Abstract
Meiotic recombination within the proximal region of the major histocompatibility complex (MHC) of the mouse is not random but occurs in clusters at certain restricted sites, so-called recombinational hotspots. The wm7 haplotype of the MHC, derived from the wild mouse, enhances recombination specifically during female meiosis within a fragment of 1.3 kb of DNA located between the A beta 3 and A beta 2 genes in genetic crosses with laboratory haplotypes. Previous studies revealed no significant strain differences in nucleotide sequences around the hotspot, irrespective of the ability of the strain to enhance the recombination. It appeared that a distant genetic element might, therefore, control the rate of recombination. In the present study, original recombinants whose breakpoints were defined by direct sequencing of PCR-amplified DNAs were tested for the rate of secondary recombination in the crosses with laboratory strains in order to determine the location of such a genetic element. The results clearly demonstrated that the chromosomal segment proximal to the hotspot is essential for enhancement of recombination. Moreover, the male recombination is suppressed by a segment distal to the hotspot.
Full text
PDF


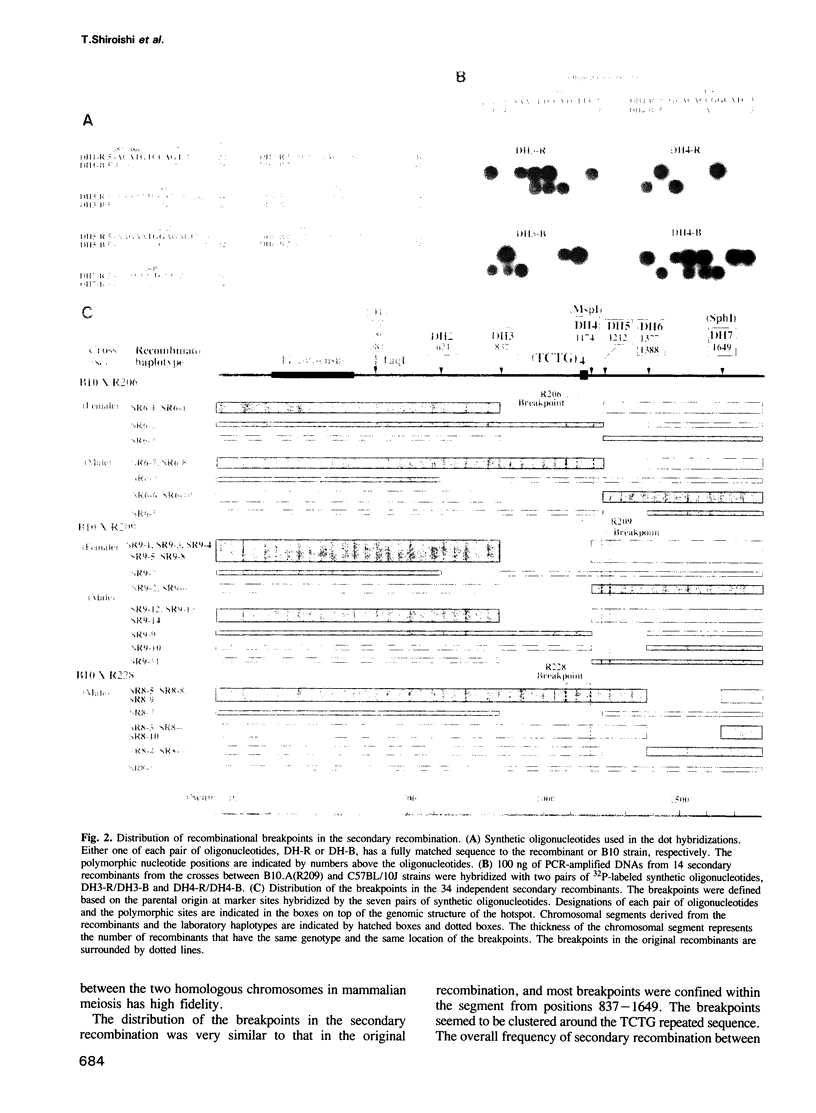


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Begovich A. B., Jones P. P. Free Ia E alpha chain expression in the E+ alpha : E- beta recombinant strain A.TFR5. Immunogenetics. 1985;22(6):523–532. doi: 10.1007/BF00430300. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Green P., Helms C., Cartinhour S., Weiffenbach B., Stephens K., Keith T. P., Bowden D. W., Smith D. R., Lander E. S. A genetic linkage map of the human genome. Cell. 1987 Oct 23;51(2):319–337. doi: 10.1016/0092-8674(87)90158-9. [DOI] [PubMed] [Google Scholar]
- Heinlein U. A., Lange-Sablitzky R., Schaal H., Wille W. Molecular characterization of the MT-family of dispersed middle-repetitive DNA in rodent genomes. Nucleic Acids Res. 1986 Aug 26;14(16):6403–6416. doi: 10.1093/nar/14.16.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobori J. A., Strauss E., Minard K., Hood L. Molecular analysis of the hotspot of recombination in the murine major histocompatibility complex. Science. 1986 Oct 10;234(4773):173–179. doi: 10.1126/science.3018929. [DOI] [PubMed] [Google Scholar]
- Lafuse W. P., Berg N., Savarirayan S., David C. S. Mapping of a second recombination hot spot within the I-E region of the mouse H-2 gene complex. J Exp Med. 1986 Jun 1;163(6):1518–1528. doi: 10.1084/jem.163.6.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuse W. P., David C. S. Recombination hot spots within the I region of the mouse H-2 complex map to the E beta and E alpha genes. Immunogenetics. 1986;24(6):352–360. doi: 10.1007/BF00377952. [DOI] [PubMed] [Google Scholar]
- Lindahl K. F. Genetic variants of histocompatibility antigens from wild mice. Curr Top Microbiol Immunol. 1986;127:272–278. doi: 10.1007/978-3-642-71304-0_31. [DOI] [PubMed] [Google Scholar]
- Lindahl K. F., Hausmann B., Chapman V. M. A new H-2-linked class I gene whose expression depends on a maternally inherited factor. Nature. 1983 Nov 24;306(5941):383–385. doi: 10.1038/306383a0. [DOI] [PubMed] [Google Scholar]
- Reeves R. H., Crowley M. R., O'Hara B. F., Gearhart J. D. Sex, strain, and species differences affect recombination across an evolutionarily conserved segment of mouse chromosome 16. Genomics. 1990 Sep;8(1):141–148. doi: 10.1016/0888-7543(90)90236-n. [DOI] [PubMed] [Google Scholar]
- Saha B. K., Raziuddin, Cullen S. E. Molecular mapping of murine I region recombinants. II. Crossing over in the E beta gene is restricted to a 4.5 kb stretch of DNA that excludes the beta 1 exon. J Immunol. 1986 Dec 15;137(12):4004–4009. [PubMed] [Google Scholar]
- Shiroishi T., Hanzawa N., Sagai T., Ishiura M., Gojobori T., Steinmetz M., Moriwaki K. Recombinational hotspot specific to female meiosis in the mouse major histocompatibility complex. Immunogenetics. 1990;31(2):79–88. doi: 10.1007/BF00661217. [DOI] [PubMed] [Google Scholar]
- Shiroishi T., Sagai T., Moriwaki K. A new wild-derived H-2 haplotype enhancing K-IA recombination. Nature. 1982 Nov 25;300(5890):370–372. doi: 10.1038/300370a0. [DOI] [PubMed] [Google Scholar]
- Shiroishi T., Sagai T., Moriwaki K. Sexual preference of meiotic recombination within the H-2 complex. Immunogenetics. 1987;25(4):258–262. doi: 10.1007/BF00404696. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Minard K., Horvath S., McNicholas J., Srelinger J., Wake C., Long E., Mach B., Hood L. A molecular map of the immune response region from the major histocompatibility complex of the mouse. Nature. 1982 Nov 4;300(5887):35–42. doi: 10.1038/300035a0. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Stephan D., Fischer Lindahl K. Gene organization and recombinational hotspots in the murine major histocompatibility complex. Cell. 1986 Mar 28;44(6):895–904. doi: 10.1016/0092-8674(86)90012-7. [DOI] [PubMed] [Google Scholar]
- Uematsu Y., Kiefer H., Schulze R., Fischer-Lindahl K., Steinmetz M. Molecular characterization of a meiotic recombinational hotspot enhancing homologous equal crossing-over. EMBO J. 1986 Sep;5(9):2123–2129. doi: 10.1002/j.1460-2075.1986.tb04475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerer E. J., Ogin E., Shreffler D. C., Passmore H. C. Molecular mapping of crossover sites within the I region of the mouse MHC. Analysis of ten recombinant chromosomes. Immunogenetics. 1987;25(4):274–276. doi: 10.1007/BF00404701. [DOI] [PubMed] [Google Scholar]






