Abstract
Small peptides of the Arabidopsis GLV/RGF/CLEL family are involved in different developmental programmes, including meristem maintenance and gravitropic responses. In addition, our previous report suggested that they also participate in the formation of lateral roots. Specifically, GLV6 is transcribed during the first stages of primordium development and GLV6 overexpression results in a strong reduction of emerged lateral roots. To investigate the cause of this phenotype we analysed primordium development in gain-of-function (gof) mutants and found that GLV6 induces supernumerary pericycle divisions, hindering the formation of a dome-shaped primordium, a prerequisite for successful emergence. The GLV6 phenotype could be reproduced by ectopic expression of the gene only in xylem-pole pericycle cells. Furthermore, GLV6 seems to function at the very beginning of lateral root initiation because GLV6 excess—either gene overexpression or peptide treatment—disrupts the first asymmetric cell divisions required for proper primordium formation. Our results suggest that GLV6 acts during lateral root initiation controlling the patterning of the first pericycle divisions.
Key words: Asymmetric division, Arabidopsis thaliana, CLE-like, GOLVEN, lateral root development, primordium initiation, root growth factors, secreted peptides.
Introduction
Unlike animals, plant organs are initiated in the meristems during post-embryonic development. Meristematic cells proliferate in an undifferentiated state but also produce differentiating cell lineages that eventually give rise to new lateral organs. This process is highly regulated and relies on a complex network of genetic and hormonal cues. In the root, specific pericycle cells at the xylem pole retain the capacity to resume meristematic activity after leaving the root apical meristem (RAM) (Beeckman et al., 2001). As the main root keeps growing, some xylem pole pericycle (XPP) cells undergo a sequence of events resulting in the formation of a novel lateral root primordium (LRP), followed by the emergence of the lateral root (LR) out of the main root tissues.
The earliest event, known to date, leading to LR formation is a regular auxin pulse occurring in the upper portion of the meristematic zone (the basal meristem) that ‘primes’ pericycle cells (De Smet et al., 2007) making them competent to produce LRs. Recently a gene transcription oscillatory mechanism was found to operate in a region of the basal meristem and the elongation zone, termed the oscillation zone, to eventually establish the so-called branching points. This oscillating transcriptional mechanism is thought to define periodic LR formation (Moreno-Risueno et al., 2010). Later on, some primed XPP cells are specified as lateral root founder cells (LRFCs). These cells are usually arranged as pairs of adjacent cells present in two or three contiguous cell files and undergo a series of divisions in the maturation zone to generate a LRP (Casimiro et al., 2001).
The lateral root initiation process starts with the first division of LRFCs. Prior to this initial division, the LRFC nuclei move towards each other, then both cells divide asymmetrically in the anticlinal direction yielding two small central cells and two large peripheral cells (Casimiro et al., 2001; De Smet et al., 2008; De Rybel et al., 2010). Auxin transport and transcriptional response components are required for this process (Benkova et al., 2003; Fukaki et al., 2005; Ditengou et al., 2008; Dubrovsky et al., 2008). A second round of anticlinal divisions yields the first primordium layer. The initial anticlinal divisions generate a recognizable hallmark that is referred to as a stage I primordium (Malamy and Benfey, 1997). The division plane then changes orientation and periclinal divisions result in primordia with an additional cell layer (stage II). Subsequent anticlinal and periclinal divisions generate a dome-shaped structure that eventually emerges from the primary root. Despite extensive studies, the LR formation process (reviewed in Van Norman et al., 2013 and Atkinson et al., 2014) remains poorly understood and the signalling network that controls patterning at early stages of primordium initiation is largely uncharacterized.
Over the last two decades it became clear that small secreted peptides carry cell-to-cell signals in a wide range of plant developmental processes, including root and shoot meristem homeostasis, defense, abscission, vascular and embryonic tissue differentiation, and stomata development (reviewed in Murphy et al., 2012 and Grienenberger and Fletcher, 2015). Several peptide families have recently been implicated specifically in LR development (reviewed in Delay et al., 2013). For example, members of the INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) family, binding to the HAESA and HAESA-LIKE2 receptors, promote the separation of the outer cell layers in the main root to open the path to the emerging LR (Kumpf et al., 2013).
Signalling peptides belonging to the GOLVEN/root growth factor/CLE-like (GLV/RGF/CLEL) family are known regulators of RAM maintenance in Arabidopsis thaliana. The GLV11/RGF1, GLV5/RGF2 and GLV7/RGF3 peptides participate in RAM homeostasis via the positive regulation of the PLETHORA (PLT) 1 and 2 transcription factors at both the transcriptional and post-transcriptional levels (Matsuzaki et al., 2010). In addition, GLV/RGF/CLEL peptides are involved in root gravitropic responses since they modulate the turnover of the auxin efflux carrier PIN2 and thus control auxin fluxes in the root tip (Whitford et al., 2012). We showed in a recent report that the overexpression of several GLV genes also results in a strong decrease in emerged lateral root (ELR) density as observed independently by another research group for GLV1/CLEL6 and GLV10/CLEL8 (Meng et al., 2012). However, the precise function of GLV peptides during LR formation has yet to be defined. Furthermore, the phenotypes resulting from constitutive gene overexpression must be interpreted with caution. For example, GLV1 is only transcribed in above-ground tissues (Whitford et al., 2012) even though its constitutive overexpression alters LR development (Meng et al., 2012; Fernandez et al., 2013a ). Because the bioactive mature peptides cleaved off the precursor proteins belonging to the same family are usually highly similar, they may be recognized by receptors to which they are normally not exposed, thereby generating ectopic effects.
Our systematic transcriptional analysis of all eleven GLV genes revealed that GLV6 is the only member of the family already expressed in stage I LRPs and its overexpression resulted in the strongest LR phenotype (Fernandez et al., 2013a ). The early GLV6 transcriptional pattern and gain-of-function (gof) phenotype suggest a function during the initial steps of primordium formation. Therefore, in this report, we investigate the role of GLV6 during LR initiation.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana seedlings were grown on half-strength Murashige and Skoog medium (MS; Duchefa Biochemie B.V.) supplemented with 1% (w/v) sucrose and 1% (w/v) agarose, at pH 5.8 and germinated in a growth chamber at 22°C under continuous light conditions (100 μmol m-2 s-1). The lines GATA23pro:NLS-GFP-GUS (De Rybel et al., 2010), 35Spro:GLV6 (Fernandez et al., 2013a ), PIN1pro:PIN1-GFP (Benkova et al., 2003) and Wave 131Y, containing a plasma membrane-localized YFP (Geldner et al., 2009) have been previously described. GLV6pro::NLS-GFP-GUS plants including the GLV6 promoter driving nuclear GFP/GUS expression (Fernandez et al., 2013a ) were crossed to Wave 131Y and F1 seedlings were used to detect GLV6 transcription in the developing LRP. Enhancer trap lines are part of the Haseloff collection and were donated or ordered from NASC.
For GLV6 overexpression in different root cell layers, an Upstream Activating Sequence (UAS) was fused to the GLV6 open reading frame (ORF) via Gateway (Geldner et al., 2009) (Invitrogen) by combining the pEN-L1-GLV6-L2, PEN-L4-UAS-R1 and pB7m24GW vectors (Geldner et al., 2009). The resulting UASpro:GLV6 construct was transformed in enhancer trap lines specific for different root cell layers and T3 single locus homozygous plants were obtained. An empty vector was used as control. All constructs were also transformed in the C24 wild-type background where no transactivation occurs.
Truncated GLV6 ORFs were amplified and cloned into pDONR221 by Gateway BP reaction (Invitrogen). The GLV6∆VR amplicon flanked by Gateway attL1 and attL2 sites was generated using overlapping PCR as previously described (Atanassov et al., 2009) (see Supplementary Table S1 for primer sequences). Overexpression constructs were generated with the FAST vector (Shimada et al., 2010) which contains a GFP seed marker to select transformants. T1 GFP positive seeds were selected and grown to quantify LR density.
GLV6 artificial microRNAs (amiRglv6) were designed using the Web MicroRNA Designer (WMD1 for amiRglv6_V1: TACTACGTTACTACAACCGAT and WMD3 for amiRglv6_V2: TAAACTACGTTACTACAACCG and V3: TCTAAACGTA CGATGATCCAT) (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi). The amiRglv6 sequences were cloned into pDONR221 as described in Whitford et al. (2012) and each resulting entry clone was assembled with the GLV6 promoter (pEN-L4-GLV6pro-R1) (Fernandez et al., 2013a ) by LR reaction into the pB7m24GW-FAST (Shimada et al., 2010) binary vector. Single locus homozygous lines where the gene was silenced (Supplementary Table S2) were obtained for each amiRglv6 construct. Two lines were used for further analysis of root length showing consistent results. Lateral root density was quantified in one line per amiRglv6 construct.
Root bending experiments
For induction of lateral root initiation GATA23pro:NLS-GFP-GUS seedlings were germinated on MS medium. Four days-after-germination (dag) seedlings were placed in a chambered coverglass (Thermo Scientific Nunc) and the root tip was mechanically bent with flat tweezers between the elongation and the maturation zone. A piece of gelled MS medium, supplemented or not with a peptide, was then immediately placed on top. After bending, roots were imaged in a Zeiss LSM5 confocal microscope every 5min for 14−16h. On average, 30min elapsed from the preparation of the sample up to the first image, therefore t0 should be considered as 30min after bending.
Peptide treatments
GLV6 peptides carrying different post-translational modifications were synthesized as previously described (Whitford et al., 2012). The following peptide sequences were assayed: GLV6p SO3, DY(SO3)RTFRRRRPVHN; rGLV6p SO3, NRRY(SO3)RHRFTVDPR and GLV6p Hyp SO3, DY(SO3)RTFRRRRHypVHN [Y(SO3), sulfonated tyrosine; Hyp, hydroxyproline]. Peptide treatments at the indicated concentrations were carried out in liquid to measure ELR density or on solid media for the analysis of the first asymmetric divisions in PIN1pro:PIN1-GFP roots.
Morphological analysis
ELRs were quantified in 8−12 dag seedlings and normalized by the root length (measured with the ImageJ software) to obtain the ELR density. Quantification of total lateral roots (non-emerged and emerged) was carried out in 8−9 dag seedlings after root clearing (Malamy and Benfey, 1997). For transactivation lines, initiation sites were considered to have restricted anticlinal divisions when the primordium border could be detected within the microscope field at 40× magnification.
Microscopic analysis
Confocal images were taken on a Zeiss LSM5 or 710 confocal microscope. GFP/YFP was detected with a 488nm filter for excitation and 520nm for detection. DIC pictures were obtained with an Olympus microscope (DIC-BX51) equipped with a CAMEDIA C-3040 zoom digital camera (Olympus).
RNA extraction and qRT-PCR
Total RNA was isolated in 3−5 dag seedlings with TriReagent (Life Technologies). Residual DNA contaminants were removed with RNase-free DNase (Roche). One microgram of RNA was used as template to synthesize the first cDNA strand with the iScript cDNA Synthesis Kit (Bio-Rad). Expression levels were analysed by qRT-PCR. Reactions were performed in 384 well-plates with a LightCycler real-time thermocycler (Roche).
Statistical analysis
Statistical differences were assessed with student t-tests. All experiments were repeated two to three times with similar results. Error bars show standard errors of the mean (±SEM). Statistical significance are indicated as: *, P<0.05; **, P<0.01; ***, P<0.001.
Results
GLV6 transcription is induced in LRFCs prior to their first division
Based on the characterization of promoter-reporter lines, we have previously shown that GLV6 is transcribed in the RAM and is the first GLV transcript detected during primordium formation (Fernandez et al., 2013a ). The observation that GLV6 transcription is already detected in stage I LRPs suggests that the encoded signalling peptide is involved in the very first steps of LR initiation.
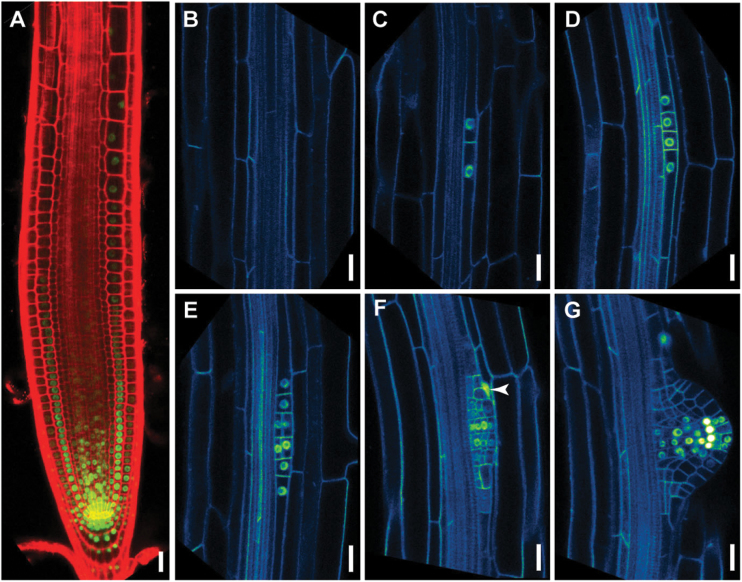
To address this hypothesis, we first examined the timing of GLV6 induction during primordium initiation in plants carrying the GLV6pro::NLS-GFP-GUS transgene, where a nuclear GFP signal indicates GLV6 transcription (Fig. 1). To visualize cell boundaries, we introduced in these plants a plasma membrane marker (Wave 131Y; Geldner et al., 2009) (Fig. 1B−G). Analysis of the reporter line in the maturation zone revealed that GLV6 is only transcribed in pericycle cells where a primordium is forming and appeared also at later stages in the endodermal cell(s) surrounding the primordium. The GFP nuclear signal was first detected in paired pericycle cells with nuclei positioned close to the common cell wall suggesting that these are LRFCs (Fig. 1C), then after the first division in all primordium stages (Fig. 1D−G). At later stages, GLV6 transcription was mainly restricted to the centre of the developing primordium (Fig. 1F, G).
Fig. 1.
GLV6 expression pattern during lateral root initiation. GLV6 transcription was detected in a GLV6pro:NLS-GFP-GUS line. (A) GLV6 transcription in the primary root. Cell walls were counterstained with propidium iodide (red). (B−G) GLV6 expression at the site of lateral root initiation in plants carrying the GLV6pro:NLS-GFP-GUS marker together with the Wave 131Y plasma membrane reporter. (B) The GLV6 signal is not detected in pericycle cells at the beginning of the differentiation zone. (C) The GLV6pro-driven GFP nuclear signal appears in founder cells prior to the first anticlinal division. (D−G) GLV6 transcription remains active throughout primordium formation and is confined to the central cells at later stages. The arrowheads point to expression in the endodermal cell adjacent to the primordium. Scale bars, 20 µm.
We conclude from this data that GLV6 transcription starts with the nuclear migration that marks LRP initiation, before the first asymmetric division, then persists throughout LRP development and extends into the overlying endodermal cells.
GLV6 mutants show defective lateral root formation
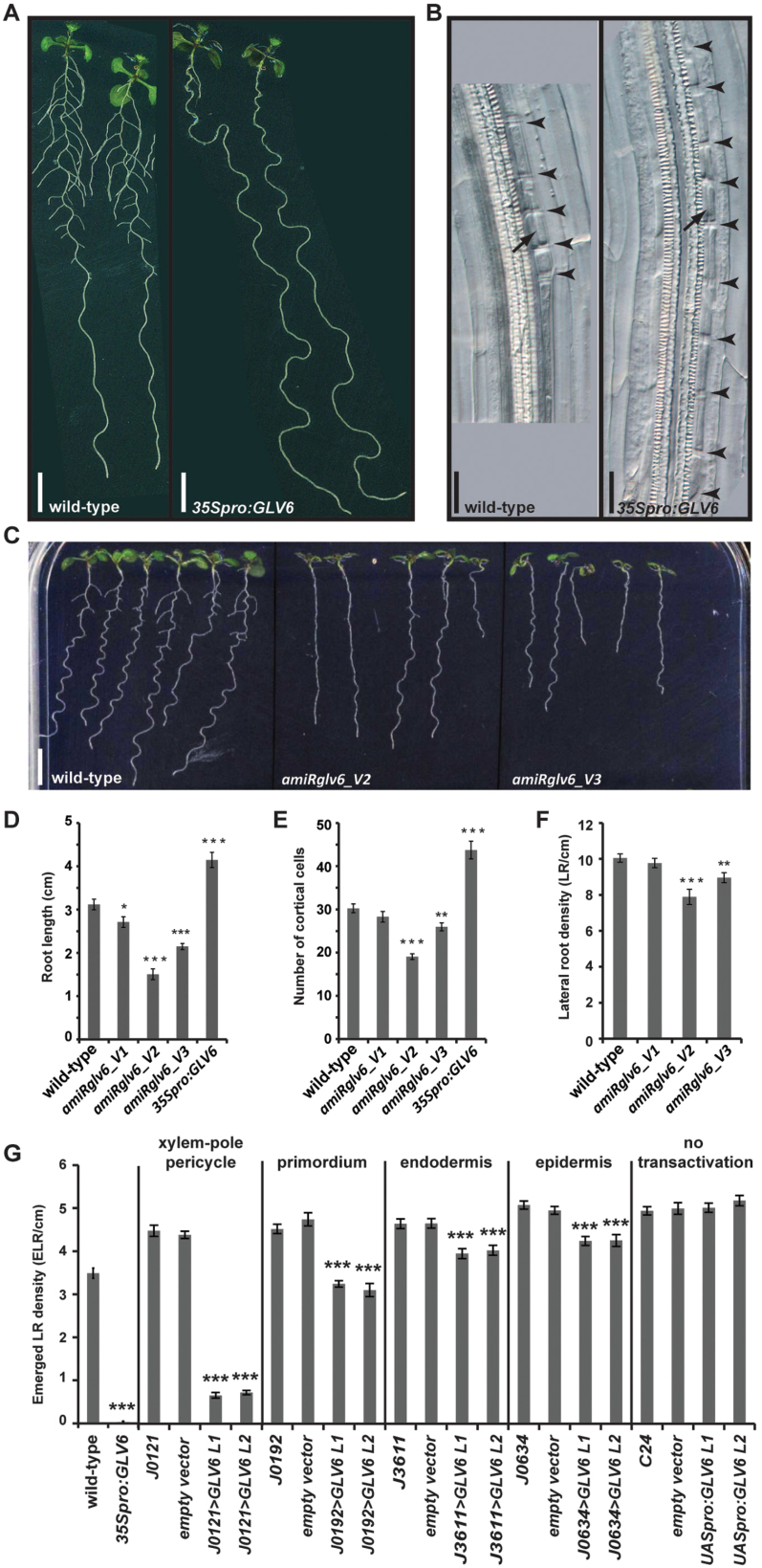
Next, we analysed the phenotype of plants that ectopically express the GLV6 gene under the control of the constitutive 35S CaMV promoter (35Spro:GLV6). We previously reported, and confirmed herein, that the 35Spro:GLV6 primary roots produced considerably fewer ELRs (Fernandez et al., 2013a ) (Fig. 2A). However, the lack of visible laterals was not caused by the absence of cellular activity. Indeed, the detailed microscopic analysis of 35Spro:GLV6 plantlets revealed that pericycle cells underwent several rounds of division at multiple sites along the differentiated primary roots (Fig. 2B). XPP cell files were characterized by excessive anticlinal divisions resulting in numerous cells along outstretched root segments. The size of these segments suggests that the borders between separate initiation sites could no longer be distinguished because the portions of the pericycle undergoing anticlinal divisions had probably expanded along the root axis and eventually merged. In addition it appeared that one single round of periclinal division produced a bilayered pericycle, but additional periclinal events were only rarely observed. Consequently, the formation of the stereotypical dome-shaped primordium did not occur, thereby preventing almost completely the normal formation of LRs and their emergence out of the primary root body.
Fig. 2.
Lateral root phenotypes in GLV6 mutants. (A) GLV6 constitutive overexpression results in high reduction of ELR. (B) GLV6 overexpression induces supernumerary pericycle cell divisions. Arrowheads and arrows point to anticlinal and periclinal divisions, respectively. (C) Phenotype of silenced GLV6 lines. Quantification of root length (D) and RAM size (E) in GLV6 silencing and overexpression lines. RAM size was determined as the number of cortical cells from the QC up to the first elongating cell (n=15−20). (F) Lateral root density in 9 dag seedlings (n=15). (G) Lateral root phenotype resulting from ectopic GLV6 expression in different root cell layers. ELR density was measured in 12 dag T3 single-locus homozygous plants where GLV6 overexpression is transactivated in different root cell layers (n=20). The UASpro:GLV6 construct or an empty vector control was transformed into different enhancer trap lines (Supplementary Fig. 1) or the C24 wild type (no transactivation). Four to seven independent GLV6 transactivation lines were obtained with similar phenotypes for each driver locus. For clarity, the data for only two lines are shown per driver (named L1 and L2). The asterisks indicate that the ELR density was significantly different (P<0.001) compared to the empty vector control. Scale bars: 0.5cm (A and C); 20 µm (B).
To further study the role of GLV6 in lateral root development, we searched for mutant lines in publicly available mutant collections. Unfortunately, no T-DNA line was found with an insertion in either the coding sequence or the promoter of the GLV6 gene. Furthermore, the tilling of an EMS-mutagenized population (Till et al., 2003; http://tilling.fhcrc.org/) did not identify alleles encoding premature stop codons or altering conserved protein sequences (data not shown).
We then silenced GLV6 expression using artificial microRNA (amiRNA) (Schwab et al., 2006). Three independent amiRNAglv6 constructs were designed and assembled under the control of the endogenous GLV6 promoter (V1−3, see ‘Materials and methods’ for details). Analysis of single locus homozygous GLV6pro:amiRglv6 lines revealed that their primary root was shorter (Fig. 2C, D). In agreement with GLV6 silencing levels (Supplementary Table S2), the loss-of-function phenotype was stronger for V2 and V3 than for V1 (Fig. 2D). The reduced root growth of amiRglv6 lines might be indicative of GLV6 function in primary root growth and is consistent with the observed GLV6 transcription pattern in QC cells and surrounding initials of the RAM (Fernandez et al., 2013a ) (Fig. 1A). Accordingly, RAM size as determined by the number of cortical cells, was significantly reduced in amiRglv6 lines compared to wild-type. In contrast, GLV6 overexpression resulted in increased root length and RAM size (Fig. 2D, E).
Considering all LRs, non-emerged and emerged, the total LR density was significantly reduced in GLV6-silenced lines V2 and V3 (Fig. 2F). Apart from that, we did not observe other defects in primordia at specific stages or in primordium shape (data not shown). That may be explained by the fact that other GLV genes are expressed at later stages (from stage II on) and could act redundantly with GLV6 (Fernandez et al., 2013a ).
Ectopic expression restricted to XPP cells is sufficient to trigger the GLV6 LR gof phenotype
The specific expression of GLV6 and the related LR overexpression phenotypes point to a function in early stages of primordium formation. However, because GLV6 is ectopically expressed in all the root tissues of 35Spro:GLV6 lines, it is difficult to conclude whether the LR gof phenotype is a direct or a secondary effect. To confirm that the observed phenotype is caused by GLV6 being ectopically transcribed in pericycle cells, we expressed the gene in different cell layers. For this purpose, the GLV6 ORF was fused to an upstream activation sequence (UAS) and the resulting UASpro:GLV6 construct was transformed in different GAL4-GFP enhancer trap lines where the GAL4 yeast transcription factor is expressed in specific tissues. The transactivation lines were selected on the basis of the GAL4-responsive green fluorescent protein (GFP) marker (Supplementary Fig. S1). Gene transcription is driven by J0121 in all XPP cells, from the elongation zone upward (Laplaze et al., 2005; Parizot et al., 2008); by J0192, only in the developing primordium, from stage I on (Laplaze et al., 2005, 2007); by J3611, in the endodermis and at a lower level in the cortex in the more mature part of the root; and by J0634, in the epidermis starting at the beginning of the differentiation zone. All lines were chosen so that no transactivation was detected in the RAM to avoid secondary effects potentially caused by expressing GLV6 in that region (Supplementary Fig. S1).
Several independent UASpro:GLV6 homozygous lines were generated for each enhancer trap line and the number of ELRs was counted as an easily quantifiable phenotype characteristic of GLV6 gof (Fig. 2G and data not shown). Transactivation within all XPP cells (J0121>GLV6) yielded a severe phenotype similar to the one previously observed in 35Spro:GLV6 roots. In J0192>GLV6 roots, the number of emerged LRs was also notably reduced although to a lesser extent than in J0121>GLV6 seedlings. J3611>GLV6 and J0634>GLV6 roots displayed a milder phenotype.
Microscopic analysis revealed massive anticlinal divisions in the J0121>GLV6 pericycle as in 35Spro:GLV6 roots. Excessive divisions were also observed in the other transactivation lines but the frequency and the length of root segments undergoing continuous anticlinal divisions decreased as GLV6 overexpression was transactivated farther away from the pericycle (Supplementary Fig S2A). This is in agreement with the observed differences in emerged lateral root number since the probability to form dome-shaped primordia that develop into mature LRs is higher when anticlinal divisions are more restricted, similar to the wild-type.
Massive anticlinal divisions prevented the distinction of neighboring initiation events that cannot thus, be easily counted, especially in J0121>GLV6 and J0192>GLV6 lines. Therefore, instead of counting initiation sites with continuous divisions we quantified those where anticlinal divisions were restricted, reasoning that these were more likely to form a functional primordium and to reflect the number of emerged LRs. Thus, fewer primordia (or initiation sites with restricted divisions) correspond to longer and more frequent regions of excessive pericycle divisions and vice versa (Supplementary Fig. S2B). As expected, only a few primordia were observed in J0121>GLV6 roots in agreement with the counted emerged LRs. J0192>GLV6 and J3611>GLV6 roots contained an intermediate number of restricted division sites, however these were usually longer in J0192>GLV6 seedlings and therefore less likely to develop into mature LRs. Excessive pericycle divisions were also observed in some J0634>GLV6 roots, but rarely compared to the other transactivation lines.
In conclusion, the phenotype was much less severe when the peptide was secreted from the epidermis, or the endodermis and cortex than from XPP cells. This observation agrees with the assumption that secreted peptides carry a molecular signal over short distances and confirms our hypothesis that the GLV6 LR gof phenotype is mostly due to ectopic expression in XPP cells. The phenotypic difference observed between J0121 and J0192 transactivation can be explained by the fact that ectopic expression is gradually turned on after LRP initiation in J0192, while it is constitutively on in XPP cells of J0121 (Supplementary Fig. S1). The ranking of the transactivation phenotypes appears to reflect a distance effect rather than differences between driver promoter activity because the phenotype strength was similar among all independent transgenic lines analysed for the same enhancer trap locus, regardless of GLV6 transcript level (Fig. 2G; Supplementary Fig S2B; Supplementary Table S2; data not shown). Consequently, we postulate that the GLV6 peptide is an autocrine signal because our data indicates that it is perceived by the LRFCs that produce it.
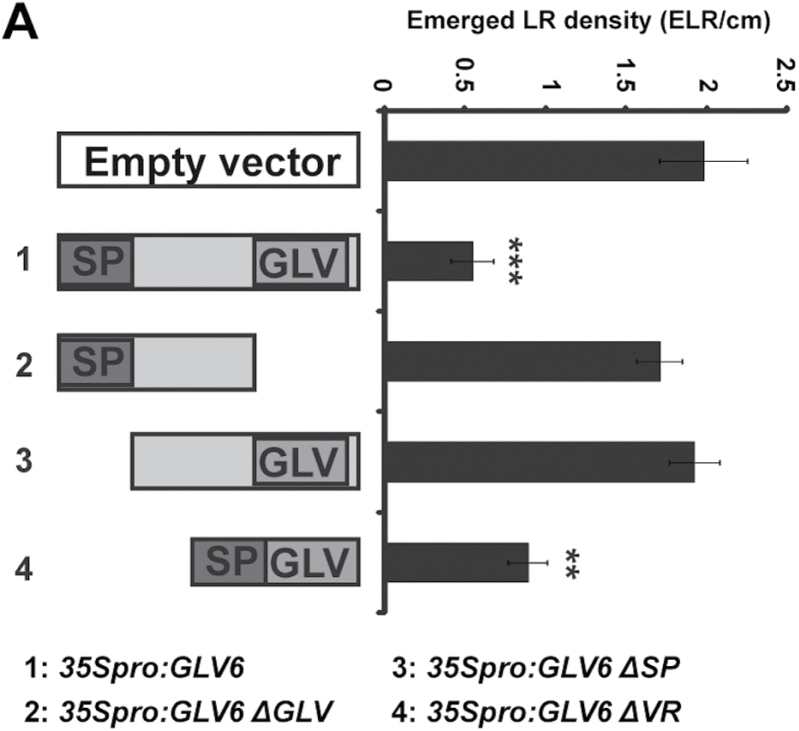
The GLV6 conserved carboxyl-terminal domain carries the bioactive secreted signal
Signalling peptides are translated as large precursors that are processed to yield the mature secreted peptides (Matsubayashi, 2011). Three domains can be distinguished in GLV precursor proteins, from the amino- to the carboxyl-terminus: a signal peptide (SP) presumably necessary for secretion, a variable region (VR), and a conserved GLV motif that defines the family (Fernandez et al., 2013b ). To confirm that the bioactive GLV6 signal peptide is encoded in the GLV motif, we overexpressed truncated versions of the GLV6 ORF in Arabidopsis plants. The tested constructs included the GLV6 sequences either (i) coding for the full precursor where the three aforementioned domains are present, (ii) lacking the region corresponding to the SP (35Spro:GLV6 Δ SP), (iii) lacking the region corresponding to the GLV motif (35Spro:GLV6 Δ GLV), or (iv) comprising a translational fusion between the SP and the GLV domains and thus where the variable region has been deleted (35Spro:GLV6 Δ VR) (Fig. 3).
Fig. 3.
The GLV6 mature peptide is contained in the C-terminus of the precursor. Figure shows the ELR density in lines overexpressing truncated GLV6 ORFs (n=12−28). SP, signal peptide; GLV, conserved C-terminal domain; VR, variable region. The asterisks indicate that the ELR density was significantly different compared to the empty vector control (**, P<0.01; ***, P<0.001).
Roots overexpressing the truncated GLV6 ORFs that lacked either the SP or GLV motif sequences produced the same number of ELRs as the control plants transformed with an empty vector (Fig. 3). However, overexpression of the GLV6 sequences corresponding to a fusion between the SP and GLV domains, without the VR, was sufficient to significantly reduce the number of ELRs. These experiments confirm that both the SP and the GLV motif are necessary and sufficient for GLV6 function during LR formation and that the mature GLV6 peptide contained in the carboxyl-terminal region of the precursor is secreted from pericycle cells to activate a signalling pathway.
GLV6 peptide application phenocopies GLV6 gof mutants
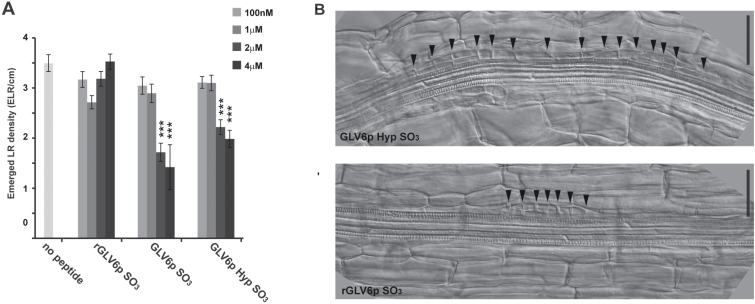
The endogenous GLV mature peptides have been identified for GLV1, GLV2, GLV3 and GLV11 (Matsuzaki et al., 2010; Whitford et al., 2012). In all four cases, the mature peptides have been found to be post-translationaly modified with sulfonation of a tyrosine and hydroxylation of a proline. In previous studies, we showed that tyrosine sulfonation is important for peptide bioactivity (Whitford et al., 2012; Fernandez et al., 2013a ).
We then investigated whether LR development can be perturbed upon treatment with a synthetic sulfonated GLV6 peptide (GLV6p SO3). A random peptide (rGLV6p SO3) containing the same amino acids but in a randomized sequence was used as control. Seedlings germinated in liquid media supplemented with GLV6p SO3 had decreased ELR density compared to untreated plants or rGLV6p SO3 controls reminiscent of the phenotype observed in GLV6 overexpression plants (Fig. 4A). Nevertheless, the reduction in emerged LR density resulting from peptide treatment, even at the highest tested concentration (4 µM), was not as strong as that of GLV6 ectopic expression (compare Fig. 2G and 4A). Since the functional role of proline hydroxylation has not been reported so far for GLV peptides, we then tested whether it could further increase peptide bioactivity. Treatments with GLV6p Hyp SO3 had the same effect as those with GLV6p SO3, indicating that, at least in exogenous synthetic GLV6 peptides, the presence of a hydroxproline residue does not enhance bioactivity (Fig. 4A). As in 35Spro:GLV6 plants, excessive anticlinal divisions were observed in early stage LRPs of roots germinated in the presence of GLV6 SO3 or GLV6p Hyp SO3 peptides (Fig. 4B and data not shown).
Fig. 4.
Treatments with GLV6 peptides phenocopy overexpression of the gene. (A) ELR density of seedlings treated with GLV6 synthetic peptides. The asterisks indicate statistically significant differences compared to the randomized rGLV6p SO3 control at the same concentration (*,P<0.05; **, P<0.01; ***, P<0.001) (n=25). (B) Treatment with GLV6 bioactive peptides induce pericycle divisions similar to those observed in GLV6 overexpression lines. Peptides were added at 1 µM. Arrowheads indicate anticlinal divisions. Scale bars, 50 µm.
Excess GLV6 activity decreases the asymmetry of the initial founder cell division
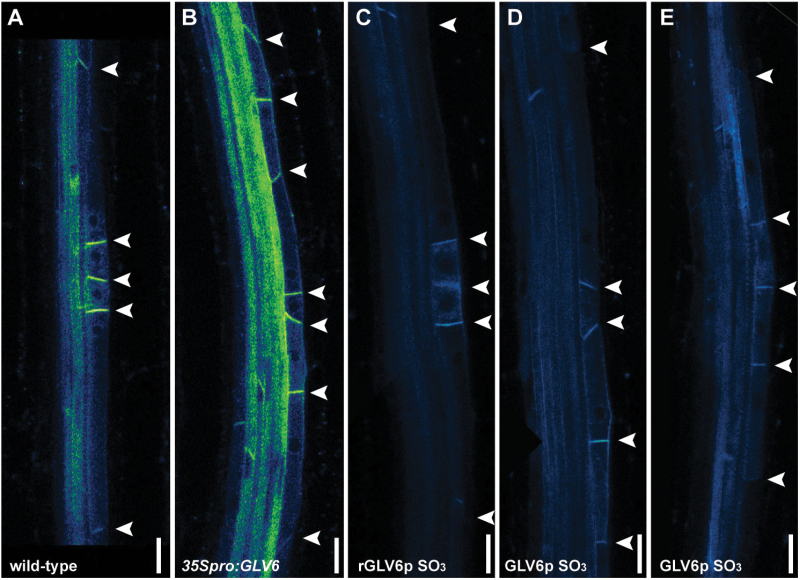
To further study GLV6 function in primordium initiation we used a PIN1pro:PIN1-GFP line as a plasma membrane marker to visualize the first divisions occurring in LRFCs since this auxin transporter is known to localize to the plasma membrane in the forming primordium (Benkova et al., 2003). In untreated PIN1pro:PIN1-GFP plants and in plants treated with a random control peptide, the first asymmetric anticlinal division was clearly visible, resulting in two small central cells and two larger flanking cells as previously reported (Fig. 5A, C). However, a remarkably different pattern was observed when these plants were either crossed to 35Spro:GLV6 (Fig. 5B) or germinated on GLV6p SO3 (Fig. 5D, E), whereby the initial anticlinal divisions yielded daughter cells of more similar sizes. Sometimes, one of the two neighbouring LRFCs divided symmetrically while the other one divided asymmetrically (Fig. 5D).
Fig. 5.
Excess GLV6 activity disrupts the first pericycle asymmetric divisions during primordium initiation. (A−E) The GFP signal in PIN1pro:PIN1-GFP plants was used as a plasma membrane marker. These plants were either crossed to GLV6 overexpression plants (A, B) and the F1 progeny was analysed or they were germinated on 1 µM of the indicated peptide (C−E). Arrowheads indicate anticlinal divisions. Scale bars, 20 µm.
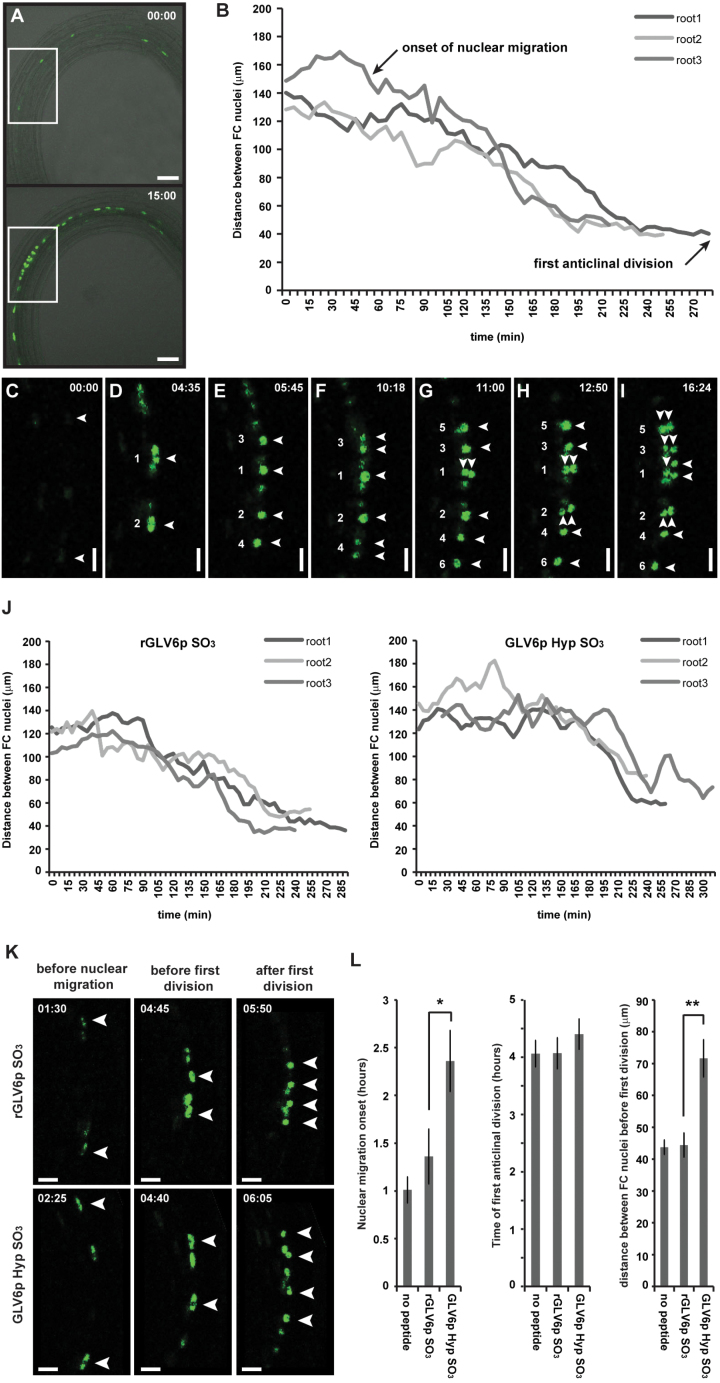
To determine how early GLV6 may function, it is necessary to detect the very first cellular events at the onset of LRP initiation. For this purpose, we used the GATA23 gene whose transcription is induced when founder cells are specified in the elongation zone. Therefore, in a GATA23pro:NLS-GFP-GUS maker line, the founder cell nuclei can already be identified before the first cellular signs of LPR initiation as marked with a GFP signal (De Rybel et al., 2010). As reported before, primordium formation can be induced by mechanical bending of the primary root (Ditengou et al., 2008; Laskowski et al., 2008). Thus, we manually bent the main root of GATA23pro:NLS-GFP-GUS seedlings and performed confocal time-lapse series for 14−16h, long enough for the appearance of a stage II primodium in the bent region (Fig. 6A).
Fig. 6.
GLV6 peptide treatment affects nuclear migration before the first division of the founder cells. (A) Stage II primordium is observed 15h after mechanical bending of the GATA23pro:NLS-GFP-GUS primary root. (B) Dynamics of LRFC nuclei represented as the distance between the two nuclei over time, from the beginning of the experiment until the first anticlinal division. Three independent roots are shown. Arrows show timing examples of the indicated events. Higher magnification of the area framed in A shows different events taking place in LRFCs during primordium initiation: (C) immediately after bending; (D) after nuclear migration; (E) first anticlinal division; (F) second anticlinal division; (G, H) first periclinal division in central nuclei; (I) second periclinal division in flanking nuclei. Notice plane change again in central nuclei. (J) Effect of peptide treatment on the onset of nuclear migration. The distance between LRFC nuclei was measured from t0 up to the first anticlinal division in the presence of 1 µM of the indicated peptides. Three independent roots are shown. (K) Representative images showing beginning and end of nuclear migration and first anticlinal division in LRFCs treated with 1 µM of the indicated peptides. (L) Quantification of events illustrated in K. Charts show average time/distance ±SEM (n=6 to 8). Time is shown as h:min. All indicated time points refer to time after the first image was taken. Nuclei were followed in six to eight movies for each condition. Scale bars: 50 µm (A); 20 µm (C−I and K).
In agreement with previous reports, we observed that during primordium initiation the nuclei of two adjacent pericycle cells migrate towards the common cell wall, then anticlinal and periclinal divisions give rise to the first stages of primordium development (Fig. 6C−I; Supplementary Movie S1). Thus, using this system, the LR initiation process can be analysed with high resolution up to stage II, including in response to peptide addition.
We then analysed the dynamics of primordium initiation without any treatment. First, we followed nuclear migration towards the common cell wall by measuring the distance between the two founder cell nuclei over time. The dynamics of nuclear migration after bending was strikingly similar between independent roots (Fig. 6B; Table 1). Parameters such as onset and end of nuclear rapprochement, as well as the final distance between the nuclei before the first anticlinal division, were highly reproducible (Fig. 6B, 6L; Table 1). Likewise, subsequent cell divisions took place at regular time intervals (Table 1). The time between the first and second anticlinal divisions was on average 4.7h (284min) (Table 1). Then, the division plane changed in central cells and the first periclinal division took place only 25min after (Fig. 6F−H; Table 1). We also remarked that the paired LRFCs are usually not exactly synchronized, and can divide a few minutes apart. In fact, this time lag is maintained in later divisions occurring in daughter cells regardless of the division plane, i.e. daughter cells of the earlier dividing founder cell keep dividing before descendants of the later dividing founder cell (Table 1). Our analysis of the initial LRP cell divisions suggests that this process is highly controlled and that it operates as a single functional module once launched.
Table 1.
Timing of cellular events following lateral root primordium initiation
| Cellular events a | No peptide | rGLV6p SO 3 | GLV6p Hyp SO 3 |
|---|---|---|---|
| Nuclei initiate migration | 61±8 | 82±18 | 142±19 |
| Nuclei stop migration | 199±20 | 206±20 | 233±13 |
| First anticlinal division | |||
| Cell position 1 | 244±14 | 244±16 | 264±16 |
| Cell position 2 | 268±15 | 272±15 | 292±22 |
| Second anticlinal division | |||
| Cell position 3 | 527±24 | 510±19 | 541±20 |
| Cell position 4 | 581±21 | 567±26 | 596±22 |
| First periclinal division | |||
| Cell position 1 | 552±28 | 551±14 | 579±25 |
| Cell position 2 | 606±28 | 614±28 | 634±34 |
a LRP initiation was triggered by root bending. Events were tracked in time-lapse image series. The position of the nuclei or cells refers to Fig. 6D, end of nuclear migration; Fig. 6E first anticlinal division; Fig. 6F, second anticlinal division; Fig. 6G and H, first periclinal division. The first dividing founder cell was given position 1. Cells with odd position numbers are descendants of the first dividing founder cell, those with even numbers of the second. No peptide (n=8); rGLV6p SO3, random sulfated GLV6 peptide (n=6); GLV6p Hyp SO3, GLV6 peptide containing a hydroxyproline and a sulfated tyrosine (n=6). Peptides were added at 1µm. Time after the first image is indicated in minutes ±SEM.
We then studied the effect of GLV6 peptide addition on the different cellular events. Seedlings were germinated on MS medium and peptides were applied to the root immediately after primary root bending (see ‘Materials and methods’). In untreated roots, the nuclei of founder cells started migrating towards each other ~1h after primary root bending and this polar movement lasted 2.3h (138min) on average (Table 1). Subsequently, nuclei stopped approaching and divided shortly after. In contrast, when LR initiation was induced in the presence of GLV6p Hyp SO3, the onset of nuclear migration was delayed by 1h compared to the rGLV6p SO3 control (Fig. 6J−L; Table 1; Supplementary Movie S2). Nevertheless, despite the late nuclear migration, founder cells in GLV6p Hyp SO3-treated roots divided approximately at the same time as in control experiments (Fig. 6K, L; Table 1). This data indicate that, at least in stages I and II, GLV6 excess does not alter the progression of the cell cycle but rather disrupts the division pattern. In addition, the distance between founder cell nuclei before the first division was larger when roots were treated with GLV6-derived peptides which is another proof of the loss of asymmetry caused by GLV6 activity (Fig. 6K, L). This result is in agreement with the defects observed in gof mutants where the wild-type division pattern is disturbed resulting in more symmetric divisions.
Discussion
LRs are formed through a process that comprises several steps, the first one known to date occurring in the basal meristem. As XPP cells differentiate, some of them undergo a series of developmental programmes to eventually give rise to a mature lateral root. Despite numerous studies on LR development, many aspects of each of these steps remain unknown. In this report, we showed that GLV6 activity could be an important factor involved in primordium initiation. GLV6 expression starts in LRFCs during the nuclear migration indicating that the GLV6 signal is produced very early, that is, before the first anticlinal division takes place. Furthermore GLV6 excess disturbs the initial asymmetric divisions.
The analysis of amiRglv6 plants revealed that the root length is shorter in agreement with a reduced number of cortical cells in the RAM, while GLV6 overexpression resulted in the opposite phenotype. This data, together with GLV6 transcription in the QC and stem cells, indicate a function for GLV6 in RAM homeostasis, possibly redundant with other GLV genes (Matsuzaki et al., 2010).
Due to the inhibition of root growth in amiRglv6 seedlings the analyses of LR formation is not straightforward. Nevertheless our results indicate a reduction in the total number of lateral roots. Our hypothesis is that a threshold level of GLV6 activity is necessary to trigger the first anticlinal asymmetric division and thus, lower GLV6 levels will result in less anticlinal divisions during LR initiation (reflected in reduced number of initiation events) while increased GLV6 levels would produce excessive anticlinal divisions and disturb the asymmetric pattern. The fact that short peptide treatments resulted in delayed nuclear migration immediately after LR induction indicates that the defects in asymmetric divisions observed in GLV6 overexpression lines is not a secondary effect. However, it is important to notice that we have no evidence of GLV6 directly affecting the nuclear movement itself and therefore the differences observed in nuclear migration upon peptide treatment can only be considered as the read out of GLV6 action in the initial events of primordium formation. The time of the first divisions after LR induction was approximately the same between untreated and GLV6p-treated roots, confirming that GLV6 addition does not disturb the onset of cell division but rather interferes with the division pattern. Based on our data, it is tempting to speculate that GLV6 might be involved in cell polarity, for example by controlling a polarizing cue. However additional insights into its signalling pathway are needed to confirm this hypothesis.
The induction of GLV6 transcription pattern coincides, spatially and temporally, with the auxin maximum that is first formed at the beginning of the LRP initiation and that is maintained at the core of the primordium throughout its development (Benkova et al., 2003). The understanding of the GLV function in LR development will therefore require a careful analysis of the potential links between peptide and auxin signalling pathways, as such connections have already been described in other developmental programmes (Whitford et al., 2012).
GLV6 is transcribed in LRFCs and GLV6 gof lines show a defect in a process taking place in the same cells. Deleting the signal peptide from the precursor results in the loss of the overexpression phenotype indicating that the peptide must be secreted. In addition, GLV6 phenotype is stronger when the gene is ectopically expressed in the pericycle compared to other cell layers. Therefore, we conclude that GLV6 is an autocrine signal. Furthermore, GLV6 could be part of a communication mechanism taking place between the two founder cells to coordinate their division pattern. Interestingly GLV6 overexpression from the adjacent endodermal cells results in a much milder gof phenotype. This could be explained in several ways: (i) the Casparian strip may filter out (part of) the peptide before it reaches the pericycle cells; (ii) the peptide secreted from the endodermis may partially diffuse away as only one side of the endodermal cells is in direct proximity with the pericycle; (iii) a GLV6 peptide gradient resulting from its secretion by the neighbouring LRFCs controls a polarity cue and thus, the asymmetry of the initial divisions in the wild type. In this case, excess of peptide perceived at one cell side (secreted from endodermis), as opposed to all cell sides (secreted from XPP), will perturb cell polarization less.
Two post-translational modifications have been described in mature GLV peptides: tyrosine sulfonation and proline hydroxylation. It is known that sulfonation increases peptide bioactivity (Whitford et al., 2012; Fernandez et al., 2013a ). In this report we showed that peptide bioactivity is not further increased by the presence of a hydroxylated proline in synthetic peptides. One possibility is that the hydroxyproline in GLV peptides is further modified with glycosylations as has been shown for other signalling peptides such as CLV3 and PSY (Amano et al., 2007; Ohyama et al., 2009). Nevertheless, despite repeated attempts to purify the native GLV6 peptide from media conditioned with GLV6 overexpression plants (Ohyama et al., 2008), we have been unable to detect any GLV6-related sequence (data not show). One reason for this failure could be that the high arginine content in the predicted mature GLV6 peptide sequence prevents peptide detection by mass spectrometry (Foettinger et al., 2006).
As we studied the role of GLV6 in the early divisions of lateral root formation, we developed a new system to track primordium initiation with high time and space resolution. Combining root mechanical bending and the GATA23pro:NLS-GFP-GUS reporter line, we were able to follow and describe the dynamics of nuclear migration and the initial divisions after LR induction. Our results indicate that the first divisions leading to primordium initiation follow a regular pattern. In another study, Lucas et al. (2013) found that cell divisions during primordium formation are not stereotypical. Although we agree that after stage II, some cell divisions may become more randomly oriented, our exhaustive analysis of LRP initiation indicates that nuclear migration, the initial anticlinal asymmetric and the first periclinal divisions are stereotypical in the wild type, at least after induction by primary root bending. Furthermore using nuclear movement as a read out, we showed that GLV6 peptide treatment interferes with the first steps of primordium initiation. We foresee that this experimental setup will also be suitable to study the influence of other factors involved in early steps of primordium development.
To conclude, we have described new aspects of the dynamics of primordium initiation and presented evidence indicating that the GLV6 peptide is an important factor controlling this process. We hope our work will form the basis for future studies of LR development and can contribute to the understanding of the role of signalling peptides in the control of plant developmental processes.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Figure S1. Enhancer trap lines used for transactivation of GLV6 expression in different root cell layers.
Supplementary Figure S2. Pericycle divisions caused by ectopic GLV6 expression in different root cell layers.
Supplementary Movie S1. Early events during primordium formation visualized with the GATA23:NLS-GFP-GUS line.
Supplementary Movie S2. Effect of GLV6 peptide addition on lateral root initiation.
Supplementary Table S1. Primers used to generate truncated GLV6 open reading frames.
Supplementary Table S2. GLV6 RNA fold change in gain- and loss-of-function mutants.
Acknowledgements
This work was funded by grants of the Interuniversity Attraction Poles Programme (IUAP P7/29 ‘MARS’) initiated by the Belgian Science Policy Office and the Research Foundation-Flanders (FWO), as well as an FWO postdoctoral fellowship (application number 1293814N).
Glossary
Abbreviations:
- dag
days-after-germination
- ELR
emerged lateral root
- gof
gain-of-function
- LRFC
lateral root founder cell
- LRP
lateral root primordium
- ORF
open reading frame
- RAM
root apical meristem
- SP
signal peptide
- UAS
Upstream Activating Sequence
- VR
variable region
- XPP
xylem pole pericycle.
References
- Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y. 2007. Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis . Proceedings of the National Academy of Sciences, USA 104, 18333–18338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassov II, Atanassov II, Etchells JP, Turner SR. 2009. A simple, flexible and efficient PCR-fusion/Gateway cloning procedure for gene fusion, site-directed mutagenesis, short sequence insertion and domain deletions and swaps. Plant Methods 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson JA, Rasmussen A, Traini R, Voss U, Sturrock C, Mooney SJ, Wells DM, Bennett MJ. 2014. Branching out in roots: uncovering form, function, and regulation. Plant Physiology 166, 538–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman T, Burssens S, Inze D. 2001. The peri-cell-cycle in Arabidopsis . Journal of Experimental Botany 52, 403–411. [DOI] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J. 2003. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. [DOI] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, et al. 2001. Auxin transport promotes Arabidopsis lateral root initiation. The Plant Cell 13, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, et al. 2010. A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Current Biology 20, 1697–1706. [DOI] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, et al. 2007. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis . Development 134, 681–690. [DOI] [PubMed] [Google Scholar]
- De Smet I, Vassileva V, De Rybel B, et al. 2008. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322, 594–597. [DOI] [PubMed] [Google Scholar]
- Delay C, Imin N, Djordjevic MA. 2013. Regulation of Arabidopsis root development by small signaling peptides. Frontiers in Plant Science 4, 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditengou FA, Teale WD, Kochersperger P, et al. 2008. Mechanical induction of lateral root initiation in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 105, 18818–18823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benkova E. 2008. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proceedings of the National Academy of Sciences, USA 105, 8790–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A, Drozdzecki A, Hoogewijs K, Nguyen A, Beeckman T, Madder A, Hilson P. 2013. a Transcriptional and functional classification of the GOLVEN/ROOT GROWTH FACTOR/CLE-like signaling peptides reveals their role in lateral root and hair formation. Plant Physiology 161, 954–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A, Hilson P, Beeckman T. 2013. b GOLVEN peptides as important regulatory signalling molecules of plant development. Journal of Experimental Botany 64, 5263–5268. [DOI] [PubMed] [Google Scholar]
- Foettinger A, Leitner A, Lindner W. 2006. Derivatisation of arginine residues with malondialdehyde for the analysis of peptides and protein digests by LC-ESI-MS/MS. Journal of Mass Spectrometry 41, 623–632. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M. 2005. Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis . The Plant Journal 44, 382–395. [DOI] [PubMed] [Google Scholar]
- Geldner N, Denervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J. 2009. Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. The Plant Journal 59, 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger E, Fletcher JC. 2015. Polypeptide signaling molecules in plant development. Current Opinion in Plant Biology 23, 8–14. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kumpf RP, Shi CL, Larrieu A, Sto IM, Butenko MA, Peret B, Riiser ES, Bennett MJ, Aalen RB. 2013. Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proceedings of the National Academy of Sciences, USA 110, 5235–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L, Benkova E, Casimiro I, et al. 2007. Cytokinins act directly on lateral root founder cells to inhibit root initiation. The Plant Cell 19, 3889–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L, Parizot B, Baker A, Ricaud L, Martiniere A, Auguy F, Franche C, Nussaume L, Bogusz D, Haseloff J. 2005. GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana . Journal of Experimental Botany 56, 2433–2442. [DOI] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, Hove CA, Hogeweg P, Maree AF, Scheres B. 2008. Root system architecture from coupling cell shape to auxin transport. PLoS Biology 6, e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Kenobi K, von Wangenheim D, et al. 2013. Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proceedings of the National Academy of Sciences, USA 110, 5229–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. 1997. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y. 2011. Small post-translationally modified peptide signals in Arabidopsis . The Arabidopsis Book. American Society of Plant Biologists. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. 2010. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis . Science 329, 1065–1067. [DOI] [PubMed] [Google Scholar]
- Meng L, Buchanan BB, Feldman LJ, Luan S. 2012. CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis . Proceedings of the National Academy of Sciences, USA 109, 1760–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN. 2010. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329, 1306–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Smith S, De Smet I. 2012. Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. The Plant Cell 24, 3198–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K, Ogawa M, Matsubayashi Y. 2008. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. The Plant Journal 55, 152–160. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. 2009. A glycopeptide regulating stem cell fate in Arabidopsis thaliana . Nature Chemical Biology 5, 578–580. [DOI] [PubMed] [Google Scholar]
- Parizot B, Laplaze L, Ricaud L, et al. 2008. Diarch symmetry of the vascular bundle in Arabidopsis root encompasses the pericycle and is reflected in distich lateral root initiation. Plant Physiology 146, 140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. 2006. Highly specific gene silencing by artificial microRNAs in Arabidopsis . The Plant Cell 18, 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Hara-Nishimura I. 2010. A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana . The Plant Journal 61, 519–528. [DOI] [PubMed] [Google Scholar]
- Till BJ, Reynolds SH, Greene EA, et al. 2003. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Research 13, 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Norman JM, Xuan W, Beeckman T, Benfey PN. 2013. To branch or not to branch: the role of pre-patterning in lateral root formation. Development 140, 4301–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford R, Fernandez A, Tejos R, et al. 2012. GOLVEN secretory peptides regulate auxin carrier turnover during plant gravitropic responses. Developmental Cell 22, 678–685. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.