Abstract
Programmed cell death in bacteria is generally associated with two-component toxin-antitoxin systems. The SpoIIS toxin-antitoxin system, consisting of a membrane-bound SpoIISA toxin and a small, cytosolic antitoxin SpoIISB, was originally identified in Bacillus subtilis. In this work we describe the Bacillus cereus SpoIIS system which is a three-component system, harboring an additional gene spoIISC. Its protein product serves as an antitoxin, and similarly as SpoIISB, is able to bind SpoIISA and abolish its toxic effect. Our results indicate that SpoIISC seems to be present not only in B. cereus but also in other Bacilli containing a SpoIIS toxin-antitoxin system. In addition, we show that B. cereus SpoIISA can form higher oligomers and we discuss the possible role of this multimerization for the protein's toxic function.
Keywords: Bacillus cereus, toxin-antitoxin system, SpoIIS, programmed cell death, Bacillus subtilis
Introduction
Programmed cell death (PCD) is a genetically regulated system in which a bacterial cell is able to commit suicide in response to a variety of different stresses. This response includes cell lysis or growth inhibition induced by harsh environmental conditions such as starvation or antibiotic treatment, active mother cell lysis during sporulation to release the spore, or altruistic suicide to release cell content to provide the nutrients required for the normal development of the remaining bacterial population (Engelberg-Kulka et al., 2006). PCD is usually mediated by a pair of toxin/antitoxin (TA) genes. Toxins are always highly stabile proteins. Their antidotes, the antitoxins, are usually labile proteins or small RNAs. TA systems are classified according to the nature of the antitoxin. Type I and III are small RNAs which either inhibit the synthesis of the toxin or capture it. Examples include the type I hok-sok system (Pedersen and Gerdes, 1999) and the type III ToxIN system (Fineran et al., 2009). Types II, IV, and V, on the other hand, are all proteins. They include the type II mazEF TA system (Gerdes et al., 2005), the type IV yeeU-yeeV system (Masuda et al., 2012), and the type V ghoT-ghoS system (Wang et al., 2012). These three types are distinguished based on their mode of action. The type II antitoxin is a small protein with an N-terminal DNA-binding domain and a C-terminal toxin-bonding domain, the type IV antitoxin is an antagonist of its cognate toxin and competes with it in binding to its target, and the type V antitoxin is an endoribonuclease that degrades the toxin-encoding mRNA (Goeders and Van Melderen, 2014).
Many bacteria harbor genes for TA systems on plasmids (Ruiz-Echevarría et al., 1995; Gerdes et al., 1997; Sayeed et al., 2000; Van Melderen, 2001; Camacho et al., 2002). These genes are part of a mechanism called post-segregational killing, which ensures that their host plasmids are retained in the daughter cells of a growing bacterial population. In this process, the stable, long-lived toxin kills those daughter cells which do not inherit the plasmid encoding the labile antitoxin (Gerdes et al., 1986; Lehnherr and Yarmolinsky, 1995; Hayes, 2003). Other bacterial species contain numerous toxin-antitoxin genes on their chromosome (Hayes, 2003; Tsilibaris et al., 2007; Van Melderen and Saavedra De Bast, 2009). Chromosomal TA systems may serve to prevent the spread of mobile genetic elements such as phages or plasmids; they are typically involved in the general stress response and in guarding against DNA loss (reviewed in Schuster and Bertram, 2013).
The spoIIS locus was originally identified on the Bacillus subtilis chromosome during a study of the genetic mutants that block sporulation after the formation of the polar septum (Adler et al., 2001). Formerly, the locus was thought to consist of two genes, spoIISA coding for the toxin and spoIISB for proteic antitoxin (Adler et al., 2001), thus classifying as type II TA system. A condition-dependent analysis of the transcription of all B. subtilis genes indicated that a third transcriptionally active region, S458, might be present in the spoIIS operon (Nicolas et al., 2012), which we name spoIISC. Inactivation of the spoIISA toxin gene has no effect on sporulation, but inactivation of the spoIISB antitoxin gene decreases sporulation efficiency by four orders of magnitude. Furthermore, disruption of spoIISA in a spoIISB null mutant restores sporulation. Thus, SpoIISB is required for sporulation only if SpoIISA is present in the cell (Adler et al., 2001). The morphological consequence of an artificially induced higher level of toxin expression is the formation of plasmolysis zones in the cytoplasmic membrane, leading to the death of the cell. The transcription of spoIISA, spoIISB, and spoIISC is upregulated during sporulation from four to up to eight hours (Nicolas et al., 2012); however, the expression of SpoIISA is independent of the crucial sporulation initiation transcription factor, Spo0A (Rešetárová et al., 2010). Production of the SpoIISA toxin is also induced during ethanol stress and nutrient deprivation. During starvation, the production of SpoIISB was detected, which suggests that SpoIISB is able to diminish the toxic effect of SpoIISA. Moreover, SpoIISB is also produced during swarming and at times of high cell density. There is presently only a little information about spoIISC, but it is known that its transcription is activated during both sporulation and biofilm formation (Nicolas et al., 2012). The SpoIISA toxin is neutralized by the formation of a tight complex with the SpoIISB antitoxin. The crystal structure of this complex revealed that SpoIISB and the cytoplasmic domain of SpoIISA form a heterotetrameric complex with C-SpoIISA2:SpoIISB2 stoichiometry (Florek et al., 2011).
Homologs of SpoIISA and SpoIISB proteins have also been identified among other Bacillus species, but they display only a low level of homology. Both B. subtilis and B. cereus SpoIISA inhibit the growth of E. coli cells, and the SpoIISB antitoxin is able to neutralize SpoIISA toxicity in E. coli (Florek et al., 2008).
In the present study we analyze the spoIIS operon in B. cereus ATCC 14579. Even though a third trancriptionally active region in the spoIIS operon of B. subtilis was identified, it is unclear whether its product is really part of this TA system. We have found that both B. subtilis and B. cereus spoIISC encode an antitoxin that is able to diminish SpoIISA toxicity in E. coli. As in B. subtilis, the B. cereus spoIIS operon consists of three genes: spoIISA, spoIISB, and spoIISC. Using a bacterial two hybrid system we show that B. cereus C-SpoIISA interacts with other C-SpoIISA molecules, as well as with SpoIISB and SpoIISC. These new positive interactions, identified in vivo, were also confirmed in vitro using a pull-down assay. In vitro analysis of the oligomeric states of B. cereus C-SpoIISA revealed that the soluble C-SpoIISA exists in monomeric, dimeric and trimeric forms.
Materials and methods
Bacterial strains, growth conditions, and media
The bacterial strains E. coli XL1-BLUE, DH5α, and MM294 were used for routine DNA manipulations. The E. coli BTH101 reporter strain was employed in the bacterial adenylate cyclase-based two-hybrid system. E. coli BL21 (λDE3) cells were employed in expression of recombinant protein. E. coli cells were grown at 37°C, 28°C or room temperature in LB (Ausubel et al., 1987) or SOC medium (Hanahan, 1983) or on agar plates. When required, the medium was supplemented with appropriate antibiotics and other additives. E. coli transformation and DNA manipulations were performed using standard protocols (Sambrook et al., 1989).
The kill/rescue assay cultivation
To evaluate the effect of the expression of B. cereus spoIIS genes on the growth of E. coli MM294, a single colony of bacterial cells was resuspended in 100 μl of LB and grown overnight on LB agar plates. The bacterial lawn was washed off with 1 ml LB and this primary culture was used to inoculate a second cell generation in LB containing 100 μg ml−1 ampicillin and 0.5% glucose (w/v). The starting optical density (OD600) of the cell cultures was 0.05–0.06. The cells were cultivated at 37°C in an orbital shaker at 150 rpm and growth was monitored by measuring the OD600 in 1-h intervals. When the OD600 reached 0.4, spoIIS expression was induced by the addition of l-arabinose to a final concentration of 0.02% (w/v).
Recombinant plasmid construction
All bacterial strains and plasmids used in this study are listed in Table 1. All primers for cloning were designed for the PCR amplification of specific genes and regulatory regions and are listed in Table 2. Chromosomal DNA of B. subtilis PY79 (Youngman et al., 1984) and Bacillus cereus ATCC 14579 was used for amplification of spoIIS genes.
Table 1.
Strains and plasmids used in this study.
| Strain | Genotype or description | Reference or origin |
|---|---|---|
| E. coli | ||
| MM294 | F− endA1 hsdR17 (rk−, mk) supE44 thi-1 recA+ | Meselson and Yuan, 1968 |
| BTH101 | F− cya-99 araD139 galE15 galK16 rpsL1(StrR) hsdR2 mcrA1 mcrB1 | Karimova et al., 1998 |
| DH5α | F' Iq supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Meselson and Yuan, 1968 |
| XL1-BLUE | Δ(mcrA)183 (mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA lac (F' proAB lacIq ΔM15Tn5 kan') | Stratagene |
| IB890 | pBAD24 in MM294 | Florek et al., 2008 |
| IB926 | pBAD24-BCIISA in MM294 | Florek et al., 2008 |
| PLASMIDS USED IN KILL/RESCUE ASSAY | ||
| pBAD24 | AmpR araC; PBAD promoter | Guzman et al., 1995 |
| pBADCIISA | AmpR araC; PBAD promoter, B. cereus spoIISA-like gene | Florek et al., 2008 |
| pBADIISAB Bc | AmpR araC; PBAD promoter, B. cereus spoIISAB-like genes | This study |
| pBADIISC Bc | AmpR araC; PBAD promoter, B. cereus spoIISC-like gene | This study |
| pBADIISAC Bc | AmpR araC; PBAD promoter, B. cereus spoIISAC-like genes | This study |
| pBADIISA Bs | AmpR araC; PBAD promoter, B. subtilis spoIISA | This study |
| pBADIISAB Bs | AmpR araC; PBAD promoter, B. subtilis spoIISAB | This study |
| pBADIISC Bs | AmpR araC; PBAD promoter, B. subtilis spoIISC-like gene | This study |
| pBADIISAC Bs | AmpR araC; PBAD promoter, B. subtilis spoIISAC | This study |
| PLASMIDS FOR TESTING PROTEIN-PROTEIN INTERACTIONS IN VITRO | ||
| pET15b | AmpR; T7lac promoter | Novagen |
| pETDuet-1 | AmpR; T7lac promoter | Novagen |
| pET15b-Bc-CIISA | AmpR; T7lac promoter, B. cereus C-spoIISA | Laboratory stock |
| pET15b-Bc-HCIISA | AmpR; T7lac promoter, His6 tag fused with B. cereus C-spoIISA | This study |
| pETDuet-Bc-IISC | AmpR; T7lac promoter, B. cereus spoIISC | This study |
| pETDuet-Bc-HCIISAC | AmpR; T7lac promoter, His6 tag fused with B. cereus C-spoIISA, T7lac promoter, B. cereus spoIISC | This study |
| pETDuetCIISA Bc | AmpR; T7lac promoter, His6 tag fused with B. cereus C-spoIISA | This study |
| pETDuetIISB Bc | AmpR; T7lac promoter, B. cereus spoIISB fused with S-tag | This study |
| pETDuetCIISAB Bc | AmpR; T7lac promoter His6-tag fused with B. cereus C-spoIISA, T7lac promoter, B. cereus spoIISB fused with S-tag | This study |
| PLASMIDS FOR THE BACTERIAL TWO-HYBRID SYSTEM | ||
| pKT25 | KanR; Plac promoter, T25 | Karimova et al., 1998 |
| pKNT25 | KanR; Plac promoter, T25 | Karimova et al., 1998 |
| pUT18 | AmpR; Plac promoter, T18 | Karimova et al., 1998 |
| pUT18C | AmpR; Plac promoter, T18 | Karimova et al., 1998 |
| pKT25-zip | KanR; Plac promoter, T25 fused with zip | Karimova et al., 1998 |
| pUT18C-zip | AmpR; Plac promoter, T18 fused with zip | Karimova et al., 1998 |
| pKTCIISA Bc | KanR; Plac promoter, T25 fused with B. cereus C-SpoIISA | This study |
| pKNTCIISA Bc | KanR; Plac promoter, B. cereus C-spoIISA fused with T25 | This study |
| pUTCIISA Bc | AmpR; Plac promoter, B. cereus C-spoIISA fused with T18 | This study |
| pUTCCIISA Bc | AmpR; Plac promoter, T18 fused with B. cereus C-spoIISA | This study |
| pUTIISB Bc | AmpR; Plac promoter, B. cereus spoIISB fused with T18 | This study |
| pUTCIISB Bc | AmpR; Plac promoter, T18 fused with B. cereus spoIISB | This study |
| pUTIISC Bc | AmpR; Plac promoter, B. cereus spoIISC fused with T18 | This study |
| pUTCIISC Bc | AmpR; Plac promoter, T18 fused with B. cereus spoIISC | This study |
| pKTIISC Bc | KanR; Plac promoter, T25 fused with B. cereus spoIISC | This study |
| pKNTIISC Bc | KanR; Plac promoter, B. cereus spoIISC fused with T25 | This study |
| pKTCIISA Bs | KanR; Plac promoter, T25 fused with B. subtilis C-spoIISA | This study |
| pKNTCIISA Bs | KanR; Plac promoter, B. subtilis C-spoIISA fused with T25 | This study |
| pUTCIISA Bs | AmpR; Plac promoter, B. subtilis C-spoIISA fused with T18 | This study |
| pUTCCIISA Bs | AmpR; Plac promoter, T18 fused with B. subtilis C-spoIISA | This study |
| pUTIISB Bs | AmpR; Plac promoter, B. subtilis spoIISB fused with T18 | This study |
| pUTCIISB Bs | AmpR; Plac promoter, T18 fused with B. subtilis spoIISB | This study |
| pUTIISC Bs | AmpR; Plac promoter, B. subtilis spoIISC fused with T18 | This study |
| pUTCIISC Bs | AmpR; Plac promoter, T18 fused with B. subtilis spoIISC | This study |
| pKTIISC Bs | KanR; Plac promoter, T25 fused with B. subtilis spoIISC | This study |
| pKNTIISC Bs | KanR; Plac promoter, B. subtilis spoIISC fused with T25 | This study |
Table 2.
Primers used in this study.
| Primer | Sequence (5′–3′), restriction sites are in bold | Final construct |
|---|---|---|
| SP/Bc-CIISA/XhoI | TCATCATCACTCGAGGAAATATGGGGTGCGAAATT | pET15b-Bc-HCIISA |
| ASP/Bc-CIISABamE | TCATCATCAGGATCCTTTACTAAAATAACTATGAT | |
| SP/BcIISB/NdeI | TCATCATCACATATGGTGATTGTAGTGGTAAAAGA | pETDuet-Bc-IISC |
| ASP/BcIISB/XhoI | TCATCATCACTCGAGTACACTTATGATTTTCTTTT | |
| SP/IISA/NcoI | TCATCATCACCATGGATGATCTCTAACATTCGAAT | pBADIISAB Bc |
| ASP/IISB/HindIII | TCATCATCAAAGCTTGCAAATGTAGAAAGAGTGTA | |
| SP/IISCBc/PstI | TCATCATCACTGCAGTGAAAAGGGGGAGAAGAGATG | pBADIISC Bc |
| ASP/IISCBc/HindIII | TCATCATCAAAGCTTATGCTCTATGCATTTTCTTT | |
| SP/IISABc/EcoRI | TCATCATCAGAATTCATGATCTCTAACATTCGAAT | pBADIISAC (via pBADIISC Bc) |
| ASP/IISABc/NcoI | TCATCATCACCATGGTAGAAGAAAAGGACAGAAAA | |
| SP/CIISABc/BamHI | TCATCATCAGGATCCCGAAATATGGGGTGCGAAATT | All four BACTH vectors carrying CIISA Bc |
| ASP/CIISABcSTOP/EcoRI | TCATCATCAGAATTCGATTCTGTCCTTATTTACTA | pUTCCIISA Bc, pKTCIISA Bc |
| ASP/CIISABcNOSTOP/EcoRI | TCATCATCAGAATTCGATTTACTAAAATAACTATGA | pUTCIISA Bc, pKNTCIISA Bc |
| SP/IISBBc/BamHI | TCATCATCAGGATCCCGTGATTGTAGTGGTAAAAGA | pUTIISB Bc, pUTCIISB Bc |
| ASP/IISBBcNOSTOP/EcoRI | TCATCATCAGAATTCGATGATTTTCTTTTTAATTCTT | pUTIISB Bc |
| ASP/IISBBcSTOP/EcoRI | TCATCATCAGAATTCGAGCAAATGTAGAAAGAGTGTA | pUTCIISB Bc |
| SP/IISCBc/BamHI | TCATCATCAGGATCCCATGGCTGAAGTCAATGTGCA | All four BACTH vectors carrying SpoIISC Bc |
| ASP/IISCBcNOSTOP/EcoRI | TCATCATCAGAATTCGATGCATTTTCTTTTGTTCTTT | pUTIISC Bc, pKNTIISC Bc |
| ASP/IISCBcSTOP/EcoRI | TCATCATCAGAATTCGACTATGCATTTTCTTTTGTTC | pUTCIISC Bc, pKTIISC Bc |
| SP/CIISABc/BamHI2 | TCATCATCAGGATCCGATTTCAGAAATATGGGG | pETDuetCIISA Bc |
| ASP/CIISABc/EcoRI | TCATCATCAGAATTCGATTCTGTCCTTATTTACTAA | |
| SP/IISBBc/KpnI | TCATCATCAGGTACCGTGATTGTAGTGGTA | pETDuetIISB Bc, pETDuetCIISAB Bc (via pETDuetCIISA Bc) |
| ASP/IISBBc/XhoI | TCATCATCACTCGAGTGATTTTCTTTTTAA | |
| SP/IISABs/EcoRI | TCATCATCAGAATTCATGGTTTTATTCTTTCAGATCATGGTCTGG | pBADIISA Bs, pBADIISAB Bs |
| ASP/IISABs/NcoI | TCATCATCACCATGGTTCCATTATCCTTCACCTTC | pBADIISA Bs |
| ASP/IISBBs/NcoI | TCATCATCACCATGGTTTAGTGTGATCATGCTTTT | pBADIISAB Bs |
| SP/IISCBs/PstI | TCATCATCACTGCAGAGAGGATAATGTCAGGTGAT | pBADIISAC Bs |
| ASP/IISCBs/HindIII | TCATCATCAAAGCTTCAAAGACCATAAAAATCCCGGAGCCGCTCC | |
| SP/CIISABs/BamHI | TCATCATCAGGATCCCAAAAAACTGGCCGGCAGCGAGCTTGAAACA | All four BACTH vectors carrying CIISA Bs |
| ASP/CIISABsSTOP/EcoRI | TCATCATCAGAATTCTTATCCTTCACCTTCCTCCT | pUTCCIISABs, pKTCIISABs |
| ASP/CIISABsNOSTOP/EcoRI | TCATCATCAGAATTCGATCCTTCACCTTCCTCCTCAA | pUTCIISABs, pKNTCIISABs |
| SP/IISBBs/BamHI | TCATCATCAGGATCCCATGGAACGTGCGTTTCAAAACAGATGCGAG | pUTIISB Bs, pUTCIISB Bs |
| ASP/IISBBsNOSTOP/EcoRI | TCATCATCAGAATTCGATCCTTCACCTTCCTCCTCAA | pUTIISB Bs |
| ASP/IISBBsSTOP/EcoRI | TCATCATCAGAATTCTCATGCTTTTTTTCGTTTAT | pUTCIISB Bs |
| SP/IISCBs/BamHI | TCATCATCAGGATCCCGTGACATATAATAAATACAA | All four BACTH vectors carrying SpoIISC Bs |
| ASP/IISCBsNOSTOP/EcoRI | TCATCATCAGAATTCGATGCTTTTTTTCGTTTATACT | pUTIISC Bs, pKNTIISC Bs |
| ASP/IISCBsSTOP/EcoRI | TCATCATCAGAATTCGATTATTTTTTCTTCTTCAACT | pUTCIISC Bs, pKTIISC Bs |
Bacterial two-hybrid system
Fragments T25 and T18 from the adenylate cyclase bacterial two-hybrid system (Karimova et al., 1998) were fused with the C-terminal domain of SpoIISA, full-length SpoIISB and SpoIISC, all from both B. cereus and B. subtilis. Chromosomal DNA from B. subtilis PY79 and B. cereus ATCC 14579 were used as PCR templates. E. coli BTH101 was used as a host for testing protein-protein interactions. Cells were co-transformed with the relevant plasmid combinations and plated onto LB plates supplemented with 100 μg ml−1 ampicillin, 30 μg ml−1 kanamycin, 40 μg ml−1 X-Gal and 0.1 mM IPTG and grown for 48 h at room temperature.
SDS-PAGE analysis
One dimensional SDS-PAGE was performed according to Laemmli (1970). Samples of whole cell lysates of recombinant-protein expressing E. coli BL21 (λDE3) cells, protein complexes, or purified protein samples were resuspended in sample buffer [4% SDS (w/v); 10% β-mercaptoethanol (v/v); 20% glycerol (v/v); 0.25 M Tris-Cl, pH 8] and boiled for 10 min. Denatured proteins were separated in 12% polyacrylamide gels. Due to the low molecular weight of B. cereus SpoIISC (6.6 kDa), this protein was analyzed using 16.5% Tricine–SDS-PAGE (Schägger and von Jagow, 1987), which better resolves such small proteins. As for the SDS-PAGE, samples of whole cell lysates of E. coli BL21 (λDE3) cells expressing recombinant SpoIISC and purified protein samples were resuspended in Novex sample buffer (Invitrogen, USA), then heated for 5 min in a boiling water bath and briefly spun down. The gels were run at 25 mA and stained with Coomassie brilliant blue R-250.
Pull-down assay
Pull-down assays were used to confirm in vitro the interactions between B. cereus C-SpoIISA and SpoIISB, SpoIISC and B. subtilis C-SpoIISA. In order to investigate the interaction of His6-tagged B. cereus C-SpoIISA with S-tagged SpoIISB, the following proteins were isolated: His6-tagged C-SpoIISA, S-tagged SpoIISB and His6-tagged C-SpoIISA expressed together with S-tagged SpoIISB. E. coli BL21 (λDE3) competent cells were transformed with the pETDuetCIISA Bc and pETDuetIISB Bc plasmids (Table 1) for the overexpression of His6-tagged C-SpoIISA and S-tagged SpoIISB, respectively. Transformation with pETDuetCIISAB Bc was performed to obtain co-expression of His6-tagged C-SpoIISA with S-tagged SpoIISB. The resulting cell cultures were grown at 28°C in LB medium supplemented with 100 μg ml−1 ampicillin and 0.5% glucose. Recombinant protein expression was induced by the addition of IPTG to a final concentration of 0.5 mM, when the culture reached an OD600 of ~0.6. Cells were harvested 5 h after induction, centrifuged, and resuspended in solubilization buffer [20 mM Tris-Cl, pH 8; 150 mM NaCl; 10% glycerol (v/v); 10 mM MgCl2; 1 mM AEBSF]. Proteins were solubilized by overnight incubation at 14°C in the presence of 10 mM CHAPS (Sigma Aldrich). Samples were centrifuged for 30 min at 60 000 × g and 4°C. Soluble fractions were loaded onto a Ni Sepharose HP column (Amersham Bioscience) and washed; bound proteins were eluted with an imidazole step gradient from 0.2 M, to 0.4 M, 0.6 M and 1 M. The most concentrated fraction of the His6-tagged C-SpoIISA, that with 1M imidazole, was used in further experiments. The S-tagged B. cereus SpoIISB 0.2 M imidazole fraction was used as a control for non-specific binding to the Ni column. Finally, the 0.4 M imidazole fraction of SpoIISB was used in the assay to confirm that His6-tagged C-SpoIISA interacts with S-tagged SpoIISB. These proteins and the C-SpoIISA–SpoIISB protein complex were fractionated by 16.5% Tricine–SDS–PAGE. The fractioned proteins were transferred onto a nitrocellulose membrane and subsequently Western blotted.
The pull-down assay of His6-tagged B. cereus C-SpoIISA with untagged SpoIISC was performed similarly as described above. In this case, E. coli BL21 (λDE3) cells were transformed with pETDuet-Bc-HCIISAC for the interaction study and pETDuet-Bc-IISC (Table 1) to control for the non-specific binding of B. cereus SpoIISC to the Ni column.
Glutaraldehyde crosslinking
The oligomeric state of recombinant B. cereus His6-C-SpoIISA was assessed by glutaraldehyde crosslinking. E. coli BL21 (λDE3) competent cells were transformed with pETDuetCIISA Bc, and protein expression was induced with 0.5 mM IPTG for 5 h at 28°C. Cells were then harvested and resuspended in a buffer containing 20 mM HEPES pH 7.5 and 150 mM NaCl and sonicated. The soluble fractions were centrifuged for 30 min at 60 000 × g and 4°C and then loaded onto a Ni Sepharose HP column (Amersham Bioscience). Proteins were eluted with an imidizole step gradient from 0.1 M to 0.2 M, 0.3 M and 1 M. For the crosslinking, 80 μg of protein was mixed with 5 μl of a freshly prepared solution of 2.3% glutaraldehyde to make a total volume of 100 μl. This reaction mixture was incubated for 30 min at 37°C and the reaction was then stopped by the addition of 10 μl of 1 M Tris-HCl, pH 8.0. The crosslinked molecules of B. cereus C-SpoIISA were loaded onto a 12% SDS-PAGE gel and detected by Western blotting.
Western blotting
To visualize the interaction of B. cereus C-SpoIISA with the heterologous B. subtilis C-SpoIISA as well as the interaction of B. cereus C-SpoIISA with SpoIISB, we performed Western blotting using the general protocol of Ausubel et al. (1987). Briefly, proteins were fractionated by either 12% SDS-PAGE or 16.5% Tricine-SDS-PAGE and transferred onto a nitrocellulose membrane (Hybond ECL; Amersham Bioscience). To prevent non-specific binding, the membrane was treated using 5% non-fat milk in Tris-buffered saline with 0.05% Tween 20 (v/v). His6-tagged B. cereus C-SpoIISA was probed with an anti His6-tag monoclonal antibody (Novagen; catalog no. 70796-3) while S-tagged B. subtilis C-SpoIISA and S-tagged B. cereus SpoIISB were probed with an anti S-tag monoclonal antibody (Novagen; catalog no. 71549-3). Protein interactions were detected using antimouse horseradish peroxidase-conjugated secondary antibodies (Promega; catalog no. W402B).
Gel filtration
To analyze the oligomerization of B. cereus C-SpoIISA using gel filtration, we developed a procedure for purifying untagged B. cereus C-SpoIISA. First, E. coli BL21 (λDE3) cells were transformed with the plasmid pET15b-Bc-CIISA. Next, the cell culture was grown at 28°C in LB medium supplemented with 100 μg ml−1 ampicillin. When the culture reached an OD600 of 0.6, the expression of untagged C-SpoIISA was induced with 0.5 mM IPTG. The cells were harvested 5 h after induction, centrifuged and resuspended in a resuspension buffer containing 50 mM glycine, pH 10; 50 mM NaCl; 10 mM MgCl2; 10% glycerol (v/v); and 1 mM AEBSF. The protein was solubilized by incubating at 14°C overnight in the presence of 10 mM CHAPS (Sigma Aldrich). The soluble fractions were cleared by centrifugation for 30 min at 60 000 × g and 4°C and loaded onto a HiPrep DEAE Sepharose FF 16/10 column (GE Healthcare Life Sciences), which had previously been equilibrated with a resuspension buffer containing 10 mM CHAPS. The protein eluted in the flow-through fraction and was loaded onto a HiPrep Q Sepharose HP 16/10 column (GE Healthcare Life Sciences), previously equilibrated with the same solution. The protein was eluted from this column with a continuous salt gradient ranging from 0.2 to 1 M NaCl. The purified protein was applied to a Superose 6 10/300 GL column (GE Healthcare Life Sciences) connected to an FPLC (GE Healthcare Life Sciences) instrument controlled by UNICORN 5.11 software, at a flow rate of 0.4 ml min−1. The elution was followed using UV absorbance at 280 nm.
Dynamic light scattering measurements
DLS experiments were performed at 20°C on a Zetasizer Nano ZS instrument (Malvern Instrument) controlled by DTS software (version 5.1, Malvern Instruments Ltd). The instrument has a 90° scattering angle. The purified protein, at a concentration of 100 μM in a resuspension buffer at pH 8 containing 10 mM CHAPS, was filtered through 20 nm filters into a 40 μl cuvette. A single measurement consisted of 20 runs of 12 s each. All measurements were done in triplicate. The samples gave a clear signal (the y-intercept was 0.95) and required only moderate attenuation (set at 7).
Bioinformatics analysis
Promoter analysis was performed using BPROM (Solovyev and Salamov, 2011). Identification of Rho-independent bacterial terminators was done using was done using ARNold web tool (Naville et al., 2011; http://rna.igmors.u-psud.fr/toolbox/arnold/). B. cereus SpoIISA membrane topology prediction was done using the MEMSAT3 and MEMSAT-SVM algorithms (http://bioinf.cs.ucl.ac.uk/psipred/; Nugent and Jones, 2009).
Results and discussion
The SpoIISABC toxin-antitoxin system
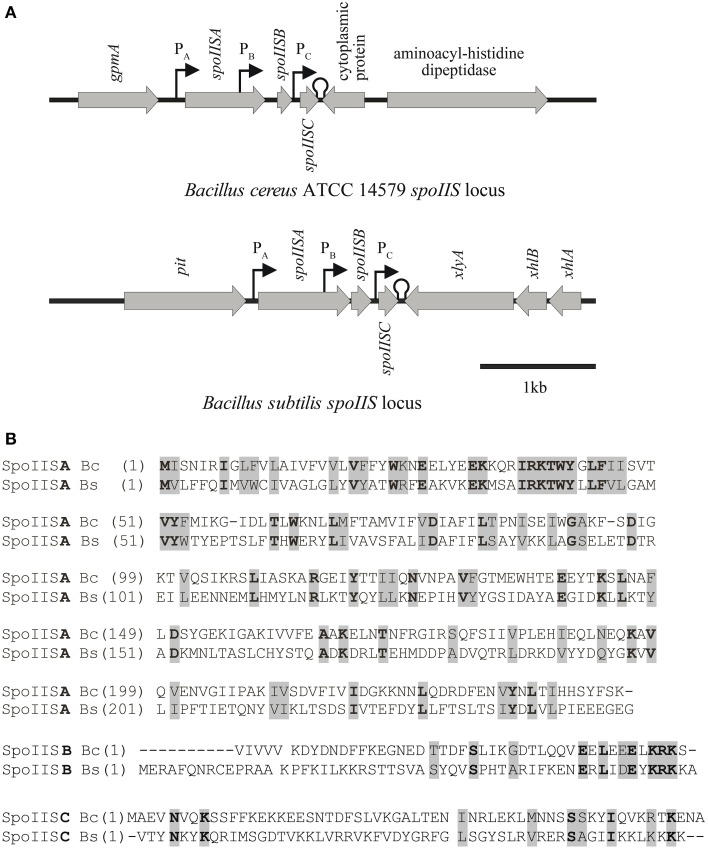
The SpoIIS toxin-antitoxin system in Bacillus subtilis consists of a SpoIISA toxin that is neutralized by a SpoIISB antitoxin (Adler et al., 2001; Florek et al., 2008). However, profiling of the condition-dependent transcription of B. subtilis revealed the presence of a third transcriptionally active region, denoted as S458 (Nicolas et al., 2012), located 55 bp downstream of spoIISB in the spoIIS operon, which we named spoIISC. Adler et al. (2001) identified two promoters in the B. subtilis spoIIS operon. The first promoter (PA) is located upstream of spoIISA and is important for regulating the expression of both spoIISA and spoIISB. The second promoter (PB) is located within the spoIISA gene and serves to regulate the expression of spoIISB. A promoter search using BPROM (see Materials and Methods) revealed a possible additional promoter (PC) downstream of spoIISB which could be used to regulate the expression of spoIISC. Its -35 sequence is 5′-TTCCTT-3′ and its -10 sequence is 5′-ACATATAAT-3′. In addition, a search for Rho-independent bacterial terminators using the ARNold tool identified the terminator (5′- GAAAAAATAAATCCCGGAGCGGCTCCGGGATTTTTATGGTCT -3′; letters in bold indicates bases contributing to the loop structure, underlined letters are bases forming the stem of terminator hairpin) immediately after the spoIISC STOP codon.
We previously found that a two-component SpoIIS system also exists in B. cereus (Florek et al., 2008). The position of its locus on the chromosome is completely different from that of the spoIIS operon in B. subtilis. While the B. subtilis spoIIS operon is 115° away from the origin of replication, the B. cereus spoIIS locus is 158° away. The B. cereus spoIIS operon consists of spoIISA (BC_2436), which encodes a 245-residue SpoIISA-like protein, and BC_2437, which encodes a hypothetical protein with 58 residues. As shown in Florek et al. (2008), BC_2437 is found 316 bp downstream of the spoIISA-like gene and was named spoIISB since its SpoIISB-like product was able to neutralize the toxicity of the SpoIISA-like protein in E. coli. Prompted by the identification of a putative third transcript in the B. subtilis spoIIS operon (Nicolas et al., 2012), we revisited the bioinformatics analysis of the B. cereus spoIIS operon and found that the B. cereus spoIIS operon also likely contains three genes: the BC_2436 ORF encoding a 245-residue SpoIISA-like protein; a 138-bp ORF 103 bp downstream of this gene, which encodes a 45-residue, putative SpoIISB; and a further 72 bp downstream of that, the BC_2437 ORF, which encodes the 58-residue protein we had previously called SpoIISB, but which we now call SpoIISC (Figure 1; Florek et al., 2008). As in the B. subtilis analysis, BPROM identified putative promoters in this operon. B. cereus spoIISA appears to be driven by the putative promoter PA, the putative PB promoter for controlling spoIISB gene expression is found within the spoIISA gene, and the putative PC promoter that likely regulates the expression of spoIISC is located downstream of the spoIISB gene. ARNold tool predicts that a Rho-independent bacterial transcription terminator, with the sequence 5′-AA AGAACAAAAGAAAATGCATAGAGCATTTTCTTTTGTTTTTTT A-3′ (letters in bold indicates bases contributing to the loop structure, underlined letters are bases forming the stem of terminator hairpin). This sequence overlaps with the end of B. cereus spoIISC gene (Figure 1A).
Figure 1.
Comparison of the spoIIS loci of Bacillus cereus and Bacillus subtilis. (A) Genomic organization of the spoIIS locus in B. cereus and B. subtilis. (B) Alignment of the SpoIIS proteins of B. cereus (Bc) and B. subtilis (Bs). Amino acids printed in normal weight on a gray background indicate similar amino acids, while bold weight on a gray background indicates identical amino acids.
The presence of three promoters in the spoIIS locus may be due to the different conditions under which the expression of individual genes is induced. The transcription of all three B. subtilis spoIIS genes is clearly induced during sporulation, but during nutrient deprivation only the spoIISA and spoIISB genes are transcribed (Nicolas et al., 2012). Moreover, there are conditions which induce transcription of only one of these genes: spoIISA is transcribed during ethanol stress, spoIISB during swarming and at high cell density, and spoIISC during biofilm formation (Nicolas et al., 2012).
Both B. subtilis and B. cereus SpoIISA-SpoIISB systems are clear examples of type II TA systems (Adler et al., 2001; Florek et al., 2011). The spoIIS operon has been identified only in Bacilli, and only a low level of homology can be detected between the SpoIIS proteins of B. subtilis and B. cereus (Florek et al., 2008). SpoIISA proteins display 17.3% identity and 30.2% similarity, while the SpoIISB proteins have only 12.5% identity and 17.9% similarity. The SpoIISC proteins have the lowest homology, with only 8.6% identity and 15.5% similarity. On the other hand, the SpoIISB and SpoIISC proteins from one of these organisms exhibit a higher level of homology with each other than with their counterparts in the other organism. Thus the B. subtilis SpoIISB and SpoIISC proteins show 37.5% homology and 12.5% identity while B. cereus SpoIISB and SpoIISC have 35.6% similarity and 27.1% identity (Figure 1B).
Bacterial type II TA systems are normally organized so that the first gene in the operon codes for the antitoxin and the toxin is positioned farther downstream; both genes are usually preceded by their own promoters. This arrangement ensures an abundance of antitoxin is produced to prevent toxin activity when it is undesirable. One exception to this arrangement is the higBA TA module in pathogenic Proteus species (Hurley and Woychik, 2009). As noted above, the spoIIS system is another, with the toxin preceding its two putative antitoxin genes. The SpoIIS TA system is unusual in another way as well. The typical type II TA system is a two-component system, but the SpoIIS TA system consists of three components: the SpoIISA toxin, the SpoIISB antitoxin and the third component SpoIISC (a likely antitoxin). Other three-component type II TA systems have previously been reported, including the ω-ε-ζ TA module encoded by the Streptococcus pyogenes plasmid pSM19035, the paaR-paaA-parE TA module encoded by E. coli O157:H7, and the pasA/pasB/pasC module of the Thiobacillus ferrooxidans plasmid pTF-FC2 (reviewed in Unterholzner et al., 2013). In all of these systems, at least one of the three components is involved in autoregulating the operon. There is presently no information about whether the expression of the spoIIS operon in Bacilli is autoregulated.
The spoIISB and spoIISC genes encode antitoxins in Bacillus cereus
B. subtilis transcription analysis by Nicolas et al. (2012) and in this study have revealed that the spoIIS operon is formed by the spoIISA, spoIISB, and spoIISC genes. In E. coli, B. subtilis SpoIISA inhibited bacterial growth and SpoIISB was able to neutralize SpoIISA toxicity (Florek et al., 2008). Previously, we observed that B. cereus SpoIISA, like B. subtilis SpoIISA, has a toxic effect on E. coli growth (Florek et al., 2008), but at that time, we had incorrectly designated ORF BC_2437 as spoIISB. A new bioinformatics analysis, prompted by the likely existence of a third gene in the B. subtilis spoIIS operon by Nicolas et al. (2012), shows that BC_2437 indeed contains spoIISC and that SpoIISB is a 45-residue protein of unknown function encoded by a small ORF (only 138 bp) located between the spoIISA and spoIISC genes.
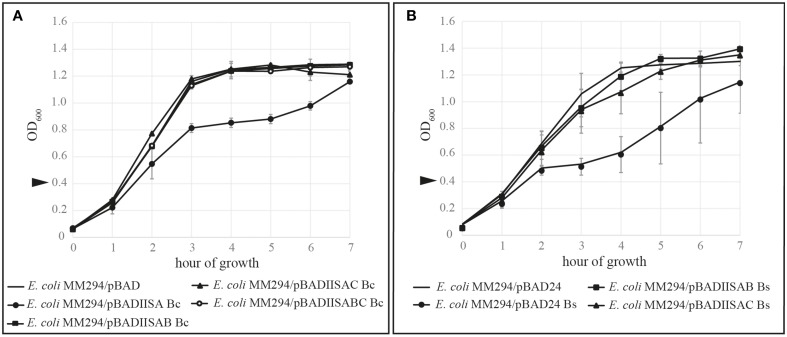
To determine if B. cereus SpoIISB and SpoIISC are both able to neutralize the toxicity of B. cereus SpoIISA in E. coli, the corresponding genes spoIISAB and spoIISAC were cloned into pBAD24 vectors under the control of arabinose-inducible PBAD promoters to generate pBADIISAB Bc and pBADIISAC Bc. These plasmids were subsequently introduced into E. coli MM294 cells. The growth of these transformed cells, together with the control strains IB890 (E. coli MM294 / pBAD24) and IB926 (E. coli MM294/pBAD-BCIISA) (Florek et al., 2008), was monitored after the induction of protein expression. As found previously (Florek et al., 2008), the growth of E. coli cells expressing only B. cereus SpoIISA was inhibited. On the other hand, both SpoIISB and SpoIISC were able to neutralize the toxicity of SpoIISA: the growth curves of those strains which expressed both SpoIISA and either the SpoIISB antitoxin or SpoIISC were similar to that of the wild-type IB890 E. coli cells (Figure 2A). Because B. cereus SpoIISB and SpoIISC disturb SpoIISA toxicity when expressed in E. coli cells, it can be concluded that both spoIISB and spoIISC encode antitoxins and that they are likely to have similar functions as the antitoxins in B. subtilis. Indeed, an identical set of experiments using the B. subtilis genes rather than the B. cereus ones gives very similar results (Figure 2B).
Figure 2.
Kill/rescue assay in E. coli MM294. In order to test the ability of SpoIISC to act as an antitoxin for SpoIISA, SpoIIS proteins were expressed alone or in combination in E. coli cells. All results are mean values of three independent replicates and the bars represent 1 SD. The growth of E. coli cells expressing the SpoIISA toxin (circle) was inhibited while those cells expressing either the SpoIISAB complex (square) or the SpoIISAC complex (triangle) had wild-type growth (no marker). Arrows indicate the addition of 0.02% arabinose to induce expression. (A) The effect of the B. cereus SpoIIS proteins on the growth of E. coli MM294 cells. (B) The effect of the B. subtilis SpoIIS proteins on the growth of E. coli MM294 cells.
The interactions of SpoIIS proteins in a bacterial two hybrid system
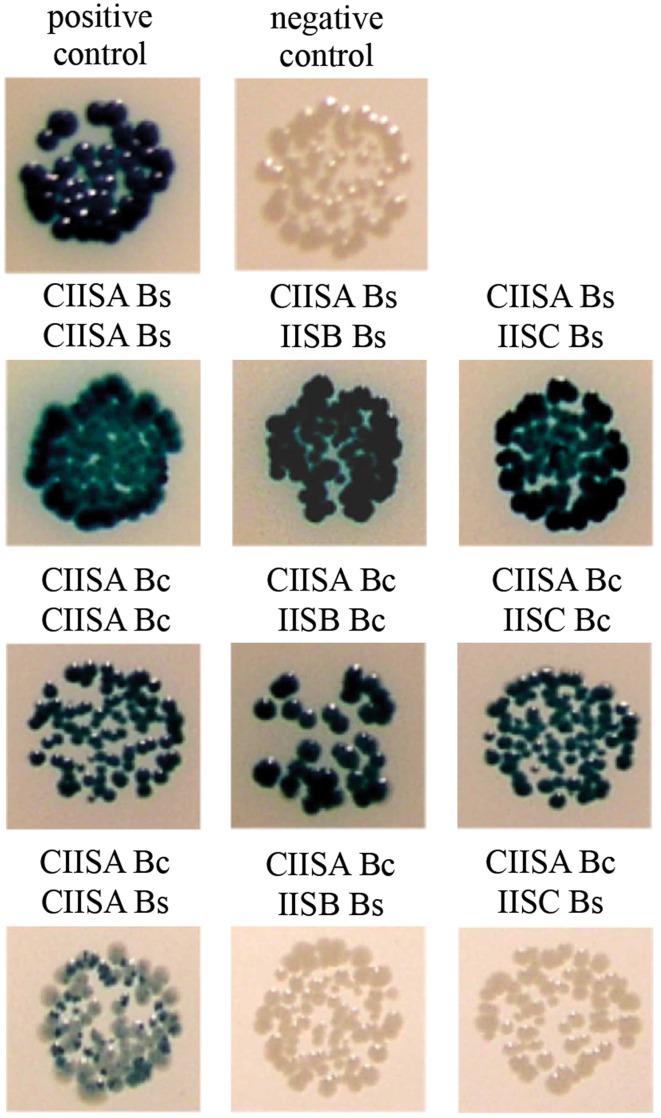
The clearest evidence that B. subtilis SpoIISA and SpoIISB directly interact can be found in the crystal structure of the C-terminal domain of SpoIISA in complex with SpoIISB (Florek et al., 2011). To analyze the protein–protein interactions of the B. cereus SpoIIS proteins in vivo, we made use of the bacterial adenylate cyclase two hybrid system (Karimova et al., 1998). Like B. subtilis SpoIISA, B. cereus SpoIISA is predicted to be a membrane protein with three membrane-spanning segments. However, we decided to use only the cytoplasmic domains in this protein–protein interaction study, since the whole SpoIISA protein is toxic for E. coli as we have shown previously. We prepared fusions of the C-terminal domain of B. cereus SpoIISA, SpoIISB, and SpoIISC with the adenylate cyclase fragments T25 and T18. All possible interactions were tested and compared with those of similar SpoIIS fusion proteins from B. subtilis (Figure 3).
Figure 3.
Interaction study of the SpoIIS proteins using the BACTH system. Since fusions with SpoIIS proteins in both orientations were positive in some cases, only representative ones were selected. A strain expressing a pair of leucine zipper proteins, T25-Zip and T18-Zip, served as the positive control; the negative control was a strain expressing the pair T25-CIISA Bc and T18-Zip. Abbreviations: Bc, B. cereus; Bs, B. subtilis; CIISA, C-terminal domain of SpoIISA; IISB, SpoIISB; IISC, SpoIISC.
Our results confirmed the dimerization of B. subtilis C-SpoIISA as well as the interaction of B. subtilis C-SpoIISA with SpoIISB described in Florek et al. (2011). A positive interaction was also observed for B. subtilis C-SpoIISA with SpoIISC (Figure 3). Finally, we found that the B. cereus C-terminal domain of SpoIISA can interact with another C-SpoIISA protomer, with SpoIISB and with SpoIISC (Figure 3).
B. cereus SpoIISB and SpoIISC are able to bind the C-terminal domain of SpoIISA in vitro
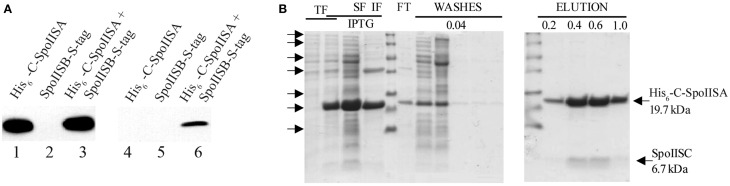
To analyze these protein–protein interactions in vitro, we prepared three derivatives of the pETDuet recombinant expression plasmid, each containing one of the following genes, all under the control of an IPTG-inducible T7 promoter: a gene coding for a His6-tagged B. cereus C-SpoIISA, an S-tagged SpoIISB and an untagged SpoIISC (Table 1). We found that His6-tagged C-SpoIISA binds the Ni column and that S-tagged SpoIISB and untagged SpoIISC creates a tight complex with C-SpoIISA which can be eluted by a solubilization buffer step gradient containing 0.1–1 mM imidazole (Figure 4).
Figure 4.
Pull-down assays of B. cereus SpoIISB and SpoIISC with C-SpoIISA. The soluble fractions of lysed bacterial cells were applied to a Ni Sepharose HP column. The eluted proteins were identified by Western blotting (A) and Coomassie brilliant blue R-250 staining (B). (A) In the Western blot, the eluted proteins were probed with an anti-His6 monoclonal antibody (lanes 1–3) or with an anti-S monoclonal antibody (lanes 4–6). Lanes 1 and 4 contain purified His6-tagged C-SpoIISA, lanes 2 and 5, purified S-tagged SpoIISB. S-tagged SpoIISB does not bind a Ni Sepharose HP column. Lanes 3 and 6 show that His6-tagged C-SpoIISA can pull down S-tagged SpoIISB and therefore that there is an interaction between them. (B) A pull-down assay showing an interaction between His6-tagged C-SpoIISA and SpoIISC when both proteins are co-expressed. TF, total fraction; SF, soluble fraction; IF, insoluble fraction; FT, flow-through fraction, 0.04; 0.2; 0.4, 0.6, and 1.0—molarity of imidazole used in washing and elution. The arrows mark the following positions on the protein ladder from top to bottom: 116, 66.2, 45, 35, 25, 18.4, and 14.4 kDa.
The interaction of B. cereus His6-tagged C-SpoIISA with S-tagged SpoIISB was confirmed in a pull-down assay by the co-elution of both proteins from a Ni column. When SpoIISB is co-expressed in E. coli together with His6-tagged C-SpoIISA, the His6-tagged C-SpoIISA binds the Ni column, and since S-tagged SpoIISB binds C-SpoIISA, the two are pulled down together as a complex during elution with 0.4 M imidazole (Figure 4A). This complex could be detected by Western blotting using an anti-His6-tag monoclonal antibody to identify His6-tagged C-SpoIISA (Figure 4A, lane 3) and an anti-S-tag monoclonal antibody to identify the S-tagged SpoIISB (Figure 4A, lane 6).
A similar approach was used to test the interaction of untagged B. cereus SpoIISC with His6-tagged C-SpoIISA in vitro (Figure 4B). B. cereus SpoIISC expressed in E. coli BL21 (DE3) appears in the insoluble fraction of the cell lysate according to 16.5% Tricine/SDS–PAGE (data not shown). However, when co-expressed with B. cereus His6-tagged C-SpoIISA in the same cells, they form a complex which is able to pull SpoIISC out of the insoluble fraction. The whole complex can then be solubilized and purified from the soluble fraction by affinity chromatography.
B. cereus C-terminal domain of SpoIISA forms an oligomer
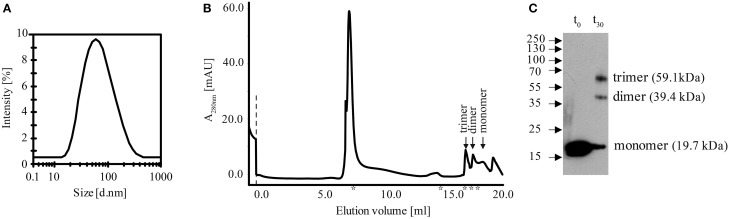
The crystal structure of the B. subtilis SpoIISA C-terminal domain shows that the protein dimerizes by forming a four-helix bundle using the first and last α-helices of each molecule (Florek et al., 2011). Our bacterial two-hybrid experiments showed that B. cereus C-SpoIISA interacts with other B. cereus C-SpoIISA molecules (Figure 3), suggesting that this molecule also forms oligomers. The oligomeric form of C-SpoIISA was examined by measuring the hydrodynamic radius of dissolved particles using dynamic light scattering. A cumulant analysis showed that the sample was monomodal (i.e., had only one peak, Figure 5A), and was polydisperse, with a polydispersity index of 0.255 and an overall polydispersity of 50.32%. The polydispersity indicates broader particle size distribution, and thus the hydrodynamic radius and corresponding molecular mass cannot be reliably calculated.
Figure 5.
Analysis of the multimeric state of B. cereus C-SpoIISA. (A) Dynamic light scattering analysis of C-SpoIISA oligomer. Size distribution (by intensity) of B. cereus C-SpoIISA, at 20°C, average hydrodynamic radius = 55 nm. (B) Gel filtration of C-SpoIISA. The stars indicate the positions at which the following protein standards eluted from the column (left to right): 2000, 450, 66, 45, and 29 kDa. (C) Western blot analysis of glutaraldehyde-crosslinked His6-tagged C-SpoIISA.
The SpoIISA oligomerisation was examined further by size-exclusion chromatography of C-SpoIISA using a Superose 6 10/300 GL column. In this analysis, most of the protein appeared in the void volume fraction of the column, which was determined from the elution of Blue dextran 2000 (~2000 kDa, Pharmacia) (Figure 5B). Three small peaks were detected, however, and likely correspond to the 59.1 kDa trimer, the 39.4 kDa dimer and the 19.7 kDa monomer of C-SpoIISA. The existence of monomeric, dimeric and trimeric states was confirmed by glutaraldehyde crosslinking (Figure 5C), but the existence of higher oligomeric forms could not be confirmed because such large species would not have been able to enter the crosslinking gel.
Taken together, the above results indicate that B. cereus C-SpoIISA is able to form higher multimers, even if their nature is unclear. In this respect, its behavior differs from that of B. subtilis C-SpoIISA, which formed only dimers (Florek et al., 2011). Whole B. subtilis SpoIISA does seem to form higher oligomers, but this seems to require its N-terminal transmembrane domain rather than just its C-terminal cytosolic domain (Makroczyová et al., 2014). Earlier studies also suggested that whole SpoIISA oligomerizes, and moreover suggested that it forms holin-like pores (Adler et al., 2001). Whether either the B. subtilis or B. cereus proteins actually do form such pores remains unknown, however.
Finally, this study describes the SpoIISC protein, a third component of the spoIIS locus. This protein serves as an antitoxin and shows similarity to SpoIISB. The presence of two antitoxin genes in the spoIIS locus of both B. subtilis and B. cereus naturally poses the question of the role of such duplication. One possibility is that the different proteins are linked to different conditions under which they might be expressed, as was shown for B. subtilis SpoIIS system (Nicolas et al., 2012). They may also act as transcription regulators, as some other antitoxins are known to. It is also possible that their different amino-acid compositions could affect their affinity for SpoIISA, leading to different degrees of inhibition. In any case, our results show that the SpoIIS TA system is much more complex than had previously been thought.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Daniela Krajčíková for critical reading of the manuscript, Katarína Muchová for help in designing the pull-down experiments and for critically evaluating the results. The authors would also like to thank Emília Chovancová for technical assistance and Jacob Bauer for helpful comments. This work was supported by Grant 2/0009/13 from the Slovak Academy of Sciences, by a Grant from the Slovak Research and Development Agency under contract APVV-00335-10; APVV-14-0181 and by the Research and Development Operational Programme funded by the ERDF (ITMS code: 26240220044).
References
- Adler E., Barák I., Stragier P. (2001). Bacillus subtilis locus encoding a killer protein and its antidote. J. Bacteriol. 183, 3574–3581. 10.1128/JB.183.12.3574-3581.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., et al. (1987). Protocols in Molecular Biology. New York, NY: Greene Publishing Associates and Wiley-Interscience John Wiley and Sons. [Google Scholar]
- Camacho A. G., Misselwitz R., Behlke J., Ayora S., Welfle K., Meinhart A., et al. (2002). In vitro and in vivo stability of the 2ζ2 protein complex of the broad host-range Streptococcus pyogenes pSM19035 addiction system. Biol. Chem. 383, 1701–1713. 10.1515/bc.2002.191 [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H., Amitai S., Kolodkin-Gal I., Hazan R. (2006). Bacterial programmed cell Death and multicellular behavior in bacteria. PLoS Genet. 2:e135. 10.1371/journal.pgen.0020135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineran P. C., Blower T. R., Foulds I. J., Humphreys D. P., Lilley K. S., Salmond G. P. C. (2009). The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. U.S.A. 106, 894–899. 10.1073/pnas.0808832106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florek P., Levdikov V. M., Blagova E., Lebedev A. A., Škrabana R., Rešetárová S., et al. (2011). The structure and interactions of SpoIISA and SpoIISB, a toxin-antitoxin system in Bacillus subtilis. J. Biol. Chem. 286, 6808–6819. 10.1074/jbc.M110.172429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florek P., Muchová K., Pavelčíková P., Barák I. (2008). Expression of functional Bacillus SpoIISAB toxin-antitoxin modules in Escherichia coli. FEMS Microbiol. Lett. 278, 177–184. 10.1111/j.1574-6968.2007.00984.x [DOI] [PubMed] [Google Scholar]
- Gerdes K., Christensen S. K., Løbner-Olesen A. (2005). Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3, 371–382. 10.1038/nrmicro1147 [DOI] [PubMed] [Google Scholar]
- Gerdes K., Gultyaev A. P., Franch T., Pedersen K., Mikkelsen N. D. (1997). Antisense RNA-regulated programmed cell death. Annu. Rev. Genet. 31, 1–31. 10.1146/annurev.genet.31.1.1 [DOI] [PubMed] [Google Scholar]
- Gerdes K., Rasmussen P. B., Molin S. (1986). Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. U.S.A. 83, 3116–3120. 10.1073/pnas.83.10.3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeders N., Van Melderen L. (2014). Toxin-antitoxin systems as multilevel interaction systems. Toxins (Basel). 6, 304–324. 10.3390/toxins6010304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. 10.1016/S0022-2836(83)80284-8 [DOI] [PubMed] [Google Scholar]
- Hayes F. (2003). Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301, 1496–1499. 10.1126/science.1088157 [DOI] [PubMed] [Google Scholar]
- Hurley J. M., Woychik N. A. (2009). Bacterial toxin HigB associates with ribosomes and mediates translation-dependent mRNA cleavage at A-rich sites. J. Biol. Chem. 284, 18605–18613. 10.1074/jbc.M109.008763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G., Pidoux J., Ullmann A., Ladant D. (1998). A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756. 10.1073/pnas.95.10.5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Lehnherr H., Yarmolinsky M. B. (1995). Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 92, 3274–3277. 10.1073/pnas.92.8.3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makroczyová J., Rešetárová S., Florek P., Barák I. (2014). Topology of the Bacillus subtilis SpoIISA protein and its role in toxin–antitoxin function. FEMS Microbiol. Lett. 358, 180–187. 10.1111/1574-6968.12531 [DOI] [PubMed] [Google Scholar]
- Masuda H., Tan Q., Awano N., Wu K.-P., Inouye M. (2012). YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol. Microbiol. 84, 979–989. 10.1111/j.1365-2958.2012.08068.x [DOI] [PubMed] [Google Scholar]
- Meselson M., Yuan R. (1968). DNA restriction enzyme from E. coli. Nature 217, 1110–1114. 10.1038/2171110a0 [DOI] [PubMed] [Google Scholar]
- Naville M., Ghuillot-Gaudeffroy A., Marchais A., Gautheret D. (2011). ARNold: a web tool for the prediction of Rho-independent transcription terminators. RNA Biol. 8, 11–13. 10.4161/rna.8.1.13346 [DOI] [PubMed] [Google Scholar]
- Nicolas P., Maeder U., Dervyn E., Rochat T., Leduc A., Pigeonneau N., et al. (2012). Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335, 1103–1106. 10.1126/science.1206848 [DOI] [PubMed] [Google Scholar]
- Nugent T., Jones D. T. (2009). Transmembrane protein topology prediction using support vector machines. BMC Bioinformatics 10:159. 10.1186/1471-2105-10-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K., Gerdes K. (1999). Multiple hok genes on the chromosome of Escherichia coli. Mol. Microbiol. 32, 1090–1102. 10.1046/j.1365-2958.1999.01431.x [DOI] [PubMed] [Google Scholar]
- Rešetárová S., Florek P., Muchová K., Wilkinson A. J., Barák I. (2010). Expression and localization of SpoIISA toxin during the life cycle of Bacillus subtilis. Res. Microbiol. 161, 750–756. 10.1016/j.resmic.2010.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Echevarría M. J., Giménez-Gallego G., Sabariegos-Jareño R., Díaz-Orejas R. (1995). Kid, a small protein of the parD stability system of plasmid R1, is an inhibitor of DNA replication acting at the initiation of DNA synthesis. J. Mol. Biol. 247, 568–577. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual, 2nd Edn. New York, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Sayeed S., Reaves L., Radnedge L., Austin S. (2000). The stability region of the large virulence plasmid of Shigella flexneri encodes an efficient postsegregational killing system. J. Bacteriol. 182, 2416–2421. 10.1128/JB.182.9.2416-2421.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379. 10.1016/0003-2697(87)90587-2 [DOI] [PubMed] [Google Scholar]
- Schuster C. F., Bertram R. (2013). Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol. Lett. 340, 73–85. 10.1111/1574-6968.12074 [DOI] [PubMed] [Google Scholar]
- Solovyev V., Salamov A. (2011). Automatic annotation of microbial genomes and metagenomic sequences, in Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies, ed Li R. W. (New York, NY: Nova Science Publishers; ), 61–78. [Google Scholar]
- Tsilibaris V., Maenhaut-Michel G., Mine N., Van Melderen L. (2007). What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J. Bacteriol. 189, 6101–6108. 10.1128/JB.00527-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner S. J., Poppenberger B., Rozhon W. (2013). Toxin–antitoxin systems. Mob. Genet. Elements 3:e26219. 10.4161/mge.26219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melderen L. (2001). Molecular interactions of the CcdB poison with its bacterial target, the DNA gyrase. Int. J. Med. Microbiol. 291, 537–544. 10.1078/1438-4221-00164 [DOI] [PubMed] [Google Scholar]
- Van Melderen L., Saavedra De Bast M. (2009). Bacterial toxin–antitoxin systems: more than selfish entities? PLoS Genet. 5:e1000437. 10.1371/journal.pgen.1000437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Lord D. M., Cheng H.-Y., Osbourne D. O., Hong S. H., Sanchez-Torres V., et al. (2012). A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat. Chem. Biol. 8, 855–861. 10.1038/nchembio.1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P., Perkins J. B., Losick R. (1984). Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 12, 1–9. 10.1016/0147-619X(84)90061-1 [DOI] [PubMed] [Google Scholar]