Abstract
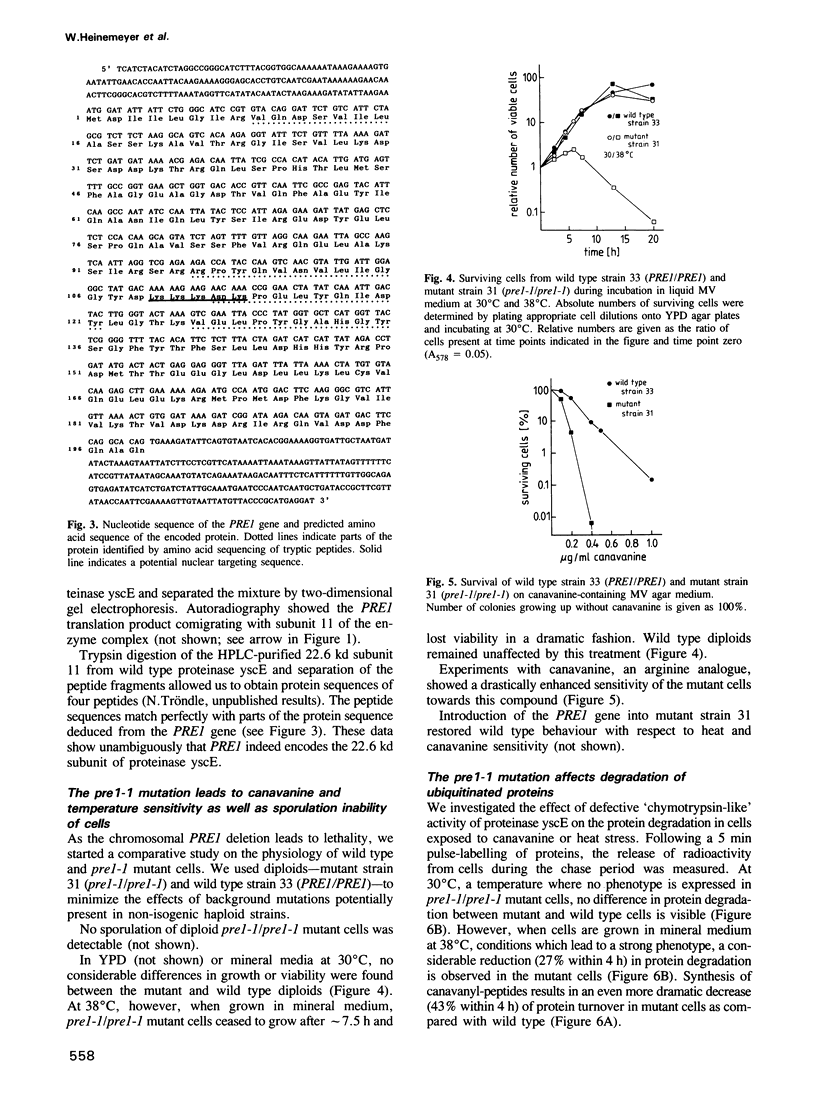
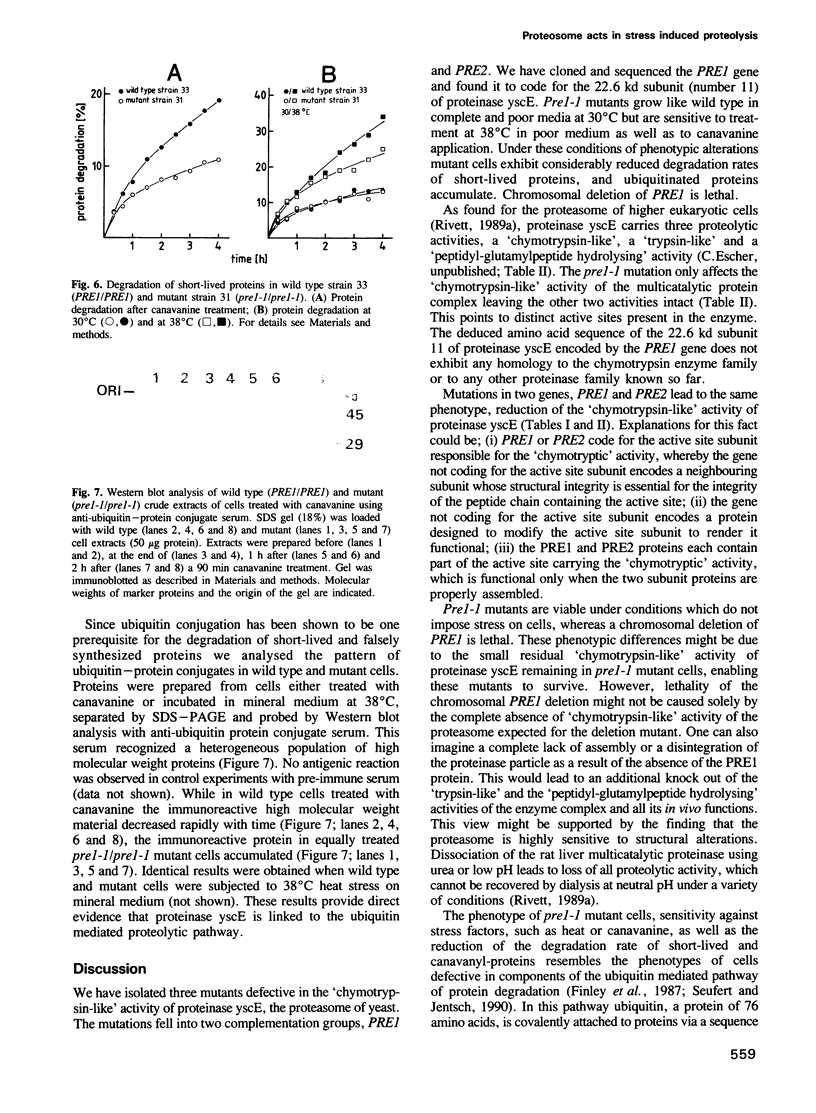
Proteinase yscE is the yeast equivalent of the proteasome, a multicatalytic-multifunctional proteinase found in higher eukaryotic cells. We have isolated three mutants affecting the proteolytic activity of proteinase yscE. The mutants show a specific reduction in the activity of the complex against peptide substrates with hydrophobic amino acids at the cleavage site and define two complementation groups, PRE1 and PRE2. The PRE1 gene was cloned and shown to be essential. The deduced amino acid sequence encoded by the PRE1 gene reveals weak, but significant similarities to proteasome subunits of other organisms. Two-dimensional gel electrophoresis identified the yeast proteasome to be composed of 14 different subunits. Comparison of these 14 subunits with the translation product obtained from PRE1 mRNA synthesized in vitro demonstrated that PRE1 encodes the 22.6 kd subunit (numbered 11) of the yeast proteasome. Diploids homozygous for pre1-1 are defective in sporulation. Strains carrying the pre1-1 mutation show enhanced sensitivity to stresses such as incorporation of the amino acid analogue canavanine into proteins or a combination of poor growth medium and elevated temperature. Under these stress conditions pre1-1 mutant cells exhibit decreased protein degradation and accumulate ubiquitin-protein conjugates.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achstetter T., Ehmann C., Osaki A., Wolf D. H. Proteolysis in eukaryotic cells. Proteinase yscE, a new yeast peptidase. J Biol Chem. 1984 Nov 10;259(21):13344–13348. [PubMed] [Google Scholar]
- Achstetter T., Emter O., Ehmann C., Wolf D. H. Proteolysis in eukaryotic cells. Identification of multiple proteolytic enzymes in yeast. J Biol Chem. 1984 Nov 10;259(21):13334–13343. [PubMed] [Google Scholar]
- Achstetter T., Wolf D. H. Proteinases, proteolysis and biological control in the yeast Saccharomyces cerevisiae. Yeast. 1985 Dec;1(2):139–157. doi: 10.1002/yea.320010203. [DOI] [PubMed] [Google Scholar]
- Arrigo A. P., Tanaka K., Goldberg A. L., Welch W. J. Identity of the 19S 'prosome' particle with the large multifunctional protease complex of mammalian cells (the proteasome). Nature. 1988 Jan 14;331(6152):192–194. doi: 10.1038/331192a0. [DOI] [PubMed] [Google Scholar]
- Barrett A. J. A new assay for cathepsin B1 and other thiol proteinases. Anal Biochem. 1972 May;47(1):280–293. doi: 10.1016/0003-2697(72)90302-8. [DOI] [PubMed] [Google Scholar]
- Betz H., Weisner U. Protein degradation and proteinases during yeast sporulation. Eur J Biochem. 1976 Feb 2;62(1):65–76. doi: 10.1111/j.1432-1033.1976.tb10098.x. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Schwartz A. L. How are substrates recognized by the ubiquitin-mediated proteolytic system? Trends Biochem Sci. 1989 Dec;14(12):483–488. doi: 10.1016/0968-0004(89)90180-1. [DOI] [PubMed] [Google Scholar]
- Dahlmann B., Kopp F., Kuehn L., Niedel B., Pfeifer G., Hegerl R., Baumeister W. The multicatalytic proteinase (prosome) is ubiquitous from eukaryotes to archaebacteria. FEBS Lett. 1989 Jul 17;251(1-2):125–131. doi: 10.1016/0014-5793(89)81441-3. [DOI] [PubMed] [Google Scholar]
- Driscoll J., Goldberg A. L. The proteasome (multicatalytic protease) is a component of the 1500-kDa proteolytic complex which degrades ubiquitin-conjugated proteins. J Biol Chem. 1990 Mar 25;265(9):4789–4792. [PubMed] [Google Scholar]
- Emter O., Wolf D. H. Vacuoles are not the sole compartments of proteolytic enzymes in yeast. FEBS Lett. 1984 Jan 30;166(2):321–325. doi: 10.1016/0014-5793(84)80104-0. [DOI] [PubMed] [Google Scholar]
- Eytan E., Ganoth D., Armon T., Hershko A. ATP-dependent incorporation of 20S protease into the 26S complex that degrades proteins conjugated to ubiquitin. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7751–7755. doi: 10.1073/pnas.86.20.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenburg P. E., Haass C., Kloetzel P. M., Niedel B., Kopp F., Kuehn L., Dahlmann B. Drosophila small cytoplasmic 19S ribonucleoprotein is homologous to the rat multicatalytic proteinase. Nature. 1988 Jan 14;331(6152):190–192. doi: 10.1038/331190a0. [DOI] [PubMed] [Google Scholar]
- Finley D., Ozkaynak E., Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987 Mar 27;48(6):1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Tanaka K., Kumatori A., Shin S., Yoshimura T., Ichihara A., Tokunaga F., Aruga R., Iwanaga S., Kakizuka A. Molecular cloning of cDNA for proteasomes (multicatalytic proteinase complexes) from rat liver: primary structure of the largest component (C2). Biochemistry. 1989 Sep 5;28(18):7332–7340. doi: 10.1021/bi00444a028. [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Tanaka K., Orino E., Yoshimura T., Kumatori A., Tamura T., Chung C. H., Nakai T., Yamaguchi K., Shin S. Proteasomes are essential for yeast proliferation. cDNA cloning and gene disruption of two major subunits. J Biol Chem. 1990 Sep 25;265(27):16604–16613. [PubMed] [Google Scholar]
- Fuller R. S., Sterne R. E., Thorner J. Enzymes required for yeast prohormone processing. Annu Rev Physiol. 1988;50:345–362. doi: 10.1146/annurev.ph.50.030188.002021. [DOI] [PubMed] [Google Scholar]
- García-Alvarez N., Teichert U., Wolf D. H. Proteinase yscD mutants of yeast. Isolation and characterization. Eur J Biochem. 1987 Mar 2;163(2):339–346. doi: 10.1111/j.1432-1033.1987.tb10805.x. [DOI] [PubMed] [Google Scholar]
- Haass C., Kloetzel P. M. The Drosophila proteasome undergoes changes in its subunit pattern during development. Exp Cell Res. 1989 Jan;180(1):243–252. doi: 10.1016/0014-4827(89)90228-0. [DOI] [PubMed] [Google Scholar]
- Haass C., Pesold-Hurt B., Kloetzel P. M. The Drosophila PROS-29 gene is a new member of the PROS-gene family. Nucleic Acids Res. 1990 Jul 11;18(13):4018–4018. doi: 10.1093/nar/18.13.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C., Pesold-Hurt B., Multhaup G., Beyreuther K., Kloetzel P. M. The PROS-35 gene encodes the 35 kd protein subunit of Drosophila melanogaster proteasome. EMBO J. 1989 Aug;8(8):2373–2379. doi: 10.1002/j.1460-2075.1989.tb08366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Eytan E., Ciechanover A., Haas A. L. Immunochemical analysis of the turnover of ubiquitin-protein conjugates in intact cells. Relationship to the breakdown of abnormal proteins. J Biol Chem. 1982 Dec 10;257(23):13964–13970. [PubMed] [Google Scholar]
- Hershko A. Ubiquitin-mediated protein degradation. J Biol Chem. 1988 Oct 25;263(30):15237–15240. [PubMed] [Google Scholar]
- Hershko A. Ubiquitin: roles in protein modification and breakdown. Cell. 1983 Aug;34(1):11–12. doi: 10.1016/0092-8674(83)90131-9. [DOI] [PubMed] [Google Scholar]
- Hough R., Pratt G., Rechsteiner M. Purification of two high molecular weight proteases from rabbit reticulocyte lysate. J Biol Chem. 1987 Jun 15;262(17):8303–8313. [PubMed] [Google Scholar]
- Hough R., Pratt G., Rechsteiner M. Ubiquitin-lysozyme conjugates. Identification and characterization of an ATP-dependent protease from rabbit reticulocyte lysates. J Biol Chem. 1986 Feb 15;261(5):2400–2408. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S., Seufert W., Sommer T., Reins H. A. Ubiquitin-conjugating enzymes: novel regulators of eukaryotic cells. Trends Biochem Sci. 1990 May;15(5):195–198. doi: 10.1016/0968-0004(90)90161-4. [DOI] [PubMed] [Google Scholar]
- Jones E. W. The synthesis and function of proteases in Saccharomyces: genetic approaches. Annu Rev Genet. 1984;18:233–270. doi: 10.1146/annurev.ge.18.120184.001313. [DOI] [PubMed] [Google Scholar]
- Klebe R. J., Harriss J. V., Sharp Z. D., Douglas M. G. A general method for polyethylene-glycol-induced genetic transformation of bacteria and yeast. Gene. 1983 Nov;25(2-3):333–341. doi: 10.1016/0378-1119(83)90238-x. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt J. A., Escher C., Wolf D. H. Proteinase yscE of yeast shows homology with the 20 S cylinder particles of Xenopus laevis. FEBS Lett. 1988 Oct 24;239(1):35–40. doi: 10.1016/0014-5793(88)80540-4. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt J. A., Hügle B., Grund C., Franke W. W. The 22 S cylinder particles of Xenopus laevis. I. Biochemical and electron microscopic characterization. Eur J Cell Biol. 1983 Nov;32(1):143–156. [PubMed] [Google Scholar]
- Krebs H. O., Hoffschulte H. K., Müller M. In vitro studies on the translocation of acid phosphatase into the endoplasmic reticulum of the yeast Saccharomyces cerevisiae. Eur J Biochem. 1989 May 1;181(2):323–329. doi: 10.1111/j.1432-1033.1989.tb14727.x. [DOI] [PubMed] [Google Scholar]
- Kuehn L., Dahlmann B., Kopp F. Effect of the multicatalytic proteinase (prosome) on translational activity in rabbit reticulocyte lysates. FEBS Lett. 1990 Feb 26;261(2):274–278. doi: 10.1016/0014-5793(90)80570-9. [DOI] [PubMed] [Google Scholar]
- Kumatori A., Tanaka K., Tamura T., Fujiwara T., Ichihara A., Tokunaga F., Onikura A., Iwanaga S. cDNA cloning and sequencing of component C9 of proteasomes from rat hepatoma cells. FEBS Lett. 1990 May 21;264(2):279–282. doi: 10.1016/0014-5793(90)80267-m. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Ohlsson B. G., Weström B. R., Karlsson B. W. Enzymoblotting: a method for localizing proteinases and their zymogens using para-nitroanilide substrates after agarose gel electrophoresis and transfer to nitrocellulose. Anal Biochem. 1986 Feb 1;152(2):239–244. doi: 10.1016/0003-2697(86)90404-5. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M. Ubiquitin-mediated pathways for intracellular proteolysis. Annu Rev Cell Biol. 1987;3:1–30. doi: 10.1146/annurev.cb.03.110187.000245. [DOI] [PubMed] [Google Scholar]
- Rivett A. J. The multicatalytic proteinase of mammalian cells. Arch Biochem Biophys. 1989 Jan;268(1):1–8. doi: 10.1016/0003-9861(89)90558-4. [DOI] [PubMed] [Google Scholar]
- Rivett A. J. The multicatalytic proteinase. Multiple proteolytic activities. J Biol Chem. 1989 Jul 25;264(21):12215–12219. [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer K. Prosomes, subcomplexes of untranslated mRNP. Mol Biol Rep. 1990 Feb;14(1):1–9. doi: 10.1007/BF00422709. [DOI] [PubMed] [Google Scholar]
- Schmid H. P., Akhayat O., Martins De Sa C., Puvion F., Koehler K., Scherrer K. The prosome: an ubiquitous morphologically distinct RNP particle associated with repressed mRNPs and containing specific ScRNA and a characteristic set of proteins. EMBO J. 1984 Jan;3(1):29–34. doi: 10.1002/j.1460-2075.1984.tb01757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengstag C., Hinnen A. The sequence of the Saccharomyces cerevisiae gene PHO2 codes for a regulatory protein with unusual aminoacid composition. Nucleic Acids Res. 1987 Jan 12;15(1):233–246. doi: 10.1093/nar/15.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert W., Jentsch S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990 Feb;9(2):543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T., Tanaka K., Kumatori A., Yamada F., Tsurumi C., Fujiwara T., Ichihara A., Tokunaga F., Aruga R., Iwanaga S. cDNA cloning and sequencing of component C5 of proteasomes from rat hepatoma cells. FEBS Lett. 1990 May 7;264(1):91–94. doi: 10.1016/0014-5793(90)80773-c. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Fujiwara T., Kumatori A., Shin S., Yoshimura T., Ichihara A., Tokunaga F., Aruga R., Iwanaga S., Kakizuka A. Molecular cloning of cDNA for proteasomes from rat liver: primary structure of component C3 with a possible tyrosine phosphorylation site. Biochemistry. 1990 Apr 17;29(15):3777–3785. doi: 10.1021/bi00467a026. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Yoshimura T., Kumatori A., Ichihara A., Ikai A., Nishigai M., Kameyama K., Takagi T. Proteasomes (multi-protease complexes) as 20 S ring-shaped particles in a variety of eukaryotic cells. J Biol Chem. 1988 Nov 5;263(31):16209–16217. [PubMed] [Google Scholar]
- Teichert U., Mechler B., Müller H., Wolf D. H. Lysosomal (vacuolar) proteinases of yeast are essential catalysts for protein degradation, differentiation, and cell survival. J Biol Chem. 1989 Sep 25;264(27):16037–16045. [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk S., Orlowski M. Cation-sensitive neutral endopeptidase: isolation and specificity of the bovine pituitary enzyme. J Neurochem. 1980 Nov;35(5):1172–1182. doi: 10.1111/j.1471-4159.1980.tb07873.x. [DOI] [PubMed] [Google Scholar]
- Wilk S., Orlowski M. Evidence that pituitary cation-sensitive neutral endopeptidase is a multicatalytic protease complex. J Neurochem. 1983 Mar;40(3):842–849. doi: 10.1111/j.1471-4159.1983.tb08056.x. [DOI] [PubMed] [Google Scholar]





