Abstract
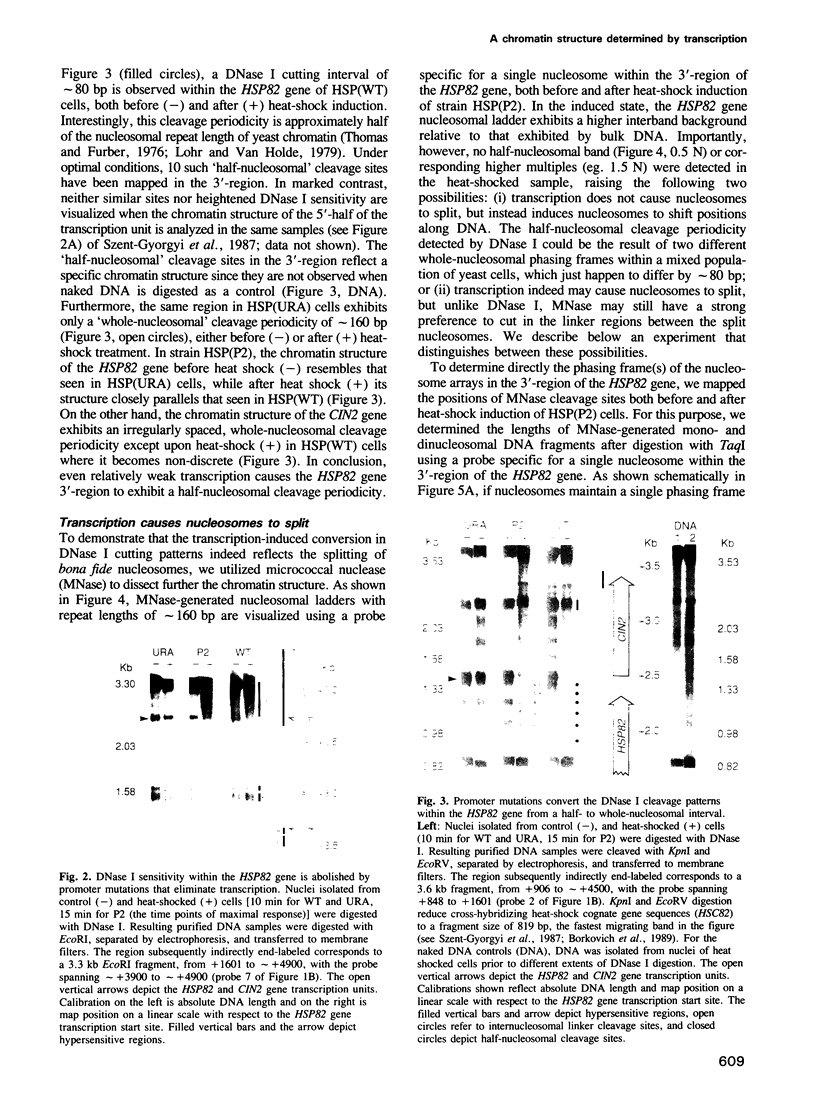
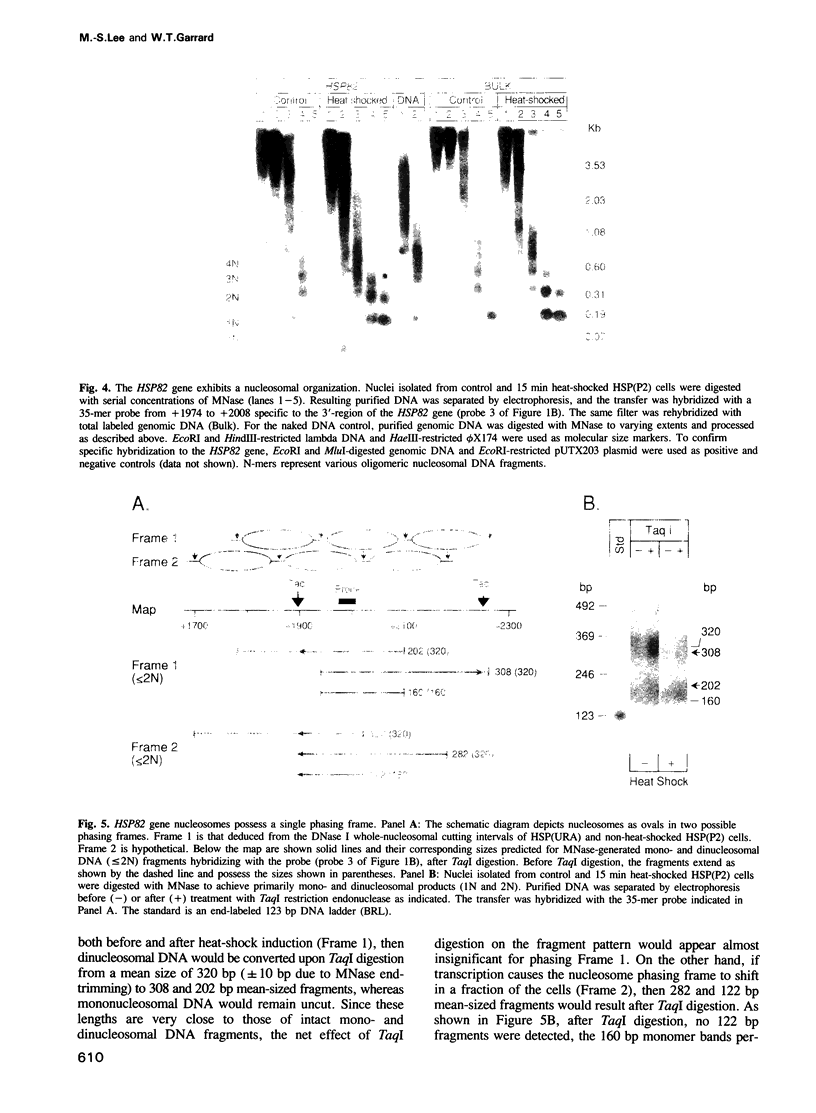
Utilizing yeast strains containing promoter mutations, we demonstrate that transcription of the HSP82 gene causes nucleosomes toward the 3'-end to become DNase I sensitive and 'split' into structures that exhibit a 'half-nucleosomal' cleavage periodicity. Splitting occurs even when only a few RNA polymerase II molecules are engaged in basal level transcription or during the first round of induced transcription. The split nucleosomal structure survives nuclear isolation suggesting that it may be stabilized by post-translational modifications or non-histone proteins, and may require DNA replication for reversal to a whole nucleosomal structure. Split nucleosomes represent a structure for DNase I sensitive chromatin and are probably of common occurrence but difficult to detect experimentally. We suggest that transient positive supercoils downstream of traversing RNA polymerase lead to nucleosome splitting.
Full text
PDF

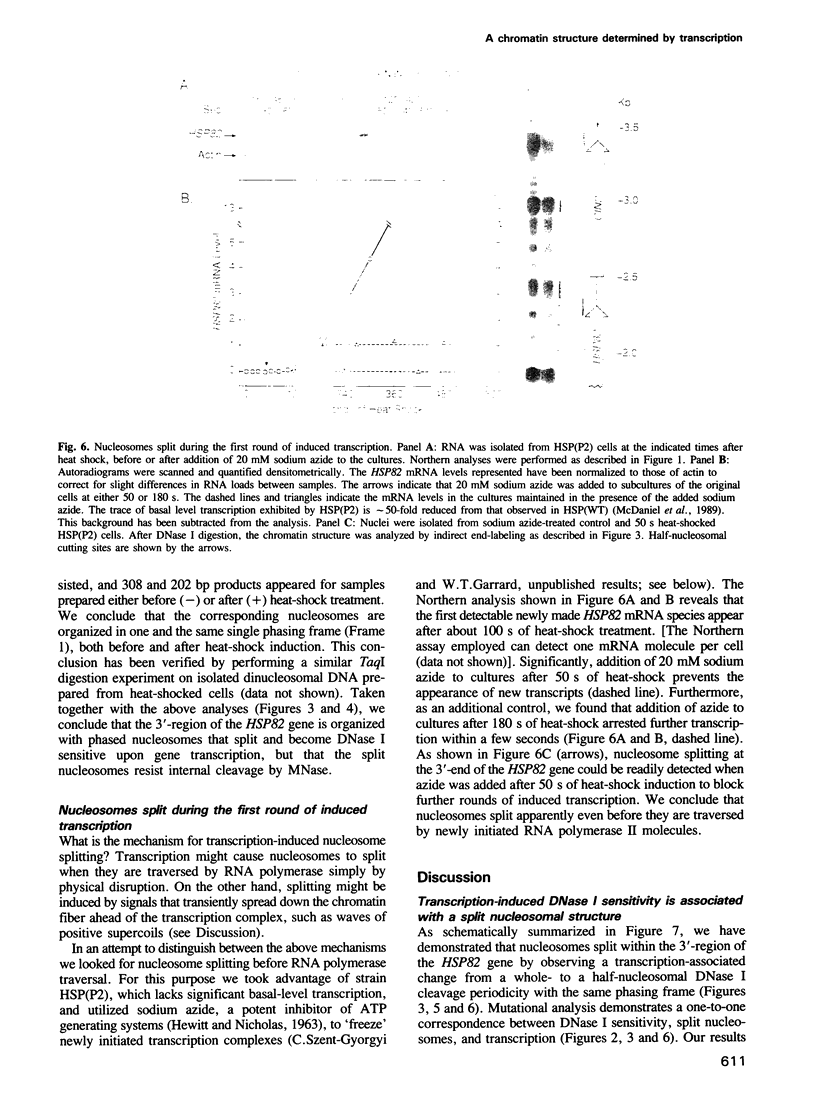
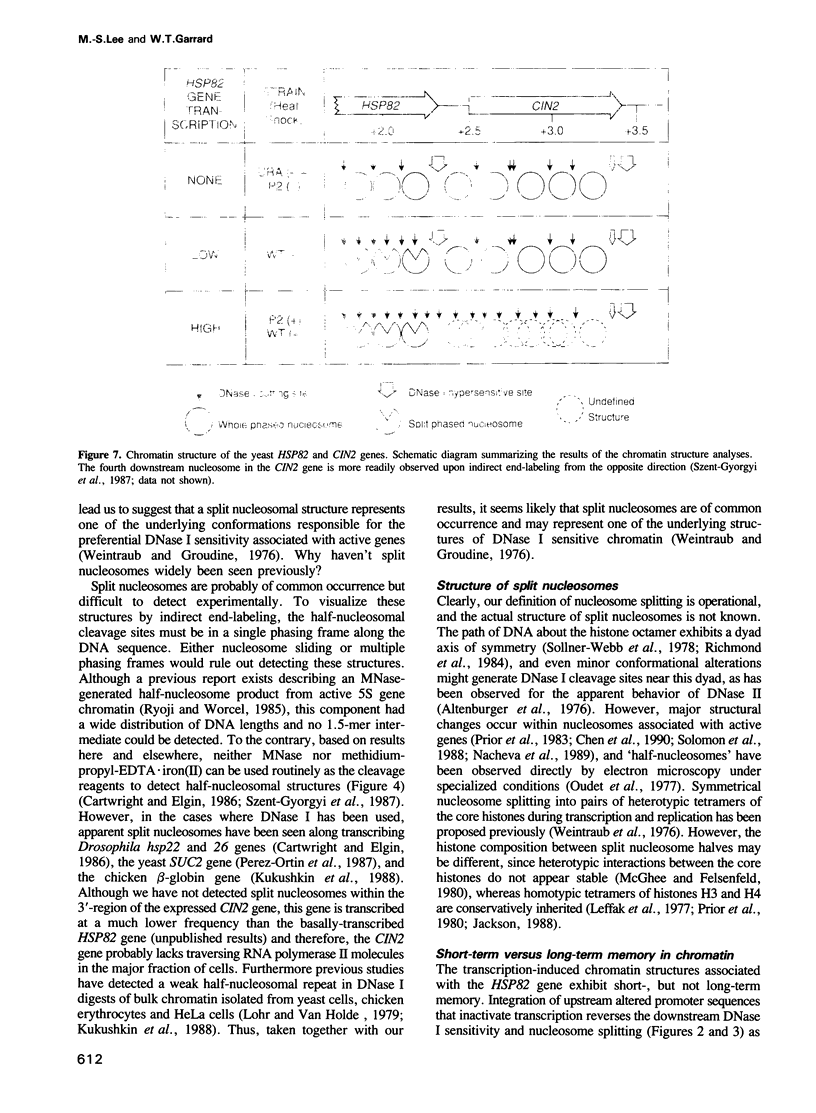



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altenburger W., Hörz W., Zachau H. G. Nuclease cleavage of chromatin at 100-nucleotide pair intervals. Nature. 1976 Dec 9;264(5586):517–522. doi: 10.1038/264517a0. [DOI] [PubMed] [Google Scholar]
- Bellard M., Dretzen G., Bellard F., Kaye J. S., Pratt-Kaye S., Chambon P. Hormonally induced alterations of chromatin structure in the polyadenylation and transcription termination regions of the chicken ovalbumin gene. EMBO J. 1986 Mar;5(3):567–574. doi: 10.1002/j.1460-2075.1986.tb04248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Moore M. W., Yancopoulos G. D., Suh H., Lutzker S., Selsing E., Alt F. W. Recombination between immunoglobulin variable region gene segments is enhanced by transcription. Nature. 1986 Dec 11;324(6097):585–589. doi: 10.1038/324585a0. [DOI] [PubMed] [Google Scholar]
- Borkovich K. A., Farrelly F. W., Finkelstein D. B., Taulien J., Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989 Sep;9(9):3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotherton T. W., Reneker J., Ginder G. D. Binding of HMG 17 to mononucleosomes of the avian beta-globin gene cluster in erythroid and non-erythroid cells. Nucleic Acids Res. 1990 Apr 25;18(8):2011–2016. doi: 10.1093/nar/18.8.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright I. L., Elgin S. C. Nucleosomal instability and induction of new upstream protein-DNA associations accompany activation of four small heat shock protein genes in Drosophila melanogaster. Mol Cell Biol. 1986 Mar;6(3):779–791. doi: 10.1128/mcb.6.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T. A., Sterner R., Cozzolino A., Allfrey V. G. Reversible and irreversible changes in nucleosome structure along the c-fos and c-myc oncogenes following inhibition of transcription. J Mol Biol. 1990 Apr 5;212(3):481–493. doi: 10.1016/0022-2836(90)90327-I. [DOI] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- De Bernardin W., Koller T., Sogo J. M. Structure of in-vivo transcribing chromatin as studied in simian virus 40 minichromosomes. J Mol Biol. 1986 Oct 5;191(3):469–482. doi: 10.1016/0022-2836(86)90142-7. [DOI] [PubMed] [Google Scholar]
- Einck L., Bustin M. Inhibition of transcription in somatic cells by microinjection of antibodies to chromosomal proteins. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6735–6739. doi: 10.1073/pnas.80.22.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrelly F. W., Finkelstein D. B. Complete sequence of the heat shock-inducible HSP90 gene of Saccharomyces cerevisiae. J Biol Chem. 1984 May 10;259(9):5745–5751. [PubMed] [Google Scholar]
- Flint S. J., Weintraub H. M. An altered subunit configuration associated with the actively transcribed DNA of integrated adenovirus genes. Cell. 1977 Nov;12(3):783–794. doi: 10.1016/0092-8674(77)90277-x. [DOI] [PubMed] [Google Scholar]
- Giaever G. N., Wang J. C. Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell. 1988 Dec 2;55(5):849–856. doi: 10.1016/0092-8674(88)90140-7. [DOI] [PubMed] [Google Scholar]
- Goodwin G. H., Nicolas R. H., Cockerill P. N., Zavou S., Wright C. A. The effect of salt extraction on the structure of transcriptionally active genes; evidence for a DNAseI-sensitive structure which could be dependent on chromatin structure at levels higher than the 30 nm fibre. Nucleic Acids Res. 1985 May 24;13(10):3561–3579. doi: 10.1093/nar/13.10.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D. S., Garrard W. T. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Huang S. Y., Barnard M. B., Xu M., Matsui S., Rose S. M., Garrard W. T. The active immunoglobulin kappa chain gene is packaged by non-ubiquitin-conjugated nucleosomes. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3738–3742. doi: 10.1073/pnas.83.11.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson V. Deposition of newly synthesized histones: hybrid nucleosomes are not tandemly arranged on daughter DNA strands. Biochemistry. 1988 Mar 22;27(6):2109–2120. doi: 10.1021/bi00406a044. [DOI] [PubMed] [Google Scholar]
- Jackson V. In vivo studies on the dynamics of histone-DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry. 1990 Jan 23;29(3):719–731. doi: 10.1021/bi00455a019. [DOI] [PubMed] [Google Scholar]
- Jantzen K., Fritton H. P., Igo-Kemenes T. The DNase I sensitive domain of the chicken lysozyme gene spans 24 kb. Nucleic Acids Res. 1986 Aug 11;14(15):6085–6099. doi: 10.1093/nar/14.15.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. E., Herskowitz I. Directionality and regulation of cassette substitution in yeast. Cold Spring Harb Symp Quant Biol. 1984;49:97–104. doi: 10.1101/sqb.1984.049.01.013. [DOI] [PubMed] [Google Scholar]
- Kukushkin A. N., Svetlikova S. B., Pospelov V. A. A structure of potentially active and inactive genes of chicken erythrocyte chromatin upon decondensation. Nucleic Acids Res. 1988 Sep 12;16(17):8555–8569. doi: 10.1093/nar/16.17.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Lindquist S. Changing patterns of gene expression during sporulation in yeast. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7323–7327. doi: 10.1073/pnas.81.23.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey R. A. The use of intensifying screens or organic scintillators for visualizing radioactive molecules resolved by gel electrophoresis. Methods Enzymol. 1980;65(1):363–371. doi: 10.1016/s0076-6879(80)65047-2. [DOI] [PubMed] [Google Scholar]
- Leffak I. M., Grainger R., Weintraub H. Conservative assembly and segregation of nucleosomal histones. Cell. 1977 Nov;12(3):837–845. doi: 10.1016/0092-8674(77)90282-3. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loc P. V., Strätling W. H. The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J. 1988 Mar;7(3):655–664. doi: 10.1002/j.1460-2075.1988.tb02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Van Holde K. E. Organization of spacer DNA in chromatin. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6326–6330. doi: 10.1073/pnas.76.12.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. On the displacement of histones from DNA by transcription. Cell. 1988 Dec 2;55(5):743–744. doi: 10.1016/0092-8674(88)90128-6. [DOI] [PubMed] [Google Scholar]
- Lutzker S., Rothman P., Pollock R., Coffman R., Alt F. W. Mitogen- and IL-4-regulated expression of germ-line Ig gamma 2b transcripts: evidence for directed heavy chain class switching. Cell. 1988 Apr 22;53(2):177–184. doi: 10.1016/0092-8674(88)90379-0. [DOI] [PubMed] [Google Scholar]
- McDaniel D., Caplan A. J., Lee M. S., Adams C. C., Fishel B. R., Gross D. S., Garrard W. T. Basal-level expression of the yeast HSP82 gene requires a heat shock regulatory element. Mol Cell Biol. 1989 Nov;9(11):4789–4798. doi: 10.1128/mcb.9.11.4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Bustin M., Miller O. L., Jr Electron microscopic analysis of chromosome metabolism in the Drosophila melanogaster embryo. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):741–754. doi: 10.1101/sqb.1978.042.01.075. [DOI] [PubMed] [Google Scholar]
- Nacheva G. A., Guschin D. Y., Preobrazhenskaya O. V., Karpov V. L., Ebralidse K. K., Mirzabekov A. D. Change in the pattern of histone binding to DNA upon transcriptional activation. Cell. 1989 Jul 14;58(1):27–36. doi: 10.1016/0092-8674(89)90399-1. [DOI] [PubMed] [Google Scholar]
- Osheim Y. N., Miller O. L., Jr, Beyer A. L. RNP particles at splice junction sequences on Drosophila chorion transcripts. Cell. 1985 Nov;43(1):143–151. doi: 10.1016/0092-8674(85)90019-4. [DOI] [PubMed] [Google Scholar]
- Oudet P., Spadafora C., Chambon P. Nucleosome structure II: structure of the SV40 minichromosome and electron microscopic evidence for reversible transitions of the nucleosome structure. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):301–312. doi: 10.1101/sqb.1978.042.01.032. [DOI] [PubMed] [Google Scholar]
- Parikh V. S., Conrad-Webb H., Docherty R., Butow R. A. Interaction between the yeast mitochondrial and nuclear genomes influences the abundance of novel transcripts derived from the spacer region of the nuclear ribosomal DNA repeat. Mol Cell Biol. 1989 May;9(5):1897–1907. doi: 10.1128/mcb.9.5.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior C. P., Cantor C. R., Johnson E. M., Allfrey V. G. Incorporation of exogenous pyrene-labeled histone into Physarum chromatin: a system for studying changes in nucleosomes assembled in vivo. Cell. 1980 Jul;20(3):597–608. doi: 10.1016/0092-8674(80)90306-2. [DOI] [PubMed] [Google Scholar]
- Prior C. P., Cantor C. R., Johnson E. M., Littau V. C., Allfrey V. G. Reversible changes in nucleosome structure and histone H3 accessibility in transcriptionally active and inactive states of rDNA chromatin. Cell. 1983 Oct;34(3):1033–1042. doi: 10.1016/0092-8674(83)90561-5. [DOI] [PubMed] [Google Scholar]
- Pérez-Ortín J. E., Estruch F., Matallana E., Franco L. Fine analysis of the chromatin structure of the yeast SUC2 gene and of its changes upon derepression. Comparison between the chromosomal and plasmid-inserted genes. Nucleic Acids Res. 1987 Sep 11;15(17):6937–6956. doi: 10.1093/nar/15.17.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984 Oct 11;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Rose M., Grisafi P., Botstein D. Structure and function of the yeast URA3 gene: expression in Escherichia coli. Gene. 1984 Jul-Aug;29(1-2):113–124. doi: 10.1016/0378-1119(84)90172-0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Ryoji M., Worcel A. Structure of the two distinct types of minichromosomes that are assembled on DNA injected in Xenopus oocytes. Cell. 1985 Apr;40(4):923–932. doi: 10.1016/0092-8674(85)90352-6. [DOI] [PubMed] [Google Scholar]
- Schlissel M. S., Baltimore D. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell. 1989 Sep 8;58(5):1001–1007. doi: 10.1016/0092-8674(89)90951-3. [DOI] [PubMed] [Google Scholar]
- Smerdon M. J., Thoma F. Site-specific DNA repair at the nucleosome level in a yeast minichromosome. Cell. 1990 May 18;61(4):675–684. doi: 10.1016/0092-8674(90)90479-x. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Melchior W., Jr, Felsenfeld G. DNAase I, DNAase II and staphylococcal nuclease cut at different, yet symmetrically located, sites in the nucleosome core. Cell. 1978 Jul;14(3):611–627. doi: 10.1016/0092-8674(78)90246-5. [DOI] [PubMed] [Google Scholar]
- Solomon M. J., Larsen P. L., Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988 Jun 17;53(6):937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Stearns T., Hoyt M. A., Botstein D. Yeast mutants sensitive to antimicrotubule drugs define three genes that affect microtubule function. Genetics. 1990 Feb;124(2):251–262. doi: 10.1093/genetics/124.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szent-Györgyi C., Finkelstein D. B., Garrard W. T. Sharp boundaries demarcate the chromatin structure of a yeast heat-shock gene. J Mol Biol. 1987 Jan 5;193(1):71–80. doi: 10.1016/0022-2836(87)90628-0. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Furber V. Yeast chromatin structure. FEBS Lett. 1976 Jul 15;66(2):274–280. doi: 10.1016/0014-5793(76)80521-2. [DOI] [PubMed] [Google Scholar]
- Villeponteau B., Martinson H. G. Gamma rays and bleomycin nick DNA and reverse the DNase I sensitivity of beta-globin gene chromatin in vivo. Mol Cell Biol. 1987 May;7(5):1917–1924. doi: 10.1128/mcb.7.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K., Keil R. L., Roeder G. S. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987 Mar 27;48(6):1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- Weintraub H. Assembly and propagation of repressed and depressed chromosomal states. Cell. 1985 Oct;42(3):705–711. doi: 10.1016/0092-8674(85)90267-3. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Worcel A., Alberts B. A model for chromatin based upon two symmetrically paired half-nucleosomes. Cell. 1976 Nov;9(3):409–417. doi: 10.1016/0092-8674(76)90085-4. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Xu M., Barnard M. B., Rose S. M., Cockerill P. N., Huang S. Y., Garrard W. T. Transcription termination and chromatin structure of the active immunoglobulin kappa gene locus. J Biol Chem. 1986 Mar 15;261(8):3838–3845. [PubMed] [Google Scholar]
- Yancopoulos G. D., Alt F. W. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985 Feb;40(2):271–281. doi: 10.1016/0092-8674(85)90141-2. [DOI] [PubMed] [Google Scholar]