Abstract
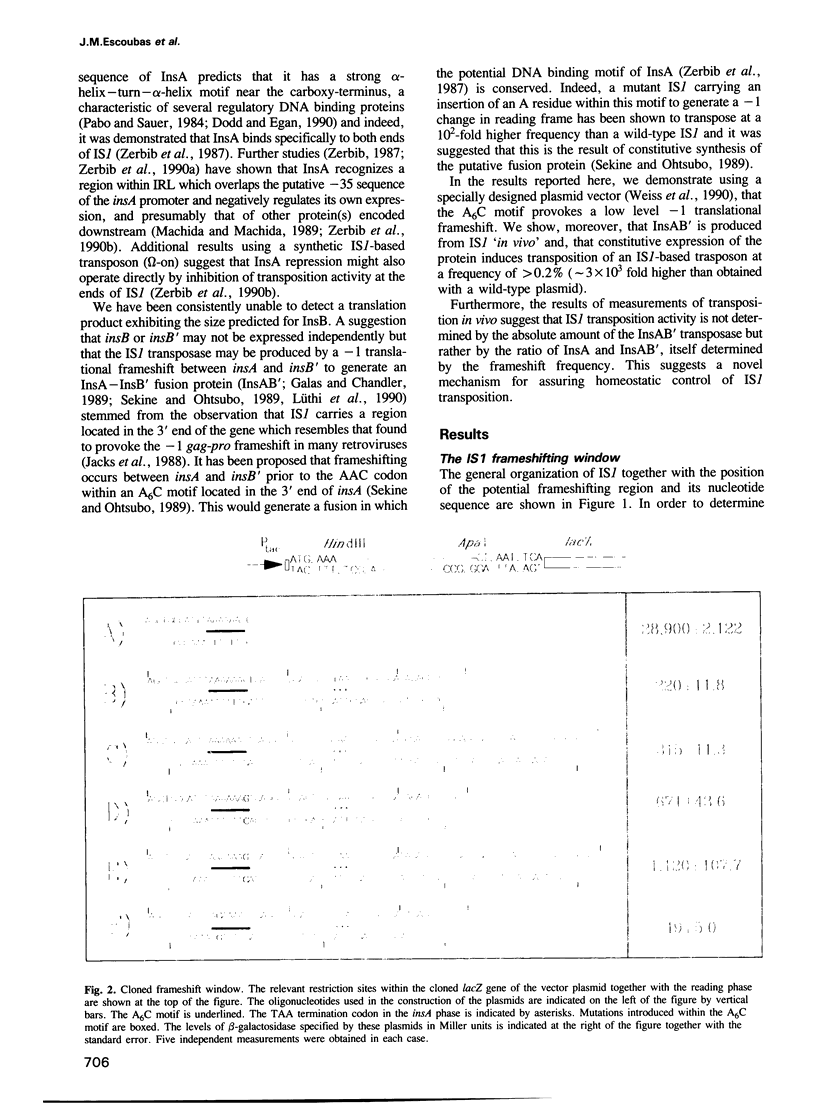
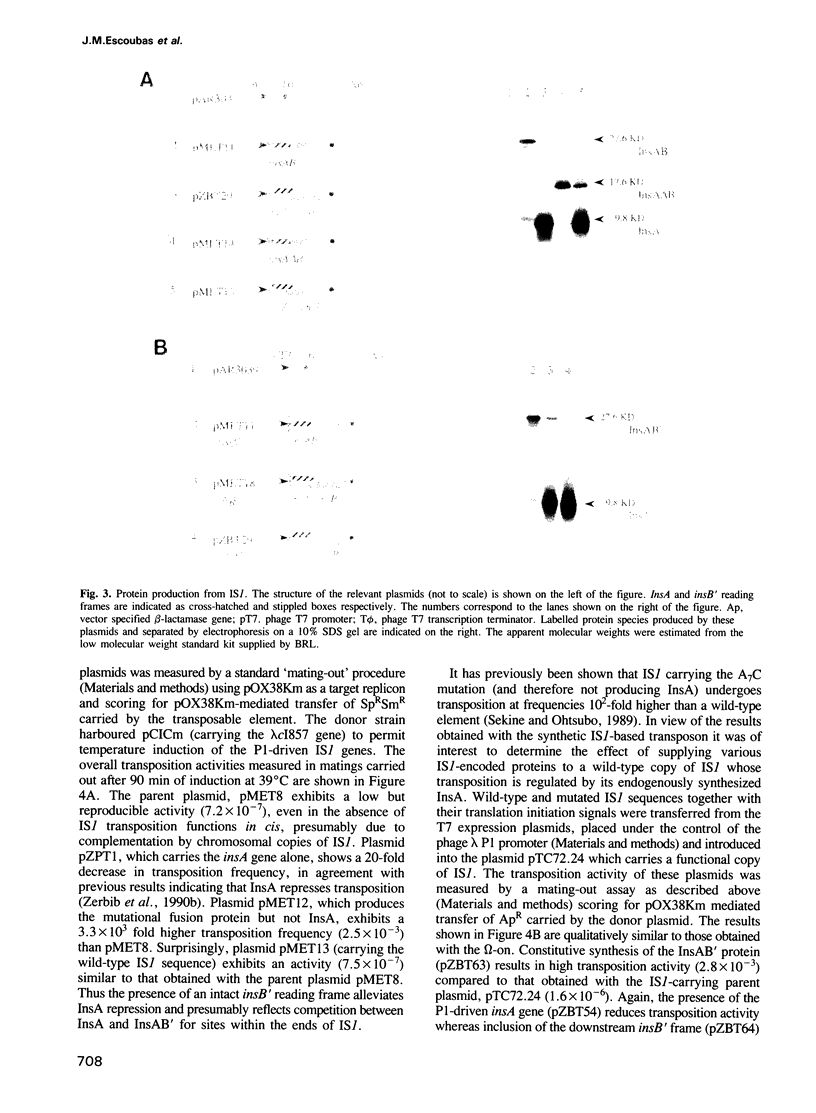
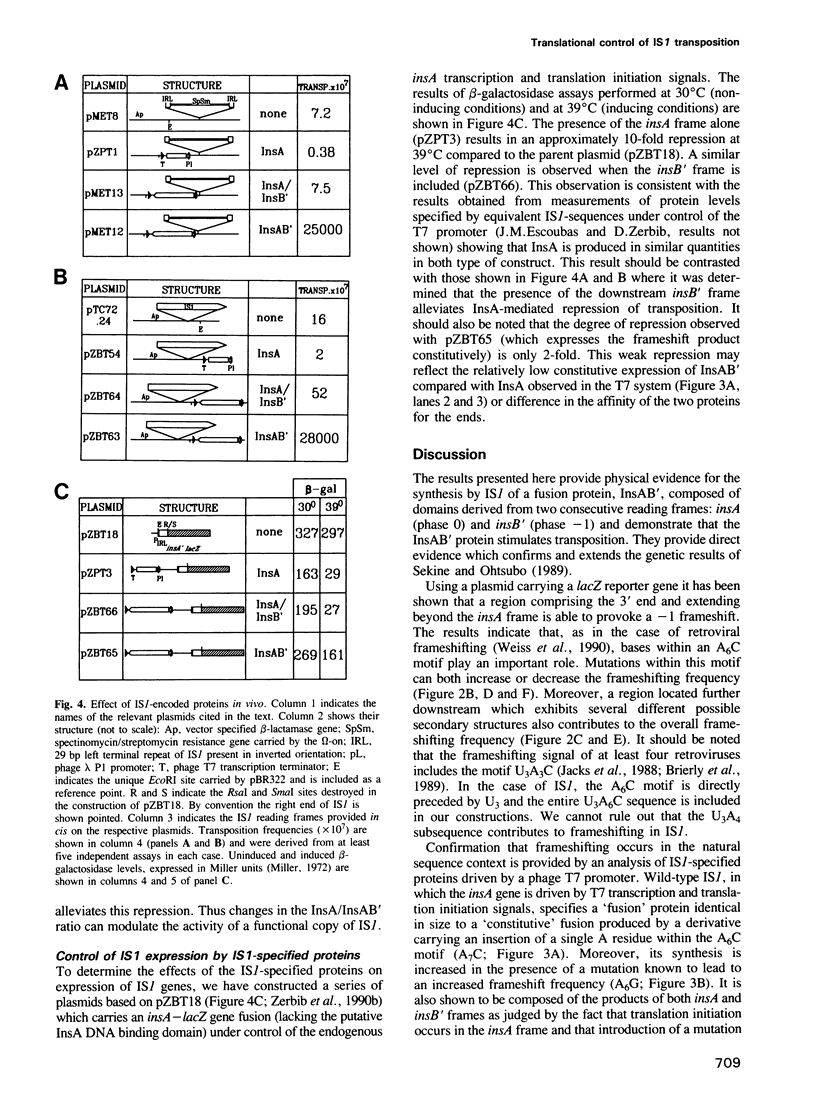
The experiments reported here provide strong evidence indicating that the transposition frequency of the bacterial insertion sequence IS1 is determined principally by two IS1-specified proteins. The first, InsA, was previously shown to bind to the ends of the element and to act as a repressor. We present both physical and genetic evidence which reveals that the second, the InsAB' transposase, is a fusion of InsA with the product of a downstream reading frame, InsB'. Synthesis of this protein occurs by a -1 frameshift between the insA and insB' frames. It requires the presence of an intact retroviral-like frameshift signal composed of an A6C motif and a downstream region able to form several alternative secondary structures. In vivo studies show that IS1 transposition activity depends on the relative rather than on the absolute levels of InsA and InsAB'. The ratio is determined primarily at the translational level by frameshifting and appears to be relatively insensitive to large variations in levels of transcription. This novel homeostatic control could therefore protect IS1 from activation as a consequence of insertion into active transcription units.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brierley I., Digard P., Inglis S. C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989 May 19;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam K., Béjar S., Gil D., Bouché J. P. Identification and sequence of gene dicB: translation of the division inhibitor from an in-phase internal start. Nucleic Acids Res. 1988 Jul 25;16(14A):6327–6338. doi: 10.1093/nar/16.14.6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chan P. T., Lebowitz J. Mapping of RNA polymerase binding sites in R12 derived plasmids carrying the replication-incompatibility region and the insertion element IS1. Nucleic Acids Res. 1982 Nov 25;10(22):7295–7311. doi: 10.1093/nar/10.22.7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M., Galas D. J. Cointegrate formation mediated by Tn9. II. Activity of IS1 is modulated by external DNA sequences. J Mol Biol. 1983 Oct 15;170(1):61–91. doi: 10.1016/s0022-2836(83)80227-7. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I. B., Egan J. B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990 Sep 11;18(17):5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Fellay R., Frey J., Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52(2-3):147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- Fowler A. V., Zabin I. Purification, structure, and properties of hybrid beta-galactosidase proteins. J Biol Chem. 1983 Dec 10;258(23):14354–14358. [PubMed] [Google Scholar]
- Galas D. J., Chandler M. Structure and stability of Tn9-mediated cointegrates. Evidence for two pathways of transposition. J Mol Biol. 1982 Jan 15;154(2):245–272. doi: 10.1016/0022-2836(82)90063-8. [DOI] [PubMed] [Google Scholar]
- Gamas P., Chandler M. G., Prentki P., Galas D. J. Escherichia coli integration host factor binds specifically to the ends of the insertion sequence IS1 and to its major insertion hot-spot in pBR322. J Mol Biol. 1987 May 20;195(2):261–272. doi: 10.1016/0022-2836(87)90648-6. [DOI] [PubMed] [Google Scholar]
- Hübner P., Iida S., Arber W. A transcriptional terminator sequence in the prokaryotic transposable element IS1. Mol Gen Genet. 1987 Mar;206(3):485–490. doi: 10.1007/BF00428889. [DOI] [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988 Nov 4;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakowec M., Prentki P., Chandler M., Galas D. J. Mutational analysis of the open reading frames in the transposable element IS1. Genetics. 1988 Sep;120(1):47–55. doi: 10.1093/genetics/120.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lüthi K., Moser M., Ryser J., Weber H. Evidence for a role of translational frameshifting in the expression of transposition activity of the bacterial insertion element IS1. Gene. 1990 Mar 30;88(1):15–20. doi: 10.1016/0378-1119(90)90054-u. [DOI] [PubMed] [Google Scholar]
- Machida C., Machida Y., Ohtsubo E. Both inverted repeat sequences located at the ends of IS1 provide promoter functions. J Mol Biol. 1984 Aug 5;177(2):247–267. doi: 10.1016/0022-2836(84)90455-8. [DOI] [PubMed] [Google Scholar]
- Machida C., Machida Y. Regulation of IS1 transposition by the insA gene product. J Mol Biol. 1989 Aug 20;208(4):567–574. doi: 10.1016/0022-2836(89)90148-4. [DOI] [PubMed] [Google Scholar]
- Machida Y., Machida C., Ohtsubo E. Insertion element IS1 encodes two structural genes required for its transposition. J Mol Biol. 1984 Aug 5;177(2):229–245. doi: 10.1016/0022-2836(84)90454-6. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Nucleotide sequence of an insertion element, IS1. Proc Natl Acad Sci U S A. 1978 Feb;75(2):615–619. doi: 10.1073/pnas.75.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Prentki P., Krisch H. M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984 Sep;29(3):303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Prentki P., Pham M. H., Gamas P., Chandler M., Galas D. J. Artificial transposable elements in the study of the ends of IS1. Gene. 1987;61(1):91–101. doi: 10.1016/0378-1119(87)90368-4. [DOI] [PubMed] [Google Scholar]
- Remaut E., Tsao H., Fiers W. Improved plasmid vectors with a thermoinducible expression and temperature-regulated runaway replication. Gene. 1983 Apr;22(1):103–113. doi: 10.1016/0378-1119(83)90069-0. [DOI] [PubMed] [Google Scholar]
- Schimmel P. RNA pseudoknots that interact with components of the translation apparatus. Cell. 1989 Jul 14;58(1):9–12. doi: 10.1016/0092-8674(89)90395-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine Y., Ohtsubo E. Frameshifting is required for production of the transposase encoded by insertion sequence 1. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4609–4613. doi: 10.1073/pnas.86.12.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Shuh M., Atkins J. F., Gesteland R. F. E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol. 1989 Nov;1(2):159–169. [PubMed] [Google Scholar]
- Zerbib D., Jakowec M., Prentki P., Galas D. J., Chandler M. Expression of proteins essential for IS1 transposition: specific binding of InsA to the ends of IS1. EMBO J. 1987 Oct;6(10):3163–3169. doi: 10.1002/j.1460-2075.1987.tb02627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbib D., Polard P., Escoubas J. M., Galas D., Chandler M. The regulatory role of the IS1-encoded InsA protein in transposition. Mol Microbiol. 1990 Mar;4(3):471–477. doi: 10.1111/j.1365-2958.1990.tb00613.x. [DOI] [PubMed] [Google Scholar]
- Zerbib D., Prentki P., Gamas P., Freund E., Galas D. J., Chandler M. Functional organization of the ends of IS1: specific binding site for an IS 1-encoded protein. Mol Microbiol. 1990 Sep;4(9):1477–1486. [PubMed] [Google Scholar]





