Highlight
An 11 aa peptide derived from one of the cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 (CAP) superfamily is salt regulated, conferring salt susceptibility through suppression of salt-tolerance genes.
Key words: Environmental regulation, CAP, negative regulator of salt resistance, plant peptide, proteolytic process, salinity.
Abstract
High salinity has negative impacts on plant growth through altered water uptake and ion-specific toxicities. Plants have therefore evolved an intricate regulatory network in which plant hormones play significant roles in modulating physiological responses to salinity. However, current understanding of the plant peptides involved in this regulatory network remains limited. Here, we identified a salt-regulated peptide in Arabidopsis. The peptide was 11 aa and was derived from the C terminus of a cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins (CAP) superfamily. This peptide was found by searching homologues in Arabidopsis using the precursor of a tomato CAP-derived peptide (CAPE) that was initially identified as an immune signal. In searching for a CAPE involved in salt responses, we screened CAPE precursor genes that showed salt-responsive expression and found that the PROAtCAPE1 (AT4G33730) gene was regulated by salinity. We confirmed the endogenous Arabidopsis CAP-derived peptide 1 (AtCAPE1) by mass spectrometry and found that a key amino acid residue in PROAtCAPE1 is critical for AtCAPE1 production. Moreover, although PROAtCAPE1 was expressed mainly in the roots, AtCAPE1 was discovered to be upregulated systemically upon salt treatment. The salt-induced AtCAPE1 negatively regulated salt tolerance by suppressing several salt-tolerance genes functioning in the production of osmolytes, detoxification, stomatal closure control, and cell membrane protection. This discovery demonstrates that AtCAPE1, a homologue of tomato immune regulator CAPE1, plays an important role in the regulation of salt stress responses. Our discovery thus suggests that the peptide may function in a trade-off between pathogen defence and salt tolerance.
Introduction
Excessive salinity has a highly negative impact on plant growth due to the increased osmotic potential of soil and the toxicity of salt ions to cells (Bernstein, 1975; Munns and Tester, 2008). Accordingly, plants have developed various mechanisms for growth and survival in harsh saline environments. Extensive research has revealed an array of salinity regulatory mechanisms at the molecular level (Kreps et al., 2002; Taji et al., 2004), including changes in a vast number of genes encoding ion channels, transporters, transcription factors, signalling molecules, detoxification enzymes, chaperones, and osmolytes (Wang et al., 2003; Munns and Tester, 2008; Turan et al., 2012), that can be utilized to induce salt tolerance in crop plants (Turan et al., 2012; Roy et al., 2014). A systems-level study also revealed that the salt response involves an intricate web of regulatory networks that crosstalk with other stress (Wang et al., 2003; Mahajan and Tuteja, 2005) and developmental (Bernstein, 1975) responses. Among the regulation signals for high salt, phytohormones are thought to be the most important endogenous substances for modulating physiological responses that eventually lead to adaptation to salinity (Pearce et al., 2001b). Recently, plant peptides have been found to function as hormones in a diverse array of roles in plant growth, development, and signalling to environmental stresses (Matsubayashi, 2014). However, to date, little has been reported on plant peptides that regulate the salt response (Matsubayashi, 2014).
Recent research based on studies of precursor genes has suggested a functional link between plant peptides and modulation of the salt-stress response. A knockout mutant of one of the C-TERMINALLY ENCODED PEPTIDE (CEP) family proteins exhibited more tolerance to salinity than wild-type Arabidopsis, and external application of the peptide to seedlings suppressed root growth (Ohyama et al., 2008; Delay et al., 2013). This suggests the relevance of plant peptides functioning in root development in the regulation of underground stresses such as salinity. Additional evidence showed that tomato with overexpression of PROSYSTEMIN, the precursor of the anti-herbivore systemin peptide signal (Pearce et al., 1991), displayed more tolerance to salt stress than wild-type tomato (Pearce et al., 1991; Orsini et al., 2010). This finding suggests that defence-related peptides may also participate in the modulation of salt-stress tolerance.
We recently identified a peptide derived from tomato pathogenesis-related protein 1 (PR1) that is wound induced in the same way as systemin and regulates immune responses (Chen et al., 2014). Since PR1 protein was classified as one of the cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins (CAP) superfamily (Gibbs et al., 2008), the peptide derived from the protein in this superfamily was designated CAP-derived peptide (CAPE) (Chen et al., 2014). Notably, CAPE was found to be wide spread throughout the plant kingdom, being found in dicots (e.g. Solanoideae, Nicotianoideae, Vitaceae, Brassicaceae, and Fabaceae) and monocots (e.g. Poaceae), with a conserved ·PxGNxxxxxPY- motif (Chen et al., 2014). It has been demonstrated that application of a synthetic peptide predicted to be a CAPE derived from Arabidopsis PR1 can lead to resistance against Pseudomonas syringae pv. tomato strain DC3000 (Pst DC3000) infection (Chen et al., 2014). This finding indicates that Arabidopsis CAP proteins may produce functional CAPEs. Moreover, overexpression of Arabidopsis PR1 and tomato PROSYSTEMIN showed drought and salt-stress tolerance, respectively (Seo et al., 2008; Wang et al., 2011; Liu et al., 2013), which led us to test the hypothesis that Arabidopsis CAPEs may function not only in regulation of innate immunity but also in the response to salt-stress tolerance. In this report, we aimed to identify the endogenous Arabidopsis CAPEs that are involved in salt responses and we showed that a CAPE negatively regulates the salt-tolerance response by downregulating salt-tolerance genes upon high-saline challenge. The phenotypic evidence and the CAPE-mediated salt-responsive mechanism were studied and are discussed.
Materials and methods
Details of the seedling germination assay, RNA extraction and quantitative reverse transcription PCR (qRT-PCR) analysis, and western blotting are provided in the Supplementary materials and methods, available at JXB online.
Plant materials and growth conditions
A T-DNA mutant of AT4G33730 (GT_5_110620; named proatcape1 here) was obtained from the Nottingham Arabidopsis Stock Centre (NASC) for phenotypic investigation. Arabidopsis ecotype Landsberg erecta (Ler) was used in all experiments as the background control for proatcape1 and PROATCAPE1ox in proatcape1 transgenic lines generated in a proatcape1 background. In addition, Arabidopsis ecotype Columbia-0 (Col-0) was used as the background control for CAPE1oxCNYD and CAPE1oxCNAD transgenic plants. A schematic representation of the target site of PROAtCAPE1 by T-DNA is shown in Supplementary Fig. S2A (available at JXB online). Homozygous proatcape1 was determined by genotyping (Supplementary Fig. S2B). qRT-PCR results indicated that proatcape1 was a knockout mutant (Supplementary Fig. S2C). The primers used for PCR-based genotyping are listed in Supplementary Table S2, available at JXB online.
Seeds were surface sterilized with 30% bleach (CLOROX) for 8min and then washed with sterilized ddH2O five times. Seeds were germinated on half-strength Murashige and Skoog (1/2 MS) medium under a 16h photoperiod (80–100 μmol m−2 s−1 illumination) at 22 °C.
Plasmid constructions and generation of transgenic lines
Total RNA extracted from Ler was utilized as the template for reverse transcription to generate cDNA. The cDNA was used as template for PCR using primers (Supplementary Table S2) corresponding to the 5′ and 3′ ends of the PROAtCAPE1 coding sequence. The amplified DNA fragment was cloned into pCR8/GW/TOPO (Invitrogen) according to the manufacturer’s protocol. The validated PROAtCAPE1 sequence was then cloned into pMDC32 (Curtis and Grossniklaus, 2003), a constitutive expression vector harbouring a dual 35S promoter, by site-specific recombination (LR clonase; Invitrogen). Similarly, for the generation of CAPE1oxCNYD transgenic plants, the PROAtCAPE1 sequence was recombined into pK7YWG2 (Karimi et al., 2002). For the generation of pPROAtCAPE1:GUS lines, the sequence upstream of the transcription start site of PROAtCAPE1 (2kb) was cloned into pCR8/GW/TOPO and then recombined into pMDC164 (Curtis and Grossniklaus, 2003). Transgenic Arabidopsis was generated via an Agrobacterium tumefaciens-mediated transformation system (Zhang et al., 2006).
Microarray analysis
Quality control of total RNA was determined using an Agilent Bioanalyzer 2100 (Agilent Technologies, St Clara, CA, USA). Total RNA (0.2 μg) was amplified by a Low Input Quick-Amp Labeling kit (Agilent Technologies). Preparation of fluorescence-labelled cDNA and microarray experiments were performed at the DNA Microarray Core Facility, Institute of Plant and Microbial Biology, Academia Sinica, Taiwan. Agilent Arabidopsis (V4) Gene Expression Microarray 4×44k chips were used in this study. Labelling of cDNA probes and hybridization experiments were performed according to the single-colour microarray protocols provided by the manufacturer. The fluorescence signals were detected by an Agilent DNA Microarray Scanner G2565CA and Agilent Feature Extraction 10.7.1.1 software. Three biological repeats were conducted using cDNAs obtained from Ler and proatcape1 in 1/2 MS medium and with 12h of 125mM NaCl treatment. Raw data from hybridization were imported into microarray analysis software GeneSpring 11.5 (Agilent Technologies), and the data of Ler subjected to medium alone was used as a control for normalization. As a quality control, we kept the genes whose raw signal intensity was >100 and coefficient of variation (CV) was <50% (default value) at any time point for further analyses. The coefficient of variation is a statistical measurement to compare the degree of variation from one data set to another, even if the means differ dramatically from each other.
The microarray data discussed in this publication have been deposited in the Gene Expression Omnibus of the National Center for Biotechnology Information (NCBI) (Edgar et al., 2002) and are accessible through GEO Series accession number GSE66946.
Endogenous peptide isolation
Ten-day-old Ler seedlings grown vertically on 1/2 MS medium were transferred to a hydroponic system culture of 1/2 MS liquid medium without sucrose. The medium was changed every 4 d. Three weeks after planting, the plant tissue was harvested. For salt treatments, fresh 1/2 MS medium in the presence or absence of 125mM NaCl was exchanged and the treatments were prolonged to 24h. Samples were homogenized with 200ml of 1% (v/v) chilled trifluoroacetic acid (TFA; Sigma-Aldrich) in a blender for 2min. To normalize the variations in sample preparation and for better quantification accuracy by liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis, 20 μl of 1 pmole μl–1 of internal standard (synthetic peptide: PAAAYIGARAY) with two amino acid substitutions of G3A and N4A of AtCAPE1 was added to the extraction buffer while crude peptide was isolated. The extracts were then filtered through four layers of Miracloth (Calbiochem, San Diego, CA, USA) to remove plant debris. The procedure followed a previously described protocol (Pearce et al., 2001a, b ). After centrifuging at 8500rpm for 20min at 4 °C (Beckman Coulter, Avanti J-26 XP), the pH value of the supernatant was adjusted to 4.5. The extracts were then centrifuged again at 8500rpm for 20min at 4 °C and the pH value of supernatant was adjusted to 2.5. Subsequently, the supernatant was bound with a customized Sep-Pak C18 solid phase extraction cartridge (Waters, Milford, MA, USA) according to the following steps: the C18 cartridge was conditioned by 0.1% (v/v) TFA first, bound with the supernatant, and then washed with 0.1% (v/v) TFA. Finally, the polypeptides were eluted with 60% (v/v) methanol/0.1% (v/v) TFA. The peptides in the final eluate were evaporated to dryness by rotary evaporation under high vacuum, and the pellets were resuspended in 0.1% (v/v) TFA. The polypeptides were further fractionated by fast protein liquid chromatography (ÄKTA purifier) to remove protein contaminates with size exclusion columns (Superdex peptide 10/300 GL; GE Healthcare, Little Chalfont, UK). Fractions containing polypeptides or small proteins with similar sizes to AtCAPE1 were collected and combined. The combined eluates were rotary evaporated under high vacuum and resuspended in 0.1% (v/v) TFA for ZipTip (Millipore, Billerica, MA, USA) to remove the contamination of salts. After final evaporation and resuspension in 0.1% (v/v) formic acid (Fluka), targeted LC-MS/MS analysis was performed.
Targeted LC-MS/MS analysis
An LTQ Velos PRO mass spectrometer (Thermo Scientific, Waltham, MA, USA) coupled with an online capillary nanoUHPLC system (Waters) was utilized for peptide identification and quantification. The capillary LC system was equipped with a homemade C18 trap cartridge (5 μm particles, Symmetry C18; Waters), and a homemade C18 reversed-phase analytical column (1.7 μm particles, BEH130 C18; Waters) (Chen et al., 2012) was used to deliver the solvent and target peptide with a linear gradient from 8 to 90% (v/v) acetonitrile in 0.1% (v/v) formic acid for 95min at a nanoflow rate (approx. 300 nl min–1). The analytical column was coupled to a nanoelectrospray ionization source, and acquisition of the data was performed with a full MS scan followed by MS/MS scans of the targeted precursor ions. Precursor ions of AtCAPE1 (PAGNYIGARPY; m/z 589.8) and the internal standard (PAAAYIGARAY; m/z 562.4) were selected for subsequent targeted MS/MS scans. The fragment ions m/z 563.2, 676.3, and 900.5 and m/z 537.2, 650.3, and 813.34 were used for further identification and quantification of AtCAPE1 and the internal standard, respectively.
Relative quantification of endogenous AtCAPE1
Quantitative analysis of AtCAPE1 was carried out using a label-free approach. Values of the peak area extracted from the selected fragment ions in the extracted ion current chromatogram were calculated. The sum of three peak values represented the expression of the target peptide. After normalizing AtCAPE1 to the internal standard in each sample, the relative area ratio for AtCAPE1 displayed a comparison of AtCAPE1 expression in different samples with that in shoots under normal conditions.
Results
Identification of PROAtCAPE as specifically responding to salt stress in Arabidopsis
Putative CAP proteins in Arabidopsis were searched using the precursor of tomato CAP-derived peptide (CAPE1), the first peptide derived from the CAP protein superfamily in plants (Chen et al., 2014), as a query protein for a homology search against the TAIR10 protein database. Twenty-two Arabidopsis CAPs were discovered that displayed sequence similarity (E-value <1) with the tomato CAPE1 precursor. To further refine the candidate proteins that may produce CAPEs, the 22 potential Arabidopsis CAP proteins were searched by the C-terminal conserved motif, CNYx.PxGNxxxxxPY- (Chen et al., 2014) (Table 1). Nine potential CAPs were identified as precursor candidates for CAPEs. These proteins were named PROAtCAPEs and their putative CAPEs, containing the ·PxGNxxxxxPY- motif, were named AtCAPEs (Table 1). The nomenclature followed the order of percentage identity between the putative AtCAPEs and tomato CAPE1 (Chen et al., 2014). Thus, the putative AtCAPE derived from AT4G33730 was designated AtCAPE1 and its precursor as PROAtCAPE1. The putative AtCAPEs derived from the known pathogenesis-related proteins PRB1 and PR1 were named AtCAPE7 and AtCAPE9, respectively (Table 1).
Table 1.
List of putative CAPEs in Arabidopsis
| Peptide namea | Putative CAPE sequenceb | Gene symbolc |
|---|---|---|
| AtCAPE1 | .PAGNYIGARPY- | AT4G33730 |
| AtCAPE2 | .PPGNWVGEWPY- | AT4G25780 |
| AtCAPE3 | .PPGNWVGEWPY- | AT4G33720 |
| AtCAPE4 | .PPGNYVGEKPY- | AT4G25790 |
| AtCAPE5 | .PPGNYVGEKPY- | AT5G57625 |
| AtCAPE6 | .PPGNFLGRKPY- | AT4G30320 |
| AtCAPE7 | .PPGNYANQKPY- | PRB1 |
| AtCAPE8 | .PPGNYRGRWPY- | AT5G26130 |
| AtCAPE9 | .PRGNYVNEKPY- | PR1 |
a Peptide name given in the present work.
b Putative CAPEs were defined by the conserved sequence. ·PxGNxxxxxPY- at the C terminus of the CAP proteins.
c The gene symbol refers to NCBI nomenclature.
To search potential salt-responsive CAPEs, the expression of the nine PROAtCAPE genes that responded to salt stress were examined by public microarray data analysis (Hruz et al., 2008). A probe for PROAtCAPE5 was not available in the Affymetrix GeneChip (Supplementary Table S1, available at JXB online). Excluding PROAtCAPE5, the eight PROAtCAPE transcripts were identified as responding to salt and salt-related stresses according to our criteria of expression fold change of ≥1.5 (up- or downregulated) (P≤0.05; Supplementary Table S1). In the roots, PROAtCAPE1 transcripts were decreased upon salinity, the PROAtCAPE3 gene was decreased upon salinity, osmotic, drought, and cold stresses, the PROAtCAPE4 gene was downregulated by salt and osmotic stresses, and the PROAtCAPE6 gene was suppressed by salt, osmotic, and drought stresses, while PROAtCAPE7 (also known as PRB1) was upregulated upon oxidative and cold stresses (Supplementary Table S1). In the shoots, PROAtCAPE1 transcripts were increased upon salinity, the PROAtCAPE2 gene was decreased upon saline and drought stresses but increased under cold stress, the PROAtCAPE3 gene was upregulated by salt; PROAtCAPE6 transcripts were suppressed by oxidative stress, and PROAtCAPE9 (also known as PR1) was downregulated upon salinity, oxidative, drought and cold stresses (Supplementary Table S1). Notably, PROAtCAPE1 was regulated mainly by salt, while the transcripts of the other seven PROAtCAPEs were regulated by more than two abiotic stresses (Supplementary Table S1). This analysis implied that AtCAPE1 derived from PROAtCAPE1 may be more specific to the salt-stress response than the other eight AtCAPEs. We therefore focused our further investigation on the role of the potential AtCAPE1 in salt-stress responses.
AtCAPE1 is endogenous in Arabidopsis
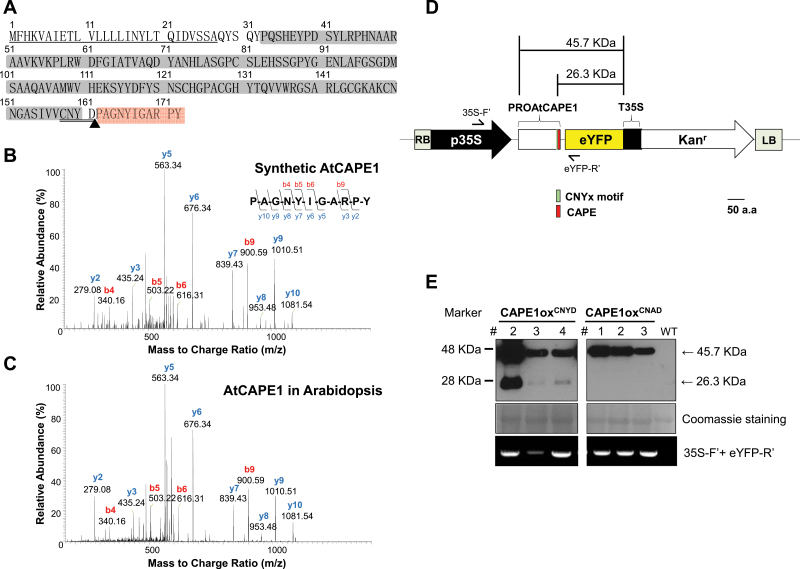
The deduced coding region of the PROAtCAPE1 product was 172 aa (Fig. 1A). A predicted signal peptide was found at the N terminus with a cleavage site between aa 27 and 28 (SignalP 4.1 Server, http://www.cbs.dtu.dk/services/SignalP/), suggesting that PROAtCAPE1 is a secretory protein that is synthesized in the endoplasmic reticulum (ER) and secreted into the extracellular space. The CAP domain (Gibbs et al., 2008) conserved among CAPs from various species was located in the middle of the sequence (aa 43–160), while the putative AtCAPE1 was at the C-terminal end (Fig. 1A).
Fig. 1.
Identification of AtCAPE1 in Arabidopsis. (A) Deduced amino acid sequence of the PROAtCAPE1 product. The predicted secretion signal peptide is underlined. The CAP domain is shaded in grey. The putative AtCAPE1 peptide is shaded in red. The cleavage site predicted to produce AtCAPE1 is indicated with an arrowhead. The putative cleavage signal motif is double-underlined. (B) LC-MS/MS spectrum of the synthetic AtCAPE1. The y-ion is the C-terminal fragments after peptide bond cleavage while the b-ion is the N-terminal fragments. (C) LC-MS/MS spectrum of the identified AtCAPE1 in Arabidopsis. (D) Schema representing the construct used for constitutive overexpression of enhanced yellow fluorescent protein (eYFP)-tagged PROAtCAPE1. The green box shows the CNYx motif. The putative CAPE is shown in red. The numbers indicate the predicted molecular weight of precursor protein tagged with eYFP (45.7kDa) and the cleaved precursor tagged with eYFP (26.3kDa). (E) Production of the precursor PROAtCAPE1 and the cleaved PROAtCAPE1 in CAPE1oxCNYD and CAPE1oxCNAD transgenic plants, where eYFP was fused to PROAtCAPE1 containing wild type (CNYD) and the mutated (CNAD) junction sequence, respectively. T3 seedlings derived from independent transgenic lines were sampled for western blotting with anti-GFP antibody. Coomassie blue staining was used for protein loading control. The lower panel shows the presence of the T-DNA insertions in the transgenic plants by genomic DNA PCR with the primer pair 35S-F’ and eYFP-R’ shown in (D).
To find endogenous evidence of AtCAPE1 in Arabidopsis, we performed nanoflow LC-MS/MS (nanoLC-MS/MS) operated in MS/MS full scan mode (Chen et al., 2012). After optimizing the collision-induced dissociation energy using synthetic AtCAPE1 as the standard, the condition attained a detection limit of ~10 attomole in our system. For the synthetic standard, 80% y-ion and 40% b-ion series coverage for the peptide fragments were observed in the MS/MS spectrum (Fig. 1B). In the analysis of endogenous peptides extracted from wild type (Ler), one MS/MS spectrum showed all the b- and y-ions matched to the standard. The ID confidence for a full-scan MS/MS spectrum is above the criteria for ID assignment that was proposed by the European Commission and noted in the ‘COMMISSION DECISION of 12 August 2002 implementing Council Directive 96/23/EC concerning the performance of analytical methods and the interpretation of results’. Therefore, the result indicated the existence of endogenous AtCAPE1 (Fig. 1C).
Among the PROAtCAPE proteins, the short conserved CNYx motif juxtaposing the CAPE sequence was assumed to be the cleavage signal that generates the CAPEs (Fig. 1A) (Chen et al., 2014). To confirm this proteolytic process of the AtCAPE1 precursor, we generated a transgenic plant, CAPE1oxCNYD, where wild-type PROAtCAPE1 fused with C-terminal enhanced yellow fluorescent protein (eYFP) was constitutively overproduced by the 35S promoter (Fig. 1D). Expression of PROAtCAPE1 was examined in three independent CAPE1oxCNYD transgenic lines by probing with anti-green fluorescent protein (GFP) antibody (Fig. 1E). In all of the three transgenic lines, we observed the presence of two bands with a molecular weight (MW) close to 48 and 28kDa, indicated by the protein marker (Fig. 1E, left-hand panel). The MWs of the two bands were close to that of the precursor protein tagged with eYFP (45.76kDa) and the putative AtCAPE1 fused to eYFP (26.3kDa), respectively (Fig. 1E, left-hand panel), when the cleavage occurred at the predicted cleavage site (Fig. 1A). We then generated transgenic lines, named CAPE1oxCNAD, where eYFP was fused with PROAtCAPE1 but with a mutation (Y160A, Fig. 1A) in the conserved cleavage motif. We then examined expression of the mutated PROAtCAPE1–eYFP. Only a single band with a MW of 45.76kDa was detected in all three independent transgenic lines (Fig. 1E, right-hand panel). Taken together, these results suggested that the identified AtCAPE1 was derived from its precursor, PROAtCAPE1, through cleavage at the conserved CNYx motif, and that an aromatic amino residue, tyrosine, is important for the process.
Expression of the PROAtCAPE1 gene is downregulated by salt stress
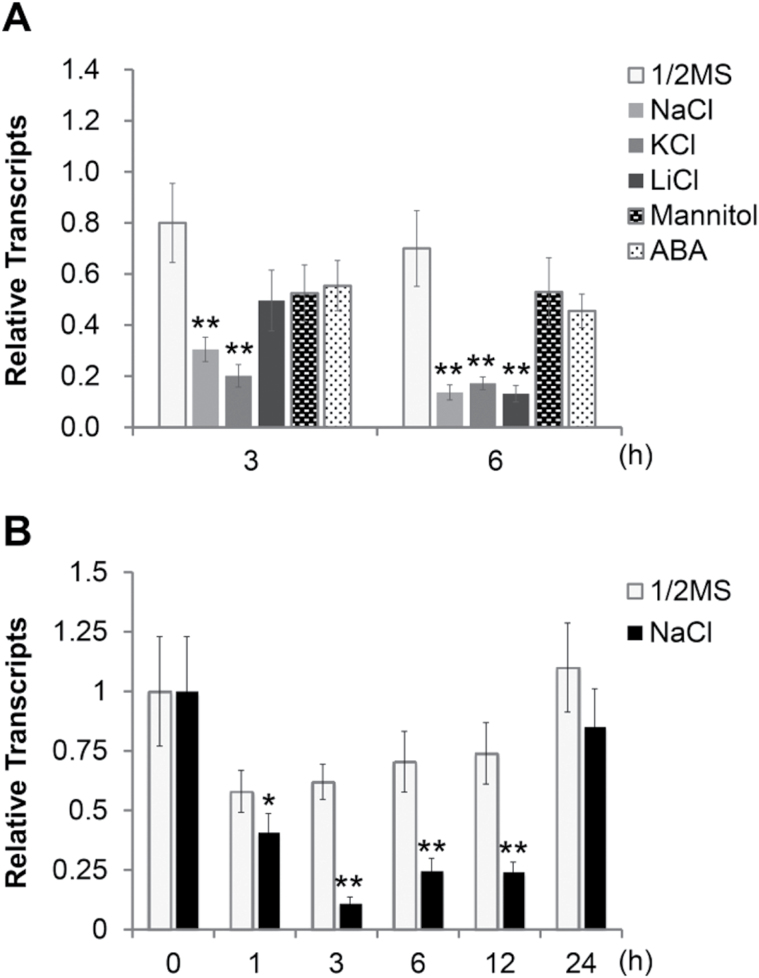
Having confirmed the endogenous presence of AtCAPE1, we further validated that expression of the precursor gene, PROAtCAPE1, was regulated by salt stress via qRT-PCR (Fig. 2A). In comparison with normal growth condition, the transcripts of PROAtCAPE1 were significantly reduced by around 3- to 4- fold (P≤0.01) after 3 and 6h treatment with 150mM NaCl and KCl, as well as 6h treatment with 30mM LiCl (Fig. 2A). However, incubation with 300mM mannitol or 100 μM abscisic acid (ABA) for 3 and 6h showed no significant effect on the expression of the PROAtCAPE1 gene (Fig. 2A). As expression of the PROAtCAPE1 gene was significantly reduced upon incubation with three different salts, NaCl, KCl, and LiCl, but with not mannitol, we propose that the reduced expression of PROAtCAPE1 under high salinity is mainly due to ionic stress, not osmotic stress. In addition, ABA showed much less effect on expression of PROAtCAPE1 in comparison with the effect derived from salt ions, and the reduction in expression of the PROAtCAPE1 gene was still observed in the ABA biosynthesis mutants (Supplementary Fig. S1, available at JXB online) nced3, aba2, and aba3 (Nambara and Marion-Poll, 2005). These findings indicated that the PROAtCPAE1 gene may not be regulated by an ABA-dependent pathway under salinity. A comparison of the expression level at 3 and 6h of incubation with 150mM NaCl revealed that the expression of PROAtCAPE1 showed a kinetic response to salt stress. Therefore, more detailed kinetics of the expression of PROAtCAPE1 were examined upon incubation with a slightly reduced salt concentration (125mM NaCl) (Fig. 2B). We found that the transcript level of PROAtCAPE1 started to show a noticeable decrease in the first 1h, reaching a minimum at 3h (Fig. 2B). This result showed that the downregulation of PROAtCAPE1 is an early cellular response to salt stress. Interestingly, the expression level remained low up to 12h but recovered to a level close to that of wild type at 24h (Fig. 2B; see Discussion).
Fig. 2.
Levels of PROAtCAPE1 transcripts under salinity. (A) Ten-day-old wild-type (Ler) seedlings were subjected to 150mM NaCl, 150mM KCl, 30mM LiCl, 300mM mannitol, and 100 μM abscisic acid (ABA). Relative transcripts indicate the normalized PROAtCAPE1 level (PROAtCAPE1/ACTIN2) from each sample compared with that in wild type in medium alone (1/2MS) at 0h. The bar shown here is the mean of three biological repeats. Error bars indicate means±SEM (stress-treated wild-type versus untreated wild-type at different time points; Student’s t-test: **P≤0.01, *P≤0.05). (B) Ten-day-old wild-type seedlings were treated without (1/2MS) or with 125mM NaCl. Zero hours means that the seedlings were subjected to medium alone and harvested immediately. The bars shown here are the means from four biological repeats. Error bars indicate means±SEM (salt-treated wild-type versus untreated wild-type at different time points; Student’s t-test: **P,≤0.01, *P,≤0.05).
AtCAPE1 negatively regulates salt tolerance
To test the physiological role of AtCAPE1 in response to salt stress, the growth of wild-type (Ler) and proatcape1 mutant (T-DNA inserted knockout line with Ler background; Supplementary Fig. S2) in normal growth medium and medium containing NaCl for 20 d was compared (Supplementary Fig. S3, available at JXB online). The proatcape1 mutant seedlings did not show a noticeable defect under normal growth conditions. In 100mM NaCl, more surviving seedlings with green and large-sized leaves were observed in proatcape1 than in the wild type (Supplementary Fig. S3). In 125mM NaCl, neither wild-type nor proatcape1 seedlings survived. We also tested whether exogenously supplied synthetic AtCAPE1 could restore the salt response of the proatcape1 mutant (Supplementary Fig. S3). The presence of 1 μM synthetic AtCAPE1 partially restored the salt response, as shown by the reduced growth in 100mM NaCl to a level close to that in wild type (Supplementary Fig. S3). These results indicated that AtCAPE1 confers salt sensitivity on Arabidopsis and thus acts negatively on the salt-tolerance response.
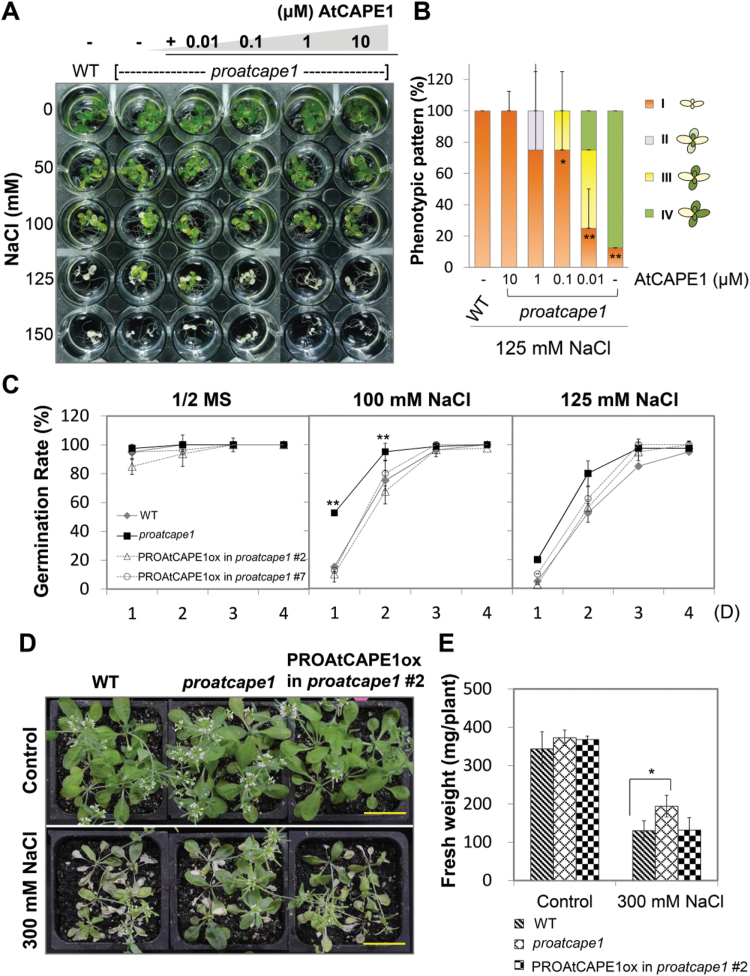
Salt stress retards plant development and impairs seed germination (Greenway and Munns, 1980). We analysed these two responses in more detail in the proatcape1 mutant. The growth response of the mutants was measured by subjecting 5-d-old wild-type and proatcape1 seedlings to various concentrations of NaCl for a further 10 d (Fig. 3A). Although the treatment with 50 and 100mM NaCl caused the leaves of the wild-type and proatcape1 seedlings to turn light green, no significant difference was observed between the two (Fig. 3A). Upon treatment with 125mM NaCl, proatcape1 seedlings displayed greenish cotyledons and generated true leaves, whereas wild-type seedlings exhibited yellowish cotyledons and stunted growth (Fig. 3A). Neither Ler nor proatcape1 seedlings survived following 150mM NaCl treatment (Fig. 3A). We further investigated the phenotype of salt-treated Ler and proatcape1 subjected to various concentrations of peptides. Introduction of various concentrations (0, 0.01, 0.1, 1, and 10 μM) of synthetic AtCAPE1 into the medium did not cause any visible growth retardation. The effect of the peptide became clearly noticeable upon treatment with 125mM NaCl. Under this condition, a semi-quantitative analysis of the phenotypic severity observed with various concentrations of peptide was conducted by categorizing the plants into four classes. As shown in Fig. 3B, upon 125mM NaCl treatment, none of the wild-type seedlings survived (class I) after treatment for 10 d; similarly, the leaves of all the proatcape1 seedlings turned totally yellow and stopped growing (class I) when subjected to an additional 10 μM synthetic AtCAPE1. In the presence of 1 and 0.1 μM peptide, 75% of the proatcape1 seedlings died (class I) and 25% displayed either four yellow to pale green (class II) or four green (class III) leaves out of a total of six leaves, respectively (Fig. 3B). The peptide had a visible effect at concentrations as low as 0.01 μM (25% of seedlings in class I); when no peptide was supplemented, nearly 90% of the proatcape1 mutants displayed five out of six green leaves (class IV) (Fig. 3B). The quantitative phenotype indicated that the negative effect caused by AtCAPE1 is dosage dependent.
Fig. 3.
AtCAPE1 negatively regulates the salt-tolerance response. (A) Growth response of proatcape1 mutants to salt stress and to AtCAPE1. Five-day-old seedlings were treated with various concentrations of NaCl in the absence (–) and presence (+) of various concentrations of synthetic AtCAPE1. The photograph was taken after treatment for another 10 d. (B) Semi-quantitative analysis of the phenotype of plants shown in (A). Each bar represents the mean percentage of the phenotypic pattern from two independent experiments. Error bars indicate means±SD. The phenotypic pattern according to leaf number and colour of seedlings was defined into categories I–IV as shown in the diagram (salt- and peptide- treated proatcape1 versus salt-treated wild-type Ler; Student’s t-test: **P≤0.01, *P≤0.05). (C) Germination rates of proatcape1 mutant and PROAtCAPE1ox in proatcape1 transgenic plant seeds compared with the corresponding wild-type (Ler) seeds. Each value represents the percentage of germination (with 40 seeds) for four independent tests. Error bars indicate the means±SEM. (Student’s t-test: **P≤0.01, *P≤0.05). (D) Three-week-old plants were irrigated without (Control) and with 300mM NaCl three times for every second day. After that, plants were recovered with water for another one week. The photograph was taken then. (E) Shoot fresh weight was measured after treatments from (D). Data are shown as an average fresh weight from 36 plants. Error bars indicate means±SD. An asterisk indicates significant differences (*P≤0.05) between proatcape1 mutant and wild-type Ler plants upon 300mM NaCl treatment.
We next examined the germination rates of wild-type and proatcape1 mutant seeds on medium containing various concentrations of NaCl (Fig. 3C). When grown on a medium supplemented with 100mM NaCl, proatcape1 seeds displayed significantly higher germination rates than wild-type (Ler) seeds at early time points (1–2 d, Fig. 3C). No significant difference was found when both types of seeds were sown on medium containing 125mM NaCl (Fig. 3C). To complement the phenotype of proatcape1 under salinity, we generated a transgenic line of proatcape1 harbouring a constitutive overexpression cassette for PROAtCAPE1 (named PROATCAPE1ox in proatcape1; Supplementary Fig. S2D). Two PROATCAPE1ox in proatcape1 transgenic lines displayed a slower germination rate than proatcape1 mutant seeds in the presence of 100mM NaCl for 1–2 d. Moreover, adult PROATCAPE1ox in proatcape1 transgenic lines also displayed reduced resistance to salinity as compared with proatcape1 mutants, with less fresh weight of shoots (Fig. 3D, E). These results indicated that overexpression of PROAtCAPE1 could counteract the salt resistance of the proatcape1 mutant.
Taken together, the knockout mutant of PROAtCAPE1 showed a reduced response in growth retardation and induced germination under high-salt conditions, whereas exogenous application of synthetic AtCAPE1 (Fig. 3A, B; Supplementary Fig. S3) or overexpression of peptide precursor (Fig. 3C–E) restored the salt response of proatcape1 mutants. These results demonstrated that AtCAPE1 functions as a negative regulator of salt-stress tolerance in Arabidopsis.
Transcriptome analysis reveals that AtCAPE1 negatively regulates the transcripts of salt-inducible genes
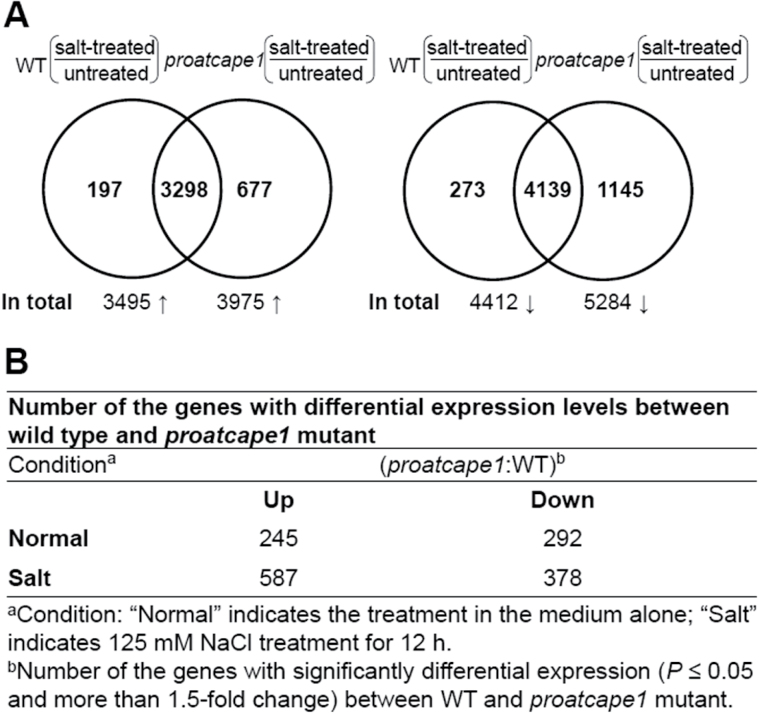
To gain insight into the mechanism by which AtCAPE1 regulates salt responses, we investigated the gene expression profiles of wild-type (Ler) and proatcape1 mutant seedlings in the presence and absence of 125mM NaCl by microarray analysis (Fig. 4). First, we identified 3495 and 4412 genes that were up- and downregulated, respectively, by over 1.5-fold in the wild type after 12h of treatment in 125mM NaCl (Fig. 4A; Supplementary dataset I, available at JXB online). This observation was in agreement with previous reports that salt treatment leads to a large change in the gene expression profile and is involved in many physiological processes (Kreps et al., 2002; Seki et al., 2002; Jiang and Deyholos, 2006). In the mutant, 3975 and 5284 genes were up- and downregulated, respectively, by over 1.5-fold under salinity (Fig. 4A; Supplementary dataset II, available at JXB online). The result showed that the overall sensitivity of plants to salt stress was increased in the mutant, as more genes were affected in the mutant than in the wild type. We then compared the genes that were differentially expressed in the wild type and mutant. Under normal conditions, 245 and 292 genes were up- and downregulated by over 1.5-fold in the mutants (Fig. 4B; Supplementary dataset III, available at JXB online). In contrast, after 12h of treatment with 125mM NaCl, 587 and 378 genes were up- and downregulated by over 1.5-fold in the mutants (Fig. 4B; Supplementary dataset IV, available at JXB online). The result showed that more genes were upregulated under high salinity. As the mutant displayed salt tolerance, it is likely that these upregulated genes in the mutant under high salinity contribute to the salt-tolerant phenotype. As analysed by Gene Ontology (GO) (Huang et al., 2009a, b ), the gene products of these genes could be described by GO terms including ‘response to water deprivation’, ‘response to abscisic acid stimulus’, and ‘response to salt stress’ (Table 2).
Fig. 4.
Number of genes differentially expressed in wild type (Ler) and the proatcape1 mutant under high salt. (A) Numbers indicate the genes with significantly differential expression (P≤0.05 and a greater than 1.5-fold change) between the indicated data sets derived from microarray analysis. (B) Number of genes with differential expression levels between wild type and the proatcape1 mutant under normal and salt (12h of treatment in 125mM NaCl) conditions (P≤0.05 and a greater than 1.5-fold change).
Table 2.
GO analysis a of the 587 differentially upregulated genes in proatcape1 under salt conditions
| Biological functionb | Number of genesc | P value |
|---|---|---|
| Cellular amino acid derivative metabolic process | 21 | 2.51E–06 |
| Phenylpropanoid metabolic process | 20 | 2.61E–09 |
| Lipid biosynthetic process | 19 | 3.61E–03 |
| Cellular lipid metabolic process | 19 | 1.48E–02 |
| Cellular amino acid derivative biosynthetic process | 18 | 9.05E–07 |
| Aromatic compound biosynthetic process | 18 | 3.73E–06 |
| Phenylpropanoid biosynthetic process | 17 | 1.36E–08 |
| Response to salt stress | 16 | 1.32E–02 |
| Response to fungus | 16 | 4.95E–02 |
| Response to abscisic acid stimulus | 15 | 3.32E–03 |
| Response to water deprivation | 13 | 4.05E–04 |
| Response to reactive oxygen species | 12 | 2.08E–04 |
| Response to bacterium | 12 | 1.60E–02 |
| Innate immune response | 12 | 3.47E–02 |
| Response to cold | 11 | 2.85E–02 |
a The selected salt-responsive genes were uploaded into DAVID Bioinformatics Resources 6.7 (Huang et al., 2009a, b ) for GO analysis. The biological functions categorized by GO analysis were selected by level four.
b The top 15 most abundant biological process categories are shown.
c Number of genes (from a total of 587 genes) displayed for each category.
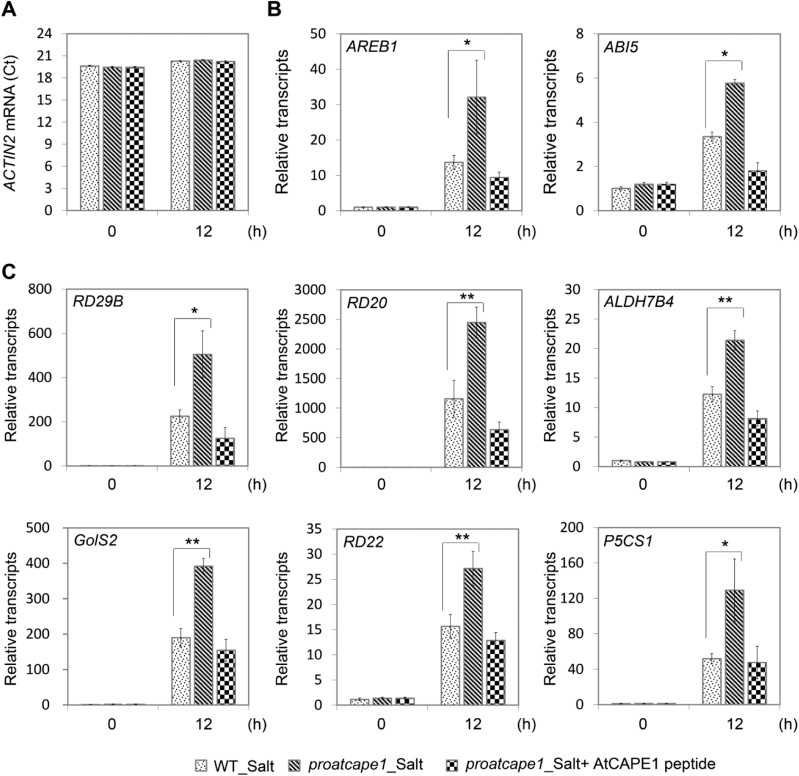
We further confirmed some of the salt-inducible genes regulated by AtCAPE1. The ACTIN2 transcript was used as an internal control in this experiment as its expression did not respond to salinity (Fig. 5A). ABA-responsive element binding protein 1 (AREB1) (Fujita et al., 2005) and ABA-INSENSITIVE 5 (ABI5) (Finkelstein and Lynch, 2000) are well-characterized basic leucine zipper transcription factors involved in ABA signalling under drought and high-salinity conditions (Uno et al., 2000; Nakashima and Yamaguchi-Shinozaki, 2013 m). The expression level of the AREB1 gene was highly upregulated by salt by around 13-fold in the wild type (Fig. 5B), which is in agreement with previous reports (Uno et al., 2000; Fujita et al., 2005). The proatcape1 mutation resulted in a further increase in AREB1 of around 2-fold compared with that in the wild type under salinity. However, the increased expression of AREB1 in the mutant was completely restored to the wild-type level by exogenous treatment with AtCAPE1 (Fig. 5B). A similar trend was found in the regulation of the ABI5 gene (Fig. 5B), although the ABI5 expression was low after seedling establishment (Nakashima and Yamaguchi-Shinozaki, 2013). AtCAPE1 also negatively regulated the expression of high-salt-inducible downstream genes, including the genes for the enzymes involved in osmoprotectant biosynthesis [Δ 1 -PYRROLINE-5-CARBOXYLATE SYNTHASE 1 (P5CS1) (Yoshiba et al., 1999); and GALACTINOL SYNTHASE (GolS2) (Taji et al., 2002)], for detoxification [ALDEHYDE DEHYDROGENASES 7B4 (ALDH7B4) (Kirch et al., 2004; Kotchoni et al., 2006)], and for the dehydration response [RD22, RD20 (also known as CLO3), and RD29B (Yamaguchi-Shinozaki and Shinozaki, 1993; Ingram and Bartels, 1996; Takahashi et al., 2000)] (Fig. 5C). Taken together, these results indicated that the negative role of AtCAPE1 in salt tolerance is through downregulation of the salt-tolerance genes involved in salt-stress resistance. A further investigation of the levels of RD29B transcripts regulated by various concentrations of AtCAPE1 (0.5, 5, 50, and 500nM and 5 μM) in proatcape1 mutants upon salinity indicated that, when introducing 50–500nM AtCAPE1 to the mutants, the induced RD29B genes in mutant lines were suppressed to the same level as that of the wild type upon salt stress (Supplementary Fig. S4, available at JXB online). However, an increasing peptide concentration (5 μM) did not show more suppression (Supplementary Fig. S4). This result suggested that the AtCAPE1 level in wild-type plants in response to salinity may have reached the maximum suppression efficacy for RD29B expression.
Fig. 5.
Expression of various salt-inducible genes is downregulated by AtCAPE1. Ten-day-old seedlings were treated without and with salt for 12h. The transcript levels of the selected salt-inducible genes in wild-type (Ler) and proatcape1 mutant seedlings were determined by qRT-PCR: ACTIN2 (A), AREB1 and ABI5 (B), and RD29B, RD20, ALDH7B4, GolS2, RD22, and P5CS1 (C). Zero hours means that the seedlings were subjected to the medium indicated and harvested immediately. For proatcape1, the mutants were subjected to either 125mM NaCl (proatcape1_Salt) or 125mM NaCl in the presence of 500nM AtCAPE1 peptide (proatcape1_Salt+AtCAPE1 peptide). The mean values from four biological repeats are shown. Error bars are means±SEM (proatcape1 versus Ler upon salt treatment; Student’s t-test: **P≤0.01, *P≤0.05).
Production of AtCAPE1 is post-translationally regulated under saline conditions
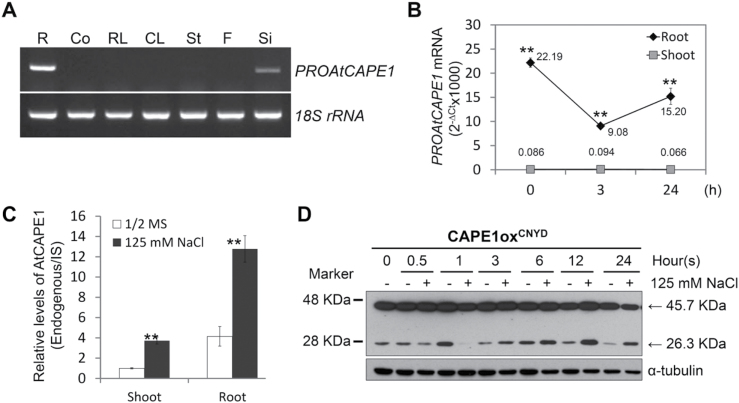
A series of molecular mechanisms converting the signal into phenotypic resistance to salinity in plants is initiated in the root and systemically transmitted to the shoot (Munns and Tester, 2008; Roy et al., 2014). Thus, the expression of PROAtCAPE1 in different tissues was examined to reveal how PROAtCAPE1 functions in response to salinity. Based on RT-PCR analysis, the PROAtCAPE1 gene was found to be expressed most highly in the roots and was barely detected in above-ground tissues except for the silique (Fig. 6A). In comparison with the wild type under normal conditions, slightly shorter primary roots were observed in proatcape1 seedlings (Supplementary Fig. S5E, available at JXB online). This phenotype suggested the possibility of a functional trade-off of AtCAPE1 between root growth and salt tolerance. When grown to the stage of inflorescence, no growth retardation and no abnormal morphology were observed in siliques in a comparison between the wild type and mutants (Supplementary Fig. S5, available at JXB online). A further investigation on roots by β-glucuronidase (GUS) staining showed that the promoter activity was observed in stelar cells from the elongation zone to the maturation zone and the trichoblast at the maturation zone (Supplementary Fig. S6, available at JXB online).
Fig. 6.
Production of AtCAPE1 is mainly derived from root tissues and is regulated by salt. (A) Transcriptional levels of PROAtCAPE1 in different tissues were determined by RT-PCR. Total RNA was extracted from root (R), cotyledon (Co), rosette leaf (RL), cauline leaf (CL), stem (St), flower (F) and silique (Si). 18S rRNA transcripts were used as internal control. (B) Salt response of the transcriptional levels of PROAtCAPE1 in shoots and roots. Ten-d-old seedlings of wild type (Ler) were treated with 125mM NaCl for 0, 3, and 24h. The transcripts of PROAtCAPE1 from the harvested roots and shoots were determined by RT-qPCR. Shown are the average values of (2-∆Ctx1000) from four biological repeats. Error bars, means±SE (PROAtCAPE1 transcripts in shoots versus PROAtCAPE1 transcripts in roots at different time points; Student’s t-test, **P≤0.01). (C) Relative level of endogenous AtCAPE1 in shoots and roots. Seedlings grown for 24h without (1/2 MS) and with 125mM NaCl were subjected to quantitative LC-MS/MS analysis. IS, internal standard. The average values from two biological repeats are shown. Error bars, means±SE. Asterisks indicate statistically significant differences between salt-treated and untreated samples (Student’s t-test; **P≤0.01). (D) Post-translational regulation of AtCAPE1 production. Protein extracts from the transgenic lines (CAPE1oxCNYD) harbouring the AtCAPE1-eYFP fusion grown with (+) and without (-) 125mM NaCl for the indicated times were subjected to western blot analysis. The upper and lower bands with approximate size of 45.7 KDa and 26.3 KDa represent the expected size of the PROAtCAPE1-eYFP fusion protein and the AtCAPE1-eYFP fusion protein, respectively. The fusion proteins were detected by anti-GFP antibody. α-tubulin, loading control.
To elucidate whether PROAtCAPE1 expressed in the roots was the main signal in response to salt stress, PROAtCAPE1 transcripts were further examined separately in the shoots and roots of Arabidopsis seedlings in 125mM NaCl (Fig. 6B). In roots, the transcript level of PROAtCAPE1 was downregulated within 3h, in agreement with our previous observation shown in Fig. 2. In shoots, the transcript was barely detected under the same conditions (Fig. 6B). GUS staining of 10-d-old pPROAtCAPE1:GUS transgenic lines in the absence and presence of salt for 24h reflected the mRNA expression pattern in the shoots and roots shown by qRT-PCR (Fig. 6B and Supplementary Fig. S7, available at JXB online). PROAtCAPE1 was expressed mainly in the roots, yet mediated salt-tolerant phenotypic effects in the leaves (Fig. 3). We thus examined whether AtCAPE1 is produced in the roots and systemically transferred to shoots resulting in salt-stress responses throughout the whole seedling. Targeted LC-MS/MS analysis was performed (Chen et al., 2012, 2014) to quantify the level of endogenous AtCAPE1 in shoots and roots (Fig. 6C). We found that, under normal conditions, AtCAPE1 was present in the shoots at 25% of the level in the roots (Fig. 6C). Interestingly, in seedlings treated with salt for 24h, the level of AtCAPE1 was 4-fold and 3-fold higher in the shoots and in roots, respectively, than in the seedlings without salt treatment (Fig. 6C). Thus, the presence of AtCAPE1 in both tissues suggests that AtCAPE1 may function systemically.
As the transcription of PROAtCAPE1 displayed the wild-type level after 24h of treatment in 125mM NaCl (Figs 2B and 6B), but AtCAPE1 was increased 3-fold (Fig. 6C), we tested whether the production of AtCAPE1 was regulated at the post-translational level. For this test, the transgenic plant CAPE1oxCNYD (Fig. 1D, E) was utilized. In Fig. 6D, the putative AtCAPE1–eYFP (indicated at a MW of 26.3kDa) was reduced rapidly upon 125mM NaCl treatment, reaching a minimum at 1h, but then increased, being relatively higher than that without salt treatment. The western blot clearly demonstrated that the proteolytic process of PROAtCPE1 was dynamically regulated under saline conditions.
Discussion
Here, we attempted to identify a salt-responsive peptide from Arabidopsis. We found a small peptide, AtCAPE1, by analysing the structure and expression patterns of the CAP superfamily proteins, that contained a CNYx.PxGNxxxxxPY- motif at the C terminus (Table 1). The predicted peptide existed endogenously and was 11 aa, making it the smallest currently known unmodified peptide signal in plants. The substitution of Y160A in the conserved CNYx motif completely blocked the processing of PROAtCAPE1 (Fig. 1E), demonstrating that the aromatic residue tyrosine is critical for the cleavage of PROAtCAPE1. A dibasic -RR- or -GR- motif is the only currently well-known protease recognition site located upstream of mature peptides in plants (Pearce and Ryan, 2003; Liu et al., 2007; Matos et al., 2008; Srivastava et al., 2008). However, no -RR- or -GR- motif was observed preceding the CAPEs, suggesting that such motifs are not utilized for the cleavage of CAP proteins. This work, therefore, uncovered a unique protease recognition motif important for regulation of the proteolytic process. The signal peptide is predicted to be at the N-terminal PROAtCAPE1 protein sequence (Fig. 1A). Thus, PROAtCAPE1 may be secreted into the extracellular space and cleaved into mature AtCAPE1 in the apoplast (Delaunois et al., 2013).
As the knockout mutant of PROAtCAPE1 conferred salt tolerance to Arabidopsis seedlings, and exogenous application of AtCAPE1 to the mutant rendered the seedlings salt sensitive, this peptide is identified as a negative regulator of salt tolerance or a positive regulator of the salt-sensitive response in Arabidopsis. Based on our data showing the existence of the peptide (Fig. 6C, D), we envision that AtCAPE1, under normal conditions, suppresses the expression of salt-tolerance-related genes. This may be to avoid unnecessary expression of salt-tolerance genes in the absence of salt.
The level of the peptide was regulated dynamically upon salt treatment. Initially, following salt treatment, expression of PROAtCAPE1 gene was rapidly downregulated and was recovered at later time points (Fig. 2B). Furthermore, processing of PROAtCAPE1 into mature peptides was also rapidly diminished (Fig. 6D). Together, these results showed that the reduction of AtCAPE1 level is an early response of plants to salt stress. This leads to temporary relief of expression of the salt-tolerance genes suppressed by AtCAPE1. However, after a prolonged period of salt treatment, expression of the PROAtCAPE1 gene, proteolytic processing, and accordingly production of AtCAPE1 was increased to an even higher level than that in the wild type (Figs 2B, and 6C, D). The increased amount of the peptide then led to suppression of salt-tolerance genes and resulted in susceptibility to salt stress (Figs 3 and 5). However, in the proatcape1 mutant with no AtCAPE1 production, the salt tolerance-related genes may be expressed constitutively, leading to better salt tolerance than in the wild type. Therefore, treatment of the mutant with exogenous AtCAPE1 suppressed expression of the salt-tolerance genes, recovering the salt sensitivity. Nonetheless, the expression level of AtCAPE1 may have reached maximum efficacy in terms of salt susceptibility in the wild type, because the salt-tolerance gene expression (Supplementary Fig. S4) and the salt susceptibility phenotype (Fig. 3A, B) in the mutant lines were only suppressed to the wild-type level, even when high concentrations (micromolar level) of AtCAPE1 were applied. In addition, overexpression of PROAtCAPE1 in transgenic lines only displayed a similar germination rate to the wild type under high salinity (Fig. 3C). This can be explained by the threshold effect of the peptide, and it is probable that protease activity for processing PROAtCAPE1 into mature AtCAPE1 is also a limiting effector.
Endogenous AtCAPE1 was present in the shoots at 25% of the level in the roots. AtCAPE1 detected in the shoots is less likely to be derived from its precursor since PROAtCAPE1 gene expression was undetectable or extremely low in the shoots (Fig. 6B). Given the large discrepancy between the level of PROAtCAPE1 transcripts in shoots and roots (Fig. 6A, B), AtCAPE1 detected in the shoots might have originated from the roots. The shoots of wild-type seedlings were more sensitive to salt than the mutant seedlings. In addition, the leaf-specific RD20/CLO3 gene encoding a calcium-binding protein that signals the regulation of stomatal closure under stress (Aubert et al., 2010) was regulated by AtCAPE1. We propose that, at least in part, the salt sensitivity of the wild-type shoot is mediated by the systemic peptide signal.
Our study further shed light on how AtCAPE1 mediates salt sensitivity or negatively regulates salt tolerance at the molecular level. As the microarray comparison between the wild type and the proatcape1 mutant revealed, expression of hundreds of salt-responsive genes was altered in the mutants. A part of the molecular mechanism of AtCAPE1-mediated salt sensitivity can be explained as follows. The genes downregulated by AtCAPE1 included the transcription factors ABI5 and AREB1, which control the transcription of downstream ABA-dependent and salt-responsive genes. Both ABI5 and AREB1 are basic leucine zipper transcription factors; however, ABI5 functions in response to stresses mainly during germination, while AREB1 functions mainly at the vegetative stage (Nakashima and Yamaguchi-Shinozaki, 2013). Therefore, the delayed germination caused by AtCAPE1 may in part be governed by the ABI5-dependent signalling pathway, while the growth defects observed in seedlings are in part mediated by the AREB1-dependent regulon. AREB1 activates the downstream gene expression by binding to a conserved cis-acting ABA-responsive element (ABRE) in their promoter regions (Uno et al., 2000). The salt-responsive genes RD29B and RD20/CLO3 containing the ABRE element in the promoter region (Uno et al., 2000; Fujita et al., 2005; Aubert et al., 2010) were found to be a direct target of AREB1 (Uno et al., 2000; Fujita et al., 2005). Since P5CS1 and ALDH7B4 also contain the cis ABRE (Yamaguchi-Shinozaki and Shinozaki, 1994; Yoshiba et al., 1999; Missihoun et al., 2014), it is highly likely that they are regulated by AREB1 as well. Thus, reduced expression of these downstream genes by AtCAPE1 may have been, in part, the result of reduced expression of AREB1 by AtCAPE1. The reduced expression of P5CS1 may result in the reduced accumulation of proline, functioning as an osmolyte to balance water potential in plant cells (Ross et al., 2004; Denoux et al., 2008). Reduced expression of ALDH7B4 would lead to a lesion in the detoxification processes of aldehydes that overaccumulate in plants when exposed to abiotic stresses (Kotchoni et al., 2006) and to the susceptibility to NaCl treatments as observed in the aldh7b4 mutant (Kotchoni et al., 2006). We also found that AtCAPE1 downregulated the transcription of GolS2, which encodes a biosynthesis enzyme for the production of raffinose and galactinol, which function as osmoprotectants against high salinity (Taji et al., 2002). Since there is no ABRE element in the promoter region of GolS2 (Taji et al., 2002), AtCAPE1-mediated downregulation of this gene is unlikely through AREB1.
We have unequivocally shown that AtCAPE1 negatively regulates salt tolerance or mediates salt sensitivity. Thus, what would be the possible advantage of having this endogenous peptide? From the phenotypic assays, we showed that proatcape1 seedlings have slightly shorter primary roots than wild-type seedlings. This phenotype may indicate the possible involvement of the peptide in a trade-off between root growth and salt tolerance. We reported previously that a CAP-derived peptide could mediate an anti-pathogenic response in tomato (Chen et al., 2014). It is thus also feasible that AtCAPE1 may be involved in a possible trade-off between pathogenic defence and salt tolerance. In addition, the knowledge that an endogenous peptide is a negative regulator of salt stress and information about the regulation of the proteolysis of a precursor protein into peptides can be utilized in strategies to introduce salt stress tolerance in plants by blocking the cleavage of precursor proteins.
Supplementary data
Supplementary data are available at JXB online.
Supplementary materials and methods.
Supplementary Fig. S1. Levels of PROAtCAPE1 transcripts in the mutants of ABA biosynthesis genes under salinity.
Supplementary Fig. S2. Transcripts of PROAtCAPE1 in transgenic plants.
Supplementary Fig. S3. AtCAPE1 negatively regulates salt tolerance response.
Supplementary Fig. S4. Application of AtCAPE1 peptide reduces the transcript levels of RD29B under salinity.
Supplementary Fig. S5. Phenotypic investigation of wild-type Ler and proatcape1 mutant.
Supplementary Fig. S6. Expressions of PROAtCAPE1 in roots.
Supplementary Fig. S7. GUS-staining assay revealed the PROAtCAPE1 promoter activity in the shoots and roots upon salt stress.
Supplementary Table S1. Effect of selected abiotic stresses on expressions of PROAtCAPEs.
Supplementary Table S2. Primers used in this study.
Supplementary dataset I. Differentially expressed genes in wild type upon the treatment in the presence and absence of 125mM NaCl for 12h.
Supplementary dataset II. Differentially expressed genes in proatcape1 mutants upon the treatment in the presence and absence of 125mM.
Supplementary dataset III. Differentially expressed genes between proatcape1 mutants and wild type in the normal condition.
Supplementary dataset IV. Differentially expressed genes between proatcape1 mutants and wild type in the presence of 125mM NaCl for 12h.
Acknowledgements
This work was supported by the Ministry of Science and Technology of the Republic of China (grant no. NSC103-2113-M-001-002), the Agricultural Biotechnology Research Center (ABRC), Academia Sinica, Taiwan, and the Center for Plant Aging Research, Institute for Basic Science (IBS), Republic of Korea (IBS-R013-D1-2015-a00). We acknowledge Ms. Shu-Jen Chou in the DNA Microarray Core Laboratory, Institute of Plant and Microbial Biology, Academia Sinica, Taiwan, for assistance with microarray analysis and Dr. Choun-Sea Lin and Mr. Chen-Chuan Hsu in the Plant Tech Core Facility, ABRC, Academia Sinica, Taiwan, for cross-section imaging of GUS staining in plants. We also thank Ms. Miranda Loney for English editing of the manuscript.
Glossary
Abbreviations:
- ABA
abscisic acid
- Col-0
Columbia-0
- ER
endoplasmic reticulum
- eYFP
enhanced yellow fluorescent protein
- GFP
green fluorescent protein
- GO
Gene Ontology
- GUS
β-glucuronidase
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- Ler
Landsberg erecta
- MS
Murashige and Skoog
- MW
molecular weight
- qRT-PCR
quantitative reverse transcription PCR
- TFA
trifluoroacetic acid.
References
- Aubert Y, Vile D, Pervent M, Aldon D, Ranty B, Simonneau T, Vavasseur A, Galaud JP. 2010. RD20, a stress-inducible caleosin, participates in stomatal control, transpiration and drought tolerance in Arabidopsis thaliana . Plant and Cell Physiology 51, 1975–1987. [DOI] [PubMed] [Google Scholar]
- Bernstein L. 1975. Effects of salinity and sodicity on plant growth. Annual Reviews of Phytopathology 13, 295–312. [Google Scholar]
- Chen CJ, Chen WY, Tseng MC, Chen YR. 2012. Tunnel frit: a nonmetallic in-capillary frit for nanoflow ultra high-performance liquid chromatography-mass spectrometry applications. Analytical Chemistry 84, 297–303. [DOI] [PubMed] [Google Scholar]
- Chen YL, Lee CY, Cheng KT, Chang WH, Huang RN, Nam HG, Chen YR. 2014. Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signaling in tomato. The Plant Cell 26, 4135–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunois B, Colby T, Belloy N, Conreux A, Harzen A, Baillieul F, Clément C, Schmidt J, Jeandet P, Cordelier S. 2013. Large-scale proteomic analysis of the grapevine leaf apoplastic fluid reveals mainly stress-related proteins and cell wall modifying enzymes. BMC Plant Biology 13, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delay C, Imin N, Djordjevic MA. 2013. CEP genes regulate root and shoot development in response to environmental cues and are specific to seed plants. Journal of Experimental Botany 64, 5383–5394. [DOI] [PubMed] [Google Scholar]
- Denoux C, Galletti R, Mammarella N, Gopalan S, Werck D, De Lorenzo G, Ferrari S, Ausubel FM, Dewdney J. 2008. Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Molecular Plant 1, 423–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash A. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Lynch T. 2000. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. The Plant Cell 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K. 2005. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis . The Plant Cell 17, 3470–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs GM, Roelants K, O’Bryan MK. 2008. The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins--roles in reproduction, cancer, and immune defense. Endocrine Reviews 29, 865–897. [DOI] [PubMed] [Google Scholar]
- Greenway H, Munns R. 1980. Mechanisms of salt tolerance in nonhalophytes. Annual Reviews of Plant Physiology 31, 149–190. [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. 2008. Genevestigator V3: A reference expression database for the meta-analysis of transcriptomes. Advances in Bioinformatics 2008, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009a. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocol 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009b. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram J, Bartels D. 1996. The molecular basis of dehydration tolerance in plants. Annual Review of Plant Physiology and Plant Molecular Biology 47, 377–403. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Deyholos MK. 2006. Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biology 6, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY™ vectors for agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kirch HH, Bartels D, Wei Y, Schnable PS, Wood AJ. 2004. The ALDH gene superfamily of Arabidopsis . Trends in Plant Science 9, 371–377. [DOI] [PubMed] [Google Scholar]
- Kotchoni SO, Kuhns C, Ditzer A, Kirch HH, Bartels D. 2006. Over-expression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant, Cell and Environment 29, 1033–1048. [DOI] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF. 2002. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130, 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Che P, Howell SH. 2007. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. The Plant Journal 51, 897–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WX, Zhang FC, Zhang WZ, Song LF, Wu WH, Chen YF. 2013. Arabidopsis Di19 functions as a transcription factor and modulates PR1, PR2, and PR5 expression in response to drought stress. Molecular Plant 6, 1487–1502. [DOI] [PubMed] [Google Scholar]
- Mahajan S, Tuteja N. 2005. Cold, salinity and drought stresses: an overview. Archives of Biochemistry and Biophysics 444, 139–158. [DOI] [PubMed] [Google Scholar]
- Matos JL, Fiori CS, Silva-Filho MC, Moura DS. 2008. A conserved dibasic site is essential for correct processing of the peptide hormone AtRALF1 in Arabidopsis thaliana . FEBS Letters 582, 3343–3347. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y. 2014. Posttranslationally modified small-peptide signals in plants. Annual Reviews of Plant Biology 65, 385–413. [DOI] [PubMed] [Google Scholar]
- Missihoun TD, Hou Q, Mertens D, Bartels D. 2014. Sequence and functional analyses of the aldehyde dehydrogenase 7B4 gene promoter in Arabidopsis thaliana and selected Brassicaceae: regulation patterns in response to wounding and osmotic stress. Planta 239, 1281–1298. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. 2008. Mechanisms of salinity tolerance. Annual Reviews of Plant Biology 59, 651–681. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K. 2013. ABA signaling in stress-response and seed development. Plant Cell Reports 32, 959–970. [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. 2005. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol . 56, 165–185. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Ogawa M, Matsubayashi Y. 2008. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. The Plant Journal 55, 152–160. [DOI] [PubMed] [Google Scholar]
- Orsini F, Cascone P, De Pascale S, Barbieri G, Corrado G, Rao R, Maggio A. 2010. Systemin-dependent salinity tolerance in tomato: evidence of specific convergence of abiotic and biotic stress responses. Physiologia Plantarum 138, 10–21. [DOI] [PubMed] [Google Scholar]
- Pearce G, Moura DS, Stratmann J, Ryan CA. 2001a. Production of multiple plant hormones from a single polyprotein precursor. Nature 411, 817–820. [DOI] [PubMed] [Google Scholar]
- Pearce G, Moura DS, Stratmann J, Ryan CAJ. 2001b. RALF, a 5kDa ubiquitous polypeptide in plants, arrests root growth and development. Proceedings of the National Academy of Sciences 98, 12843–12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Ryan CA. 2003. Systemic signaling in tomato plants for defense against herbivores. Isolation and characterization of three novel defense-signaling glycopeptide hormones coded in a single precursor gene. The Journal of Biological Chemistry 278, 30044–30050. [DOI] [PubMed] [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan CA. 1991. A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253, 895–897. [DOI] [PubMed] [Google Scholar]
- Ross AR, Ambrose SJ, Cutler AJ, Feurtado JA, Kermode AR, Nelson K, Zhou R, Abrams SR. 2004. Determination of endogenous and supplied deuterated abscisic acid in plant tissues by high-performance liquid chromatography-electrospray ionization tandem mass spectrometry with multiple reaction monitoring. Analytical Biochemistry 329, 324–333. [DOI] [PubMed] [Google Scholar]
- Roy SJ, Negrao S, Tester M. 2014. Salt resistant crop plants. Current Opinions in Biotechnology 26, 115–124. [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, Satou M, Akiyama K, Taji T, Yamaguchi-Shinozaki K, Carninci P, Kawai J, Hayashizaki Y, Shinozaki K. 2002. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. The Plant Journal 31, 279–292. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Lee AK, Xiang F, Park CM. 2008. Molecular and functional profiling of Arabidopsis pathogenesis-related genes: insights into their roles in salt response of seed germination. Plant and Cell Physiology 49, 334–344. [DOI] [PubMed] [Google Scholar]
- Srivastava R, Liu JX, Howell SH. 2008. Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilisin serine protease in Arabidopsis . The Plant Journal 56, 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. 2002. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana . Plant J 29, 417–426. [DOI] [PubMed] [Google Scholar]
- Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K, Narusaka Y, Narusaka M, Zhu JK, Shinozaki K. 2004. Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiology 135, 1697–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Katagiri T, Yamaguchi-Shinozaki K, Shinozaki K. 2000. An Arabidopsis gene encoding a Ca2+-binding protein is induced by abscisic acid during dehydration. Plant and Cell Physiology 41, 898–903. [DOI] [PubMed] [Google Scholar]
- Turan S, Cornish K, Kumar S. 2012. Salinity tolerance in plants: Breeding and genetic engineering. Australian Journal of Crop Science 6, 1337–1348. [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. 2000. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proceedings of the National Academy of Sciences 97, 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Xiao B, Xiong L. 2011. Identification of a cluster of PR4-like genes involved in stress responses in rice. Journal of Plant Physiology 168, 2212–2224. [DOI] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Altman A. 2003. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218, 1–14. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. 1993. The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana . Molecular Genetics and Genomics 238, 17–25. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. 1994. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. The Plant Cell 6, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiba Y, Nanjo T, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. 1999. Stress-Responsive and Developmental Regulation of Δ1-Pyrroline-5-carboxylate Synthetase 1 (P5CS1) Gene Expression in Arabidopsis thaliana . Biochemical and Biophysical Research Communications 261, 766–772. [DOI] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. 2006. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nature Protocol 1, 641–646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.