Highlight
Using a genome-wide approach, the complete CLE peptide-encoding gene families of soybean and common bean were identified, characterized, and compared with those of Arabidopsis.
Key words: Autoregulation of nodulation, nitrate regulation of nodulation, plant development, plant hormone, plant peptide signalling, symbiosis
Abstract
CLE peptides are key regulators of cell proliferation and differentiation in plant shoots, roots, vasculature, and legume nodules. They are C-terminally encoded peptides that are post-translationally cleaved and modified from their corresponding pre-propeptides to produce a final ligand that is 12–13 amino acids in length. In this study, an array of bionformatic and comparative genomic approaches was used to identify and characterize the complete family of CLE peptide-encoding genes in two of the world’s most important crop species, soybean and common bean. In total, there are 84 CLE peptide-encoding genes in soybean (considerably more than the 32 present in Arabidopsis), including three pseudogenes and two multi-CLE domain genes having six putative CLE domains each. In addition, 44 CLE peptide-encoding genes were identified in common bean. In silico characterization was used to establish all soybean homeologous pairs, and to identify corresponding gene orthologues present in common bean and Arabidopsis. The soybean CLE pre-propeptide family was further analysed and separated into seven distinct groups based on structure, with groupings strongly associated with the CLE domain sequence and function. These groups provide evolutionary insight into the CLE peptide families of soybean, common bean, and Arabidopsis, and represent a novel tool that can aid in the functional characterization of the peptides. Transcriptional evidence was also used to provide further insight into the location and function of all CLE peptide-encoding members currently available in gene atlases for the three species. Taken together, this in-depth analysis helped to identify and categorize the complete CLE peptide families of soybean and common bean, established gene orthologues within the two legume species, and Arabidopsis, and provided a platform to help compare, contrast, and identify the function of critical CLE peptide hormones in plant development.
Introduction
CLAVATA/embryo surrounding region (ESR) peptide hormones (CLE peptides) are a group of post-translationally modified signal molecules involved in the regulation and differentiation of meristematic plant tissues. They have been shown to control cell divisions in the shoot apical meristem (SAM), root apical meristem (RAM), vasculature, and legume nodules (Matsubayashi, 2014; Ferguson and Mathesius, 2014; Grienenberger and Fletcher, 2015; Hastwell et al., 2015). They arise from a structurally conserved gene family and are named after the first identified CLE peptide (AtCLV3 in Arabidopsis thaliana; Fletcher et al., 1999), and the structurally and functionally similar, but unrelated, ESR peptides (first identified in Zea mays; Opsahl-Ferstad et al., 1997; Cock and McCormick, 2001).
Mature CLE peptides are typically 12–13 amino acids in length and are located at or near the C-terminus of their pre-propeptide. CLE pre-propeptides are cysteine-poor and have a tripartite domain structure, consisting of an N-terminal signal peptide, a central variable domain, and a highly conserved and functional CLE peptide domain (Matsubayashi, 2014; Hastwell et al., 2015). Some also have a fourth domain, called a C-terminal extension, which is not highly conserved, except between orthologous genes. Multi-CLE domain-containing pre-propeptides have also been identified in several plant species (Kinoshita et al., 2007; Oelkers et al., 2008), but little is known about their processing in plants. There is also a group of CLE-Like (CLEL) peptides, whose functional domain shares a similar structure but exhibits unrelated activity (Meng et al., 2012). Interestingly, one gene identified in Arabidopsis (AtCLE18) contains both a CLE and a CLEL domain (Meng et al., 2012).
The mature CLE peptide ligand is post-translationally cleaved and modified from its pre-propeptide. Hydroxylatation of proline residues is common, with one central hydroxyproline having a tri-arabinose moiety attached (Matsubayashi, 2014); however, it is important to note that all arabinose post-translational modifications identified in plants to date are limited to three peptides in A. thaliana (AtCLV3, AtCLE2, and AtCLE9) and one in Lotus japonicus (LjCLE-RS2) (Ohyama et al., 2009; Okamoto et al., 2013; Shinohara and Matsubayashi, 2013; Matsubayashi, 2014). Mature CLE peptides are ligands for leucine-rich repeat receptor kinases (LRR-RKs), with the first identified ligand receptor pair being CLV3 and CLV1 of Arabidopsis (Fletcher et al., 1999), which has since expanded to include a number of additional binding partners and associated factors (Shinohara and Matsubayashi, 2015). A comprehensive list of putative CLE ligand–LRR-RK pairs was recently presented (Endo et al., 2014).
The role of many CLE peptides remains unknown, with the majority that have been functionally characterized found in Arabidopsis. The most widely studied is AtCLV3, which acts in the SAM to regulate stem cell numbers (Fletcher et al., 1999; Gaillochet et al., 2015). Additional Arabidopsis CLE peptides acting in the root have also been characterized, including AtCLE40 (Hobe et al., 2003; Sharma et al., 2003; Stahl et al., 2009), which regulates cell proliferation in the RAM as part of a mechanism mirroring that acting in the SAM (van der Graff et al., 2009). Other root-acting CLE peptides of Arabidopsis include AtCLE1, 2, 3, 4, and 7, which are involved in nitrate-responsive mechanisms, with some also involved in lateral root development (Scheible et al., 2004; Araya et al., 2014). Additional CLE peptide-encoding genes involved in cell proliferation and differentiation include AtCLE8, which acts in embryogenesis (Fiume and Fletcher, 2012), and AtCLE45, which has been implicated in both root protophloem and pollen development (Depuydt et al., 2013; Endo et al., 2013; Rodriguez-Villalon et al., 2014). Three CLE peptides, known as tracheary element differentiation factors (TDIFs), control vascular meristematic tissue proliferation and differentiation (encoded by AtCLE41, AtCLE42, and AtCLE44; Sawa et al., 2006; Ito et al., 2006; Hirakawa et al., 2010). This group has the highest conservation amongst gymnosperms and angiosperms (Strabala et al., 2014), and consists of the only CLE peptides to begin with a histidine, rather than the archetypical arginine residue that is characteristic of all other CLE peptides (with the sole exception of AtCLE46, whose CLE domain begins with a histidine, and whose function remains unknown; Hirakawa et al., 2011).
In addition to those identified in Arabidopsis, a number of CLE peptides have been identified in various legume species. This includes CLE peptides acting to control the highly important nodulation process, which is a symbiotic relationship legumes enter into with nitrogen-fixing rhizobia bacteria (Okamoto et al., 2009, 2013; Mortier et al., 2010, 2012; Reid et al., 2011a, 2013; Ferguson et al., 2014; reviewed in Hastwell et al., 2015). By regulating nodulation, these CLE peptides essentially enable the host plant to balance nitrogen uptake from the bacteria with resource allocation to form and maintain nodules (Ferguson et al., 2010). Prominent pathways involved in this regulation are the systemic autoregulation of nodulation (AON) and the local nitrogen regulation pathways, both of which commence with the induction of CLE peptide signals (reviewed in Ferguson et al., 2010; Reid et al., 2011b ). Similarly, a number of legume CLE peptides have also been shown to respond to phosphate application (Funayama-Noguchi et al., 2011) and more recently mycorrhiza infection (Handa et al., 2015).
Aside from plants, cyst nematodes are the only other known organism to have CLE peptide-encoding genes (Mitchum et al., 2013). These genes have multiple CLE domains that are processed into a single mature peptide ligand (Chen et al., 2015). The peptides are thought to assist in nematode infection, possibly by manipulating the host to gain entry into the plant (Olsen and Skriver, 2003; Wang et al., 2005; reviewed in Mitchum et al., 2013). They are post-translationally modified and processed by the host plant’s machinery, and are perceived by plant receptors (Replogle et al., 2011; Chen et al., 2015), suggesting that they may have evolved through horizontal gene transfer.
Here, advantage was taken of recent advances in genomics and bioinformatics to identify, categorize, and functionally characterize the highly important CLE peptide families of soybean and common bean, two agriculturally important crop species. Soybean and common bean share a common ancestor whose genome duplicated ~59 million years ago (MYA), from which soybean subsequently diverged (19 MYA) and duplicated again 13 MYA (Lavin et al., 2005; Schmutz et al., 2010, 2014). As a result, 75% of soybean genes have more than one copy across the genome (a homeologous or duplicate copy; Schmutz et al., 2010, 2014; Roulin et al., 2013), whereas common bean does not. Indeed, for these reasons, soybean and common bean are commonly used for comparative and evolutionary studies in genomics and genetics (e.g. McClean et al., 2008; Lin et al., 2010; Ferguson et al. 2014; Schmutz et al., 2014).
The present investigations identified a total of 84 CLE peptide-encoding genes in soybean and 44 in common bean. In-depth sequence analyses enabled the identification of all homeologous copies within soybean, in addition to all orthologous copies existing between soybean, common bean, and Arabidopsis. Transcriptional analysis of all CLE peptide-encoding genes available in gene atlases of soybean, common bean, and Arabidopsis were evaluated to provide further insight into the localization and function of the genes. Moreover, using the complete family in soybean, seven distinct CLE peptide groups were defined based on both sequence similarity and phylogenetic analysis, with consensus sequences subsequently derived for each. Collectively, the findings provide new insight into the sequence, structure, and evolution of critical CLE peptide hormones of plants.
Materials and methods
Gene identification
To identify CLE peptide-encoding genes, multiple TBLASTN and BLASTN searches using known soybean sequences were conducted in Phytozome against the Glycine max Wm82.a2.v1 and Phaseolus vulgaris v1.0 genomes (http://www.phytozome.net/; Schmutz et al., 2010, 2014; Goodstein et al., 2012). Searches were conducted using less stringent parameters [expected threshold (E)=10] to enhance the identification of genes of interest. Results were then manually validated to confirm the presence of a CLE domain in an open reading frame. Subsequent searches based on the preliminary findings were performed using BLASTN to identify additional genes, including common bean orthologues and soybean duplicates, particularly where no duplicate/orthologue was identified in the initial queries. These subsequent searches were conducted using a slightly more stringent parameter of E=1. The open reading frames of homologous chromosome regions were also examined for potential unannotated or truncated duplicates. Additional BLASTP searches of mycorrhizal (http://genome.jgi.doe.gov/) and rhizobia genomes (Rhizobase; http://genome.microbedb.jp/rhizobase; Fujisawa et al., 2014), using both whole CLE pre-propeptide sequences and also CLE domain consensus sequences from soybean, were also performed using very low stringency (E=100) to identify CLE peptide encoding genes in these species.
Genomic environments
Synteny between genomic environments was individually obtained for each gene of interest. This was achieved using Phytozome JBrowse of the Glycine max Wm82.a2.v1, Phaseolus vulgaris v1.0, Arabidopsis thaliana TAIR10, Oryza sativa v7.0 and Medicago truncatula Mt4.0v1 genomes (http://www.phytozome.net/; Ouyang et al., 2007; Schmutz et al., 2010, 2014; Young et al., 2011; Goodstein et al., 2012; Lamesch et al., 2012). For each genomic environment investigated, the five genes located directly up- and downstream of the gene of interest were assessed for their orientation, gene family, and predicted homologues.
Sequence characterization
Clustal Omega, hosted on EMBL-EBI (http://www.ebi.ac.uk/Tools/msa/clustalo/), was used to generate multiple sequence alignments (Goujon et al., 2010; Sievers et al., 2011; McWilliam et al., 2013). Manual adjustments were subsequently made to some of the sequences predicted in Phytozome, particularly in regards to their start codon. This was based on sequence similarity to duplicate genes, similarly clustering genes, and/or likely orthologous genes, in addition to signal peptide domain prediction results.
Logo diagrams used to define consensus sequences were obtained using multiple sequence alignments for each CLE peptide group (I–VII) in Geneious Pro v6.1.8 (Kearse et al., 2012). Signal peptides were identified using the SignalP prediction program v4.1 (http://www.cbs.dtu.dk/services/SignalP/; Petersen et al., 2011). Hydrophobicity values were determined from amino acid scale values on ProtScale (http://web.expasy.org/protscale/; Gasteiger et al., 2005) using the Kyte and Doolittle (1982) hydrophobicity scale.
Phylogenetic analyses
Phylogenetic trees were constructed from multiple sequence alignments using the PHYML plugin in Geneious Pro v6.1.8 (Guindon and Gascuel, 2003). They were derived using the maximum likelihood approach with 1000 bootstraps to support a branch, with the exception of the tree designed using all soybean, common bean, and Arabidopsis sequences, where 100 bootstraps were used. Multiple trees were constructed to identify homeologous soybean genes. Those appearing to lack a homeologous copy were identified and used to re-search the genome for a potential duplicate. All trees presented here include each distinct gene identified in the numerous searches made. A similar approach was used to identify all soybean gene orthologues in common bean and Arabidopsis.
Meta-analyses of transcriptome data
Transcriptional data for the meta-analysis was collected from publicly available data sets from the Soybean RNA-Seq Atlas (http://www.soybase.org/soyseq/; Severin et al., 2010); the Soybean eFP Browser (http://bar.utoronto.ca/efpsoybean/cgi-bin/efpWeb.cgi; Libault et al., 2010a, b); A Common Bean Gene Expression Atlas (http://plantgrn.noble.org/PvGEA/index.jsp; Jamie et al. 2014); and the Arabidopsis eFP Browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Schmid et al., 2005). The entire list of gene identifiers for each species was searched in their respective databases, and only those with transcriptional data are presented. Normalized RPKM (reads per kilobase per million) values were taken where possible.
Results
Identification of CLE peptide-encoding genes in soybean and common bean, in addition to mycorrhiza and rhizobia species
To identify CLE peptide-encoding genes in soybean and common bean, a genome-wide analysis was performed involving multiple BLAST queries, followed by manual validation and the removal of false positives (i.e. no CLE domain). This resulted in the identification of 84 distinct soybean genes and 44 distinct common bean genes (Figs 1, 2; Tables 1, 2). BLAST queries were based on all known soybean CLE genes, and some Arabidopsis genes, and involved searching with both pre-propeptide and CLE domain sequences to enhance the likelihood of detecting all CLE peptide-encoding genes in the two genomes.
Fig. 1.
Multiple sequence alignment of soybean (Glycine max) CLE pre-propeptides. Homeologous copies consistently align together, as do other closely related sequences. Shading of amino acid residues represents conservation, with the darker the shading the more highly conserved the residues. The CLE domain and the leucine-rich region of the signal peptide domain exhibit the greatest degree of conservation across the entire pre-propeptide family. (This figure is available in colour at JXB online.)
Table 1.
Features of the soybean (Glycine max) CLE genes
| Name | Chromosome location | Orientation | Pre-propeptide lengtha | Predicted intron | SP cleavage siteb | Homeologue similarity (%) | Common bean orthologue | Soybean and common bean pairwise identity (%) |
|---|---|---|---|---|---|---|---|---|
| GmCLE1a | Chr11:10740675..10741635 | Reverse | 84 | Y | 23 | 82.1 | PvCLE1 | 74.6 |
| GmCLE1b | Chr12:4724973..4727049 | Reverse | 83 | Y | 23 | |||
| GmCLE2a | Chr20:46634836..46635799 | Reverse | 76 | N | 30 | 92.1 | – | – |
| GmCLE2b | Chr10:38974407..38975417 | Forward | 74 | N | 28 | |||
| GmCLE3a | Chr03:43793053..43794104 | Forward | 81 | N | 27 | 89.5 | PvCLE3 | 80.2 |
| GmCLE3b | Chr19:48528559..48529545 | Forward | 75 | N | 27 | |||
| GmCLE4a | Chr01:53094482..53095085 | Forward | 67 | N | 21 | 92.5 | PvCLE4 | 82.6 |
| GmCLE4b | Chr11:3319115..3320325 | Reverse | 67 | N | 21 | |||
| GmCLE5 | Chr08:46805591..46806636 | Reverse | 99 | N | 25 | - | PvCLE5 | 69.9 |
| GmCLE6a | Chr20:35756760..35757955 | Reverse | 97 | N | 26 | 91.8 | PvCLE6 | 76.3 |
| GmCLE6b | Chr10:49704427..49706416 | Forward | 96 | N | 26 | |||
| GmCLE7a | Chr01:5559528..5560353 | Forward | 108 | N | 23 | 89.8 | PvCLE7 | 85.8 |
| GmCLE7b | Chr02:10245905..10246706 | Reverse | 108 | N | 23 | |||
| GmCLE8a | Chr06:17294801..17295629 | Reverse | 96 | N | 21 | 83.9 | PvCLE8 | 85.4 |
| GmCLE8b | Chr04:42380768..42381923 | Forward | 95 | N | 28 | |||
| GmCLE9a | Chr05:2299498..2299782 | Forward | 79 | Y | 19 | 93.8 | PvCLE9 | 80.3 |
| GmCLE9b | Chr17:7902958..7904070 | Reverse | 79 | Y | 19 | |||
| GmCLE10a | Chr01:4182744..4185349 | Reverse | 108 | Y | 42 | 83.3 | PvCLE10 | 79.7 |
| GmCLE10b | Chr02:2311001..2311717 | Forward | 102 | Y | 40 | |||
| GmCLE11a | Chr14:7781256..7782013 | Reverse | 82 | N | 27 | 89.3 | PvCLE11 | 65.4 |
| GmCLE11b | Chr17:39269471..39270222 | Forward | 84 | N | 27 | |||
| GmCLE12a | Chr13:16671710..16673786 | Forward | 97 | Y | 34 | 94.8 | PvCLE12 | 93.1 |
| GmCLE12b | Chr19:1819967..1821863 | Rreverse | 97 | Y | 34 | |||
| GmCLE13 | Chr13:36676213..36676962 | Forward | 86 | Y | 24 | – | PvCLE13 | 73.8 |
| GmCLE14 | Chr10:46589943..46590137 | Forward | 83 | N | 25 | – | PvCLE14 | 72.7 |
| GmCLE15a | Chr10:46586624..46587350 | Forward | 86 | N | 25 | 51.1 | PvCLE15a, PvCLE15b, PvCLE15c, PvCLE15d | 48.3, 47.9, 45.4, 45.6 |
| GmCLE15b | Chr06:27528956..27529216 | Forward | 86 | N | 26 | |||
| GmCLE16a | Chr09:34804635..34806006 | Forward | 86 | N | 27 | 90.7 | PvCLE16 | 85.7 |
| GmCLE16b | Chr16:35643819..35644747 | Forward | 86 | N | 27 | |||
| GmCLE17a | Chr05:38846465..38847260 | Reverse | 87 | N | 28 | 86.2 | PvCLE17 | 85.1 |
| GmCLE17b | Chr08:969117..970012 | Reverse | 87 | N | 24 | |||
| GmCLE18a | Chr13:21801637..21802409 | Forward | 85 | N | 19 | 85.9 | PvCLE18 | 80.8 |
| GmCLE18b | Chr17:4258185..4258436 | Reverse | 83 | N | 19 | |||
| GmCLE19a | Chr07:39333907..39334972 | Forward | 119 | N | 32 | 83.9 | PvCLE19 | 67.0 |
| GmCLE19b | Chr20:1750676..1751787 | Forward | 114 | N | 32 | |||
| GmCLE20a | Chr03:33954213..33955592 | Forward | 100 | N | 36 | 91.0 | PvCLE20 | 78.9 |
| GmCLE20b | Chr19:38764138..38765477 | Forward | 94 | N | 31 | |||
| GmCLE21a | Chr02:46067116..46071548 | Forward | 81 | N | 26 | 88.9 | PvCLE21 | 75.4 |
| GmCLE21b | Chr14:2730030..2731670 | Reverse | 80 | N | 26 | |||
| GmCLE22a | Chr07:41652868..41653137 | Reverse | 89 | N | 27 | 91.0 | PvCLE22 | 74.0 |
| GmCLE22b | Chr20:7721313..7721576 | Reverse | 87 | N | 27 | |||
| GmCLE23a | Chr02:45459965..45460989 | Reverse | 73 | N | 23 | 85.9 | PvCLE23 | 79.0 |
| GmCLE23b | Chr14:3533265..3534446 | Forward | 71 | N | 21 | |||
| GmCLE24a | Chr10:43660111..43661108 | Forward | 110 | N | 23 | 88.4 | PvCLE24 | 82.9 |
| GmCLE24b | Chr20:42379994..42380805 | Reverse | 111 | N | 23 | |||
| GmCLE25a | Chr05:1295698..1296578 | Forward | 118 | N | 29 | 80.0 | PvCLE25 | 68.8 |
| GmCLE25b | Chr17:9746590..9748712 | Forward | 114 | N | 29 | |||
| GmCLE26 | Chr20:2984627..2986271 | Forward | 99 | N | 27 | – | PvCLE26 | 52.6 |
| GmCLE27a | Chr02:11156483..11156827 | Reverse | 114 | N | 30 | 83.3 | PvCLE27 | 78.6 |
| GmCLE27b | Chr01:7300791..7302992 | Reverse | 107 | N | 30 | |||
| GmCLE28a | Chr13:37349043..37349282 | Reverse | 83 | N | 27 | 80.3 | PvCLE28 | 69.4 |
| GmCLE28b | Chr12:38835186..38835383 | Reverse | 65 | N | 26 | |||
| GmCLE29a | Chr12:27615321..27615566 | Forward | 82 | N | 26 | 92.8 | PvCLE29 | 84.3 |
| GmCLE29b | Chr06:36330866..36331117 | Reverse | 83 | N | 26 | |||
| GmCLE30a | Chr06:36324860..36325095 | Reverse | 78 | N | 22 | 61.5 | PvCLE30 | 60.5 |
| GmCLE30b | Chr06:36255159..36255402 | Reverse | 81 | N | 26 | |||
| GmCLE31a | Chr07:37351348..37351668 | Forward | 106 | N | 22 | 92.5 | – | – |
| GmCLE31b | Chr13:28570341..28570661 | Reverse | 106 | N | 22 | |||
| GmCLE32 | Chr13:28559073..28559703 | Reverse | 68 | N | 23 | – | – | – |
| GmCLE33a | Chr06:36402219..36402452 | Reverse | 78 | N | 23 | 84.4 | PvCLE33 | 66.3 |
| GmCLE33b | Chr12:27380684..27380911 | Forward | 76 | N | 24 | |||
| GmCLE34a | Chr12:38840660..38840902 | Reverse | 81 | N | 22 | 88.9 | PvCLE34 | 78.6 |
| GmCLE34b | Chr13:37353930..37354172 | Reverse | 81 | N | 22 | |||
| GmCLE35 | Chr13:28564185..28564418 | Reverse | 78 | N | 23 | – | PvCLE35 | 70.5 |
| GmCLE36a | Chr13:34350525..34350935 | Reverse | 76 | N | 24 | 83.1 | – | – |
| GmCLE36b | Chr15:6162182..6162415 | Forward | 77 | N | 25 | |||
| GmCLE37a | Chr16:4533525..4534140 | Forward | 185 | Y | 18 | 40.8 | – | – |
| GmCLE37b | Chr19:35239153..35240209 | Reverse | 190 | Y | 24 | |||
| GmCLE40a | Chr12:3979297..3980162 | Forward | 82 | Y | 23 | 40.0 | PvCLE40 | 47.9 |
| GmCLE40b | Chr11:9961342..9961800 | Forward | 35 | N | - | |||
| GmCLV3a | Chr12:34902722..34903650 | Forward | 105 | Y | 28 | 93.3 | PvCLV3 | 91.1 |
| GmCLV3b | Chr13:40867356..40867942 | Reverse | 105 | Y | 29 | |||
| GmNIC1a | Chr12:36837550..36838464 | Forward | 80 | N | 22 | 86.3 | PvNIC1 | 75.9 |
| GmNIC1b | Chr13:39224711..39225630 | Reverse | 79 | N | 22 | |||
| GmRIC1a | Chr13:39215403..39216108 | Reverse | 95 | N | 28 | 77.3 | PvRIC1 | 68.8 |
| GmRIC1b | Chr12:36848528..36849475 | Forward | 96 | N | 27 | |||
| GmRIC2a | Chr06:47247215..47248215 | Reverse | 93 | N | 26 | 87.2 | PvRIC2 | 74.5 |
| GmRIC2b | Chr12:13187190..13187511 | Forward | 94 | N | 26 | |||
| GmTDIF1a | Chr07:41652868..41653137 | Reverse | 104 | N | 42 | 92.4 | PvTDIF1 | 82.5 |
| GmTDIF1b | Chr18:40563162..40564249 | Reverse | 104 | N | 41 | |||
| GmTDIF2a | Chr05:32724420..32724761 | Reverse | 113 | N | 28 | 92.2 | PvTIDF2 | 87.9 |
| GmTDIF2b | Chr08:6781787..6783296 | Reverse | 113 | N | 28 | |||
| GmTDIF3a | Chr09:4193781..4194815 | Forward | 125 | N | 31 | 76.7 | PvTDIF3 | 68.6 |
| GmTDIF3b | Chr15:13038523..13039541 | Forward | 127 | N | 29 |
a Number of amino acid residues.
b After amino acid number listed.
Listed are the genetic location, pre-propeptide length, predicted intron presence, gene orientation, soybean and common bean homologue, pre-propeptide similarity (%). and SignalP signal peptide (SP) cleavage site.
Table 2.
Features of the common bean (Phaseolus vulgaris) CLE genes
| Name | Phytozome v10 ID | Pre-propeptide lengtha | Predicted intron | Chromosome location | Orientation | Oelkers et al. (2008) | uniprot.org |
|---|---|---|---|---|---|---|---|
| PvCLE1 | Phvul.011G065200 | 96 | Y | Chr11:5675757..5676469 | Reverse | – | XP_007132079 |
| PvCLE3 | Phvul.006G092600 | 99 | Y | Chr06:21113605..21114127 | Forward | PvCLE169 | XP_007147057 |
| PvCLE4 | Phvul.002G008500 | 67 | N | Chr02:960456..961284 | Reverse | – | XP_007156683 |
| PvCLE5 | Phvul.003G035700 | 121 | N | Chr03:3588969..3589711 | Forward | – | XP_007153443 |
| PvCLE6 | Phvul.007G027300 | 94 | Y | Chr07:2049797..2054614 | Reverse | PvCLE176 | XP_007142910 |
| PvCLE7 | Phvul.002G085300 | 108 | N | Chr02:13297480..13297806 | Forward | – | XP_007157625 |
| PvCLE8 | Phvul.009G187200 | 95 | N | Chr09:27684592..27685489 | Forward | – | XP_007138182 |
| PvCLE9 | Phvul.003G190100 | 95 | N | Chr03:40210422..40210709 | Forward | – | XP_007155310 |
| PvCLE10 | Phvul.002G079000 | 101 | Y | Chr02:11819569..11820862 | Reverse | – | XP_007157554 |
| PvCLE11 | Phvul.001G025500 | 77 | N | Chr01:2309373..2309606 | Reverse | – | XP_007160889 |
| PvCLE12 | Phvul.004G023800 | 108 | Y | Chr04:2459046..2460734 | Reverse | – | XP_007151170 |
| PvCLE13 | Phvul.005G069900 | 102 | Y | Chr05:11484552..11485119 | Reverse | – | XP_007149431 |
| PvCLE14 | Phvul.007G068800 | 88 | N | Chr07:6196473..6196739 | Reverse | – | XP_007143392 |
| PvCLE15a | Phvul.007G068400 | 85 | N | Chr07:6165176..6165433 | Reverse | – | XP_007143388 |
| PvCLE15b | Phvul.007G068500 | 83 | N | Chr07:6181155..6181406 | Forward | – | XP_007143389 |
| PvCLE15c | Phvul.007G068600 | 87 | N | Chr07:6184216..6184479 | Reverse | – | XP_007143390 |
| PvCLE15d | Phvul.007G068700 | 84 | N | Chr07:6189914..6190168 | Forward | – | XP_007143391 |
| PvCLE16 | Phvul.004G117600 | 86 | N | Chr04:38385127..38385862 | Forward | – | XP_007152295 |
| PvCLE17 | Phvul.002G287300 | 97 | N | Chr02:45090923..45091742 | Reverse | – | XP_007160038 |
| PvCLE18 | Phvul.003G137800 | 85 | N | Chr03:33013056..33013313 | Reverse | – | XP_007154669 |
| PvCLE19 | Phvul.002G095900 | 104 | Y | Chr02:17549689..17550064 | Forward | – | XP_007157755 |
| PvCLE20 | Phvul.001G120900 | 92 | N | Chr01:34104465..34105721 | Forward | – | XP_007162068 |
| PvCLE21 | Phvul.008G203000 | 88 | N | Chr08:51319273..51319539 | Forward | – | XP_007141519 |
| PvCLE22 | Phvul.006G016000 | 90 | N | Chr06:7671543..7672241 | Reverse | - | XP_007146145 |
| PvCLE23 | Phvul.008G211300 | 74 | N | Chr08:52313956..52316136 | Forward | – | XP_007141620 |
| PvCLE24 | Phvul.007G101800 | 109 | N | Chr07:11339237..11339566 | Reverse | – | XP_007143789 |
| PvCLE25 | Phvul.003G177600 | 110 | N | Chr03:38979082..38979719 | Forward | – | XP_007155150 |
| PvCLE26 | Phvul.002G168200 | 85 | Y | Chr02:31082684..31084138 | Reverse | – | XP_007158622 |
| PvCLE27 | Phvul.002G081400 | 106 | N | Chr02:12270950..12272253 | Reverse | – | XP_007157583 |
| PvCLE28 | Phvul.005G067900 | 83 | N | Chr05:10636536..10636787 | Reverse | – | XP_007149409 |
| PvCLE29 | Phvul.011G160600 | 81 | N | Chr11:42316953..42317385 | Forward | – | XP_007133207 |
| PvCLE30 | Phvul.011G160700 | 82 | N | Chr11:42325813..42326352 | Forward | – | XP_007133208 |
| PvCLE31 | Chr01: 14906066..14906353 | 95 | N | Chr01: 14906066..14906353 | Forward | – | – |
| PvCLE33 | Chr11:42291102..42291350 | 82 | N | Chr11:42291102..42291350 | Reverse | - | - |
| PvCLE34 | Chr05:10644869..10645097 | 75 | N | Chr05:10644869..10645097 | Reverse | – | – |
| PvCLE35 | Phvul.003G057900 | 75 | N | Chr03:7610340..7610764 | Forward | – | XP_007153705 |
| PvCLE40 | Phvul.011G056800 | 114 | Y | Chr11:4877577..4878010 | Forward | – | XP_007131981 |
| PvCLV3 | Phvul.005G120600 | 104 | Y | Chr05:34343926..34344486 | Reverse | – | XP_007150035 |
| PvNIC1 | Phvul.005G097000 | 80 | N | Chr05:28793851..28794118 | Reverse | – | XP_007149764 |
| PvRIC1 | Phvul.005G096900 | 115 | Y | Chr05:28775368..28775758 | Reverse | – | – |
| PvRIC2 | Phvul.011G135900 | 93 | N | Chr11:30985821..30986626 | Reverse | – | XP_007132915 |
| PvTDIF1 | Phvul.008G124100 | 118 | N | Chr08:17187233..17187933 | Forward | – | XP_007140575 |
| PvTDIF2 | Phvul.002G187400 | 108 | N | Chr02:34265616..34266385 | Forward | – | XP_007158853 |
| PvTDIF3 | Phvul.009G244400 | 115 | N | Chr09:35772334..35773004 | Reverse | – | XP_007138869 |
a Number of amino acid residues.
Listed are the genetic location, pre-propeptide length, and predicted intron presence.
The identified genes are scattered across the genomes, with at least one located on every chromosome, except for chromosome 10 of common bean. Chromosome 13 of soybean contains the most CLE peptide-encoding genes, with a total of 12. Most of the identified genes lack predicted introns, with the exception of 12 soybean genes and nine common bean genes (Tables 1, 2).
Many of the genes identified here had not been discovered previously and therefore had not yet been assigned a name. In contrast, those which were previously reported had as many as five different aliases. To unify the nomenclature, designations were assigned based on the names of all previously characterized soybean CLE peptides (e.g. Cock and McCormick, 2001; Reid et al., 2011a ; Wong et al., 2013), and the Arabidopsis phylogenetic approach was used for all non-characterized genes (Cock and McCormick, 2001). The duplicated nature of the soybean genome was also accounted for by identifying a and b copies of homeologous gene pairs (described below). In common bean, the gene names were assigned based on their orthologue in soybean (Table 1; Supplementary Fig. S1 available at JXB online). A comprehensive list of all soybean and common bean names, including all previous identifiers, is provided in Supplementary Table S1.
Aside from plants, cyst nematodes are the only known organisms to possess CLE peptide-encoding genes (Mitchum et al., 2013). These peptides appear to assist in parasitism of the host. To determine whether mutualistic symbiotic organisms also encode for CLE peptides that assist in infection, a protein search of mycorrhiza (http://genome.jgi.doe.gov/) and rhizobia (Rhizobase; http://genome.microbedb.jp/rhizobase; Fujisawa et al. 2014) species was conducted using CLE domain consensus sequences and also pre-propeptide sequences. This thorough search yielded the identification of no CLE peptide-encoding genes in these organisms.
Identification of homeologues and orthologues in soybean and common bean
To characterize their amino acid sequences, all identified CLE peptide-encoding genes were translated and successive multiple sequence alignments were conducted using entire CLE pre-propeptide sequences. Despite having large variable domains, the pre-propeptides grouped strongly according to their CLE domain sequence in both soybean (Fig. 1) and common bean (Fig. 2). This helped in identifying likely homeologous (duplicate) copies of genes in the palaeopolyploid genome of soybean, with 39 pairs identified compared with only six genes having no duplicate (Fig. 1; Table 1). The six genes lacking a duplicate were re-blasted against the soybean genome to confirm their lack of a duplicate, and their homeologous chromosome region was checked for unannotated genes. The presence of a common bean orthologue confirmed they were not triplicated within the soybean genome.
Fig. 2.
Multiple sequence alignment of common bean (Phaseolus vulgaris) CLE pre-propeptides. Related sequences tend to align closer together. Shading of amino acid residues represents conservation, with the darker the shading the more highly conserved the residues. As with the soybean prepropeptides shown in Fig. 1, the CLE domain and the leucine-rich region of the signal peptide domain exhibit the greatest degree of conservation across the entire pre-propeptide family. (This figure is available in colour at JXB online.)
To identify likely orthologues between soybean and common bean, an additional multiple sequence alignment was produced using the CLE peptide-encoding gene families of both species (data not shown). This alignment was also useful in confirming the 39 homeologous gene pairs of soybean. As expected, all previously reported gene orthologues of soybean and common bean clustered together (e.g. RIC, NIC; Ferguson et al., 2014). Additional orthologue candidates also clustered; however, soybean has four homeologous gene pairs and one individual gene lacking an apparent duplicate that appear to have no orthologue in common bean (GmCLE2a and b; GmCLE31a and b; GmCLE32; GmCLE36a and b; and GmCLE37a and b; Table 1).
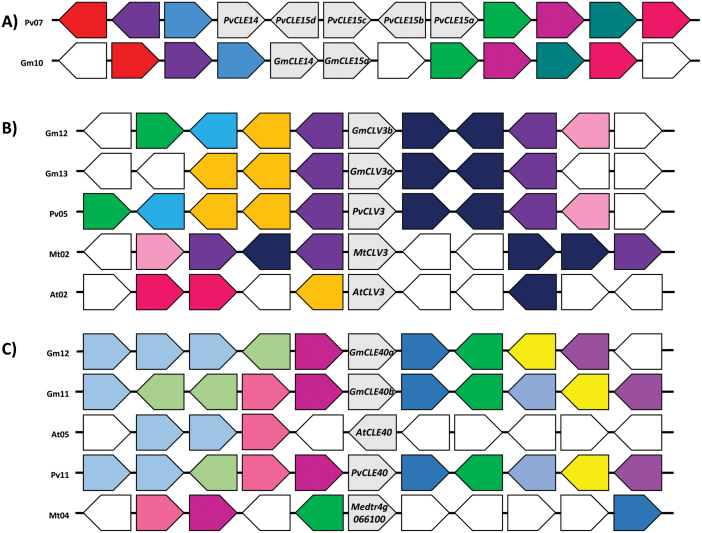
When identifying gene orthologues, it was noticed that three of the 44 genes identified in common bean did not have an apparent orthologue in soybean (Table 1; Supplementary Fig. S1 at JXB online). These genes are all part of a group of four tandemly duplicated genes located on chromosome 7, called PvCLE15a, b, c, and d, and thus can all be considered orthologous to the same genes in soybean, GmCLE15a and b. This indicates that the tandem duplication occurred in common bean after it diverged ~19 MYA from soybean. Directly upstream of these tandemly duplicated genes and adjacent to PvCLE15d is another CLE peptide-encoding gene, PvCLE14 (Fig. 3A). This tandem duplication also occurs in soybean (GmCLE14 and GmCLE15a) and thus must have occurred prior to the two species diverging.
Fig. 3.
Genomic environment of PvCLE15 tandemly duplicate genes of common bean, and the CLV3 and CLE40 genes of different species. The genes of interest are positioned centrally and shaded in grey. Species and chromosome number are indicated to the left of each genomic segment. Surrounding genes similar in putative function are indicated by the same colour and genes with unrelated putative functions are uncolored. The direction of the arrow represents the orientation of the gene compared with that of the CLE gene. (A) Common bean chromosome 7 containing a tandem gene duplication not found on the orthologous region of soybean on chromosome 10. Orthologues of (B) CLV3 and (C) CLE40 in soybean, common bean, Arabidopsis, and M. truncatula. A high level of genetic synteny is shown here for each of these CLE genes.
Two additional sets of genes occur in tandem in common bean: PvCLE29 and PvCLE30, and PvNIC1 and PvRIC1. In soybean, the NIC1 and RIC1 genes also occur in tandem, suggesting that this duplication occurred prior to the divergence of soybean and common bean. However, due to the whole-genome duplication, soybean has homeologous regions that include these genes, resulting in two tandem repeats: GmNIC1a and GmRIC1b on chromosome 12 and GmNIC1b and GmRIC1a on chromosome 13.
Manual adjustments were made to some coding sequences predicted in Phytozome regarding the placement of their start codon. These adjustments were based on sequence similarity to their duplicate gene, to clustering sequences in common bean (i.e. probable orthologues), and/or to signal peptide domain prediction results (described below). In total, eight soybean sequences were trimmed slightly to place their start codon downstream of where it was predicted in Phytozome (GmCLE10b, GmCLE16b, GmCLE21b, GmCLV3b, GmTDIF1a, GmTDIF1b, GmRIC1a, and GmRIC2b). An additional five sequences were extended to include a start codon slightly upstream of that predicted in Phytozome (GmCLE3a, GmCLE16a, GmCLE20a, GmCLE27a, and GmCLE28a).
Characterization of CLE pre-propeptides in soybean and common bean
CLE pre-propeptides typically consist of a signal peptide, a variable domain, and a CLE domain, with some also having a C-terminal extension (Hastwell et al., 2015). All of the CLE pre-propeptides identified here have this structure. Moreover, they are rich in lysine (11.4%) and serine (11.3%), and are notably poor in cysteine (1.3%), tyrosine (1.3%), and tryptophan (0.7%; often poorly represented in plants) (Supplementary Table S2 at JXB online), which is typical amongst CLE peptides (Hastwell et al., 2015). The length of the CLE pre-propeptides varies, with the smallest being 67 residues in both soybean and common bean (excluding likely pseudogenes reported below), and the longest being 127 and 121 residues, respectively. Some contain histidine repeats in their variable domain, but this does not correlate with sequence length.
The signal peptide located at the N-terminus of the pre-propeptide is typically hydrophobic and is responsible for exporting the propeptide from the cell (Rojo et al., 2002). Hydrophobicity analysis confirmed that the signal peptide is the most hydrophobic region of the CLE pre-propeptides investigated here, whereas the remaining propeptide is more hydrophilic, as determined by Kyte and Doolittle (1982) scores (Supplementary Fig. S2 at JXB online). Indeed, 61.4% of the amino acid residues occurring in the signal peptide domain are hydrophobic (Supplementary Fig. S2). SignalP prediction software was used to determine the putative cleavage site of the signal peptide (Table 1). Using these predicted signal peptide sequences, a multiple sequence alignment and phylogenetic tree was constructed that showed less conserved and confident groupings (data not shown) compared with entire pre-propeptides. One pre-propeptide, GmCLE40b, is not predicted to have a signal peptide, as it is truncated and only 34 amino acids in length (Table 1; Fig. 1).
Directly following the signal peptide domain in the pre-propeptide is the variable domain. This region only shows conservation between homeologous and/or orthologous genes (Figs 1, 2). However, the final residue of the variable domain positioned directly before the CLE domain is commonly a lysine (48.4%), with asparagine (13.9%), glutamic acid (9.0%), alanine (7.4%), and histidine (5.7%) as the next four highest represented amino acids at this position.
The CLE domain represents the region of the pre-propeptide that is cleaved and modified to become the functional CLE peptide product. Of the 126 CLE peptide-encoding genes of soybean and common bean, there are 54 unique CLE domain sequences that are 12 amino acids in length (with 44 of 82 in soybean and 40 of 44 in common bean). This number increases to 60 sequences if 13 amino acids are taken into account. All mature CLE peptides that have been biochemically confirmed to date have been 13 amino acids in length (Ohyama et al., 2009; Shinohara et al., 2012; Okamoto et al., 2013; Chen et al., 2015); however, only 54.8% of the pre-propeptide CLE sequences of soybean and common bean have a residue in position 13, with the others having a stop codon preventing them from being any more than 12 amino acids in length.
Sequence similarity within the CLE pre-propeptides of soybean and common bean is highest in the CLE domain (Figs 1, 2). There is no 100% conserved residue, although position 12 has a highly conservative histidine/asparagine substitution. The least conserved residues are at position 2 (15.8% pairwise identity) and position 5 (19.7% pairwise identity). Of the critical residues previously identified in the CLE domain (e.g. Ni et al., 2011; Reid et al., 2013), position 1 is predominantly arginine, or, in some cases, histidine (i.e. TDIF peptides). An additional group has threonine at position 1 (GmCLE16a, GmCLE16b, and PvCLE16). Three others that group together have valine, lysine, and leucine residues at this position (PvCLE15a, PvCLE15d, and GmCLE15b, respectively; Figs 1, 2), which includes two of the four common bean genes that are tandemly duplicated (described above). Position 7, which is often post-translationally modified, is predominately a proline. However, there are 10 soybean homeologues and five associated common bean orthologues where a serine (CLE7; CLE8; CLE11 and CLE23 orthologous) or alanine (CLE4 orthologues) is in that position. Interestingly, soybean has six pairs (i.e. 12 genes) of homeologous CLE peptide-encoding genes that have a mismatch within their CLE domain as a result of naturally occurring mutations (Fig. 1). The impact of amino acid changes on the function and activity of various Arabidopsis and legume CLE pre-propeptides was recently reviewed (Hastwell et al., 2015).
Some CLE pre-propeptides contain a fourth domain directly following the CLE domain, called the C-terminal extension. The precise function of this domain remains unclear. Only 32.5% of the CLE pre-propeptides in soybean and common bean have this domain, similar to the CLE pre-propeptide family of A. thaliana (31.3%; Cock and McCormick, 2001). The only prevalent feature of the C-terminal extension appears to be the common presence of proline (19.5%). Indeed, the sequence is highly variable in length and amino acid residues, except between homeologous and/or orthologous genes (Fig. 1). Interestingly, the domain is present in 83.3% of the CLE genes that contain a predicted intron. It is also present in CLV3 orthologues and in almost all rhizobia-induced nodulation-suppressing CLE peptides (with the exception of MtCLE12; Hastwell et al., 2015).
Pseudogenes and multi-CLE peptide-encoding genes of soybean and common bean
Due to insertion, duplication, and deletion events, some of the CLE peptide-encoding genes identified here do not fit the common tripartite domain structure. For example, in soybean, GmCLE28b, GmCLE30b, and GmCLE40b are all probably pseudogenes. GmCLE28b and GmCLE40b have nonsense mutations that result in a truncation prior to the CLE domain. However, the sequences downstream of these mutations align closely to GmCLE28a and GmCLE40a, respectively. GmCLE30b has low conservation in the CLE domain after residue five, when compared with its duplicate, GmCLE30a. This appears to be due to a deletion event causing a frameshift directly in the CLE domain. It is likely that none of these three pseudogenes genes produces a functional CLE peptide. They have been denoted as the b copy, consistent with the RIC, NIC, and CLV3 genes, where the b copy may not be transcribed/functional (Reid et al., 2011a ; Wong et al., 2013).
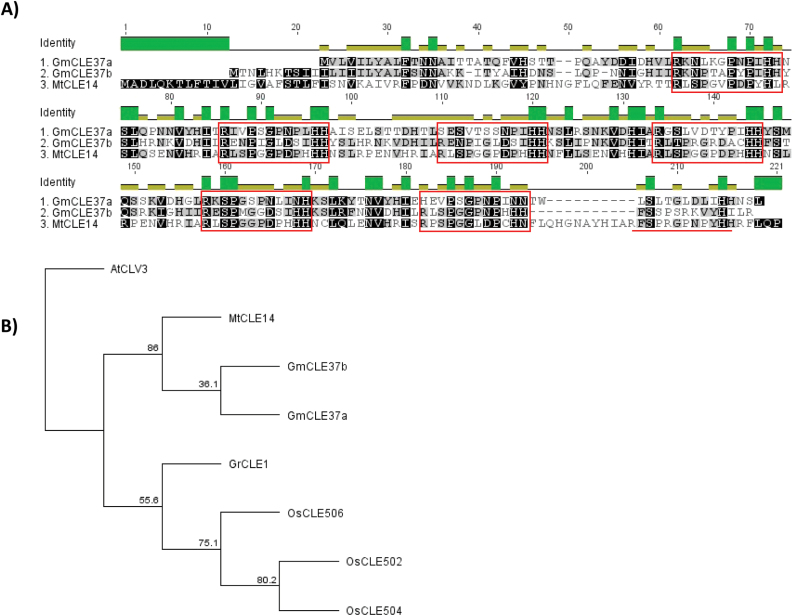
Genes encoding pre-propeptides that contain multi-CLE domains were also identified. This includes GmCLE37a and GmCLE37b, which have six possible CLE domains each (Fig. 4A). These were excluded from the alignment in Fig. 1 as they do not have the archetypical domain structure. There are only two identical CLE domains within the soybean multi-CLE domain pre-propeptides and they both occur in GmCLE37b (Fig. 4A). A multi-CLE domain-containing pre-propeptide previously reported in Medicago truncatula by Oelkers et al. (2008) was identified here as MtCLV3 (MtCLV3 was previously discovered by Chen et al., 2009, but was not reported to encode a multi-CLE domain). Although MtCLV3 encodes three CLE domains, only one is actually translated due to the presence of a previously undetected intron identified here. An additional pre-propeptide of M. truncatula, called MtCLE14, contains a multi-CLE domain with seven CLE peptide domains (Fig 4A; Mortier et al., 2011). MtCLE14 contains four identical 12 amino acid CLE domains in tandem, each followed by an asparagine residue (possible representing a 13th residue in the CLE peptide), and each preceded by the same two hydrophobic residues (Fig. 4A).
Fig. 4.
Multi-CLE domain pre-propeptides. (A) Multiple sequence alignment of the soybean and M. truncatula multi-CLE domain pre-propeptides, with putative 13 amino acid residue CLE domains highlighted by a red box. An additional CLE domain of MtCLE14 that is not detected in the two soybean pre-propeptides is underlined in red. Four MtCLE14 CLE domains are identical in sequence (CLE domains 2–5) while there are no 100% conserved 13 amino acid residue CLE domains in soybean. However, there are two fully conserved 12 residue CLE domains in GmCLE37b (CLE domains 1 and 2). (B) Phylogenetic tree of known multi-CLE domain-containing pre-propeptides of rice (Oryza sativa), potato cyst nematode (Globodera rostochiensis), MtCLE14 of M. truncatula, and the newly identified GmCLE27a and GmCLE37b of soybean, including AtCLV3 as an outgroup. The multi-CLE domain pre-propeptides identified here cluster separately from those that were previously identified. The tree is shown with bootstrap confidence values expressed as a percentage from 1000 bootstrap replications.
In A. thaliana, AtCLE18 encodes both a CLE and a CLEL domain (Meng et al., 2012). TBLASTN and BLASTN searches of the soybean and common bean genomes failed to identify a similar gene. Multi-CLE domain-encoding genes of nematodes are processed into single functional CLE peptide ligands (Chen et al., 2015). TBLASTN searches of the soybean and common bean genomes using the known multi-CLE domain-encoding gene of nematode and three others of rice (Olsen and Skriver, 2003; Oelkers et al., 2008) identified no orthologues. A phylogenetic analysis (Fig. 4B) also shows that the legume multi-CLE domain pre-propeptides cluster separately from the nematode and rice pre-propeptides.
Categorization and functional predictions of soybean CLE peptides
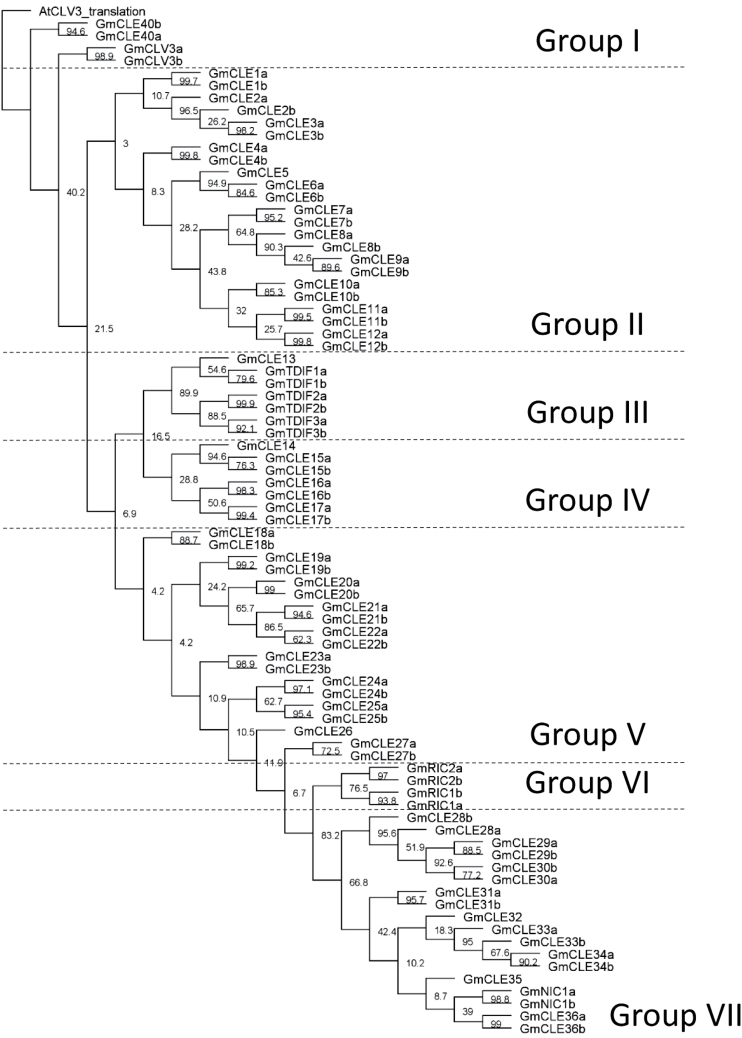
The function of many CLE peptides can be predicted based on sequence. The Arabidopsis CLE peptides are currently categorized into two groups: type-A affecting root and shoot meristem development, and type-B affecting vasculature development (Matsubayashi, 2014). The soybean CLE peptides were assigned into different categories based on the sequence alignment, phylogenetic grouping of their pre-propeptides, and their functional roles where known. The groups were initially defined based on phylogenetic analysis, and were then further refined following examination of their CLE domain and adjacent residues. In total, seven groups (Groups I–VII) were identified (Fig. 5). Logo alignments (Fig. 6) were subsequently constructed to establish the level of conservation within the 13 amino acid CLE domain of each group, with highly conserved residues probably critical to their function.
Fig. 5.
Soybean CLE pre-propeptide phylogenetic tree illustrating the seven distinct identity groups. Phylogenetic analysis was performed using the multiple sequence alignment generated with entire pre-propeptide sequences (Fig. 1), including AtCLV3 as an outgroup. Homeologous genes consistently cluster together with high confidence (indicated by high bootstrap values). The seven groups (Group I–VII) were assigned based on clustering in the tree, in addition to sequence similarity. The tree is shown with bootstrap confidence values expressed as a percentage from 1000 bootstrap replications.
Fig. 6.
CLE domain consensus sequences from the seven soybean pre-propeptide groups. Logo diagrams illustrate the 13 amino acid CLE domain consensus sequences for soybean CLE Groups I–VII, as determined from multiple sequence alignments generated for each group. The 13th amino acid is a consensus of only those sequences that have a residue at that position. Group IV does not have any residues at that position and hence the logo diagram for this group is 12 residues only.
Group I is small, consisting of only four members. It contains CLV3, CLE40, and their homeologous duplicates (Fig. 5). CLV3 and CLE40 are well characterized and are responsible for apical meristem regulation in the shoot and root, respectively (Grienenberger and Fletcher, 2015). The CLE domain of this group is highly conserved (Fig. 6), particularly for amino acid residues reported to be critical for function (Song et al., 2013).
Group II contains the least conserved CLE domain of all the established groups. It is also the largest group, with 23 members, which may account for it having the lowest degree of conservation (Figs 5, 6). The group cannot be divided further with any degree of confidence using a phylogenetic approach. Interestingly, it has low conservation at residue six, which is generally considered to be critical for function, possibly having a role in enabling the CLE peptide to rotate or bend (Hastwell et al., 2015). Most of the CLE peptides in this group remain poorly characterized in any species; however, some of the soybean CLE pre-propeptides show similarity to, and group closely with, AtCLE45 (Supplementary Fig. S3 at JXB online).
Group III contains seven members, including the three TDIF pre-propeptides and their homeologues, in addition to one other member of unknown function that lacks a duplicate copy (Fig. 5). This group is orthologous to the Arabidopsis type-B CLE pre-propeptides that influence vasculature development, including AtCLE41, ACLE42, and AtCLE44 (Fig. 5; Supplementary Fig. S3 at JXB online; Matsubayashi, 2014). A defining feature of this soybean group is that all of the CLE peptides begin with a histidine residue, as opposed to the classical arginine (Fig. 6). Interestingly, with the exception of the non-TDIF peptide (GmCLE13), the 12 amino acid CLE domain is 100% conserved. Also of note is that the members of this group are the only CLE peptides to have a serine residue at position 11, rather than the characteristic histidine (Fig. 6).
Group IV consists of seven members and notably does not encode any CLE peptides that are 13 amino acids in length (Fig. 6). It is also the group that is least conserved at residue one. The function of the group members remains poorly defined.
Group V is another large group, having 19 members (Fig. 5). Of the CLE peptides encoded by this group, all but one contain an acidic amino acid (glutamic acid or aspartic acid) and a lysine residue immediately preceding the CLE domain (Fig. 1). The CLE peptides encoded by this group also predominantly have a threonine at position 5, which is not characteristic of any of the other groups (Fig. 6).
Group VI is a small group consisting entirely of the rhizobia-induced CLE peptides (RICs) and their homeologous copies (Fig. 5). This group has been well characterized for their role in regulating legume nodule development (reviewed in Hastwell et al., 2015), including the identification of amino acid residues in the CLE domain that are critical for function (Reid et al., 2013).
Group VII consists of 18 members, and, like Group I, has two histidine residues located at positions 11 and 12 (Figs 5, 6). It contains the majority of the genes that were unpredicted in Phytozome (Table 1). The function of most remains unknown; however, it does include the nitrate-induced CLE peptide (NIC1a) and its homeologue, NIC1b (Reid et al., 2011a ; referred to as NIC2 in Lim et al., 2014), that is well known for its role in controlling legume nodulation in response to the nitrogenous content of the rhizosphere (reviewed in Hastwell et al., 2015).
These groupings hold true when the common bean CLE pre-propeptides are added to the phylogenetic analysis with soybean (Supplementary Fig. S1 at JXB online). When Arabidopsis is also included (Supplementary Fig, S3), the groupings are still conserved generally, but are supported by lower bootstrap proportions, especially Group II. This is not surprising when dealing with >150 pre-propeptides from three different species and, even though some groups are divided further when a non-legume is included, the larger groups cannot be confidently split further based on the low bootstrap proportions. In all instances, Group III is supported by very high bootstrap proportions (>88).
A C-terminal extension is encoded by one-third of the genes identified here, spanning across the various groups, but predominantly being found in Groups I, II, and VI (Figs 1, 5). GmCLE31a and b, and GmCLE13, also contain a C-terminal extension. The presence of a predicted intron correlates slightly with the groupings, as all of the genes in Group I contain a predicted intron, as do some in Group II, but none in Groups III–VII, with the exception of GmCLE13 (Group III), which incidentally also contains the only CLE domain sequence divergence of its group, as noted above (Table 2; Figs 1, 5, 6).
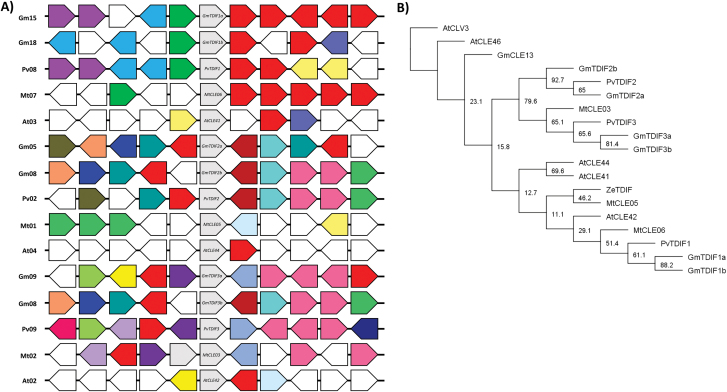
The groupings described here could help in elucidating the function of CLE peptides where a function is yet to be assigned. Indeed, these groupings, together with genomic environment analyses, were used to identify previously unknown soybean and/or common bean orthologues of AtCLV3-, AtCLE40-, and TDIF-encoding genes, as well as likely M. truncatula orthologues. AtCLV3 was the first CLE gene to be identified in any species (Fletcher et al., 1999) and has since been identified in soybean and M. truncatula (GmCLV3a, GmCLV3b, and MtCLV3; Chen et al., 2009; Wong et al., 2013). Investigations into the genomic environment and pre-propeptide sequence similarity (Fig. 3B) led to the identification of a CLV3 orthologue in common bean. Similar approaches were used to identify AtCLE40 orthologues (Fig. 3C) in common bean and M. truncatula, in addition to GmCLE40b, the homeologue of GmCLE40a. Moreover, all TDIF orthologues in soybean, common bean, and M. truncatula were established (Fig. 7). In contrast, despite AtCLE46 and GmCLE13 sharing a high level of sequence similarity in the CLE domain, they do not show synteny to the TDIF genes, or to each other, and cluster separately (Fig. 7). Thus, these genes are unlikely to be true TDIF peptides.
Fig. 7.
TDIF genes in soybean, common bean, Arabidopsis, Zinnia elegans, and M. truncatula. (A) Genomic environments of the TDIF-encoding genes highlight the genetic synteny between the genes identified here in soybean, common bean, and M. truncatula with previously characterized TDIF genes of A. thaliana, AtCLE41, AtCLE42, and AtCLE44. TDIF-encoding genes are shown positioned centrally and shaded in grey. Species and chromosome number are indicated to the left of each genomic segment. Surrounding genes similar in putative function are indicated by the same colour and genes with unrelated putative functions are uncoloured. The direction of the arrow represents the orientation of the gene compared with that of the CLE gene. A high level of genetic synteny is shown here for each of the predicted TDIF-encoding genes, but was not found for AtCLE46 and GmCLE13 (data not shown), whose CLE domain begins with a histidine residue but is not a TDIF peptide. (B) Phylogenetic tree of TDIF-encoding pre-propeptides, including ZeTDIF, and also AtCLV3 as an outgroup. Two pre-propeptides, AtCLE46 and GmCLE13, are also included that have CLE domains beginning with a histidine residue, but are not true TDIF CLE peptides and did not group with the TDIF pre-propeptides. The tree is shown with bootstrap confidence values expressed as a percentage from 1000 bootstrap replications.
Expression analysis of CLE peptide-encoding genes of soybean, common bean, and Arabidopsis
A meta-analysis of the publicly available transcriptome data was conducted in soybean, common bean, and Arabidopsis (Supplementary Tables S3–S5 at JXB online). The transcriptomic expression of functionally characterized soybean and common bean CLE peptide-encoding genes was consistent with the literature (i.e. RICs and NIC1, Reid et al., 2011a ; Ferguson et al., 2014). Interestingly, there were no transcriptional data available for CLV3 orthologues in soybean and common bean (Supplementary Tables S3, S4).
Trends observed in the expression of CLE peptide-encoding gene orthologues across different tissues of soybean and common bean were also consistent (Supplementary Tables S3, S4 at JXB online). For example: PvCLE10, GmCLE10a, and GmCLE10b showed varying levels of expression across all tissue types, in a similar trend; PvCLE17 and GmCLE17a are expressed in all tissue types except seeds, flowers, and early pod growth; and PvCLE19 and GmCLE19a show expression in all tissues except mature nodules. These three orthologous gene groups (CLE10, CLE17, and CLE19) also show high (>93) bootstrap values in the phylogenetic analyses (Supplementary Fig. S2). In contrast, CLE24 showed different expression patterns between soybean and common bean orthologues. GmCLE21a and GmCLE21b show the same expression trends, but PvCLE21 transcripts were only detected in the early seed development stage. In soybean, where data were available for both the a and b copy, the general trend of expression was consistent but in most cases the level or the time of expression varied. There is no consistent expression pattern between pre-propeptides belonging to soybean Groups I–VII, but closely related peptides probably perform a similar role in different developmental tissues as with the TDIF orthologues (Supplementary Tables S3–S5; Matsubayahsi, 2014).
To determine if expression trends are similar between orthologues of soybean, common bean, and Arabidopsis, and to see how orthologues clusters, a phylogenetic tree of the pre-propeptides from the three species was produced (Supplementary Fig. S3 at JXB online). Branches that were supported by >50 bootstrap proportions include AtCLE46 and CLE1; AtCLE21 and CLE4; AtCLE27 and CLE6; AtCLE20 and CLE23; AtCLE12 and CLE24; and the cluster containing the TDIF orthologous genes, as established previously in Fig. 7.
As expected, the legume orthologues show a similar expression trend for each of these branches and, in the case of AtCLE12, a similar trend was observed with GmCLE24a and PvCLE24 (Supplementary Tables S3–S5 at JXB online). Interestingly, AtCLE27 and AtCLE21 were not expressed in any tissues, similar to the case of their respective and related legume pre-propeptides (Supplementary Fig. S3). All the TDIF orthologues with available expression profiles show a highly similar pattern (Supplementary Tables S3–S5).
Within the meta-analysis of the transcriptomes, interesting candidates were identified as targets for future functional characterization. PvCLE29 was found only in the flower at a very high level; PvCLE24 shows very high root and nodule expression (Supplementary Table S4 at JXB online); and GmCLE25a is only expressed in root tissue (Supplementary Table S3).
The meta-analysis shows similar trends for orthologous genes. However, to date, only one-third of the CLE peptide-encoding genes of soybean, and less than half from Arabidopsis, are represented. It is also likely that some genes that respond to external stimuli (e.g. rhizobia for RIC1 and 2 and nitrate for the NIC1 orthologues) were not induced if the required treatment was not part of the study.
Feeding studies were not attempted here because the precise size and modification of each of the novel peptides is completely unknown. Although feeding unmodified or semi-modified synthetic peptides could be attempted, the peptides being fed would be designed based on prediction (in terms of both length and modifications). Furthermore, they would be applied in unnaturally high concentrations, without regard to temporal or spatial regulation, to a broad range of tissues and cell types to which they might not normally localize. These issues would be further exacerbated in feeding studies using roots grown on agar containing high levels of sucrose and nitrate, and exposed to light. Such studies would result in an extremely high frequency of false-positive outcomes that are of little biological value. For comparison sake, an ecologist investigating the impact of wild boars on the environment would not flood a forest with hams. Indeed, it has readily been shown that CLE peptides altered from their correct modification, size, and location can induce a phenotypic effect in feeding (e.g. Fiers et al., 2005; Whitford et al., 2008; Ohyama et al., 2009; Mortier et al., 2010; Kondo et al., 2011) or site-directed mutagenesis and domain-swap studies (e.g. Ni and Clark, 2006; Song et al., 2012; Reid et al., 2013). CLE peptides unlikely to come into contact with a given receptor can be forced to bind to that receptor in vitro (as elegantly demonstrated by Shinohara and Matsubayashi, 2015). Thus, results from peptide feeding studies may not be biologically relevant, and any phenotypic changes observed would need to be interpreted with extreme caution. For these reasons, the focus here was to use alternative approaches to help determine the role of novel peptides of unknown structure and function.
Discussion
CLE peptides are widely recognized as important contributors to plant signalling and development; however, a lot remains to be understood about these critical signal molecules. Here, this emerging field was enhanced by the discovery and categorization of the CLE peptide families of soybean and common bean, two of the world’s most agriculturally important crops. A total of 84 CLE peptide-encoding genes in soybean and 44 in common bean were identified, and subsequently an array of bioinformatic approaches were conducted for comparative genomic and molecular evolution analyses. Doing so led to the identification of three pseudogenes, two multi-CLE domain-encoding genes in soybean, and a tandem gene duplication event in common bean. It also enabled the establishment of all homeologous gene copies within soybean, and orthologous copies amongst soybean, common bean, and Arabidopsis. Searches using rhizobia and mycorrhiza genomes were also performed, but revealed no CLE peptide-encoding genes in these organisms. Thus, to date, CLE peptides appear to be exclusive to plants and nematodes.
The function of most CLE peptides remains completely unknown. However, phylogenetic analyses of the entire CLE pre-propeptide families of soybean, common bean, and Arabidopsis show that they group strongly according to their CLE domain and known/predicted function. Based on the analyses, it is demonstrated that the soybean CLE pre-propeptides (excluding multi-CLE domain-encoding genes) grouped into seven distinct categories (Groups I–VII) and that these groups are generally preserved when other species are included. This expands on the two groups reported in Arabidopsis (type-A affecting root and shoot development, and type-B affecting vasculature development; e.g. Matsubayashi, 2014). The categorization approach reported here could be a useful tool for elucidating the function of unknown CLE peptides and their closely related homeologous and orthologous sequences. As an example, all known CLE peptides of similar function were found to group together (CLV3 and CLV40 formed Group I, the TDIFs formed Group III, and the RICs formed Group VI). Moreover, the groupings revealed a number of highly conserved amino acid residues present in the peptide domains of each group, which are probably central to the activity of their ligands.
The groups identified here include peptides performing a similar developmental role in a range of different tissues, as exemplified by Group III, whose Arabidopsis orthologues are known to have the same function (Matsubayashi, 2014) but are expressed in a range of different tissues. This is also seen with the Group I and Group VI peptides. Given that the genes encoding the members of these groups do not show consistent expression patterns, it is possible that they too may have similar roles in different tissues. Furthermore, the transcriptome evidence presented here provides some insight into where the peptides function, as they often act in a local manner (Matsubayashi, 2014). Indeed, the only known CLE peptides to act systemically are those involved in the autoregulation of nodulation signalling pathway of legumes (Hastwell et al., 2015).
The ancestral genome shared by soybean and common bean duplicated ~59 MYA and subsequently reconverged (Schmutz et al., 2010). Later, following the divergence of the two species, the soybean genome duplicated again ~13 MYA and, as a result, there are typically two soybean orthologues present for every common bean gene (Lin et al., 2010; Schmutz et al., 2014). This trend is consistent with the present findings, where common bean contains approximately half the number of CLE peptide-encoding genes as soybean. The findings are also consistent with Arabidopsis, which is reported to have only 32 CLE peptide-encoding genes (Cock and McCormick, 2001), and is well known for fractionation (i.e. preferentially removing redundant and/or excess genomic information; Thomas et al., 2006). Indeed, Group VI of the soybean and common bean CLE peptide families identified here is completely absent from Arabidopsis. This category is known to be induced by rhizobia to control legume nodulation (reviewed in Hastwell et al., 2015), suggesting that either Arabidopsis has completely lost this group, or that the legume species have gained it as a means of regulating the relationship with their symbiotic partner.
Additional methods were employed here to identify conclusively soybean and common bean orthologues of a number of key CLE peptide-encoding genes of Arabidopsis. Indeed, orthologues of AtCLV3, which acts in the SAM to control stem cell numbers (Gaillochet et al., 2015), were identified in common bean, and confirmed in soybean and M. truncatula (Chen et al., 2009; Wong et al., 2013). Interestingly, it is also shown that MtCLV3 encodes three CLE peptide domains, but only one is translated due to the presence of an intron. Orthologues of AtCLE40, which acts in the RAM to control stem cell numbers (Hobe et al., 2003; Sharma et al., 2003; Stahl et al., 2009), were also identified here in these same three legume species. This includes the homeologous copy of GmCLE40a, called GmCLE40b, which is unlikely to produce a functional product due to a naturally occurring mutation that truncates the pre-propeptide prior to the CLE domain. Orthologues of the three TDIF CLE peptide-encoding genes of Arabidopsis, which act throughout the plant in vascular differentiation (Grienenberger and Fletcher, 2015), were also identified here, including six genes in soybean, three in common bean, and three in M. truncatula. The predicted TDIF-encoding genes (together with one other soybean gene of unknown function) make up Group III of the CLE pre-propeptide family. A number of additional Arabidopsis orthologue candidates were also identified throughout the other various CLE peptide groups defined here.
Genome-wide searches to identify CLE peptide-encoding genes in legumes have been conducted previously using soybean, M. truncatula, and L. japonicus (Cock and McCormick, 2001; Oelkers et al., 2008; Okamoto et al., 2009; Mortier et al., 2010, 2011; Lim et al. 2011), with a few additional genes also identified in common bean (Oelkers et al., 2008; Ferguson et al., 2014). However, many of these studies were limited by the technology and bioinformatic resources available at the time. Recent bioinformatic advances were capitalized on here to identify, and subsequently characterize, categorize, and compare thoroughly, the CLE peptide families of soybean and common bean. This also enabled unification of the nomenclature for these species, taking into account the duplicated nature of the soybean genome and the presence of orthologous genes amongst the two species.
Taken together, this research helped to assemble the complete CLE peptide families of two agriculturally important legume species, categorized them into groups to provide insight into their structure and function, identified key orthologues existing amongst them and Arabidopsis, and used transcriptional evidence to help elucidate their localization and activity. This represents one of the most in-depth studies conducted within and between any CLE peptide family to date. Future work to establish unequivocally the function of these critical peptides, identify their binding partners, and determine the precise structural modifications of their mature ligands is now needed to enhance further the understanding of these novel hormones in regulating plant development.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Soybean and common bean pre-propeptide phylogenetic tree.
Figure S2. Hydrophobicity plot of the CLE pre-propeptides of soybean, common bean, and Arabidopsis.
Figure S3. Soybean, common bean, and Arabidopsis pre-propeptide phylogenetic tree.
Table S1. CLE peptide-encoding genes of soybean.
Table S2. Frequency (%) of amino acid residues in CLE pre-propeptides of soybean, common bean, and Arabidopsis.
Table S3. Soybean CLE peptide-encoding gene expression from transcriptome databases.
Table S4. Common bean CLE peptide-encoding gene expression from A Common Bean Gene Expression Atlas (Jamie et al. 2014).
Table S5. Arabidopsis thaliana CLE peptide-encoding gene expression.
Acknowledgements
This work was funded by the Hermon Slade Foundation, and the Australian Research Council Discovery Project grants (DP130103084 and DP130102266). The Fellowship Fund Inc. is also thanked for provision of a Molly-Budtz Olsen PhD fellowship to AHH. We would like to thank Alina Tollenaere, Dongxue Li, Ong Cu, and Candice Jones for technical assistance, and Dr Dugald Reid for helping with preliminary investigations.
References
- Araya T, Miyamoto M, Wibowo J, Suzuki A, Kojima S, Tsuchiya YN, Sawa S, Fukuda H, von Wirén N, Takahashi H. 2014. CLE–CLAVATA1 peptide–receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proceedings of the National Academy of Sciences, USA 111, 2029–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SK, Kurdyukov S, Kereszt A, Wang XD, Gresshoff PM, Rose RJ. 2009. The association of homeobox gene expression with stem cell formation and morphogenesis in cultured Medicago truncatula. Planta 230, 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Lang P, Chronis D, Zhang S, De Jong WS, Mitchum MG, Wang X. 2015. In planta processing and glycosylation of a nematode CLAVATA3/ENDOSPERM SURROUNDING REGION-Like effector and its interaction with a host CLAVATA2-Like receptor to promote parasitism. Plant Physiology 167, 262–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock JM, McCormick S. 2001. A large family of genes that share homology with CLAVATA3. Plant Physiology 126, 939–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt S, Rodriguez-Villalon A, Santuari L, Wyser-Rmili C, Ragni L, Hardtke CS. 2013. Suppression of Arabidopsis protophloem differentiation and root meristem growth by CLE45 requires the receptor-like kinase BAM3. Proceedings of the National Academy of Sciences, USA 110, 7074–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo S, Betsuyaku S, Fukuda H. 2014. Endogenous peptide ligand–receptor systems for diverse signalling networks in plants. Current Opinion in Plant Biology 21, 140–146. [DOI] [PubMed] [Google Scholar]
- Endo S, Shinohara H, Matsubayashi Y, Fukuda H. 2013. A novel pollen–pistil interaction conferring high-temperature tolerance during reproduction via CLE45 signaling. Current Biology 23, 1670–1676. [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Indrasumunar A, Hayashi S, Lin M-H, Lin Y-H, Reid DE, Gresshoff PM. 2010. Molecular analysis of legume nodule development and autoregulation. Journal of Integrative Plant Biology 52, 61–76. [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Li D, Hastwell AH, Reid DE, Li Y, Jackson SA, Gresshoff PM. 2014. The soybean (Glycine max) nodulation-suppressive CLE peptide, GmRIC1, functions interspecifically in common white bean (Phaseolus vulgaris), but not in a supernodulating line mutated in the receptor PvNARK. Plant Biotechnology Journal 12, 1085–1097. [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Mathesius U. 2014. Phytohormone regulation of legume–rhizobia interactions. Journal of Chemical Ecology 40, 770–790. [DOI] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, Liu C. 2005. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. The Plant Cell 17, 2542–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiume E, Fletcher JC. 2012. Regulation of Arabidopsis embryo and endosperm development by the polypeptide signalling molecule CLE8. The Plant Cell 24, 1000–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. 1999. Signalling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. [DOI] [PubMed] [Google Scholar]
- Fujisawa T, Okamoto S, Katayama T, et al. 2014. CyanoBase and RhizoBase: databases of manually curated annotations for cyanobacterial and rhizobial genomes. Nucleic Acids Research 42, D666–D670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funayama-Noguchi S, Noguchi K, Yoshida C, Kawaguchi M. 2011. Two CLE genes are induced by phosphate in roots of Lotus japonicus. Journal of Plant Research 124, 155–163. [DOI] [PubMed] [Google Scholar]
- Gaillochet C, Daum G, Lohmann JU. 2015. O Cell, Where Art Thou? The mechanisms of shoot meristem patterning. Current Opinion in Plant Biology 23, 91–97. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins M, Appel R, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server. In: Walker J, ed. The proteomics protocols handbook . Clifton, NJ: Humana Press, 571–607. [Google Scholar]
- Goodstein DM, Shu S, Howson R, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40, D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Research 38, W695–W699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger E, Fletcher JC. 2015. Polypeptide signalling molecules in plant development. Current Opinion in Plant Biology 23, 8–14. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52, 696–704. [DOI] [PubMed] [Google Scholar]
- Handa Y, Nishide H, Takeda N, Suzuki Y, Kawaguchi M, Saito K. 2015. RNA-seq transcriptional profiling of an arbuscular mycorrhiza provides insights into regulated and coordinated gene expression in Lotus japonicus and Rhizophagus irregularis . Plant and Cell Physiology 56 (in press). [DOI] [PubMed] [Google Scholar]
- Hastwell AH, Gresshoff PM, Ferguson BJ. 2015. The structure and activity of nodulation-suppressing CLE peptide hormones of legumes. Functional Plant Biology 42, 229–238. [DOI] [PubMed] [Google Scholar]
- Hirakawa Y, Kondo Y, Fukuda H. 2010, TDIF peptide signalling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. The Plant Cell 22, 2618–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y, Kondo Y, Fukuda H. 2011. Establishment and maintenance of vascular cell communities through local signaling. Current Opinion in Plant Biology 14, 17–23. [DOI] [PubMed] [Google Scholar]
- Hobe M, Müller R, Grünewald M, Brand U, Simon R. 2003. Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Development Genes and Evolution 213, 371–381. [DOI] [PubMed] [Google Scholar]
- Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H. 2006. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313, 842–845. [DOI] [PubMed] [Google Scholar]
- Jamie A, Iniguez LP, Fu F, Bucciarelli B, Miller SS, Jackson SA, McClean PE, Li J, Dai X, Zhao PX. 2014, An RNA-Seq based gene expression atlas of the common bean. BMC Genomics 15, 866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A, Nakamura Y, Sasaki E, Kyozuka J, Fukuda H, Sawa S. (2007) Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-Related (CLE) peptides in Arabidopsis thaliana and Oryza sativa . Plant and Cell Physiology 48, 1821–1825. [DOI] [PubMed] [Google Scholar]
- Kondo T, Yokomine K, Nakagawa A, Sakagami Y. 2011. Analogs of the CLV3 peptide: synthesis and structure–activity relationships focused on proline residues. Plant and Cell Physiology 52, 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. Journal of Molecular Biology 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, et al. 2012. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Research 40, D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF. 2005. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Systematic Biology 54, 575–194. [DOI] [PubMed] [Google Scholar]
- Libault M, Farmer A, Brechenmacher L, Drnevich J, Langley RJ, Bilgin DD, Radwan O, Neece DJ, Clough SJ, May GD. 2010. a Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiology 152, 541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, He J, Xu D, May G, Stacey G. 2010. b An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. The Plant Journal 63, 86–99. [DOI] [PubMed] [Google Scholar]
- Lin J-Y, Stupar RM, Hans C, Hyten DL, Jackson SA. 2010. Structural and functional divergence of a 1-Mb duplicated region in the soybean (Glycine max) genome and comparison to an orthologous region from Phaseolus vulgaris . The Plant Cell 22, 2545–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CW, Lee YW, Hwang CH. 2011. Soybean nodule-enhanced CLE peptides in roots act as signals in GmNARK-mediated nodulation suppression. Plant and Cell Physiology 52, 1613–1627. [DOI] [PubMed] [Google Scholar]
- Lim CW, Lee YW, Lee SC, Hwang CH. 2014. Nitrate inhibits soybean nodulation by regulating expression of CLE genes. Plant Science 229, 1–9. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y. 2014. Posttranslationally modified small-peptide signals in plants. Annual Review of Plant Biology 65, 385–413. [DOI] [PubMed] [Google Scholar]
- McClean P, Lavin M, Gepts P, Jackson S. 2008. Phaseolus vulgaris: a diploid model for soybean. In: Stacey G, ed. Genetics and genomics of soybean , Vol. 2 New York: Springer, 55–76. [Google Scholar]
- McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. 2013. Analysis tool web services from the EMBL-EBI. Nucleic Acids Research 41, W597–W600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Buchanan BB, Feldman LJ, Luan S. 2012. A putative nuclear CLE-Like (CLEL) Peptide precursor regulates root growth in Arabidopsis. Molecular Plant 5, 955–957. [DOI] [PubMed] [Google Scholar]
- Mitchum MG, Hussey RS, Baum TJ, Wang X, Elling AA, Wubben M, Davis EL. 2013. Nematode effector proteins: an emerging paradigm of parasitism. New Phytologist 199, 879–894. [DOI] [PubMed] [Google Scholar]
- Mortier V, Den Herder G, Whitford R, Van de Velde W, Rombauts S, D’haeseleer K, Holsters M, Goormachtig S. 2010. CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiology 153, 222–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier V, De Wever E, Vuylsteke M, Holsters M, Goormachtig S. 2012. Nodule numbers are governed by interaction between CLE peptides and cytokinin signaling. The Plant Journal 70, 367–376. [DOI] [PubMed] [Google Scholar]
- Mortier V, Fenta B, Martens C, Rombauts S, Holsters M, Kunert K, Goormachtig S. 2011. Search for nodulation-related CLE genes in the genome of Glycine max . Journal of Experimental Botany 62, 2571–2583. [DOI] [PubMed] [Google Scholar]
- Ni J, Clark SE. 2006. Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiology 140, 726–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Guo Y, Jin H, Hartsell J, Clark S. 2011. Characterization of a CLE processing activity. Plant Molecular Biology 75, 67–75. [DOI] [PubMed] [Google Scholar]
- Oelkers K, Goffard N, Weiller G, Gresshoff PM, Mathesius U, Frickey T. 2008. Bioinformatic analysis of the CLE signalling peptide family. BMC Plant Biology 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. 2009. A glycopeptide regulating stem cell fate in Arabidopsis thaliana . Nature Chemical Biology 5, 578–580. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Ohnishi E, Sato S, Takahashi H, Nakazono M, Tabata S, Kawaguchi M. 2009. Nod Factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant and Cell Physiology 50, 67–77. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Shinohara H, Mori T, Matsubayashi Y, Kawaguchi M. 2013. Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nature Communications 4, 2191. [DOI] [PubMed] [Google Scholar]
- Olsen AN, Skriver K. 2003. Ligand mimicry? Plant-parasitic nematode polypeptide with similarity to CLAVATA3. Trends in Plant Science 8, 55–57. [DOI] [PubMed] [Google Scholar]
- Opsahl-Ferstad HG, Deunff EL, Dumas C, Rogowsky PM. 1997. ZmEsr, a novel endosperm-specific gene expressed in a restricted region around the maize embryo. The Plant Journal 12, 235–246. [DOI] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, et al. 2007. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Research 35, D883–D887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods 8, 785–786. [DOI] [PubMed] [Google Scholar]
- Reid DE, Ferguson BJ, Gresshoff PM. (2011. a) Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Molecular Plant-Microbe Interactions 24, 606–618. [DOI] [PubMed] [Google Scholar]
- Reid DE, Ferguson BJ, Hayashi S, Lin Y-H, Gresshoff PM. 2011. b Molecular mechanisms controlling legume autoregulation of nodulation. Annals of Botany 108, 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DE, Li D, Ferguson BJ, Gresshoff PM. 2013. Structure–function analysis of the GmRIC1 signal peptide and CLE domain required for nodulation control in soybean. Journal of Experimental Botany 64, 1575–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Replogle A, Wang J, Bleckmann A, Hussey RS, Baum TJ, Sawa S, Davis EL, Wang X, Simon R, Mitchum MG. 2011. Nematode CLE signalling in Arabidopsis requires CLAVATA2 and CORYNE. The Plant Journal 65, 430–440. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Villalon A, Gujas B, Kang YH, Breda AS, Cattaneo P, Depuydt S, Hardtke CS. 2014. Molecular genetic framework for protophloem formation. Proceedings of the National Academy of Sciences, USA 111, 11551–11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E, Sharma V, Kovaleva V, Raikhel N, Fletcher J. 2002. CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. The Plant Cell 14, 969–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulin A, Auer PL, Libault M, Schlueter J, Farmer A, May G, Stacey G, Doerge RW, Jackson SA. 2013. The fate of duplicated genes in a polyploid plant genome. The Plant Journal 73, 143–153. [DOI] [PubMed] [Google Scholar]
- Sawa S, Kinoshita A, Nakanomyo I, Fukuda H. 2006. CLV3/ESR-related (CLE) peptides as intercellular signalling molecules in plants. Chemical Record 6, 303–310. [DOI] [PubMed] [Google Scholar]
- Scheible W-R, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M. 2004. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiology 136, 2483–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin AJ, Woody JL, Bolon Y-T, Joseph B, Diers BW, Farmer AD, Muehlbauer GJ, Nelson RT, Grant D, Specht JE. 2010. RNA-Seq atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biology 10, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. 2005. A gene expression map of Arabidopsis thaliana development. Nature Genetics 37, 501–506. [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, et al. 2010. Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. [DOI] [PubMed] [Google Scholar]
- Schmutz J, McClean PE, Mamidi S, et al. 2014. A reference genome for common bean and genome-wide analysis of dual domestications. Nature Genetics 46, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Ramirez J, Fletcher J. 2003. The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Molecular Biology 51, 415–425. [DOI] [PubMed] [Google Scholar]
- Shinohara H, Matsubayashi Y. 2013. Chemical synthesis of Arabidopsis CLV3 glycopeptide reveals the impact of hydroxyproline arabinosylation on peptide conformation and activity. Plant and Cell Physiology 54, 369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H, Matsubayashi Y. 2015. Reevaluation of the CLV3–receptor interaction in the shoot apical meristem: dissection of the CLV3 signaling pathway from a direct ligand-binding point of view. The Plant Journal 82, 328–336. [DOI] [PubMed] [Google Scholar]
- Shinohara H, Moriyama Y, Ohyama K, Matsubayashi Y. 2012. Biochemical mapping of a ligand-binding domain within Arabidopsis BAM1 reveals diversified ligand recognition mechanisms of plant LRR-RKs. The Plant Journal 70, 845–854. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X-F, Guo P, Ren S-C, Xu T-T, Liu C-M. 2013. Antagonistic peptide technology for functional dissection of CLV3/ESR genes in Arabidopsis. Plant Physiology 161, 1076–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song XF, Yu DL, Xu TT, Ren SC, Guo P, Liu CM. 2012. Contributions of individual amino acid residues to the endogenous CLV3 function in shoot apical meristem maintenance in Arabidopsis. Molecular plant 5, 515–523. [DOI] [PubMed] [Google Scholar]
- Stahl Y, Wink RH, Ingram GC, Simon R. 2009. A signalling module controlling the stem cell niche in Arabidopsis root meristems. Current Biology 19, 909–914. [DOI] [PubMed] [Google Scholar]
- Strabala T, Phillips L, West M, Stanbra L. 2014. Bioinformatic and phylogenetic analysis of the CLAVATA3/EMBRYO-SURROUNDING REGION (CLE) and the CLE-LIKE signal peptide genes in the Pinophyta. BMC Plant Biology 14, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BC, Pedersen B, Freeling M. 2006. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Research 16, 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E, Laux T, Rensing S. 2009. The WUS homeobox-containing (WOX) protein family. Genome Biology 10, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Mitchum MG, Gao B, Li C, Diab H, Baum TJ, Hussey RS, Davis EL. 2005. A parasitism gene from a plant-parasitic nematode with function similar to CLAVATA3/ESR (CLE) of Arabidopsis thaliana . Molecular Plant Pathology 6, 187–91. [DOI] [PubMed] [Google Scholar]
- Whitford R, Fernandez A, De Groodt R, Ortega E, Hilson P. 2008. Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proceedings of the National Academy of Sciences, USA 105, 18625–18630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CE, Singh MB, Bhalla PL. 2013. Spatial expression of CLAVATA3 in the shoot apical meristem suggests it is not a stem cell marker in soybean. Journal of Experimental Botany 64, 5641–5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Debelle F, Oldroyd GED, et al. 2011. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.