Summary
The completion of human and mouse genome sequencing has confronted us with huge amount of data sequences that certainly need decades and many generations of scientists to be reasonably interpreted and assigned to physiological functions, and subsequently fruitfully translated into medical application. A means to assess the function of genes provides gene targeting in mouse embryonic stem (ES) cells that enables to introduce site-specific modifications in the mouse genome, and analyze their physiological consequences. Gene targeting enables almost any type of genetic modifications of interest, ranging from gene insertion (e.g. insertion of human-specific genes or reporter genes), gene disruption, point mutations, short and long range deletions, inversions. Site-specific modification into the genome of ES cells can be reached by homologous recombination using targeting vectors. Here, we describe a protocol to generate targeting constructs and homologous recombinant ES cells.
Keywords: ES cells, targeting vector, MultiSite Gateway Cloning
1. Introduction
The development of mice with site-specific genome modification has become possible because of the establishment of fundamental techniques that enable to culture embryonic stem (ES) cells in vitro, without altering their pluripotent potential, and to mediate homologous recombination between specific sites in the genome of ES cells and exogenously added DNA molecules.
In the following section, we describe a protocol to generate gene targeting constructs, culture ES cells and generate homologous recombinant ES cell clones. For detailed background overview about gene targeting in mice, we refer to the review chapter (Gene Targeting in Mice: a Review) in this issue of “Virus-Host-Interactions”.
2. Materials
2.1. Cloning of the targeting vector
MultiSite Gateway Three-Fragment Vector Construction Kit (Invitrogen).
Plasmids containing floxed Neomycin gene (loxP-neo-loxP) (or other resistance genes of preference, such as hygromycin or puromycin etc.). If appropriate, the resistance gene can also be flanked by FRT instead of loxP.
If required, vectors containing a reporter gene of interest and negative selection cassette (e.g. thymidine kinase (TK), diphtheria toxin fragment A (DT-A) or Cre recombinase), respectively.
Cosmid, bacteria artificial chromosome (BAC) or appropriate vector containing the mouse genomic sequence (gene) that should be targeted, and from which the homology arms for the targeting vector will be derived. Alternatively, the homology arms can be amplified directly from whole genomic DNA, isolated from the ES cell line that will be used for gene targeting (this ensures isogeny).
CcdB Survival E. coli bacteria. This CcdB-resistant strain can be used to propagate and maintain vectors containing the ccdB gene, such as Gateway Donor vector.
Restriction enzymes.
T4 DNA ligase.
Shrimp alkaline phosphatase.
PCR reagents: PfuUltra High-fidelity DNA polymerase (buffer provided with the enzyme) (Stratagene), or Q5 Hot Start High-Fidelity DNA Polymerase (New England Biolabs), dNTPs, primers.
Highly competent E. coli with highest efficiency cloning of large plasmids (see Note 1).
PCR purification kit.
Plasmid Mini- and Midi-preparation kits.
Gel DNA extraction kit (kit enabling damage-free extraction of big DNA fragments).
Tris-equilibrated phenol.
Chloroform:isoamyl alcohol (24:1) mixture.
Absolute ethanol.
3 M sodium-acetate at pH 5.2.
70% ethanol.
0.5 M EDTA.
TE-Puffer: 10 mM Tris-HCL (pH 8.0), 1 mM EDTA (pH 8.0).
2.2. Media for cell culture
MEF medium: Dulbecco’s modified Eagle medium (DMEM), containing 10% fetal bovine serum (FBS), 100 μM β-mercaptoethanol, 100 U/mL Penicillin, 100 μg /mL Streptomycin.
ESC medium: DMEM with 4.5 g/L glucose and 1 mM Na-pyruvate (Invitrogen), supplemented with 15% FBS (tested for ES culture), 2 mM L-glutamine, 100 μM ß-mercaptoethanol, 1× nonessential amino acids of 100 × stock solution (Invitrogen), 1000 U/mL leukemia inhibitory factor (LIF), and if preferred 100 units/mL Penicillin, and 100μg/mL Streptomycin. The use of Penicillin/Streptomycin can often mask low level contamination of cell culture with microorganisms such as mycoplasma.
ESC-G418 medium: ESC medium supplemented with 250-400 μg/mL G418 (Geneticin) (the concentration of G418 depends e.g. on the used ES cell line).
2.3.Isolation, culture and mitotic inactivation of mouse embryonic fibroblasts (MEFs)
Transgenic mice harbouring the same resistance gene (e.g. neomycin or hygromycin) that is used for the positive selection of transfected ES cells (can be purchased from the Jackson Laboratories).
Sterile phosphate-buffered saline (PBS).
Trypsin-EDTA solution.
MEF medium (from Subheading 2.2.).
70% ethanol.
Sterile dissecting instruments (e.g. dissection scissors, forceps etc.).
Cesium source γ irradiator.
Freeze medium: DMEM with 10% DMSO and 20% FBS.
2.4. Culture of ES cells
-
4.
ES cells: ES cell lines can be purchased from commercial sources, e.g. http://www.lgcstandards-atcc.org/
-
5.
ESC medium (from Subheading 2.2.).
-
6.
ESC-G418 medium (from Subheading 2.2.).
2.5. Electroporation of ES cells with the targeting vector
Linearized targeting vector.
PBS.
ES cells.
ESC medium.
Trypsin-EDTA.
Mitotically inactive MEFs.
Electroporation cuvette for eukaryotic cells.
Gene pulser.
2.6. Selection of positively transfected ESC clones
ESC medium (from Subheading 2.2.).
ESC-G418 medium (from Subheading 2.2.).
MEF medium (from Subheading 2.2.).
Mitotically inactive MEFs (from Subheading 2.3.).
96- and 24-well plates.
Trypsin-EDTA solution.
Laminar flow cabinet.
Gloves.
Face mask.
2.7. Freezing of and DNA preparation from ESC clones
Trypsin-EDTA.
Freeze medium: DMEM with 10% DMSO and 20% FBS.
ESC medium (from Subheading 2.2.).
Lysis buffer: 100 mM Tris-HCl (pH 8.5), 5 mM EDTA, 200 mM NaCl, 0.2% SDS and every time freshly added 100 μg/mL Proteinase K.
Isopropanol.
TE-Puffer: 10 mM Tris-HCL (pH 8,0), 1 mM EDTA (pH 8,0).
2.8.Screening by southern blot analysis for ESC clones with homologous recombination of the targeting vector
Restriction enzymes.
Proteinase K.
TBE buffer: 1.1 M Tris-base (54 g), 900 mM Borate (27.5 g), 25 mM EDTA, adjust to pH 8, and bring the final volume to 1 L with deionized water.
Depurination solution: 0.2M HCl (8 mL 37% HCl in 500 mL H2O).
Denaturing buffer: 0.5 M NaOH, 1.5 M NaCl.
Neutralization buffer: 0.5 M Tris–HCl, 1.5 M NaCl, pH 7.2.
Positive charged Nylon membrane
20× SSC: 3M NaCl, 0.3M tri-sodium citrate 2-hydrate, pH 7.2.
UV lamp for cross-linking of DNA to the membrane.
Hybridization solution: 50% formamide, 5× SSC 10 mM Tris-HCl (pH 7.5), 1% SDS, 5× denhardts (2% Ficoll 400, 2% polyvinylpyrrolidone K30, 2% BSA), 10% dextransulfate, 100 μg/mL salmon sperm DNA.
Radioactive labelled dATP or dCTP (α-32P-dATP or α-32P-dCTP) (can be purchased from e.g. Hartmann Analytic or Amersham). Labelling of DNA probe has to be performed in room designated for radioactive work with precautions against radioactive contamination.
DNA Labeling Kit (e.g. HexaLabel & trade DNA Labeling Kit from Fermentas).
Hybridization bottles.
Hybridization oven.
Wash-buffer 1: 2× SSC, 0.1% SDS.
Wash buffer 2: 0.5× SSC, 0.4% SDS.
Autoradiography film.
Phosphor imaging plates, Scanner for detection and analyser of radioactive signals (e.g., Fuji Film Scanner FLA-3000 and Aida Image Analyser v.4.00 software).
2.9. The use of Cre/loxP recombination system in gene targeting
Vectors harbouring cre recombinase gene and loxP sequences, respectively.
If appropriate, vectors harbouring a gene encoding for FLP recombinase and FRT sequences, respectively.
Mice harbouring a cre gene that can be expressed constitutively in the whole body and all cells or conditionally in a specific tissue or cell type. Collection databases of several hundreds of Cre transgenic mouse lines expressing Cre recombinase in specific tissues or cells are available: e.g. http://www.ics-mci.fr/mousecre/; http://nagy.mshri.on.ca/cre_new/index.php; http://www.creportal.org/; http://bioit.fleming.gr/crezoo/; http://creline.org/.
3. Methods
3.1.Cloning of the targeting vector
The construction of a gene targeting vector can proceed by conventional restriction enzyme-based cloning strategies. The 5’ and 3’ homology arms can be amplified by PCR from genomic DNA prepared from the ES cells to be used or from a mouse of the same strain. Alternatively, bacterial artificial chromosome (BAC) clones can be ordered that contain the gene of interest. BAC libraries from different mouse strains, including C57BL/6J, 129/Ola and 129Sv mouse strains are available (1-4) (BAC clones can be supplied e.g. from: http://bacpac.chori.org/; http://www.brc.riken.jp/lab/dna/en/NBRPB6Nbacen.html; http://www.lifesciences.sourcebioscience.com/clone-products/genomic-dna-clones/mouse-genomic-bac-library---rpci-23/mouse-genomic-bac-library-.aspx).
A targeting vector is typically composed of at least three basic units: a 5’ homology arm, a positive selectable gene marker, and a 3’ homology arm (see Note 2). Furthermore, in order to enrich for homologous recombinant ESC clones, a negative selection marker can be included in the targeting vector, outside the homology arms (see Note 2). However, the assembly of those units (fragments) together to build the targeting vector can be a highly time-consuming and complicated process. A cloning method that may help to simplify and speed the construction of targeting vectors is based on the MultiSite Gateway Technology (commercially available by Invitrogen, http://tools.invitrogen.com/content/sfs/manuals/multisite_gateway_man.pdf) (Fig. 1) (5,6). Indeed, Gateway-mediated cloning has been exploited by the European Conditional Mouse Mutagenesis (EUCOMM) and the National Institutes of Health Knockout Mouse (KOMP) programs in their quest to generate conditional knockout alleles for all protein-coding genes in the mouse genome (7). The following illustrates how this technique can be used to assemble a basic gene targeting vector (See Note 3 for other alternative methods to generate targeting vectors).
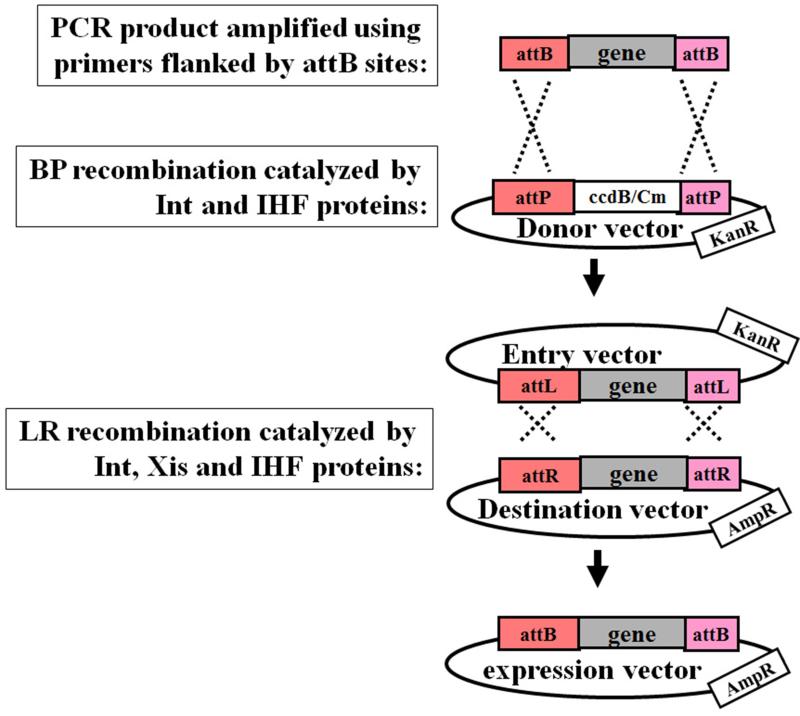
Figure 1. Gateway cloning technology.
(See description in Subheading 3.1.).
The Gateway Cloning Technology is based on the bacteriophage lambda site-specific recombination system which facilitates the integration of lambda DNA into the chromosome of E. coli, and the switch between the lytic (excision of lambda DNA from the bacterial chromosome) and lysogenic pathways (integration of lambda DNA into specific sites of the bacterial chromosome) (8-10). The phage lambda inserts its DNA into an E. coli chromosome via specific recombination sequences designated as att sites (attachment sites). The att site in the DNA of phage lambda, called attP (attachment site of phage lambda), recombines with the attachment site in the DNA of the bacteria (designated attB).
The recombination is mediated by two pairs of strand exchanges and ligation at the attB and attP sites, catalyzed by phage lambda Integrase (Int) and E. coli Integration Host Factor (IHF) proteins (supplied by Invitrogen as BP Clonase™ II enzyme mix) (Fig. 1) (8-10).
Because att sites are not palindromic, the recombination results in novel sequences designated as attL and attR (Fig. 1). The attL sites always recombine with attR resulting in the excision of lambda DNA (lytic cycle) from the bacterial chromosome (Fig. 1). This reaction is catalyzed by the Integrase (Int) and Excisionase (Xis) proteins of the phage lambda, and the E. coli Integration Host Factor (IHF) protein (Fig. 1) (supplied by the company Invitrogen as LR Clonase™ II Plus enzyme mix).
The Gateway cloning system enables efficient transfer of DNA-fragments of interest between plasmids (Fig. 1). The first step in Gateway cloning is the generation of a Gateway Entry vector. Entry vectors are usually generated in two steps: First, amplification of DNA sequence of interest using specific primers each flanked by an attB site. Second, the attB sites-flanked PCR product is then mixed with a Donor vector containing attP sites flanking a lethal ccdB gene, and BP clonase enzyme mix containing the recombinant proteins phage lambda Integrase (Int) and E. coli Integration Host Factor (IHF) (supplied by the company Invitrogen). This enzyme mix catalyzes recombination between attP and attB sites resulting in the insertion of the attB sites-flanked PCR product into attP sites to replace the lethal ccdB gene in the Donor vector (Fig. 1). The resulting vector is called Entry vector, where the inserted PCR product is now flanked by attL sites as a result of recombination between attP and attB sites. The ccdB gene is a lethal gene that serves as positive selection marker for successful BP recombination. CcdB targets DNA gyrase and thus inhibits survival and growth of E. coli strains harbouring the plasmid containing its coding sequence without the gene coding for its antitoxin, the protein CcdA, which antagonizes the toxic activity of the CcdB protein (11). Thus, bacteria can survive and propagate only if they contain the “Entry” vector, in which ccdB gene was removed by BP recombination and replaced by the attB sites-flanked PCR product (Fig. 1).
To propagate and maintain vectors containing the ccdB gene, such as Gateway Donor vector, CcdB-resistant E. coli strains (CcdB Survival strain) should be used.
The attL-flanked DNA sequence in the Entry vector can then be efficiently transferred into any Destination vector of interest that contains attR recombination sites using the enzyme mix, LR Clonase, containing the bacteriophage lambda recombination proteins Int and Excisionase (Xis), and the E. coli-encoded protein IHF (supplied by the company Invitrogen) (Fig. 1).
The MultiSite Gateway Cloning has been made possible because of the generation of modified new att sites with very high specificities enabling simultaneous, recombinational cloning of multiple DNA fragments in a single reaction. The modified att sites include among others, attB1, attB2, attB3, attB4, attB1r, of which each reacts (recombine) with the specific corresponding modified attP sites: attP1, attP2, attP3, attP4, attP1r, respectively. Furthermore, the resulting attL sites, such as attL1, attL2, etc, react specifically with the corresponding attR sites, attR1, attR2, etc. (http://tools.invitrogen.com/content/sfs/manuals/multisite_gateway_man.pdf).
3.2. Proposed strategy to generate targeting vectors using MultiSite Gateway Cloning (in combination with restriction enzymes/sites)
Gateway Cloning has been used in our laboratory. However, we didn’t yet implement the following strategy which we have designed to generate our future targeting vectors.
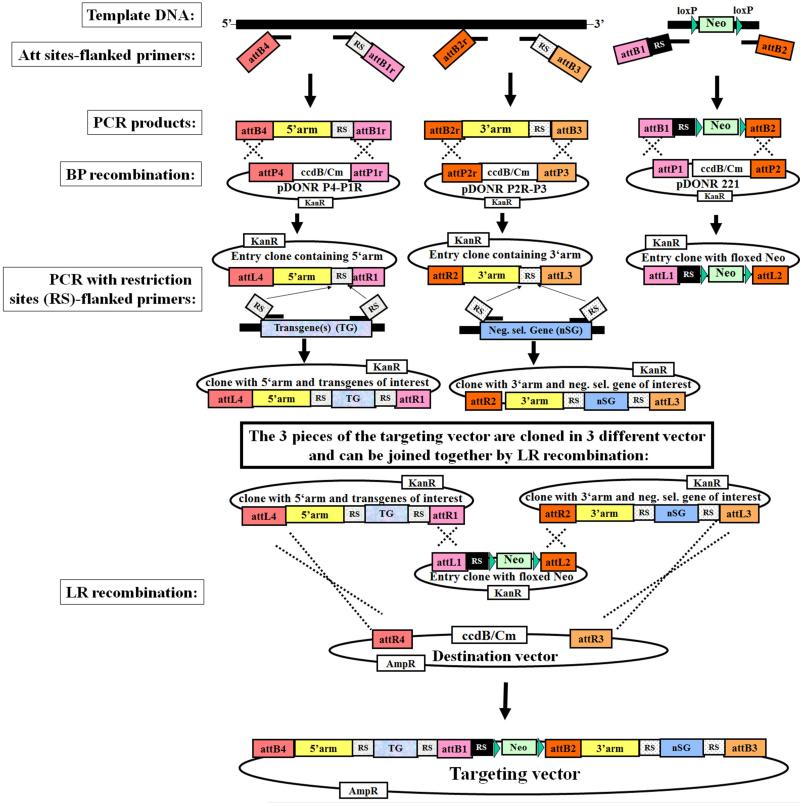
Amplification of the 5’ homology arm using specific primers flanked by attB4 site at 5’primer, and attB1r at the 3’primer. In addition, a unique restriction enzyme site (RS) can be included at the 3’primer, just upstream to the AttB1r-site (see Note 4). Such RS enables a subsequent insertion of any further DNA sequence of interest, such as a reporter gene to monitor the expression of the tagged gene, or a transgene that should be expressed in the mouse (Fig. 2).
Amplification of the 3’ homology arm using specific primers flanked by attB2r site (at 5’primer) and attB3 site (at 3’primer). In addition, a unique restriction site (RS) can be included in the 3’primer (see Note 4). Such RS enables a subsequent insertion of any further DNA sequence of interest, such as a negative selection gene (Fig. 2).
Amplification of a floxed resistance gene cassette, such as loxP-neo-loxP cassette, using specific primers flanked by attB1 and attB2 sites (Fig. 2).
BP recombination of each amplified fragment into the corresponding donor vectors (according to the manufacturer’s instructions), containing the counterpart att sites: pDONR P4-P1R (for attB4 and attB1r-flanked PCR product), pDONR P2R-P3 (for attB2r and attB3-flanked PCR product) and pDONR-221 (for attB1 and attB2-flanked PCR product), resulting in the generation of Entry vectors containing the 3 units of the targeting vector, respectively (Fig. 2) (see Note 4, 5 and 6). To propagate and maintain vectors containing the ccdB gene, such as Gateway Donor vector, ccdB Survival E. coli should be used.
Amplification of a transgene of interest using specific primers flanked by a unique restriction site (RS) identical to that introduced at the 3’end of the 5’homology arm (see Note 4). After its digestion using the appropriate restriction enzyme, the transgene PCR-product can be ligated into the corresponding restriction site (RS) of the Entry vector, downstream of the 5’arm (Fig. 2) (see Note 2, 4 and 5).
Amplification of a gene for negative selection (nSG), such as TK, DT-A or cre, using specific primers flanked by a unique restriction site (RS) identical to that introduced at the 3’end of the 3’homology arm. After its digestion using the appropriate restriction enzyme, the negative selection gene (nSG) can be ligated into the corresponding restriction site (RS) of the Entry vector, downstream of the 3’arm (Fig. 2) (see Note 4, 5 and 6).
LR recombination of the five units, which are subcloned in three different Entry vectors, into a Destination vector containing the appropriate att sites, resulting in the generation of the targeting vector composed of 5’arm, transgene of interest, positive selection cassette, 3’arm and negative selection gene (Fig. 2) (see Note 4, 5 and 6).
Figure 2. Generation of targeting vector using MultiSite Gateway Cloning (in combination with restriction enzymes/sites).
(See description in Subheading 3.2.).
3.3. Isolation, culture and mitotic inactivation of mouse embryonic fibroblasts (MEFs)
Set up breeding pair of mice expressing the same resistance gene (e.g. neomycin or hygromycin) that is used for the positive selection of transfected ES cells. Check plugs and record date of visible plug. Usually, plug is visible 0.5 day after set up breeding.
Euthanize pregnant female at 13.5-14.5 days after the appearance of the plug.
Rinse dead mouse in 70% ethanol for ~5 min.
Open the abdomen by cutting carefully the abdominal skin using sterile scissor and forceps. Care should be taken not to rip the gut to avoid contamination by gut residing bacteria.
Remove both uterine horns containing embryos. Briefly rinse them in 70% ethanol and place them in petri dish containing sterile PBS on ice.
Move to a sterile hood or laminar flow cabinet.
Open a small hole through the uterus using sterile scissors and forceps and remove the embryos through the hole.
Separate each embryo from its placenta and surrounding membranes.
Cut away head of embryos and transfer remnants to a Petri dish filled containing sterile PBS on ice.
Open abdomens of the embryos and remove intestine, liver, kidneys, and spleen.
Transfer the rest of the embryos to a small Petri dish containing 2 mL cold PBS.
Mince the embryos until they become “pipettable”, and add 2 mL of trypsin-EDTA (2ml per embryo), and incubate at 37°C for 15 min.
Break the remaining tissue pieces by up and down pipetting with a 5 mL then 2 mL pipette, and add 4 mL MEF medium per embryo.
Pipette each 2.5 mL of the MEF suspension into a 175 cm2 tissue culture flask, and add MEF medium to a final volume of ~25-30 mL.
Incubate MEFs at 37°C, 5% CO2.
Collect MEFs 2-3 days after they reached confluency (confluency is reached after a few days incubation): wash ones carefully with PBS, then add 3 mL trypsin-EDTA per 175 cm2 flask, and incubate at 37°C to detach cells from their monolayer.
Add 5 mL of MEF medium per cell culture flask to inactivate trypsin, and resuspend the cells thoroughly.
Transfer 6 mL of the cell suspension from each flask into a 50 mL tube, and add 25 ml MEF medium to the remaining 2 mL MEF suspension in each 175 cm2 flask.
Pellet the collected MEFs by centrifugation at 400×g for 3 min.
Discard supernatants and add 20 mL MEF medium to the MEFs-pellet.
γ-irradiate MEFs with 40 gray (4000 rad) using a cesium-source irradiator to mitotically inactivate them.
Pellet γ-irradiated MEFs by centrifugation at 400×g for 3 min.
Discard supernatants and add 3 mL Freeze medium to each cell pellet, resuspend the cells thoroughly, and store 1 mL aliquots at −70°C.
The steps 15-23 should be repeated for the remaining growing MEFs.
One vial frozen MEFs can be used to layer a petri dish (10 cm diameter), a 24-well plate, or a 96-well plate.
3.4. Culture of ES cells
ESCs have to be cultured in special culture conditions to maintain their pluripotency (see Note 2). A combination of mitotic inactivated MEFs and leukemia inhibitory factor (LIF) ensures an optimal culture condition to inhibit differentiation of ESCs. Furthermore, ES cells should always be kept in a low density, and thus have to be frequently split. The present protocol to culture, transfect, select and pick ES cells was used to successfully generate several knockin mice (12,13), and is based on the protocol of Talts et al. (14) with some modifications. (See Note 7 for alternative protocols using MEFs-free culture systems).
Thaw ES cells and MEFs quickly at 37°C, transfer them into a 15-mL tube and add 5-10 mL ESC medium. Centrifuge for 4 min at 400g.
Resuspend cells in appropriate ESC medium and seed ES cells and MEFs together in petri dishes (100 mm × 20 mm) (1 vial frozen MEFs per petri dish).
Change medium every day.
Trypsinize and split the ES cells before the colonies become brownish in their middle (phenotype for ongoing differentiation). Usually, ES cells have to be split every day. For trypsinization, remove carefully medium, wash one time with sterile PBS, and then add trypsin and incubate for 5 min at 37°C. Thereafter add appropriate amount of ES medium, resuspend the cells by up and down pipetting, and then split them into 2 dishes (along with new MEFs, 1 vial frozen MEFs per two petri dish of 100 mm × 20 mm size).
2-4 hours before harvesting for electroporation, ES cells should be fed (change of medium).
3.5.Electroporation of ES cells with the targeting vector
Linearization of targeting vector
Linearize at least 150 μg of the targeting construct using a unique restriction site outside the homology arms (use ~2 units enzyme per μg DNA) (see Note 8).
Purify the DNA by two-fold phenol/chloroform extraction. For each extraction add equal volume of phenol/chloroform, mix well by vigorously inverting the tube (don’t vortex), and then centrifuge for 3 min at high speed. Transfer the upper DNA- containing aqueous phase to a new vial. Then remove phenol rests by chloroform/isoamylalcohol extraction. For that add equal volume of chloroform/isoamylalcohol to the DNA-containing upper aqueous phase, mix well by vigorously inverting the tube (don’t vortex), and then centrifuge for 3 min at high speed. Transfer the upper DNA-containing aqueous phase to a new vial (see Note 9).
Precipitate DNA by addition of 0.3 M Na-acetat (pH 5.2) to upper aqueous fraction containing the DNA, mixing (don’t vortex), and then addition of 2.5 vol of absolute ethanol and mixing by inverting the tube vigorously (don’t vortex).
Transfer the DNA-precipitate with a pipette tip to a 1.5-mL containing 70% ethanol.
The DNA can be stored at this stage at −20°C until use for electroporation.
Electroporation of ES cells
Remove ethanol supernatant and air dry the DNA under sterile hood. Add 500 μl sterile PBS, resuspend by pipetting and incubate at 37°C to completely dissolve DNA.
Embryonic stem cells (~80% confluent) should be fed 2-4 hours prior to harvesting (change of medium).
Collect ES cells by trypsinization, and then wash them twice with PBS (centrifugation steps at 400×g for 4 min).
Resuspend 1×107 ES cells in 500 μl PBS in 1.5-mL tube.
Add the 500 μl PBS/DNA solution, and incubate at RT for 5 min.
Split the mixture into electroporation cuvettes. Electroporate at 800 V and 300 μF using a Gene Pulser. The time constant should be ~0.04 ms.
Carefully transfer the cells with a sterile pipette into petri dishes (~6-10 dishes) pre-layered with mitotically inactivated MEFs. Add ESC medium to a final volume of 8 mL. Shake the dishes crosswise to distribute the electroporated ES cells and incubate at 37°C, 5% CO2.
3.6. Selection and picking of positively transfected ESC clones
ESC Clones should be picked on day 6, 7 or 8 after starting selection. The number of ESC clones to be picked depends on the expected ratio of homologous recombination.
Begin selection 24 hours after electroporation. Exchange ESC medium for G418-ESC medium containing 250-400 μg/mL G418 (or appropriate concentration of alternative antibiotic, depending on the antibiotic resistance gene used). The concentration of the antibiotic depends on the ES cell line.
Change medium daily.
After day 3-4 of selection, cell death begins to be clearly observed. At those days change medium twice per day, in the morning and evening.
Several 24-well plates should be layered with mitotically inactivated MEFs in MEF medium (from subheading 2.2. and 3.3.), one day before picking ESC clones (1 vial frozen MEFs from subheading 3.3. can be used to layer one 24-well plate). Just before use, exchange MEF medium for G418-ESC medium (from subheading 2.2.).
Depending on the ES cell line and the media used, ESC colonies start to be visible ~6-8 days after beginning of selection. The optimal ESC clones to be picked have rounded or oval shape, have tight and bright borders and are closely packed, often with a weak brownish centre.
Prior to picking of ESC colonies, wear gloves and face mask, and disinfect all surfaces that will be used, including the microscope, with ethanol.
Place a petri dish containing transfected ES cells under microscope at 10× magnification (optimally, the microscope should be in a laminar flow hood, to ensure sterile work).
Push the ESCs colony loose from the MEFs layer with the tip of a pipette set to 10 μl. Aspirate the colony into the tip, transfer it into one well of a sterile 96-well plate. After filling half of a 96-well plate with 48 clones (takes approximately 60-90 minutes), add ~100 μl trypsin-EDTA per well, and incubate at 37°C for 5 min.
During the incubation of ESC clones with trypsin, exchange MEF medium of feeder cells (mitotically inactivated MEFs) in the 24-well plates for 500 μl G418-ESC medium (from subheading 2.2.).
Add 100 μL G418-ESC medium in each well of the 96-well plate containing the picked ESC clones, mix well by up and down pipetting to break up each colony, avoiding excessive foaming, and then transfer each ESC clone to a well of the prepared 24-well plate (layered with MEFs) (from this subheading 3.6. Point number 4).
Incubate at 37°C, 5% CO2.
Change the G418-ESC medium every day until a good coverage of colonies in each well is achieved (this happens ~ 5–10 days after picking ESC clones).
3.7. Freezing of and DNA preparation from ESC clones
Remove medium and wash each well with PBS.
Add ~100 μL trypsin-EDTA solution to each well and incubate at 37°C for 5 min.
Add 1 mL of ice-cold freeze medium (from subheading 2.7.) to each well.
Disperse the cells using a pipette set to 500 μL and transfer 700 μl of the cell suspension from each well to pre-labelled freeze tubes and place them immediately on dry ice.
Transfer the tubes to −80°C freezer.
Add 2 mL of G418-ESC medium to the residual cells in each well, and incubate at 37°C for ~12 h. Then replace medium by 1 mL G418-ESC medium and incubate at 37°C until the colour of the medium turns yellow.
Aspirate medium and add 500 μL of fresh lysis buffer (from subheading 2.7.) to each well that turned yellow and incubate at 37°C for at least overnight.
When a well plate is completely lysed, add 1:1 volume of isopropanol (0.5 mL) to each well and shake on an orbital shaker for at least 12 h at RT until the DNA becomes visible.
Extract the DNA by spooling it on a pipette tip, and transfer it into a pre-labelled tube containing 100-150 μl of TE-buffer (from subheading 2.7.). DNA is visible against a dark background.
Incubate for at least 8 h at 55°C to allow complete dissolving of the DNA.
Store the DNA at RT (long storage can be done at 4°C).
3.8. Screening for ESC clones with homologous recombination of the targeting vector by southern blot analysis
Homologous recombinant ESC clones can be identified by Southern blot. The genomic DNA isolated from ESC clones should be digested with an appropriate restriction enzyme that produces one cut inside the targeting vector and one cut just outside (upstream or downstream) the targeting vector, in the targeted chromosomal region. The use of an “external” probe outside of the targeting construct will produce a band with a size corresponding to the unmodified wild-type allele (see Note 2) and, if homologous recombination occurred, a second band of bigger or smaller size corresponding to the targeted allele (see Note 2). To analyse whether the targeting vector was also integrated in additional places in the genome of ES cells, an internal probe that hybridizes within the targeting construct (e.g. to the selection marker) should be used.
Digest 20-40 μl DNA solution of each ESC clone (from subheading 2.8.) (in a total volume of 50 μl) with 30-60 units of an appropriate restriction enzyme overnight.
Fractionate the digested DNA samples on 0.7 agarose gel in 1×TBE (from subheading 2.8.).
Treat the gel successively with depurination solution (0.2 M HCl), denaturing buffer and neutralization buffer (from subheading 2.8.).
Blot the gel overnight to a positive charged Nylon membrane using 20× SSC (from subheading 2.8.).
Cross-link the DNA to the membrane by irradiation of the membrane with a UV lamp for 50 sec.
Incubate the membrane with hybridization solution (from subheading 2.8) for 1 h.
During this incubation time, prepare radioactive labelled probe (see Note 10).
Add radioactive-labelled probe to the hybridization solution and incubate overnight at 42°C.
Wash the blot two times with wash-buffer 1 (2× SSC, 0.1% SDS) for 10 min at RT, then two times with wash buffer 2 (0.5× SSC, 0.4% SDS) for 20 min at 70°C.
Radioactive signals can be then detected using a phosphor imaging plate, and corresponding Scanner and software (such as Fuji Film Scanner FLA-3000 and Aida Image Analyser v.4.00 software), or by exposing the membrane with an autoradiography film.
3.9. Generation of chimeric mice from genetically modified ESC clones
The identified homologous recombinant ESC clones can be used to generate chimeric mice by injecting them into recipient pre-implantation mouse embryos (blastocysts) that are collected from female mice with coat colour different from that of the mouse strain-parent of the used ES cells (see Note 2). The injected blastocysts are then surgically transferred to a recipient pseudopregnant foster mother to allow the embryos to develop (see Note 2). Females of CD1 mouse strain make very good mothers, and are thus used by several laboratories as foster mothers. Because ESCs and recipient blastocysts are derived from mouse strains with distinguishable coat-colors, the desired chimeric offspring can be visually recognized by inspection of coat-colour chimerism (% of black and agouti hair on the mouse black-agouti). Chimeric offspring (usually males) are mated with C57BL/6 mice to produce the F1 generation. The germline transmission is then confirmed by Southern blot analysis or PCR of tail DNA from mice of the F1 generation.
Acknowledgement
Work in our laboratory is supported by grants from the BBSRC and the Wellcome Trust.
Footnotes
It is strongly recommended to use commercially available transformation competent bacteria, because they are highly competent. Transformation competency is among the important factors influencing the speed and success of cloning.
For background overview about gene targeting constructs, ES cells and gene targeting in ES cells, we refer to the review chapter (Gene Targeting in Mice: a Review) in this issue of “Virus-Host-Interactions”.
A further alternative method to generate targeting vectors uses Recombination-mediated genetic engineering ( Recombineering )-based protocols. Particularly, recombineering enables quick BAC-based construction of targeting vectors by introducing any DNA modifications of interest directly into the BAC clone containing the gene of interest, without the need for subcloning steps, restriction enzymes or DNA ligases (15-17). Recombineering is based on homologous recombination in E. coli mediated by bacteriophage recombination proteins, such as RecE, RecT, Redα, Redβ and RecA (18). Recombineering can also be used to retrieve homology arms from a BAC clone into a vector.
In addition, it is becoming increasingly affordable to generate entire or part of the targeting vector (especially complicated parts) by gene-synthesis (19,20).
Be aware that insertion of mutations by PCR site-directed mutagenesis often results in the incorporation of tandem repeats of the used complementary primer pairs at the mutated site. We actually sequence at least 5 bacteria colonies recovered from each site-directed mutagenesis reaction, and in most cases, at least one clone contained plasmid with the accurate mutation of interest, without primer tandem repeats.
According to the manufacturer’s instructions, the BP and LR reactions should be incubated at 25°C for 1 hour. However, we realized that, depending on the vectors used, the success of recombination reactions (BP and LR reactions) can be enormously increased when incubated at 25°C for at least 12 h (overnight).
The larger the plasmids (≥ ~10 kb) the more instable they are when used for ligation- or recombination-mediated DNA transfer. They often undergo unwanted recombinations resulting in “cryptic” plasmids. To reduce the occurrence of such drawbacks and increase the rate of positive colonies, we strictly incubate the transformed bacteria at maximum 27°C, and we reduce the incubation at the minimum time, just until the bacterial colonies became visible (at 27°C it takes ~20-24 h to get visible colonies).
Furthermore, we incubate the inoculated fluidic cultures with the recovered bacteria colonies at 27°C or at 37°C, and we reduce the incubation at the minimum time, just until the bacterial culture became just slightly cloudy (at 37°C it takes ~6 h to get slightly cloudy culture).
MEF-free culture systems enabling maintenance of pluripotency use glycogen synthase kinase (GSK)-3-specific inhibitors, such as 6-bromoindirubin-3’-oxime (BIO) (21), or, optimally, a combination of three inhibitors (3i medium): SU5402 (inhibits FGF receptor tyrosine kinases), PD184352 (inhibits ERK signal cascades) and CHIR99021 (a more selective inhibitor of GSK-3) (22).
In addition, recently a MEF- and inhibitors-free culture system was developed that enables maintenance of pluripotency and self-renewal by culturing ES cells on plates coated with a recombinant human extracellular matrix protein, the laminin isoform 511 (LN-511) (23). Please see also the following references for detailed protocols on how to culture ES cells without MEFs (21-23).
To ensure high efficient (almost 100%) linearization of the vector, the digestion reaction should be incubated overnight, and next day, further units of the restriction enzyme (1 unit per μg DNA) should be added, and the digestion reaction incubated for further 3-4 hours.
Do not purify the digested targeting vector by gel extraction. This can be harmful for the DNA, and will also lead to the recovery only of a small amount of the loaded DNA material.
The probe can be radioactive labelled using DNA Labeling Kits, such as HexaLabel&trade DNA Labeling Kit (Fermentas), according to manufacturer’s instructions. α-32P-dATP or α-32P-dCTP can be used as radioactive marker for the probe.
References
- 1.Ohtsuka M, Ishii K, Kikuti YY, Warita T, Suzuki D, Sato M, Kimura M, Inoko H. Construction of mouse 129/Ola BAC library for targeting experiments using E14 embryonic stem cells. Genes & genetic systems. 2006;81:143–146. doi: 10.1266/ggs.81.143. [DOI] [PubMed] [Google Scholar]
- 2.Adams DJ, Quail MA, Cox T, van der Weyden L, Gorick BD, Su Q, Chan WI, Davies R, Bonfield JK, Law F, et al. A genome-wide, end-sequenced 129Sv BAC library resource for targeting vector construction. Genomics. 2005;86:753–758. doi: 10.1016/j.ygeno.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Jansa P, Divina P, Forejt J. Construction and characterization of a genomic BAC library for the Mus m. musculus mouse subspecies (PWD/Ph inbred strain) BMC genomics. 2005;6:161. doi: 10.1186/1471-2164-6-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osoegawa K, Tateno M, Woon PY, Frengen E, Mammoser AG, Catanese JJ, Hayashizaki Y, de Jong PJ. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome research. 2000;10:116–128. [PMC free article] [PubMed] [Google Scholar]
- 5.Iiizumi S, Nomura Y, So S, Uegaki K, Aoki K, Shibahara K, Adachi N, Koyama H. Simple one-week method to construct gene-targeting vectors: application to production of human knockout cell lines. Biotechniques. 2006;41:311–316. doi: 10.2144/000112233. [DOI] [PubMed] [Google Scholar]
- 6.Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- 7.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- 9.Moitoso de Vargas L, Kim S, Landy A. DNA looping generated by DNA bending protein IHF and the two domains of lambda integrase. Science. 1989;244:1457–1461. doi: 10.1126/science.2544029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nunes-Duby SE, Matsumoto L, Landy A. Half-att site substrates reveal the homology independence and minimal protein requirements for productive synapsis in lambda excisive recombination. Cell. 1989;59:197–206. doi: 10.1016/0092-8674(89)90881-7. [DOI] [PubMed] [Google Scholar]
- 11.Maki S, Takiguchi S, Miki T, Horiuchi T. Modulation of DNA supercoiling activity of Escherichia coli DNA gyrase by F plasmid proteins. Antagonistic actions of LetA (CcdA) and LetD (CcdB) proteins. J Biol Chem. 1992;267:12244–12251. [PubMed] [Google Scholar]
- 12.Bouabe H, Moser M, Heesemann J. Enhanced selection for homologous-recombinant embryonic stem cell clones by Cre recombinase-mediated deletion of the positive selection marker. Transgenic Res. 2011 doi: 10.1007/s11248-011-9522-x. DOI 10.1007/s11248-011-9522-x. [DOI] [PubMed] [Google Scholar]
- 13.Bouabe H, Liu Y, Moser M, Bosl MR, Heesemann J. Novel Highly Sensitive IL-10-{beta}-Lactamase Reporter Mouse Reveals Cells of the Innate Immune System as a Substantial Source of IL-10 In Vivo. J Immunol. 2011;187:3165–3176. doi: 10.4049/jimmunol.1101477. [DOI] [PubMed] [Google Scholar]
- 14.Talts JF, Brakebusch C, Fassler R. Integrin gene targeting. Methods Mol Biol. 1999;129:153–187. doi: 10.1385/1-59259-249-X:153. [DOI] [PubMed] [Google Scholar]
- 15.Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol. 2003;21:652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- 16.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nature reviews. Genetics. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 17.Malureanu LA. Targeting vector construction through recombineering. Methods Mol Biol. 2011;693:181–203. doi: 10.1007/978-1-60761-974-1_11. [DOI] [PubMed] [Google Scholar]
- 18.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annual review of genetics. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 19.Hughes RA, Miklos AE, Ellington AD. Gene synthesis: methods and applications. Methods Enzymol. 2011;498:277–309. doi: 10.1016/B978-0-12-385120-8.00012-7. [DOI] [PubMed] [Google Scholar]
- 20.Matzas M, Stahler PF, Kefer N, Siebelt N, Boisguerin V, Leonard JT, Keller A, Stahler CF, Haberle P, Gharizadeh B, et al. High-fidelity gene synthesis by retrieval of sequence-verified DNA identified using high-throughput pyrosequencing. Nat Biotechnol. 2010;28:1291–1294. doi: 10.1038/nbt.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 22.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Domogatskaya A, Rodin S, Boutaud A, Tryggvason K. Laminin-511 but not -332, -111, or -411 enables mouse embryonic stem cell self-renewal in vitro. Stem Cells. 2008;26:2800–2809. doi: 10.1634/stemcells.2007-0389. [DOI] [PubMed] [Google Scholar]