1476. doi: 10.1073/pnas.89.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AM, Montagna C, Sharan S, et al. Loss of CCAAT/enhancer binding protein delta promotes chromosomal instability. Oncogene. 2004;23:1549–1557. doi: 10.1038/sj.onc.1207285. [DOI] [PubMed] [Google Scholar]
- Eltzschig HK, Carmeliet P. Hypoxia and inflammation. N Engl J Med. 2011;364:656–665. doi: 10.1056/NEJMra0910283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizet V, Johnson RS. Interdependence of hypoxic and innate immune responses. Nat Rev Immunol. 2009;9:609–617. doi: 10.1038/nri2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhausen J, Furuta GT, Tomaszewski JE, et al. Epithelial hypoxia-inducible factor-1 is protective in murine experimental colitis. J Clin Invest. 2004;114:1098–1106. doi: 10.1172/JCI21086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer T, Yamanishi Y, Clausen BE, et al. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–657. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Gilbert V, Liu Q, et al. Myeloid cell-derived hypoxia-inducible factor attenuates inflammation in unilateral ureteral obstruction-induced kidney injury. J Immunol. 2012;188:5106–5115. doi: 10.4049/jimmunol.1103377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Yokote S, Yamada A, et al. Adipose tissue-derived mesenchymal stem cells in long-term dialysis patients display downregulation of PCAF expression and poor angiogenesis activation. PLoS One. 2014;9:1–14. doi: 10.1371/journal.pone.0102311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Sivko JS, DeWille JW. Promoter methylation reduces C/EBPdelta (CEBPD) gene expression in the SUM-52PE human breast cancer cell line and in primary breast tumors. Breast Cancer Res Treat. 2006;95:161–170. doi: 10.1007/s10549-005-9061-3. [DOI] [PubMed] [Google Scholar]
- Ko CY, Hsu HC, Shen MR, et al. Epigenetic silencing of CCAAT/enhancer-binding protein delta activity by YY1/polycomb group/DNA methyltransferase complex. J Biol Chem. 2008;283:30919–30932. doi: 10.1074/jbc.M804029200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick SF, Tambuwala MM, Bruning U, et al. An intact canonical NF-κB pathway is required for inflammatory gene expression in response to hypoxia. J Immunol. 2011;186:1091–1096. doi: 10.4049/jimmunol.1002256. [DOI] [PubMed] [Google Scholar]
- Bonello S, Zähringer C, BelAiba RS, et al. Reactive oxygen species activate the HIF-1alpha promoter via a functional NFkappaB site. Arterioscler Thromb Vasc Biol. 2007;27:755–761. doi: 10.1161/01.ATV.0000258979.92828.bc. [DOI] [PubMed] [Google Scholar]
- LeClair KP, Blanar MA, Sharp PA. The p50 subunit of NF-kappa B associates with the NF-IL6 transcription factor. Proc Natl Acad Sci USA. 1992;89:8145–8149. doi: 10.1073/pnas.89.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Yano M, Liu SY, et al. A novel cis-acting element controlling the rat CYP2D5 gene and requiring cooperativity between C/EBP beta and an Sp1 factor. Mol Cell Biol. 1994;14:1383–1394. doi: 10.1128/mcb.14.2.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mink S, Haenig B, Klempnauer KH. Interaction and functional collaboration of p300 and C/EBPbeta. Mol Cell Biol. 1997;17:6609–6617. doi: 10.1128/mcb.17.11.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl JA, Hannink M. Identification of a C/EBP-Rel complex in avian lymphoid cells. Mol Cell Biol. 1994;14:6635–6646. doi: 10.1128/mcb.14.10.6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Hannink M, Ray BK. Concerted participation of NF-kappa B and C/EBP heteromer in lipopolysaccharide induction of serum amyloid A gene expression in liver. J Biol Chem. 1995;270:7365–7374. doi: 10.1074/jbc.270.13.7365. [DOI] [PubMed] [Google Scholar]
- Eto N, Miyagishi M, Inagi R, et al. Mitogen-activated protein 3 kinase 6 mediates angiogenic and tumorigenic effects via vascular endothelial growth factor expression. Am J Pathol. 2009;174:1553–1563. doi: 10.2353/ajpath.2009.080190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Yamaguchi J, Shoji K, et al. Anthracycline inhibits recruitment of hypoxia-inducible transcription factors and suppresses tumor cell migration and cardiac angiogenic response in the host. J Biol Chem. 2012;287:34866–34882. doi: 10.1074/jbc.M112.374587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio Y, Isshiki H, Kishimoto T, et al. A nuclear factor for interleukin-6 expression (NF-IL6) and the glucocorticoid receptor synergistically activate transcription of the rat alpha 1-acid glycoprotein gene via direct protein-protein interaction. Mol Cell Biol. 1993;13:1854–1862. doi: 10.1128/mcb.13.3.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.