Abstract
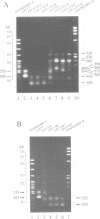
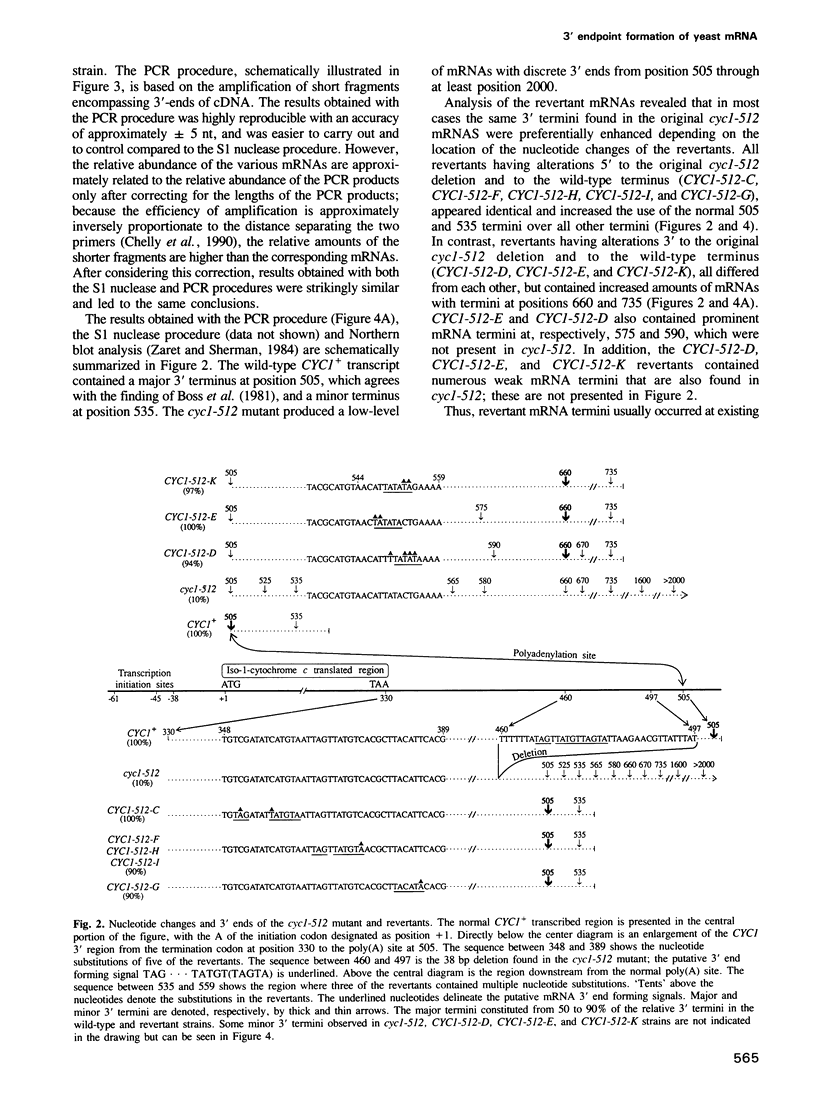
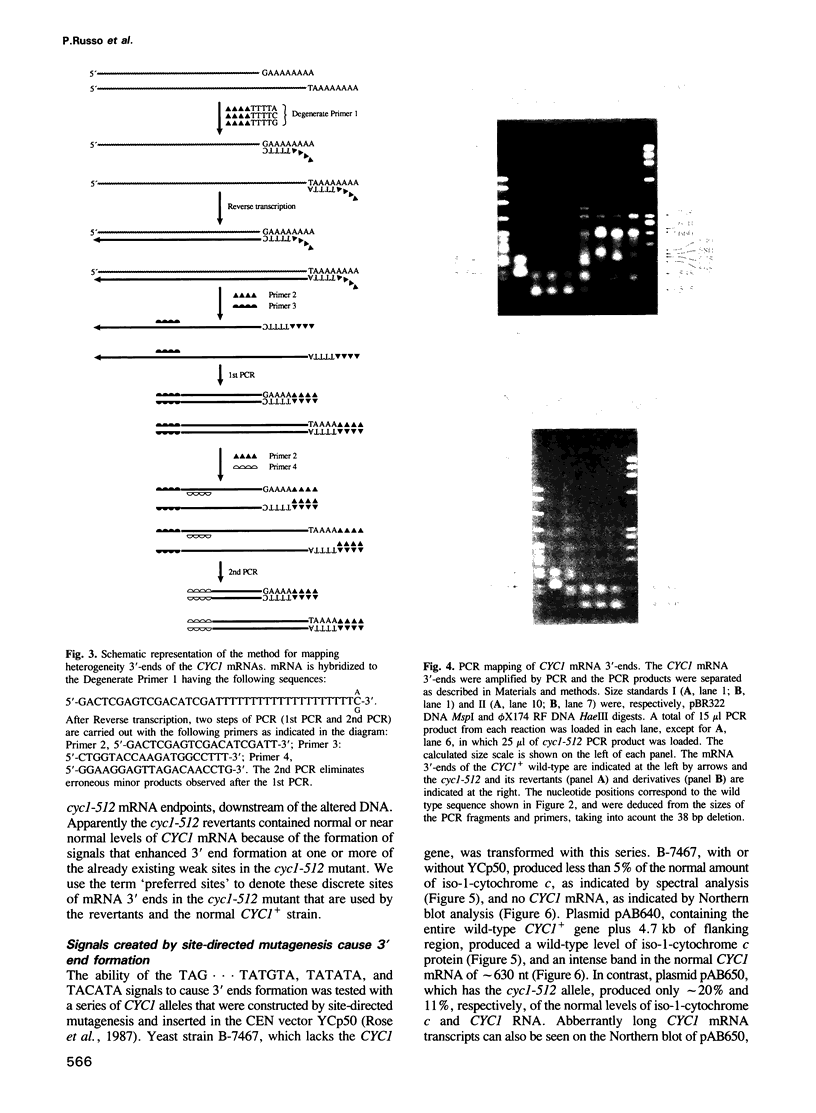
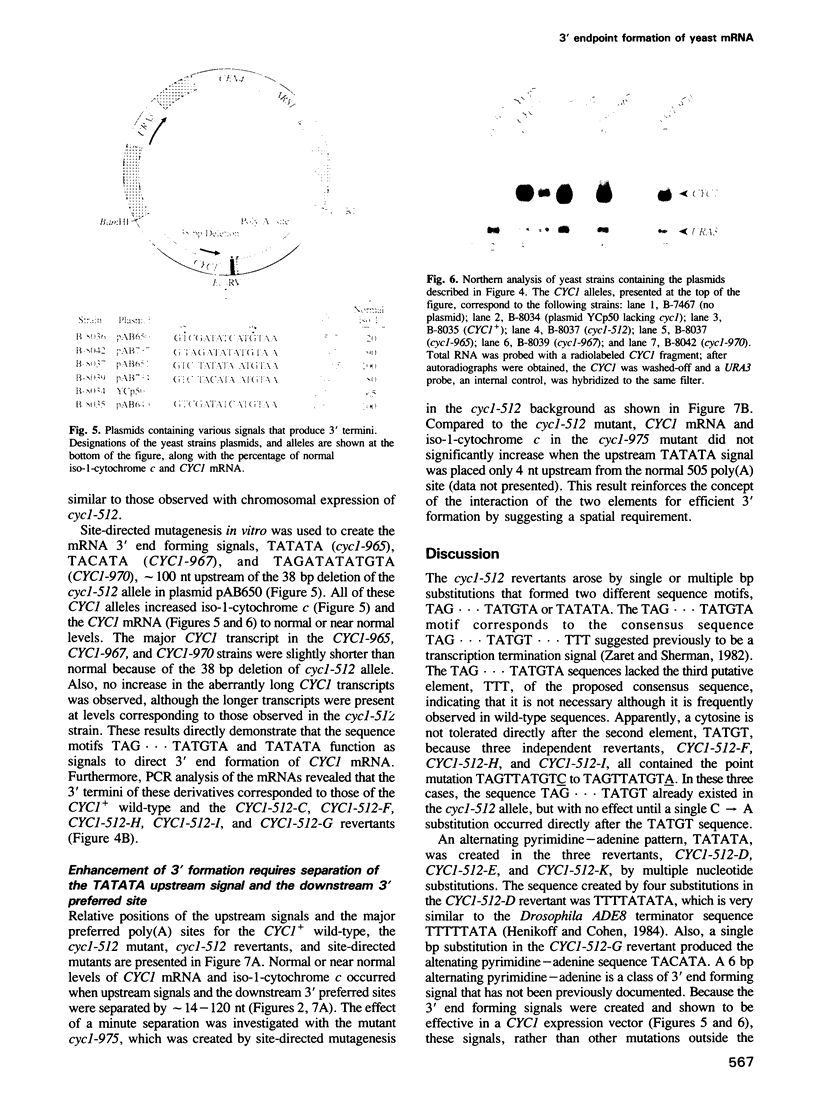
The cyc1-512 mutant of the yeast Saccharomyces cerevisiae contains a 38 bp deletion in the 3' untranslated region of the CYC1 gene, resulting in CYC1 mRNAs that are elongated, presumably labile, and reduced to 10% of the normal level. Analysis with S1 nuclease and a novel PCR procedure revealed that the low amount of cyc1-512 mRNA contained many discrete 3' termini at certain sites, ranging from the wild-type position to over 2000 nucleotides (nt) downstream. The cyc1-512 mRNA deficiency was completely or almost completely restored in eight intragenic revertants that contained six different single and multiple base-pair changes within a 300 bp region downstream from the translation terminator codon. Two of the six different reversions formed the sequence TAG...TATGTA, whereas the other four reversions created the sequences TATATA or TACATA. The positions of these revertant sequences varied, even though they caused an increased use of specific major downstream mRNA 3' endpoints, apparently identical to those seen in the cyc1-512 mRNA. However, several revertants contained minor end points not corresponding to any of the cyc1-512 mRNAs. The capacity of these three signals to form 3' ends was confirmed with sequences constructed by site-directed mutagenesis. We therefore suggest that the production of 3' termini of yeast mRNA may involve at least two functionally distinct elements working in concert. One type of element determines the sites of preferred 3' mRNA termini, as represented by the cyc1-512 termini. The second type of element, which includes TAG...TATGTA and TATATA motifs, operates at a distance to enhance the use of the downstream 3' preferred sites.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abe A., Hiraoka Y., Fukasawa T. Signal sequence for generation of mRNA 3' end in the Saccharomyces cerevisiae GAL7 gene. EMBO J. 1990 Nov;9(11):3691–3697. doi: 10.1002/j.1460-2075.1990.tb07581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Boss J. M., Gillam S., Zitomer R. S., Smith M. Sequence of the yeast iso-1-cytochrome c mRNA. J Biol Chem. 1981 Dec 25;256(24):12958–12961. [PubMed] [Google Scholar]
- Butler J. S., Platt T. RNA processing generates the mature 3' end of yeast CYC1 messenger RNA in vitro. Science. 1988 Dec 2;242(4883):1270–1274. doi: 10.1126/science.2848317. [DOI] [PubMed] [Google Scholar]
- Butler J. S., Sadhale P. P., Platt T. RNA processing in vitro produces mature 3' ends of a variety of Saccharomyces cerevisiae mRNAs. Mol Cell Biol. 1990 Jun;10(6):2599–2605. doi: 10.1128/mcb.10.6.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelly J., Montarras D., Pinset C., Berwald-Netter Y., Kaplan J. C., Kahn A. Quantitative estimation of minor mRNAs by cDNA-polymerase chain reaction. Application to dystrophin mRNA in cultured myogenic and brain cells. Eur J Biochem. 1990 Feb 14;187(3):691–698. doi: 10.1111/j.1432-1033.1990.tb15355.x. [DOI] [PubMed] [Google Scholar]
- Connelly S., Manley J. L. A CCAAT box sequence in the adenovirus major late promoter functions as part of an RNA polymerase II termination signal. Cell. 1989 May 19;57(4):561–571. doi: 10.1016/0092-8674(89)90126-8. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Frohman M. A., Dush M. K., Martin G. R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey D. M., Das G., Sherman F. Amino acid replacements in yeast iso-1-cytochrome c. Comparison with the phylogenetic series and the tertiary structure of related cytochromes c. J Biol Chem. 1986 Mar 5;261(7):3259–3271. [PubMed] [Google Scholar]
- Hampsey D. M., Ernst J. F., Stewart J. W., Sherman F. Multiple base-pair mutations in yeast. J Mol Biol. 1988 Jun 5;201(3):471–486. doi: 10.1016/0022-2836(88)90629-8. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Cohen E. H. Sequences responsible for transcription termination on a gene segment in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1515–1520. doi: 10.1128/mcb.4.8.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotval J., Zaret K. S., Consaul S., Sherman F. Revertants of a transcription termination mutant of yeast contain diverse genetic alterations. Genetics. 1983 Mar;103(3):367–388. doi: 10.1093/genetics/103.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Mann C., Davis R. W. Structure and sequence of the centromeric DNA of chromosome 4 in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jan;6(1):241–245. doi: 10.1128/mcb.6.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczynski G. T., Jaehning J. A. A transcription map of a yeast centromere plasmid: unexpected transcripts and altered gene expression. Nucleic Acids Res. 1985 Dec 9;13(23):8487–8506. doi: 10.1093/nar/13.23.8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Osborne B. I., Guarente L. Mutational analysis of a yeast transcriptional terminator. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4097–4101. doi: 10.1073/pnas.86.11.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne B. I., Guarente L. Transcription by RNA polymerase II induces changes of DNA topology in yeast. Genes Dev. 1988 Jun;2(6):766–772. doi: 10.1101/gad.2.6.766. [DOI] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Russo P., Sherman F. Transcription terminates near the poly(A) site in the CYC1 gene of the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8348–8352. doi: 10.1073/pnas.86.21.8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHERMAN F., SLONIMSKI P. P. RESPIRATION-DEFICIENT MUTANTS OF YEAST. II. BIOCHEMISTRY. Biochim Biophys Acta. 1964 Jul 15;90:1–15. doi: 10.1016/0304-4165(64)90113-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Berk A. J., Berget S. M. Transcription maps of adenovirus. Methods Enzymol. 1980;65(1):750–768. doi: 10.1016/s0076-6879(80)65071-x. [DOI] [PubMed] [Google Scholar]
- Smith A. J. DNA sequence analysis by primed synthesis. Methods Enzymol. 1980;65(1):560–580. doi: 10.1016/s0076-6879(80)65060-5. [DOI] [PubMed] [Google Scholar]
- Snyder M., Sapolsky R. J., Davis R. W. Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1988 May;8(5):2184–2194. doi: 10.1128/mcb.8.5.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M., Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3' end formation. Science. 1984 Nov 30;226(4678):1045–1051. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]
- Yarger J. G., Armilei G., Gorman M. C. Transcription terminator-like element within a Saccharomyces cerevisiae promoter region. Mol Cell Biol. 1986 Apr;6(4):1095–1101. doi: 10.1128/mcb.6.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Elder R. T. Some of the signals for 3'-end formation in transcription of the Saccharomyces cerevisiae Ty-D15 element are immediately downstream of the initiation site. Mol Cell Biol. 1989 Jun;9(6):2431–2444. doi: 10.1128/mcb.9.6.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. Mutationally altered 3' ends of yeast CYC1 mRNA affect transcript stability and translational efficiency. J Mol Biol. 1984 Jul 25;177(1):107–135. doi: 10.1016/0022-2836(84)90060-3. [DOI] [PubMed] [Google Scholar]