Abstract
When type III interferon (IFN-λ; also known as interleukin-28 [IL-28] and IL-29) was discovered in 2003, its antiviral function was expected to be analogous to that of type I IFNs (IFN-α and IFN-β) via the induction of IFN-stimulated genes (ISGs). Although IFN-λ stimulates expression of antiviral ISGs preferentially in cells of epithelial origin, recent studies have defined additional antiviral mechanisms in other cell types and tissues. Viral infection models using mice lacking IFN-λ signaling and SNP associations with human disease have expanded our understanding of the contribution of IFN-λ to the antiviral response at anatomic barriers and the immune response beyond these barriers. In this review, we highlight recent insights into IFN-λ functions, including its ability to restrict virus spread into the brain and to clear chronic viral infections in the gastrointestinal tract. We also discuss how IFN-λ modulates innate and adaptive immunity, autoimmunity, and tumor progression and its possible therapeutic applications in human disease.
Interferon-λ induces antiviral gene programs in restricted cell types, including epithelial cells. Diamond and colleagues discuss recent insights into the induction of interferon-λ, its role in barrier immunity, and its connections to human disease.
Main Text
Introduction
Interferon-λ (IFN-λ), also termed type III IFN or interleukin-28 (IL-28) and IL-29, belongs to a cytokine family that shares functional similarities with the family of type I IFNs (IFN-α and/or IFN-β [hereafter IFN-α/β]). IFN-λ and IFN-α/β are multi-gene families composed of closely related cytokines each with specific heterodimeric receptors: IFNLR (IFNLR1 and IL10Rβ) for IFN-λ and IFNAR (IFNAR1 and IFNAR2) for IFN-α/β. Humans have genes encoding four IFN-λ proteins (IFN-λ1 [IL-29], IFN-λ2 [IL-28A], IFN-λ3 [IL-28B], and IFN-λ4) as well as 17 IFN-α/β proteins (13 IFN-α subtypes, IFN-β, IFN-ω, IFN-ε, and IFN-κ). In contrast to the IFN-α/β family, which was described almost 60 years ago (Isaacs and Lindenmann, 1957), the IFN-λ family was discovered more recently. Human IFN-λ1, IFN-λ2, and IFN-λ3 were identified in 2003 (Kotenko et al., 2003, Sheppard et al., 2003), and IFNL4 at that time was considered a pseudogene. In 2013, it became clear that many humans have a functional IFNL4 and that a common SNP results in a frameshift mutation that ablates IFN-λ4 production in some populations (Hamming et al., 2013, Prokunina-Olsson et al., 2013). Genes encoding type I IFNs characteristically lack introns (with the exception of IFNK) and are syntenic in a single locus on human chromosome 9 and mouse chromosome 4. Genes encoding IFN-λ are located on human chromosome 19 and mouse chromosome 7 and share the 5-exon gene structure characteristic of IL-10 cytokine family members (Sabat, 2010).
As discussed herein and in other recent reviews (Donnelly and Kotenko, 2010, Durbin et al., 2013, Egli et al., 2014c, Griffiths et al., 2015, Hermant and Michiels, 2014, Hoffmann et al., 2015, Koch and Finotto, 2015, Kotenko, 2011, O’Brien et al., 2014, Odendall and Kagan, 2015), several aspects of IFN-λ biology differ from IFN-α/β biology. First, the effects of IFN-λ are most evident in epithelial cells, suggesting that it contributes to the specialized immune mechanisms that protect epithelial surfaces, which undergo constant exposure to commensal and pathogenic microbes (Durbin et al., 2013, Hermant and Michiels, 2014, Mahlakõiv et al., 2015). Second, because of the more focused nature of its signaling effects, IFN-λ might share the therapeutic benefits yet avoid many of the side effects that have limited the clinical use of IFN-α/β (Donnelly et al., 2011, Hermant and Michiels, 2014, Markowitz, 2007, Pagliaccetti and Robek, 2010, Pestka, 2007). Third, genome-wide association studies (GWASs) have revealed multiple IFNL polymorphisms that are linked to clearance of hepatitis C virus (HCV) infection and possibly improved outcomes with other viral infections, including hepatitis B virus (HBV), human cytomegalovirus (HCMV), and herpes simplex virus 1 (HSV-1) (Egli et al., 2014c, Galmozzi et al., 2014, Griffiths et al., 2015, Lampertico et al., 2013, Manuel et al., 2015).
In this review, we discuss new insights into the antiviral functions of IFN-λ, especially those distinguishing it from IFN-α/β. We also describe how IFN-λ modulates adaptive immunity, autoimmunity, and tumor progression, as well as possible therapeutic applications in human disease. We highlight a specialized role for IFN-λ in providing antiviral activity at anatomic barriers, including epithelial surfaces and the blood-brain barrier (BBB). Because epithelial surfaces experience constant microbial exposure, IFN-λ might provide more targeted antiviral protection at key barrier sites without activating a systemic pro-inflammatory immune response.
Induction of IFN-λ: Mechanisms Shared with and Distinct from Those of IFN-α/β
The stimuli that induce expression of IFN-λ-encoding genes, including a range of viruses, are similar to those inducing expression of genes encoding IFN-α/β (Ank et al., 2008, Ank et al., 2006, Durbin et al., 2013, Kotenko et al., 2003, Sheppard et al., 2003). Nonetheless, there are differences in transcription factor requirements between IFN-α/β and IFN-λ. Given that some of these differences have been reviewed previously (Durbin et al., 2013, Iversen and Paludan, 2010), we highlight recent advances in our understanding of transcriptional regulation of IFN-λ-encoding genes.
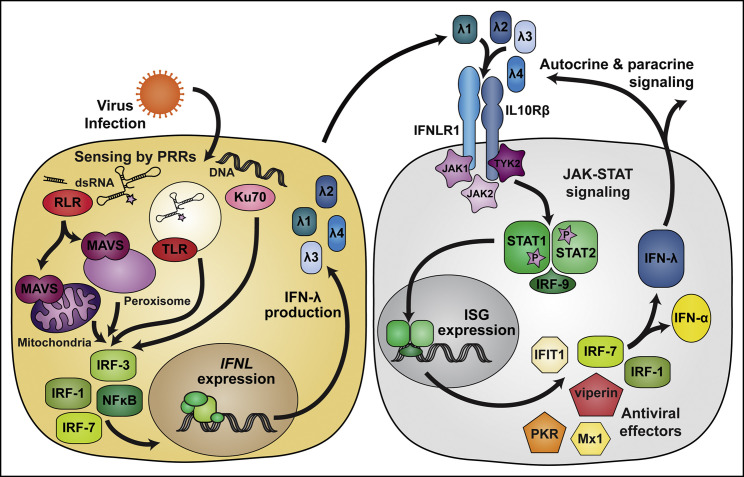
IFN expression occurs after host detection of pathogen-associated molecular patterns by specific pattern-recognition receptors (PRRs) (Swiecki and Colonna, 2011; Figure 1 ). Although most PRRs responsible for inducing expression of IFN-λ overlap those triggering IFN-α/β expression, the cytosolic DNA sensor Ku70 activates the expression of IFN-λ but not IFN-α/β (Zhang et al., 2011). Transcription factors activated downstream of PRR signaling include interferon regulatory factors (IRFs) and NF-κB. The suite of PRRs and transcription factors expressed by a cell contributes to the specific capabilities and magnitude of the IFN response following infection (Hillyer et al., 2012). For example, IFN-β is induced early after activation of PRRs because its promoter is bound by IRF-3, which is constitutively expressed, whereas most IFN-α subtypes are induced preferentially by IRF-7, which is itself encoded by an IFN-stimulated gene (ISG) (Génin et al., 2009). Initial characterization of promoter regions upstream of IFNL1 and IFNL3 identified binding elements for IRF-1, IRF-3, IRF-7, and NF-κB, and the combined activity of IRFs and NF-κB was required for maximal gene induction (Onoguchi et al., 2007, Osterlund et al., 2007, Thomson et al., 2009). Subsequent analysis of the response to TLR9 agonists showed that IFN-λ induction exhibits a greater dependence on NF-κB than does IFN-α/β induction (Iversen et al., 2010). These findings suggest that, despite similarities, there are promoter features that discriminate IFNL and IFNA/B induction.
Figure 1.
IFN-λ Induction and Signaling Pathways
IFN-λ production is induced when viral infection is sensed by pattern recognition receptors (PRRs), including members of the RIG-I-like receptor (RLR) and Toll-like receptor (TLR) families, as well as the DNA sensor Ku70. Whereas IFN-λ is induced via many of the same signaling pathways that induce IFN-α/β, Ku70 and peroxisome-localized MAVS preferentially induce IFN-λ. IFN-λ signals through its heterodimeric receptor, IFNLR, which is composed of IFNLR1 and IL10Rβ subunits. Canonical signaling through IFNLR activates JAK1 and TYK2 kinases, which phosphorylate STAT1 and STAT2. However, IFNLR signaling also can activate JAK2 and other downstream signaling pathways (not depicted). JAK-STAT signaling induces expression of IFN-stimulated genes (ISGs) and the production of effector molecules that inhibit viral infection. Among the ISGs induced by IFN-λ are IRF1 and IRF7, encoding transcription factors IRF-1 and IRF-7, respectively, which amplify IFN production.
Additional factors distinguish the regulation of IFNL and IFNA/B induction. Med23, a component of the Mediator complex, interacts with IRF-7 and enhances transcription from the IFNL1 promoter, but not the IFNB promoter; the resulting increase in IFN-λ production inhibits HSV-1 replication (Griffiths et al., 2013). A bioinformatic and biochemical analysis of the region upstream of human IFNL1 identified binding sites for the transcription factors ZEB1 and BLIMP-1. Experiments using chromatin immunoprecipitation and gene silencing established that ZEB1 and BLIMP-1 bind the IFNL1 promoter and repress transcription in airway and intestinal epithelial cell lines (Siegel et al., 2011, Swider et al., 2014). ZEB1 activity is specific to IFNL1 and does not regulate IFNB expression. BLIMP-1 functions as a repressor by displacing binding of IRF-1 (Siegel et al., 2011), which is required for IFNL1, but not IFNB, transcription (Odendall et al., 2014). Although both IFN-λ and IFN-β are induced downstream of PRR sensing and activation of mitochondrial antiviral signaling protein (MAVS), IFN-λ production is favored when activated MAVS localizes to the peroxisome, whereas IFN-β production is dominant when MAVS localizes to the mitochondria (Odendall et al., 2014). The relative abundance of peroxisomes in epithelial cells suggests a mechanism for preferential production of IFN-λ instead of IFN-β in response to viral infection at epithelial surfaces. In hepatocytes, HBV infection stimulated production of IFN-λ but not IFN-α/β, and this IFN-λ induction depended on recognition by RIG-I and signaling through MAVS (Sato et al., 2015). Similarly, HCV infection preferentially induced IFN-λ over IFN-α/β (Marukian et al., 2011, Park et al., 2012, Thomas et al., 2012), suggesting that liver-specific factors might promote IFN-λ production in response to these unrelated hepatotropic viruses.
Although epithelial cells produce IFN-λ, myeloid-lineage cells are major sources in response to double-stranded RNA (poly I:C) or viral infections (Lauterbach et al., 2010). In the small intestine, epithelial cells and immune cells both respond to poly I:C stimulation, suggesting that multiple cell types produce IFN-λ cooperatively (Mahlakõiv et al., 2015). Although gut immune cells treated with poly I:C produce IFN-α5, IFN-β, IFN-λ2, and IFN-λ3, gut epithelial cells exclusively generate IFN-λ2 and IFN-λ3 (Mahlakõiv et al., 2015), perhaps reflecting their peroxisome content (Odendall et al., 2014). There also are differences in the IFN induction profile within the myeloid cell compartment after stimulation with PRR agonists. Whereas plasmacytoid dendritic cells (DCs) produce nearly all IFNs (including IFN-λ), monocytes and myeloid DCs more selectively express IFN-β, IFN-λ1, and IFN-λ2 (Hillyer et al., 2012). Similarly, CD8α+ DCs are the major IFN-λ-producing myeloid cell type in response to poly I:C in mice (Lauterbach et al., 2010).
IFN-λ Signaling
The proximal signaling events and downstream transcriptional responses are similar between IFN-α/β and IFN-λ, even though the cytokines and their receptors are structurally and genetically distinct. The structure of IFN-λ resembles that of members of the IL-10 family, although the primary amino acid sequence is more similar to that of IFN-α/β (Gad et al., 2009, Miknis et al., 2010). Whereas all type I IFNs signal through a shared heterodimeric receptor, IFNAR (IFNAR1 and IFNAR2), type III IFNs bind to IFNLR, a unique heterodimeric receptor. IFNLR consists of one subunit that it shares with other IL-10 family cytokines (IL10Rβ) and a second that is specific to IFN-λ (IFNLR1, also called IL28Rα). Despite the structural similarities between IFN-λ and IL-10 family cytokines (i.e., IL-10, IL-19, IL-20, IL-22, IL-24, and IL-26), the crystal structure of IFN-λ1 bound to IFNLR1 revealed a distinct receptor-binding interaction, including a 1:1 stoichiometry between IFN-λ1 and IFNLR1 (compared to 2:1 between IL-10 and IL10Rα) (Miknis et al., 2010). The impact of the shared IL10Rβ chain on interactions between IFN-λ and other IL-10 family cytokines remains unclear. IFN-λ activity was enhanced by an IL-10-blocking antibody and was inhibited in the presence of IL-10 (Jordan et al., 2007a), suggesting possible competition for IL10Rβ interaction. In contrast, IL-22 (which signals through IL10Rβ and IL22Rα) enhanced the signaling and antiviral effects of IFN-λ (Hernández et al., 2015). Although IFNAR and IL10Rβ are expressed broadly on many cell types and tissues, IFNLR1 is expressed preferentially on epithelial cells (Mahlakõiv et al., 2015, Sommereyns et al., 2008). Consistent with this pattern, the antiviral effects of IFN-λ are most evident against pathogens targeting epithelial tissues.
Despite engaging different heterodimeric receptors, the post-receptor signaling events after IFN-α/β and IFN-λ binding exhibit remarkable overlap. These signaling pathways have been reviewed in detail elsewhere (Donnelly and Kotenko, 2010, Durbin et al., 2013, Hoffmann et al., 2015, Kotenko, 2011) and include activation of JAK-family kinases, phosphorylation of STAT1 and STAT2, and association between activated STAT complexes and IRF-9 to form ISGF3, which translocates to the nucleus and induces expression of hundreds of ISGs. Similar to IFN-α/β signaling, IFN-λ signaling induces JAK1 and TYK2 phosphorylation (Dumoutier et al., 2004, François-Newton et al., 2011, Ma et al., 2009). Additionally, JAK2 phosphorylation is induced specifically by IFN-λ (Odendall et al., 2014, Odendall and Kagan, 2015), suggesting that distinct upstream signaling events might differentiate IFN-λ activity from IFN-α/β activity. In addition to activating STAT1 and STAT2, IFNAR and IFNLR ligand engagement can activate other STAT family members (STAT3, STAT4, and STAT5) and STAT-independent signaling cascades (MAPK and ERK) (Cohen and Prince, 2013, Koch and Finotto, 2015).
The transcriptional responses induced by IFN-λ and IFN-α/β are similar (Bolen et al., 2014, Doyle et al., 2006, Kohli et al., 2012, Lazear et al., 2015, Marcello et al., 2006, Shindo et al., 2013, Zhou et al., 2007). No transcriptional signatures unique to IFN-λ have been identified, and the genes induced by IFN-λ typically represent a subset of the total induced by IFN-α/β. The IFN-λ transcriptional response generally is of lower magnitude than that of IFN-α/β and might exhibit a delayed peak and longer duration (Marcello et al., 2006), although different expression levels of IFNAR and IFNLR might confound comparisons. Differential effects of negative regulators might contribute to the sustained signaling pattern observed for IFN-λ in comparison to that of IFN-α. For example, the ISG USP18 desensitizes cells to further IFN-α stimulation but does not inhibit IFN-λ signaling (François-Newton et al., 2011). Among the IFN-λ subtypes, IFN-λ3 (followed by IFN-λ1 and IFN-λ2) has the most potent bioactivity (Bolen et al., 2014, Dellgren et al., 2009). These differences in potency are unexpected, given that IFN-λ2 and IFN-λ3 are nearly identical (96% amino acid identity) (Sheppard et al., 2003). IFN-α/β and IFN-λ signaling both amplify IFN production, for example, via the induction of the transcription factors IRF-1 and IRF-7 (Bolen et al., 2014, Kohli et al., 2012, Lazear et al., 2015). Because of these shared positive-feedback systems, transcriptional profiling in cells lacking IFNAR or IFNLR (or in the presence of protein-synthesis inhibitors) might be needed for distinguishing bona fide IFN-α/β- and IFN-λ-induced genes. Independent of whether there truly are IFN-λ-specific transcriptional signatures, the ability to induce a narrower set of ISGs in a more targeted set of cells suggests possible therapeutic applications in which IFN-λ could promote a more focused antiviral or immunomodulatory response with fewer side effects than IFN-α/β (Hermant and Michiels, 2014, Muir et al., 2014). Given that most IFN-λ transcriptional profiling experiments have focused on human liver cells, further studies are needed in other tissues where antiviral effects of IFN-λ have been reported.
Antiviral Effects of IFN-λ
IFN-λ exhibits antiviral activity against many viruses in vitro, but its in vivo activity has been more apparent for viruses that infect epithelial cells of the respiratory, gastrointestinal, and urogenital tracts, as well as the liver (Table 1 ). Because the antiviral effects of IFN-λ have been reviewed recently (Egli et al., 2014c, Hermant and Michiels, 2014, O’Brien et al., 2014, Sorgeloos et al., 2013), we will briefly describe the role of IFN-λ in the respiratory tract and liver and then highlight new phenotypes in the gastrointestinal tract, as well as at an unexpected site, the BBB.
Table 1.
Antiviral Effects of IFN-λ In Vivo
| Virus | Phenotype | Reference |
|---|---|---|
| Mouse Models | ||
| West Nile virus | more permeable BBB and increased neuroinvasion in Ifnlr1−/− mice; treatment with recombinant IFN-λ prevents lethality in wild-type mice | Lazear et al., 2015 |
| Norovirus | increased titers and shedding in Ifnlr1−/− mice; treatment with recombinant IFN-λ prevents infection and cures persistent infection | Nice et al., 2015 |
| Reovirus | increased growth in intestinal epithelial cells and increased viral shedding in Ifnlr1−/− mice; increased fatal liver disease in suckling Ifnlr1−/− mice | Mahlakõiv et al., 2015 |
| Rotavirus | increased titers in Ifnlr1−/− mice; treatment with recombinant IFN-λ reduces viral titers; IL-22 acts synergistically with IFN-λ to control infection | Hernández et al., 2015, Pott et al., 2011 |
| Influenza virus | increased titers in Ifnlr1−/− or Ifnlr1−/− × Ifnar1−/− mice; treatment with recombinant IFN-λ prevents lethality in wild-type mice | Crotta et al., 2013, Mordstein et al., 2008, Mordstein et al., 2010b |
| SARS coronavirus | increased titers and shedding in Ifnlr1−/− mice | Mahlakõiv et al., 2012, Mordstein et al., 2010b |
| Human metapneumovirus | increased titers in Ifnlr1−/− × Ifnar1−/− mice; treatment with recombinant IFN-λ reduces titers in wild-type mice | Baños-Lara et al., 2015, Mordstein et al., 2010b |
| Respiratory syncytial virus | increased titers in Ifnlr1−/− × Ifnar1−/− mice | Mordstein et al., 2010b |
| Herpes simplex virus 2 | treatment with recombinant IFN-λ decreases viral titers and shedding | Ank et al., 2006 |
| Lymphocytic choriomeningitis virus | no change in viral titers in Ifnlr1−/− mice; increased T cell response to acute infection and decreased T cell response to chronic infection in Ifnlr1−/− mice | Ank et al., 2008, Misumi and Whitmire, 2014 |
| Human Patients | ||
| Hepatitis C virus | SNPs rs8099917, rs12979860, rs4803217, and rs368234815 correlate with spontaneous clearance and sustained response to IFN therapy | Bibert et al., 2013, Ge et al., 2009, Griffiths et al., 2015, Hamming et al., 2013, Prokunina-Olsson et al., 2013, Suppiah et al., 2009, Tanaka et al., 2009, Thomas et al., 2009 |
| Hepatitis B virus | SNP rs12979860 correlates with response to IFN therapy | Galmozzi et al., 2014 |
| Human cytomegalovirus | SNP rs368234815 correlates with HCMV retinitis in AIDS patients; SNPs rs368234815 and rs8099917 correlate with HCMV reactivation in transplant recipients | Bibert et al., 2014, Egli et al., 2014a, Egli et al., 2014c, Griffiths et al., 2015, Manuel et al., 2015 |
| Herpes simplex virus 1 | SNP rs12979860 correlates with the severity of reactivation disease | Griffiths et al., 2013 |
| Influenza virus | increased viral titers in epithelial cells with IFNLR1 inhibition; SNP rs8099917 correlates with vaccine response in immunosuppressed patients | Egli et al., 2014b, Kim et al., 2013 |
| Rhinovirus | increased production of IFN-λ correlates with reduced viral replication in bronchial epithelial cells | Contoli et al., 2006 |
IFN-λ in the Respiratory Tract
IFNLR is expressed at relatively high levels in respiratory epithelial cells, and mice treated with IFN-λ prior to infection with human metapneumovirus (HMPV) develop lower viral titers and reduced inflammatory responses (Baños-Lara et al., 2015). Accordingly, Ifnlr1 −/− mice exhibit increased susceptibility to respiratory viral infections, including influenza virus, HMPV, respiratory syncytial virus, and SARS coronavirus (Crotta et al., 2013, Hermant and Michiels, 2014, Mahlakõiv et al., 2012, Mordstein et al., 2008, Mordstein et al., 2010b). Human myeloid and bronchial epithelial cells produce IFN-λ in response to rhinovirus infection, and IFN-λ inhibits rhinovirus replication in bronchial epithelial cells (Contoli et al., 2006). Correspondingly, production of IFN-λ correlates inversely with viral load and disease severity in humans experimentally inoculated with rhinovirus (Contoli et al., 2006). Respiratory epithelial cells respond to both IFN-λ and IFN-α/β (Mahlakõiv et al., 2015), and cultured cells respond to IFN-λ from both the apical and basolateral surfaces (Fox et al., 2015, Hamming et al., 2013). IFN-λ is the dominant IFN produced by respiratory epithelial cells after infection with influenza virus or other respiratory viruses (Crotta et al., 2013, Fox et al., 2015, Jewell et al., 2010, Okabayashi et al., 2011). This implies a key role for IFN-λ in mediating antiviral immunity in the respiratory tract (Figure 2 A).
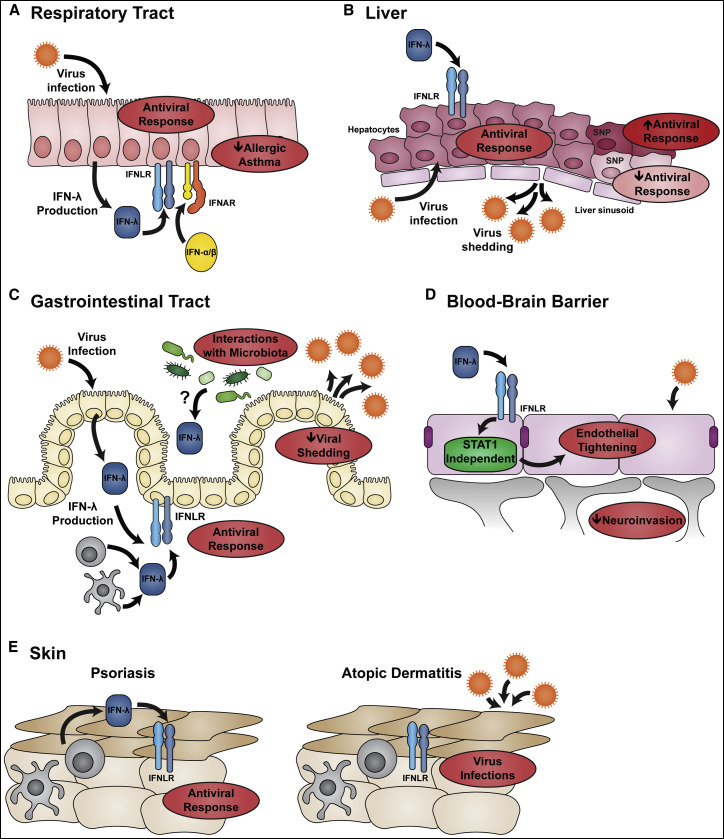
Figure 2.
Antiviral Effects of IFN-λ at Barrier Surfaces
(A) IFN-λ is a dominant IFN produced after viral infection in the respiratory tract. Respiratory epithelial cells can respond to both IFN-λ and IFN-α/β to activate an antiviral response. Th1 skewing induced by IFN-λ reduces the severity of allergic asthma.
(B) The fenestrated endothelium of the liver creates a tissue architecture in which hepatocytes directly contact blood in liver sinusoids. Hepatocytes are the primary cellular targets for HBV and HCV and are highly responsive to IFN-λ. A role for IFN-λ in controlling HCV infection is suggested by the association between HCV clinical outcome and numerous SNPs within the IFNL locus.
(C) IFN-λ has a key role in gastrointestinal tract immunity because unlike respiratory epithelial cells, gut epithelial cells do not respond to IFN-α/β and therefore rely upon IFN-λ to activate an antiviral response. The antiviral effects of IFN-λ could be especially important for restricting the shedding and transmission of enteric viruses. Immunity in the gastrointestinal tract is shaped by the bacterial microbiome; the ability of gut microbes to promote viral persistence requires IFN-λ signaling, although the mechanism of this interaction remains unclear.
(D) IFN-λ signaling tightens the endothelial junctions of the BBB, which reduces viral neuroinvasion from the circulation. The tightening activity of IFN-λ is STAT1 independent, implicating a non-canonical signaling pathway.
(E) Psoriasis and atopic dermatitis are chronic inflammatory skin conditions characterized by breakdown of the epithelial barrier. Compared to lesions from atopic dermatitis patients, psoriasis lesions exhibit elevated IFN-λ production and enhanced expression of ISGs. This might explain why disseminated viral skin infections are common in patients with atopic dermatitis but not psoriasis.
IFN-λ in the Liver
There has been great interest in defining the antiviral role of IFN-λ in the liver, particularly against HCV and HBV infections. IFNLR is expressed by human hepatocytes, the primary cellular targets of these viruses (Figure 2B), and both viruses preferentially stimulate IFN-λ production rather than IFN-α/β production (Marukian et al., 2011, Park et al., 2012, Sato et al., 2015, Thomas et al., 2012). GWASs have revealed IFNL polymorphisms that are associated with improved outcome from HCV and HBV infection, both in terms of spontaneous clearance and in response to antiviral therapy (Galmozzi et al., 2014, Ge et al., 2009, Griffiths et al., 2015, Hermant and Michiels, 2014, Lampertico et al., 2013, O’Brien et al., 2014, Russell et al., 2014, Suppiah et al., 2009, Tanaka et al., 2009, Thomas et al., 2009). The different protective alleles identified in these studies imply distinct antiviral mechanisms against HCV. One protective allele in the promoter region of IFNL3 is associated with increased IFN-λ production, which might control infection by enhancing expression of ISGs (for SNP rs8099917, the protective allele is T/T rather than T/G or G/G) (Suppiah et al., 2009, Tanaka et al., 2009). However, an unfavorable allele also is associated with elevated ISG expression and is thought to render cells refractory to further stimulation of antiviral activity (for SNP rs12979860, the protective allele is C/C rather than T/C or T/T) (Ge et al., 2009, Sheahan et al., 2014, Thomas et al., 2009, Urban et al., 2010). In addition to its association with viral hepatitis, rs12979860 predicts the severity of liver inflammation and fibrosis of other etiologies, but with the opposite association (C/C, unfavorable allele) (Eslam et al., 2015). This disparity suggests that ISG expression might differentially affect viral and non-viral liver disease. Another protective allele alters the sequence of the 3′ UTR of IFNL3 such that its transcript is no longer targeted for degradation by microRNAs induced after HCV infection (for SNP rs4803217, the protective allele is G/G rather than T/T) (McFarland et al., 2014). The SNP that is most highly associated with improved HCV outcome is one in which the protective allele has a frameshift insertion that ablates IFN-λ4 production (for SNP rs368234815, the protective allele is TT rather than -G) (Bibert et al., 2013, Hamming et al., 2013, Key et al., 2014, O’Brien et al., 2014, Prokunina-Olsson et al., 2013). It remains unclear why the loss of IFN-λ4 confers improved clinical outcome, but several explanations have been proposed: (1) a counter-regulatory role for IFN-λ4, (2) an IFN refractory state induced by high basal expression of IFN-λ4, (3) stimulation of regulatory T cell responses by IFN-λ4, and/or (4) reduced production of the more antiviral IFN-λ3. Because many of the polymorphisms within the IFNL locus exist in high linkage disequilibrium, they often segregate as haplotypes. This has made it difficult to parse out the protective functions of individual SNPs. For example, many of the protective phenotypes initially attributed to rs12979860 (C/C allele) might in fact be caused by the loss of IFN-λ4 production as a result of the highly linked and more recently identified rs368234815 (TT allele).
There was significant interest in the therapeutic potential of IFN-λ for viral hepatitis (Donnelly et al., 2011, Hayes et al., 2012), at least before the introduction of direct-acting antiviral agents to treat HCV infection (Alexopoulou and Karayiannis, 2015). Conventional pegylated IFN-α therapy is accompanied by side effects that hinder compliance. Given the more restricted expression pattern of IFNLR, and the relatively high amount of IFNLR expression in hepatocytes, IFN-λ could confer many of the antiviral benefits of IFN-α in the liver with reduced side effects (Muir et al., 2014). Although direct-acting antiviral agents are supplanting IFN therapy for HCV infection, the therapeutic advantages of IFN-λ might still have utility against HBV, other viral infections, or other hepatic conditions.
Although human hepatocytes respond to IFN-λ, mouse hepatocytes are less responsive. Instead, cholangiocytes that line the bile duct reportedly are the principal cells that respond to IFN-λ in the mouse liver (Hermant et al., 2014, Mahlakõiv et al., 2015). This observation could explain the apparent lack of an antiviral activity for IFN-λ in mouse models of hepatotropic virus infection, despite its clear role in human hepatitis (Mordstein et al., 2008). Because of their IFN-λ responsiveness, cholangiocytes could restrict excretion of hepatotropic viruses into the bile and thus control viral spread in feces.
IFN-λ in the Gastrointestinal Tract
The gastrointestinal tract is a primary site of microbial exposure, and intestinal epithelial cells form a barrier to pathogenic viruses and bacteria (Figure 2C). Initial characterization of tissue Ifnlr1 expression in mice demonstrated relatively high expression in the stomach and intestine (Sommereyns et al., 2008), and subsequent studies revealed that epithelial cells are the predominant IFN-λ-responsive cell type in the gastrointestinal tract (Mordstein et al., 2010b, Pott et al., 2011). IFN-λ and IFN-α/β responsiveness are compartmentalized within the mouse intestine. IFN-α/β has a minimal effect on intestinal epithelial cells, whereas IFN-λ has a minimal effect on cells of the lamina propria (Mahlakõiv et al., 2015, Pott et al., 2011). This differential response might be explained by the relatively high expression of IFNLR on intestinal epithelial cells (Mahlakõiv et al., 2015), as well as the low expression and apical trafficking of IFNAR on these cells (Pott et al., 2011). Although IFNLR1 transcripts are expressed in the human gastrointestinal tract (Sheppard et al., 2003), it remains to be determined whether IFN-λ and IFN-α/β responsiveness are similarly compartmentalized.
The compartmentalized response to IFN-λ in the mouse gastrointestinal tract most likely explains its activity against different viral pathogens depending on their cellular tropism. Rotavirus infects epithelial cells (Lopez and Arias, 2006), reovirus replicates in both epithelial and non-epithelial cells (Forrest and Dermody, 2003), and norovirus grows preferentially in myeloid and B cells but not in epithelial cells (Jones et al., 2014, Karst et al., 2014, Wobus et al., 2004). Rotavirus infection is controlled exclusively by IFN-λ, and there is no defined role for IFN-α/β (Angel et al., 1999, Pott et al., 2011). Reovirus is controlled cooperatively by IFN-α/β and IFN-λ, such that IFN-λ limits replication in epithelial cells, and IFN-α/β restricts infection in non-epithelial cells (Mahlakõiv et al., 2015). Norovirus infection and dissemination are controlled by IFN-α/β and IFN-λ, such that IFN-α/β prevents extra-intestinal spread, and IFN-λ inhibits the persistent shedding of virus in feces (Nice et al., 2015, Nice et al., 2013). The role for IFN-λ in norovirus infection is apparent only for strains that are persistently shed across the intestinal epithelial barrier into the gut lumen and not for strains causing only acute infection. These studies with genetically and phenotypically unrelated gastrointestinal viruses reveal unique antiviral roles for IFN-α/β and IFN-λ. In particular, IFN-λ selectively prevents viral shedding into the lumen of the gastrointestinal tract and transmission of fecal-oral viral pathogens.
The trillions of bacteria juxtaposed to the intestinal epithelial barrier are a source of numerous microbial products that are sensed by the immune system. Thus, the IFN-mediated antiviral response in the gastrointestinal tract is modulated by the bacterial microbiome. Indeed, a recent study showed that the bacterial microbiome promotes persistent norovirus infection in an IFNLR-dependent manner (Baldridge et al., 2015). Fecal shedding of norovirus, but not viral entry into Peyer’s patches or extra-intestinal spread, is reduced in mice treated with antibiotics that deplete the bacterial microbiome. This effect of antibiotics on norovirus shedding is diminished in Ifnlr1 −/− mice, but not in Ifnar1 −/−, Ifngr1 −/−, Tlr4 −/−, Myd88 −/−, or Trif −/− mice, which lack other innate immune signaling components. The specific requirement for IFNLR in bacterial promotion of norovirus infection contrasts with that in poliovirus and mouse mammary tumor virus (MMTV) infections, where bacterial promotion of viral infection depends on bacterial lipopolysaccharide and TLR4, respectively (Kane et al., 2011, Kuss et al., 2011). However, an additional role of IFN-λ in poliovirus and MMTV infections remains possible because TLR4 signaling and bacterial infection induce IFN-λ (Bierne et al., 2012, Coccia et al., 2004, Contoli et al., 2006, Odendall et al., 2014). Further experiments are required to characterize the intestinal production of and response to IFN-λ in the presence and absence of enteric bacteria. Given the importance of IFN-λ to intestinal immunity in mice, studies to test for linkage between human IFN-λ SNPs and the control of enteric viral infections appear warranted. The studies in mice also suggest that IFN-λ might have therapeutic applications for humans with persistent norovirus infection and gastroenteritis (Bok and Green, 2012).
The ability of IFN-λ to confer antiviral protection at epithelial barriers is an interesting parallel to another IL-10 family cytokine, IL-22, which promotes antibacterial immunity at epithelial surfaces (Zheng et al., 2008). Similar to IFNLR1, IL22Rα associates with IL10Rβ and is expressed preferentially by epithelial cells (Wolk et al., 2004). A recent study demonstrated that IL-22 acts synergistically with IFN-λ to control rotavirus infection and prevent intestinal tissue damage in mice (Hernández et al., 2015). Further work is needed for determining whether IL-22 contributes to the antiviral activity of IFN-λ at other epithelial surfaces and against other viruses targeting the gastrointestinal tract.
IFN-λ and Chronic Viral Infection
Surprisingly, IFN-λ stimulates clearance of persistent norovirus infection independently of the adaptive immune system (Nice et al., 2015), suggesting that innate immune responses might have a greater role in determining viral persistence than previously appreciated. Because IFNLR is expressed on epithelial cells, IFN-λ could be particularly important for restricting shedding of chronic viral infections. Consistent with this idea, IFN-λ treatment decreases HSV-2 shedding from the vaginal mucosa in mice (Ank et al., 2006). In humans, SNPs in IFNL3 and IFNL4 have been associated with the ability to clear or control multiple chronic infections, including HCV, HBV, HCMV, and HSV-1 (Bibert et al., 2014, Egli et al., 2014a, Egli et al., 2014c, Griffiths et al., 2015, Griffiths et al., 2013, Lampertico et al., 2013, Manuel et al., 2015). Although the mechanisms by which these polymorphisms affect IFN-λ production and activity remain unclear, these associations suggest that IFN-λ contributes to the control of chronic viral infections in humans. Restricted IFNLR expression within the mucosal epithelium and other key cell types might allow targeted local antiviral responses that control chronic infection and shedding without generating a potentially deleterious systemic pro-inflammatory response. A greater mechanistic understanding of how human SNPs influence IFN-λ expression or function and the study of IFN-λ in additional models of chronic viral shedding are needed.
IFN-λ at the BBB
A recent study with West Nile virus (WNV) demonstrated an antiviral effect of IFN-λ at an unexpected barrier site: the BBB (Lazear et al., 2015). The BBB protects the CNS from harmful substances in the periphery, including viruses (Koyuncu et al., 2013). Ifnlr1 −/− mice exhibited increased BBB permeability after WNV infection and sustained higher viral titers in CNS tissues. Although endothelial cells, including those composing the BBB, do not express high levels of IFNLR, administration of exogenous IFN-λ tightened the BBB, restricted viral neuroinvasion, reduced viral titers in the CNS, and protected mice from lethal viral infection (Figure 2D). These observations are consistent with those of a study demonstrating that IFN-α/β also can exert a tightening effect on the BBB (Daniels et al., 2014). IFN-λ tightening of endothelial monolayers in an in vitro model of the BBB did not require STAT1 or de novo protein synthesis, implying that the phenotype occurred via a non-canonical signaling pathway. Although further experiments are needed for determining the specific pathway by which it exerts its effects on BBB integrity, this study suggests that IFN-λ might have therapeutic activity for pathological conditions or viral infections that involve BBB breakdown or permeability.
IFN-λ Roles in Autoimmunity, Adaptive Immunity, and Anti-tumor Responses
The most direct consequence of IFN-λ signaling is the induction of ISGs, which can act in a cell-intrinsic manner to inhibit viral infection (Figure 1). However, IFN-λ has immunologic roles beyond the innate antiviral response. There is increasing evidence that IFN-λ shapes the adaptive immune response to viral infection, alters anti-tumor responses, and affects autoimmunity.
IFN-λ and Autoimmunity
Altered expression or activity of components of pathogen sensing and IFN-induction pathways is associated with autoimmunity. For example, alterations in the PRR MDA5, signaling molecule STING, or transcription factor IRF-5 can cause severe clinical syndromes known as “type I interferonopathies” (Crow, 2015, Rigby and Rehwinkel, 2015). As overlapping stimuli, sensors, and signaling mechanisms induce expression of type III IFNs, these same alterations might enhance IFN-λ production and contribute to these clinical phenotypes.
Given that it exerted a tightening effect on the BBB, IFN-λ could have a prominent barrier effect in epithelial tissues where IFNLR expression is higher. In the skin, IFNLR is expressed predominantly on keratinocytes (Witte et al., 2009). The barrier function of keratinocytes is important, as evidenced by the pathology of psoriasis and atopic dermatitis, two chronic conditions characterized by epithelial cell hyperproliferation, impaired barrier function, and inflammation (Figure 2E) (Clark, 2013, Leung, 2013, Perera et al., 2012). Patients with atopic dermatitis commonly develop disseminated bacterial (e.g., Staphylococcus aureus) and viral (e.g., HSV, human papillomavirus, and molluscum contagiosum poxvirus) infections in the skin (Christophers and Henseler, 1987, Wollenberg et al., 2003). However, these disseminated infections are not characteristic of psoriasis. Skin lesions from psoriasis patients exhibit higher amounts of IFN-λ1 and enhanced expression of ISGs than do atopic dermatitis lesions, and this occurs in the absence of differential expression of IFN-α/β (Wolk et al., 2013). Although altered epithelial barrier function is a feature of both psoriasis and atopic dermatitis, elevated IFN-λ production and ISG expression provide antiviral protection only in the context of psoriasis.
The factors that cause elevated IFN-λ production in psoriasis are not understood but might result from the Th17-driven response in psoriatic lesions rather than a Th2-driven response in atopic dermatitis. Th17 cells are the main source of IFN-λ in psoriasis lesions, and IFN-λ inhibits the production of Th2 cytokines, particularly IL-13 (Srinivas et al., 2008, Wolk et al., 2013). A protective role for IFN-λ in psoriasis is also suggested by the linkage of a SNP in the 3′ UTR of IFNLR1 to disease outcome (for SNP rs4649203, the protective allele is G rather than A), although a mechanistic basis for this association remains uncertain (Li et al., 2013, Strange et al., 2010).
A protective role for IFN-λ in allergic asthma also has been proposed (Figure 2A) (Koch and Finotto, 2015). Cells from asthma patients have impaired IFN-λ production after rhinovirus infection (Contoli et al., 2006). IFN-λ downregulates inflammatory Th2 cytokines, including IL-4, IL-5, and IL-13 (Jordan et al., 2007b, Koltsida et al., 2011, Srinivas et al., 2008), which contribute to asthma pathogenesis. In a mouse model of asthma, administration of IFN-λ reduced the production of IL-5 and IL-13, diminished eosinophil infiltration into the lung, and decreased Th17 cell responses, all of which minimized disease (Koltsida et al., 2011). The protective activity of IFN-λ in this model depended on IL-12 and IFN-γ, implicating the Th1-skewing effects of IFN-λ.
In a recent study using a mouse model of autoimmune arthritis, IFN-λ treatment reduced neutrophil recruitment into the joints and resulted in improved disease outcome (Blazek et al., 2015). This work identifies neutrophils as IFN-λ-responsive cells and suggests therapeutic applications for IFN-λ in treating autoimmune arthritis and other inflammatory conditions.
Despite their linkage to other autoimmune conditions, human SNPs near the genes encoding IFN-λ (rs8099917 and rs12979860) or IFNLR1 (rs4649203) did not show an association with multiple sclerosis (Lopez de Lapuente et al., 2012, Malhotra et al., 2011). Furthermore, the IFNLR1 allele that is protective for psoriasis was associated with exacerbated systemic lupus erythematosus (for SNP rs4649203, the protective allele is A rather than G) (Li et al., 2013), suggesting distinct contributions of IFN-λ signaling to different autoimmune conditions.
IFN-λ in Cancer
IFN-α is currently approved as a therapeutic agent for some cancers. The primary anti-tumor mechanisms of IFN-α/β are cell-intrinsic induction of apoptosis and cell-extrinsic stimulation and priming of immune cells (Booy et al., 2015, Di Trolio et al., 2015). Given that IFN-λ signaling triggers similar downstream gene expression, but in a restricted subset of IFNLR-bearing cells, it could serve as a more targeted therapeutic option. In support of this concept, IFNLR signaling induces apoptosis in colorectal cancer cells more potently than IFN-α/β or IFN-γ (Li et al., 2008) and inhibits growth and colony formation of several different tumor cell lines (Sato et al., 2006).
Induction of IFN-λ in response to viral infections could also promote anti-tumor immune responses. IFN-λ has been detected in human papillomavirus lesions, and lower levels of IFN-λ expression correlated with progression to cervical cancer (Cannella et al., 2014). Infection with a vesicular stomatitis virus strain intended for use as an oncolytic treatment also triggered IFN-λ expression in hematopoietic cells in vitro and enhanced anti-tumor natural killer (NK) cell responses in vivo (Wongthida et al., 2010). Loss of IFNLR expression on tumor cells prevented IFN-λ-stimulated expression of stimulatory ligands for the NK cell receptor NKG2D and associated anti-tumor NK cell responses (Wongthida et al., 2010).
IFN-λ is also present in the tumor microenvironment in the absence of a viral infection. IFNLR expression on mammary epithelial cells inversely correlated with tumor growth in a mouse model of breast cancer (Burkart et al., 2013). IFN-λ signaling on mammary epithelial cells promoted production of the chemokine CXCL10, which recruits CD4+ T cells into the tumor microenvironment (Burkart et al., 2013). Tumor models with melanoma, colon carcinoma, and fibrosarcoma cells also demonstrated a role of IFN-λ in triggering anti-tumor NK and T cells (Lasfar et al., 2006, Numasaki et al., 2007, Sato et al., 2006). An increased anti-tumor immune response was apparent even when the tumor cells themselves lacked the ability to respond to IFN-λ (Lasfar et al., 2006, Numasaki et al., 2007). Recent work has demonstrated that Ifnlr1 −/− mice are more susceptible to sarcoma formation induced by the carcinogen methylcholanthrene and death in transplanted tumor models (either RMA or B16F10 cells) (Souza-Fonseca-Guimaraes et al., 2015). Furthermore, IFN-λ treatment delayed lethality in the B16F10 adoptive cell transfer model and reduced sarcoma development in methylcholanthrene-treated mice (Souza-Fonseca-Guimaraes et al., 2015). Despite low levels of IFNLR expression, NK cells might respond directly to IFN-λ in vivo, resulting in enhanced IFN-γ production and anti-tumor activity (Souza-Fonseca-Guimaraes et al., 2015). Because IFN-λ can alter tumorigenesis directly and indirectly, it might have utility as an adjunctive anti-cancer therapy.
IFN-λ Modulates Adaptive Immunity
Although its innate antiviral effects have been characterized more extensively, IFN-λ also modulates adaptive immunity. During acute LCMV infection with the Armstrong strain, Ifnlr1 −/− mice had increased expansion of CD4+ and CD8+ T cells and enhanced T cell responses to LCMV rechallenge (Misumi and Whitmire, 2014). These data suggest that IFN-λ, in contrast to IFN-α/β, inhibits T cell responses during acute viral infection. This effect was cell extrinsic given that IFNLR signaling was not required on the T cells (Misumi and Whitmire, 2014). However, IFN-λ had a different effect during chronic LCMV infection: Ifnlr1 −/− mice infected with LCMV clone 13 had reduced CD8+ T cell expansion and increased weight loss (Misumi and Whitmire, 2014). In comparison, Ifnlr1 −/− mice did not show defects in the magnitude or quality of antigen-specific CD8+ T cell responses in the spleen or brain after WNV infection (Lazear et al., 2015), indicating that virus-specific host interactions might affect how IFN-λ modulates the adaptive immune response. Additional studies are required to define the role of IFN-λ in adaptive immunity to mucosal infections, where IFNLR is more abundantly expressed.
Although there are conflicting reports on whether human lymphocytes respond directly to IFN-λ (Dickensheets et al., 2013, Gallagher et al., 2010), there is consensus that IFN-λ modulates T cell responses. The addition of IFN-λ during stimulation of peripheral-blood mononuclear cells (PBMCs) with concanavalin A or during a mixed lymphocyte reaction reduced the production of Th2 cytokines (IL-4, IL-5, and IL-13) and increased the production of IFN-γ, suggesting that IFN-λ promotes a Th1 response (Dai et al., 2009, Jordan et al., 2007b, Srinivas et al., 2008). Consistent with these data, a SNP (rs8099917, with the T/T allele) in the IFNL3 locus was linked to increased IFN-λ3 production and skewing toward a Th1 response after PBMCs were stimulated with influenza virus (Egli et al., 2014b). Furthermore, administration of IFN-λ as a vaccine adjuvant resulted in reduced expression of the Th2 cytokine IL-4 by HIV gag-specific T cells and reduced numbers and activity of regulatory T cells (Morrow et al., 2009). Collectively, these observations suggest that IFN-λ skews T cell responses toward a Th1 cell bias and away from a Th2 cell bias.
The humoral immune response might be regulated by IFN-λ, although the net effect is not clear. Increased seroconversion in response to influenza or measles vaccines correlated with SNPs in the IFNL3 promoter (the T/T allele for rs8099917 and the G/G allele for rs10853727) (Egli et al., 2014b, Haralambieva et al., 2011). The T/T allele of SNP rs8099917 was linked to increased production of IFN-λ and decreased seroconversion after influenza vaccination (Egli et al., 2014b). In vitro experiments showed reduced antibody production by PBMCs stimulated with influenza virus in the presence of IFN-λ and increased antibody titers in the presence of an IFNLR-blocking peptide (Egli et al., 2014b). In contrast, a stimulatory effect on humoral response was observed in an HIV vaccine study comparing IFN-λ and IL-12 as adjuvants: the IFN-λ-adjuvanted vaccine resulted in a greater increase in IgG2a responses than did the IL-12-adjuvanted vaccine (Morrow et al., 2009). However, in the context of WNV or LCMV infection, antiviral antibody production was the same between Ifnlr1 −/− mice and wild-type animals (Lazear et al., 2015, Misumi and Whitmire, 2014). Thus, the impact of IFN-λ on humoral immunity appears to be context dependent and requires further evaluation.
More Than Just Viral Infections
Although known for their antiviral effects, IFN-α/β and IFN-λ might function in other microbial infections as well. Whereas IFN-α/β signaling protects against viral infections, it can be detrimental in the context of infection by some bacteria (e.g., Mycobacterium tuberculosis and Listeria monocytogenes) and parasites (e.g., Leishmania spp.) (Stifter and Feng, 2015). IFN-λ also is induced by bacterial infection (Bierne et al., 2012, Cohen and Prince, 2013, Lebreton et al., 2011, Love et al., 2014, Odendall et al., 2014, Pietilä et al., 2010, Travar et al., 2014) and might have consequences during Staphylococcus and Pseudomonas infections: Ifnlr1 −/− mice sustained lower bacterial loads and exhibited less pathology without differences in inflammatory cell infiltrates (Cohen and Prince, 2013). Further investigation into the role of IFN-λ in non-viral infections is warranted. The effects of IFN-λ in the context of helminth infections could be especially interesting given its ability to downregulate Th2 responses (Jordan et al., 2007b, Srinivas et al., 2008).
Antagonism of IFN-λ
Given the antiviral effects of IFN-λ, it is anticipated that some viruses will have evolved specific evasion strategies. IFN-α/β antagonist mechanisms have been described for many virus families (Hoffmann et al., 2015); because some of these target induction or signaling pathways shared between IFN-λ and IFN-α/β, these same evasion strategies most likely limit the effects of IFN-λ as well. Viruses targeting epithelial tissues might be especially likely to encode specific IFN-λ-evasion mechanisms. Yaba-like disease virus (YLDV) is a primate poxvirus that exclusively infects the skin. Similar to other orthopoxviruses, YLDV encodes a secreted glycoprotein (Y136) that binds to IFN-α/β and blocks IFNAR signaling. Unlike orthologs in vaccinia or variola viruses, Y136 also binds to and inhibits the antiviral effects of IFN-λ (Bandi et al., 2010, Huang et al., 2007). Because the structure of IFN-λ is distinct from that of IFN-α/β (Gad et al., 2009), it remains uncertain how a single viral evasion protein targets both IFN-λ and IFN-α/β. Because they infect epithelial cells and their large genomes enable them to encode a suite of immune evasion molecules (Epperson et al., 2012), other poxviruses and herpesviruses might have evolved analogous or unique mechanisms to antagonize IFN-λ responses.
IFN-λ across Species
The use of Ifnlr1 −/− mice has provided insight into the role of IFN-λ in antiviral immunity (Hermant and Michiels, 2014, Hernández et al., 2015, Lazear et al., 2015, Mahlakõiv et al., 2015, Mordstein et al., 2010a, Nice et al., 2015). Nonetheless, differences in IFN-λ between mice and humans should be considered when one interprets its array of activities. Whereas humans have three or four functional IFNL genes, mice have only two (Ifnl2 and Ifnl3), because the IFNL4 genomic region is absent in mice and mouse Ifnl1 is a pseudogene. Compared to human studies, mouse studies might underestimate some of the contributions of IFN-λ because they lack IFN-λ1. Given that most IFN-λ transcriptional profiling to date has focused on human cells, studies are needed to characterize IFN-λ responses in mouse cells to inform the interpretation of mouse models of disease.
The IFN-λ family is conserved throughout tetrapod evolution, as evidenced by synteny and structural similarity among mammalian, avian, and amphibian IFN-λ-encoding genes. In comparison, fish have genes encoding IFN-α/β but not IFN-λ (Qi et al., 2010). Antiviral effects of avian IFN-λ have been documented (Reuter et al., 2014), and in tadpoles, IFN-λ rather than IFN-α/β dominantly inhibits infection by frog virus 3 (Grayfer et al., 2015).
In contrast to the preferential epithelial expression pattern of IFNLR in humans and mice, in the bat Pteropus alecto, IFNLR is expressed in a broader range of cells and tissues (Zhou et al., 2011a). Infection of P. alecto splenocytes with the paramyxovirus Tioman virus induced the expression of IFN-λ1 but not IFN-α/β (Zhou et al., 2011b). It will be interesting to determine whether broader tissue responsiveness to IFN-λ in bats contributes to their ability to serve as reservoir hosts for diverse viruses (Baker et al., 2013). For example, enhanced IFN-λ responsiveness might enable bats to control viral infections without developing disease. Alternatively, sustained IFN-λ signaling might impede the activation of more potent antiviral responses and prevent bats from clearing viral infections.
Although the mouse and rat genomes lack IFNL4, this gene is conserved in most mammals and displays evidence of positive selection, implying functional significance (Key et al., 2014). In some human populations, however, IFNL4 is undergoing pseudogenization with the acquisition of a frameshift insertion in the IFNL4 open reading frame that ablates IFN-λ4 production (Hamming et al., 2013, Prokunina-Olsson et al., 2013). The frequency of this polymorphism varies, such that the ancestral (IFN-λ4-producing) allele dominates in African populations, and the pseudogene allele has reached near fixation in East Asian populations (Key et al., 2014). The selective pressure driving IFNL4 pseudogenization in humans remains unknown, but GWASs suggest a strong association between the pseudogene allele of IFNL4 and improved outcome after HCV infection (Griffiths et al., 2015, Hamming et al., 2013, Prokunina-Olsson et al., 2013). Because HCV infection is unlikely to provide sufficient selective pressure to drive this evolutionary process, the loss of IFN-λ4 production might be advantageous in the context of other infections or inflammatory conditions.
Conclusions
Since its discovery in 2003, many groups have identified contributions of IFN-λ to antiviral and other immune responses. These studies have prompted a growing appreciation of the unique features that distinguish IFN-λ from IFN-α/β, and these features provide possibilities for more targeted therapeutic intervention. The ability of IFN-λ to provide antiviral protection at specific tissue sites (e.g., epithelial surfaces) could explain the maintenance of a system that appears somewhat redundant with the IFN-α/β response. Epithelial surfaces could require greater innate immune protection because of their constant exposure to commensal and pathogenic microbes. Constant microbial stimulation at epithelial barriers could require local control without broadly activating a systemic immune response. Thus, IFN-λ signaling could control frequent or persistent low-level infections at epithelial barriers, and the more potent and inflammatory systemic IFN-α/β response could be reserved for severe infections. This compartmentalized tissue responsiveness to IFN-λ could be utilized therapeutically as a means of achieving antiviral benefit while minimizing systemic side effects associated with inflammation.
Acknowledgments
This work was supported by NIH grants U19 AI083019 and R01 AI074973 (M.S.D). T.J.N. was supported by NIH training grant 5T32A100716334 and postdoctoral fellowships from the Cancer Research Institute and American Cancer Society.
References
- Alexopoulou A., Karayiannis P. Interferon-based combination treatment for chronic hepatitis C in the era of direct acting antivirals. Ann. Gastroenterol. 2015;28:55–65. [PMC free article] [PubMed] [Google Scholar]
- Angel J., Franco M.A., Greenberg H.B., Bass D. Lack of a role for type I and type II interferons in the resolution of rotavirus-induced diarrhea and infection in mice. J. Interferon Cytokine Res. 1999;19:655–659. doi: 10.1089/107999099313802. [DOI] [PubMed] [Google Scholar]
- Ank N., West H., Bartholdy C., Eriksson K., Thomsen A.R., Paludan S.R. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J. Virol. 2006;80:4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ank N., Iversen M.B., Bartholdy C., Staeheli P., Hartmann R., Jensen U.B., Dagnaes-Hansen F., Thomsen A.R., Chen Z., Haugen H. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J. Immunol. 2008;180:2474–2485. doi: 10.4049/jimmunol.180.4.2474. [DOI] [PubMed] [Google Scholar]
- Baker M.L., Schountz T., Wang L.F. Antiviral immune responses of bats: a review. Zoonoses Public Health. 2013;60:104–116. doi: 10.1111/j.1863-2378.2012.01528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge M.T., Nice T.J., McCune B.T., Yokoyama C.C., Kambal A., Wheadon M., Diamond M.S., Ivanova Y., Artyomov M., Virgin H.W. Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science. 2015;347:266–269. doi: 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandi P., Pagliaccetti N.E., Robek M.D. Inhibition of type III interferon activity by orthopoxvirus immunomodulatory proteins. J. Interferon Cytokine Res. 2010;30:123–134. doi: 10.1089/jir.2009.0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baños-Lara Mdel.R., Harvey L., Mendoza A., Simms D., Chouljenko V.N., Wakamatsu N., Kousoulas K.G., Guerrero-Plata A. Impact and regulation of lambda interferon response in human metapneumovirus infection. J. Virol. 2015;89:730–742. doi: 10.1128/JVI.02897-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibert S., Roger T., Calandra T., Bochud M., Cerny A., Semmo N., Duong F.H., Gerlach T., Malinverni R., Moradpour D., Swiss Hepatitis C Cohort Study IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J. Exp. Med. 2013;210:1109–1116. doi: 10.1084/jem.20130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibert S., Wojtowicz A., Taffé P., Manuel O., Bernasconi E., Furrer H., Günthard H.F., Hoffmann M., Kaiser L., Osthoff M., Swiss HIV Cohort Study The IFNL3/4 ΔG variant increases susceptibility to cytomegalovirus retinitis among HIV-infected patients. AIDS. 2014;28:1885–1889. doi: 10.1097/QAD.0000000000000379. [DOI] [PubMed] [Google Scholar]
- Bierne H., Travier L., Mahlakõiv T., Tailleux L., Subtil A., Lebreton A., Paliwal A., Gicquel B., Staeheli P., Lecuit M., Cossart P. Activation of type III interferon genes by pathogenic bacteria in infected epithelial cells and mouse placenta. PLoS ONE. 2012;7:e39080. doi: 10.1371/journal.pone.0039080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek K., Eames H.L., Weiss M., Byrne A.J., Perocheau D., Pease J.E., Doyle S., McCann F., Williams R.O., Udalova I.A. IFN-λ resolves inflammation via suppression of neutrophil infiltration and IL-1β production. J. Exp. Med. 2015;212:845–853. doi: 10.1084/jem.20140995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok K., Green K.Y. Norovirus gastroenteritis in immunocompromised patients. N. Engl. J. Med. 2012;367:2126–2132. doi: 10.1056/NEJMra1207742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolen C.R., Ding S., Robek M.D., Kleinstein S.H. Dynamic expression profiling of type I and type III interferon-stimulated hepatocytes reveals a stable hierarchy of gene expression. Hepatology. 2014;59:1262–1272. doi: 10.1002/hep.26657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booy S., Hofland L., van Eijck C. Potentials of interferon therapy in the treatment of pancreatic cancer. J. Interferon Cytokine Res. 2015;35:327–339. doi: 10.1089/jir.2014.0157. [DOI] [PubMed] [Google Scholar]
- Burkart C., Arimoto K., Tang T., Cong X., Xiao N., Liu Y.C., Kotenko S.V., Ellies L.G., Zhang D.E. Usp18 deficient mammary epithelial cells create an antitumour environment driven by hypersensitivity to IFN-λ and elevated secretion of Cxcl10. EMBO Mol. Med. 2013;5:967–982. doi: 10.1002/emmm.201201864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannella F., Scagnolari C., Selvaggi C., Stentella P., Recine N., Antonelli G., Pierangeli A. Interferon lambda 1 expression in cervical cells differs between low-risk and high-risk human papillomavirus-positive women. Med. Microbiol. Immunol. (Berl.) 2014;203:177–184. doi: 10.1007/s00430-014-0330-9. [DOI] [PubMed] [Google Scholar]
- Christophers E., Henseler T. Contrasting disease patterns in psoriasis and atopic dermatitis. Arch. Dermatol. Res. 1987;279(Suppl):S48–S51. doi: 10.1007/BF00585919. [DOI] [PubMed] [Google Scholar]
- Clark R.A. Human skin in the game. Sci. Transl. Med. 2013;5:204ps13. doi: 10.1126/scitranslmed.3007504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia E.M., Severa M., Giacomini E., Monneron D., Remoli M.E., Julkunen I., Cella M., Lande R., Uzé G. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur. J. Immunol. 2004;34:796–805. doi: 10.1002/eji.200324610. [DOI] [PubMed] [Google Scholar]
- Cohen T.S., Prince A.S. Bacterial pathogens activate a common inflammatory pathway through IFNλ regulation of PDCD4. PLoS Pathog. 2013;9:e1003682. doi: 10.1371/journal.ppat.1003682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contoli M., Message S.D., Laza-Stanca V., Edwards M.R., Wark P.A., Bartlett N.W., Kebadze T., Mallia P., Stanciu L.A., Parker H.L. Role of deficient type III interferon-lambda production in asthma exacerbations. Nat. Med. 2006;12:1023–1026. doi: 10.1038/nm1462. [DOI] [PubMed] [Google Scholar]
- Crotta S., Davidson S., Mahlakoiv T., Desmet C.J., Buckwalter M.R., Albert M.L., Staeheli P., Wack A. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog. 2013;9:e1003773. doi: 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow Y.J. Type I interferonopathies: mendelian type I interferon up-regulation. Curr. Opin. Immunol. 2015;32:7–12. doi: 10.1016/j.coi.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Dai J., Megjugorac N.J., Gallagher G.E., Yu R.Y., Gallagher G. IFN-lambda1 (IL-29) inhibits GATA3 expression and suppresses Th2 responses in human naive and memory T cells. Blood. 2009;113:5829–5838. doi: 10.1182/blood-2008-09-179507. [DOI] [PubMed] [Google Scholar]
- Daniels B.P., Holman D.W., Cruz-Orengo L., Jujjavarapu H., Durrant D.M., Klein R.S. Viral pathogen-associated molecular patterns regulate blood-brain barrier integrity via competing innate cytokine signals. MBio. 2014;5 doi: 10.1128/mBio.01476-14. e01476–e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellgren C., Gad H.H., Hamming O.J., Melchjorsen J., Hartmann R. Human interferon-lambda3 is a potent member of the type III interferon family. Genes Immun. 2009;10:125–131. doi: 10.1038/gene.2008.87. [DOI] [PubMed] [Google Scholar]
- Dickensheets H., Sheikh F., Park O., Gao B., Donnelly R.P. Interferon-lambda (IFN-λ) induces signal transduction and gene expression in human hepatocytes, but not in lymphocytes or monocytes. J. Leukoc. Biol. 2013;93:377–385. doi: 10.1189/jlb.0812395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Trolio R., Simeone E., Di Lorenzo G., Buonerba C., Ascierto P.A. The use of interferon in melanoma patients: a systematic review. Cytokine Growth Factor Rev. 2015;26:203–212. doi: 10.1016/j.cytogfr.2014.11.008. [DOI] [PubMed] [Google Scholar]
- Donnelly R.P., Kotenko S.V. Interferon-lambda: a new addition to an old family. J. Interferon Cytokine Res. 2010;30:555–564. doi: 10.1089/jir.2010.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly R.P., Dickensheets H., O’Brien T.R. Interferon-lambda and therapy for chronic hepatitis C virus infection. Trends Immunol. 2011;32:443–450. doi: 10.1016/j.it.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S.E., Schreckhise H., Khuu-Duong K., Henderson K., Rosler R., Storey H., Yao L., Liu H., Barahmand-pour F., Sivakumar P. Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology. 2006;44:896–906. doi: 10.1002/hep.21312. [DOI] [PubMed] [Google Scholar]
- Dumoutier L., Tounsi A., Michiels T., Sommereyns C., Kotenko S.V., Renauld J.C. Role of the interleukin (IL)-28 receptor tyrosine residues for antiviral and antiproliferative activity of IL-29/interferon-lambda 1: similarities with type I interferon signaling. J. Biol. Chem. 2004;279:32269–32274. doi: 10.1074/jbc.M404789200. [DOI] [PubMed] [Google Scholar]
- Durbin R.K., Kotenko S.V., Durbin J.E. Interferon induction and function at the mucosal surface. Immunol. Rev. 2013;255:25–39. doi: 10.1111/imr.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli A., Levin A., Santer D.M., Joyce M., O’Shea D., Thomas B.S., Lisboa L.F., Barakat K., Bhat R., Fischer K.P. Immunomodulatory Function of Interleukin 28B during primary infection with cytomegalovirus. J. Infect. Dis. 2014;210:717–727. doi: 10.1093/infdis/jiu144. [DOI] [PubMed] [Google Scholar]
- Egli A., Santer D.M., O’Shea D., Barakat K., Syedbasha M., Vollmer M., Baluch A., Bhat R., Groenendyk J., Joyce M.A. IL-28B is a key regulator of B- and T-cell vaccine responses against influenza. PLoS Pathog. 2014;10:e1004556. doi: 10.1371/journal.ppat.1004556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli A., Santer D.M., O’Shea D., Tyrrell D.L., Houghton M. The impact of the interferon-lambda family on the innate and adaptive immune response to viral infections. Emerg Microbes Infect. 2014;3:e51. doi: 10.1038/emi.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epperson M.L., Lee C.A., Fremont D.H. Subversion of cytokine networks by virally encoded decoy receptors. Immunol. Rev. 2012;250:199–215. doi: 10.1111/imr.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eslam M., Hashem A.M., Leung R., Romero-Gomez M., Berg T., Dore G.J., Chan H.L., Irving W.L., Sheridan D., Abate M.L., International Hepatitis C Genetics Consortium (IHCGC) Interferon-λ rs12979860 genotype and liver fibrosis in viral and non-viral chronic liver disease. Nat. Commun. 2015;6:6422. doi: 10.1038/ncomms7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest J.C., Dermody T.S. Reovirus receptors and pathogenesis. J. Virol. 2003;77:9109–9115. doi: 10.1128/JVI.77.17.9109-9115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J.M., Crabtree J.M., Sage L.K., Tompkins S.M., Tripp R.A. Interferon Lambda Upregulates IDO1 Expression in Respiratory Epithelial Cells After Influenza Virus Infection. J. Interferon Cytokine Res. 2015 doi: 10.1089/jir.2014.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François-Newton V., Magno de Freitas Almeida G., Payelle-Brogard B., Monneron D., Pichard-Garcia L., Piehler J., Pellegrini S., Uzé G. USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon α response. PLoS ONE. 2011;6:e22200. doi: 10.1371/journal.pone.0022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gad H.H., Dellgren C., Hamming O.J., Vends S., Paludan S.R., Hartmann R. Interferon-lambda is functionally an interferon but structurally related to the interleukin-10 family. J. Biol. Chem. 2009;284:20869–20875. doi: 10.1074/jbc.M109.002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher G., Megjugorac N.J., Yu R.Y., Eskdale J., Gallagher G.E., Siegel R., Tollar E. The lambda interferons: guardians of the immune-epithelial interface and the T-helper 2 response. J. Interferon Cytokine Res. 2010;30:603–615. doi: 10.1089/jir.2010.0081. [DOI] [PubMed] [Google Scholar]
- Galmozzi E., Viganò M., Lampertico P. Systematic review with meta-analysis: do interferon lambda 3 polymorphisms predict the outcome of interferon-therapy in hepatitis B infection? Aliment. Pharmacol. Ther. 2014;39:569–578. doi: 10.1111/apt.12631. [DOI] [PubMed] [Google Scholar]
- Ge D., Fellay J., Thompson A.J., Simon J.S., Shianna K.V., Urban T.J., Heinzen E.L., Qiu P., Bertelsen A.H., Muir A.J. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- Génin P., Lin R., Hiscott J., Civas A. Differential regulation of human interferon A gene expression by interferon regulatory factors 3 and 7. Mol. Cell. Biol. 2009;29:3435–3450. doi: 10.1128/MCB.01805-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayfer L., De Jesús Andino F., Robert J. Prominent amphibian (Xenopus laevis) tadpole type III interferon response to the frog virus 3 ranavirus. J. Virol. 2015;89:5072–5082. doi: 10.1128/JVI.00051-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S.J., Koegl M., Boutell C., Zenner H.L., Crump C.M., Pica F., Gonzalez O., Friedel C.C., Barry G., Martin K. A systematic analysis of host factors reveals a Med23-interferon-λ regulatory axis against herpes simplex virus type 1 replication. PLoS Pathog. 2013;9:e1003514. doi: 10.1371/journal.ppat.1003514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S.J., Dunnigan C.M., Russell C.D., Haas J.G. The Role of Interferon-λ Locus Polymorphisms in Hepatitis C and Other Infectious Diseases. J. Innate Immun. 2015;7:231–242. doi: 10.1159/000369902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming O.J., Terczyńska-Dyla E., Vieyres G., Dijkman R., Jørgensen S.E., Akhtar H., Siupka P., Pietschmann T., Thiel V., Hartmann R. Interferon lambda 4 signals via the IFNλ receptor to regulate antiviral activity against HCV and coronaviruses. EMBO J. 2013;32:3055–3065. doi: 10.1038/emboj.2013.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralambieva I.H., Ovsyannikova I.G., Kennedy R.B., Vierkant R.A., Pankratz V.S., Jacobson R.M., Poland G.A. Associations between single nucleotide polymorphisms and haplotypes in cytokine and cytokine receptor genes and immunity to measles vaccination. Vaccine. 2011;29:7883–7895. doi: 10.1016/j.vaccine.2011.08.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes C.N., Imamura M., Aikata H., Chayama K. Genetics of IL28B and HCV--response to infection and treatment. Nat. Rev. Gastroenterol. Hepatol. 2012;9:406–417. doi: 10.1038/nrgastro.2012.101. [DOI] [PubMed] [Google Scholar]
- Hermant P., Michiels T. Interferon-λ in the context of viral infections: production, response and therapeutic implications. J. Innate Immun. 2014;6:563–574. doi: 10.1159/000360084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermant P., Demarez C., Mahlakõiv T., Staeheli P., Meuleman P., Michiels T. Human but not mouse hepatocytes respond to interferon-lambda in vivo. PLoS ONE. 2014;9:e87906. doi: 10.1371/journal.pone.0087906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández P.P., Mahlakõiv T., Yang I., Schwierzeck V., Nguyen N., Guendel F., Gronke K., Ryffel B., Hölscher C., Dumoutier L. Interferon-λ and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat. Immunol. 2015;16:698–707. doi: 10.1038/ni.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillyer P., Mane V.P., Schramm L.M., Puig M., Verthelyi D., Chen A., Zhao Z., Navarro M.B., Kirschman K.D., Bykadi S. Expression profiles of human interferon-alpha and interferon-lambda subtypes are ligand- and cell-dependent. Immunol. Cell Biol. 2012;90:774–783. doi: 10.1038/icb.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann H.H., Schneider W.M., Rice C.M. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 2015;36:124–138. doi: 10.1016/j.it.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Smirnov S.V., Lewis-Antes A., Balan M., Li W., Tang S., Silke G.V., Pütz M.M., Smith G.L., Kotenko S.V. Inhibition of type I and type III interferons by a secreted glycoprotein from Yaba-like disease virus. Proc. Natl. Acad. Sci. USA. 2007;104:9822–9827. doi: 10.1073/pnas.0610352104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs A., Lindenmann J. Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 1957;147:258–267. [PubMed] [Google Scholar]
- Iversen M.B., Paludan S.R. Mechanisms of type III interferon expression. J. Interferon Cytokine Res. 2010;30:573–578. doi: 10.1089/jir.2010.0063. [DOI] [PubMed] [Google Scholar]
- Iversen M.B., Ank N., Melchjorsen J., Paludan S.R. Expression of type III interferon (IFN) in the vaginal mucosa is mediated primarily by dendritic cells and displays stronger dependence on NF-kappaB than type I IFNs. J. Virol. 2010;84:4579–4586. doi: 10.1128/JVI.02591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell N.A., Cline T., Mertz S.E., Smirnov S.V., Flaño E., Schindler C., Grieves J.L., Durbin R.K., Kotenko S.V., Durbin J.E. Lambda interferon is the predominant interferon induced by influenza A virus infection in vivo. J. Virol. 2010;84:11515–11522. doi: 10.1128/JVI.01703-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.K., Watanabe M., Zhu S., Graves C.L., Keyes L.R., Grau K.R., Gonzalez-Hernandez M.B., Iovine N.M., Wobus C.E., Vinjé J. Enteric bacteria promote human and mouse norovirus infection of B cells. Science. 2014;346:755–759. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan W.J., Eskdale J., Boniotto M., Rodia M., Kellner D., Gallagher G. Modulation of the human cytokine response by interferon lambda-1 (IFN-lambda1/IL-29) Genes Immun. 2007;8:13–20. doi: 10.1038/sj.gene.6364348. [DOI] [PubMed] [Google Scholar]
- Jordan W.J., Eskdale J., Srinivas S., Pekarek V., Kelner D., Rodia M., Gallagher G. Human interferon lambda-1 (IFN-lambda1/IL-29) modulates the Th1/Th2 response. Genes Immun. 2007;8:254–261. doi: 10.1038/sj.gene.6364382. [DOI] [PubMed] [Google Scholar]
- Kane M., Case L.K., Kopaskie K., Kozlova A., MacDearmid C., Chervonsky A.V., Golovkina T.V. Successful transmission of a retrovirus depends on the commensal microbiota. Science. 2011;334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst S.M., Wobus C.E., Goodfellow I.G., Green K.Y., Virgin H.W. Advances in norovirus biology. Cell Host Microbe. 2014;15:668–680. doi: 10.1016/j.chom.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key F.M., Peter B., Dennis M.Y., Huerta-Sánchez E., Tang W., Prokunina-Olsson L., Nielsen R., Andrés A.M. Selection on a variant associated with improved viral clearance drives local, adaptive pseudogenization of interferon lambda 4 (IFNL4) PLoS Genet. 2014;10:e1004681. doi: 10.1371/journal.pgen.1004681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Kim C.H., Ryu J.H., Kim M.J., Park C.Y., Lee J.M., Holtzman M.J., Yoon J.H. Reactive oxygen species induce antiviral innate immune response through IFN-λ regulation in human nasal epithelial cells. Am. J. Respir. Cell Mol. Biol. 2013;49:855–865. doi: 10.1165/rcmb.2013-0003OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch S., Finotto S. Role of Interferon-λ in Allergic Asthma. J. Innate Immun. 2015;7:224–230. doi: 10.1159/000369459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli A., Zhang X., Yang J., Russell R.S., Donnelly R.P., Sheikh F., Sherman A., Young H., Imamichi T., Lempicki R.A. Distinct and overlapping genomic profiles and antiviral effects of Interferon-λ and -α on HCV-infected and noninfected hepatoma cells. J. Viral Hepat. 2012;19:843–853. doi: 10.1111/j.1365-2893.2012.01610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltsida O., Hausding M., Stavropoulos A., Koch S., Tzelepis G., Ubel C., Kotenko S.V., Sideras P., Lehr H.A., Tepe M. IL-28A (IFN-λ2) modulates lung DC function to promote Th1 immune skewing and suppress allergic airway disease. EMBO Mol. Med. 2011;3:348–361. doi: 10.1002/emmm.201100142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko S.V. IFN-λs. Curr. Opin. Immunol. 2011;23:583–590. doi: 10.1016/j.coi.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko S.V., Gallagher G., Baurin V.V., Lewis-Antes A., Shen M., Shah N.K., Langer J.A., Sheikh F., Dickensheets H., Donnelly R.P. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- Koyuncu O.O., Hogue I.B., Enquist L.W. Virus infections in the nervous system. Cell Host Microbe. 2013;13:379–393. doi: 10.1016/j.chom.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuss S.K., Best G.T., Etheredge C.A., Pruijssers A.J., Frierson J.M., Hooper L.V., Dermody T.S., Pfeiffer J.K. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampertico P., Viganò M., Cheroni C., Facchetti F., Invernizzi F., Valveri V., Soffredini R., Abrignani S., De Francesco R., Colombo M. IL28B polymorphisms predict interferon-related hepatitis B surface antigen seroclearance in genotype D hepatitis B e antigen-negative patients with chronic hepatitis B. Hepatology. 2013;57:890–896. doi: 10.1002/hep.25749. [DOI] [PubMed] [Google Scholar]
- Lasfar A., Lewis-Antes A., Smirnov S.V., Anantha S., Abushahba W., Tian B., Reuhl K., Dickensheets H., Sheikh F., Donnelly R.P. Characterization of the mouse IFN-lambda ligand-receptor system: IFN-lambdas exhibit antitumor activity against B16 melanoma. Cancer Res. 2006;66:4468–4477. doi: 10.1158/0008-5472.CAN-05-3653. [DOI] [PubMed] [Google Scholar]
- Lauterbach H., Bathke B., Gilles S., Traidl-Hoffmann C., Luber C.A., Fejer G., Freudenberg M.A., Davey G.M., Vremec D., Kallies A. Mouse CD8alpha+ DCs and human BDCA3+ DCs are major producers of IFN-lambda in response to poly IC. J. Exp. Med. 2010;207:2703–2717. doi: 10.1084/jem.20092720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear H.M., Daniels B.P., Pinto A.K., Huang A.C., Vick S.C., Doyle S.E., Gale M., Jr., Klein R.S., Diamond M.S. Interferon-λ restricts West Nile virus neuroinvasion by tightening the blood-brain barrier. Sci. Transl. Med. 2015;7:284ra59. doi: 10.1126/scitranslmed.aaa4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton A., Lakisic G., Job V., Fritsch L., Tham T.N., Camejo A., Matteï P.J., Regnault B., Nahori M.A., Cabanes D. A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response. Science. 2011;331:1319–1321. doi: 10.1126/science.1200120. [DOI] [PubMed] [Google Scholar]
- Leung D.Y. New insights into atopic dermatitis: role of skin barrier and immune dysregulation. Allergol. Int. 2013;62:151–161. doi: 10.2332/allergolint.13-RAI-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Lewis-Antes A., Huang J., Balan M., Kotenko S.V. Regulation of apoptosis by type III interferons. Cell Prolif. 2008;41:960–979. doi: 10.1111/j.1365-2184.2008.00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Cheng H., Zuo X.B., Sheng Y.J., Zhou F.S., Tang X.F., Tang H.Y., Gao J.P., Zhang Z., He S.M. Association analyses identifying two common susceptibility loci shared by psoriasis and systemic lupus erythematosus in the Chinese Han population. J. Med. Genet. 2013;50:812–818. doi: 10.1136/jmedgenet-2013-101787. [DOI] [PubMed] [Google Scholar]
- Lopez S., Arias C.F. Early steps in rotavirus cell entry. Curr. Top. Microbiol. Immunol. 2006;309:39–66. doi: 10.1007/3-540-30773-7_2. [DOI] [PubMed] [Google Scholar]
- Lopez de Lapuente A., Alloza I., Goertsches R., Zettl U.K., Urcelay E., Arroyo R., Comabella M., Montalban X., Antigüedad A., Vandenbroeck K. Analysis of the IL28RA locus as genetic risk factor for multiple sclerosis. J. Neuroimmunol. 2012;245:98–101. doi: 10.1016/j.jneuroim.2012.02.005. [DOI] [PubMed] [Google Scholar]
- Love A.C., Schwartz I., Petzke M.M. Borrelia burgdorferi RNA induces type I and III interferons via Toll-like receptor 7 and contributes to production of NF-κB-dependent cytokines. Infect. Immun. 2014;82:2405–2416. doi: 10.1128/IAI.01617-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Jiang D., Qing M., Weidner J.M., Qu X., Guo H., Chang J., Gu B., Shi P.Y., Block T.M., Guo J.T. Antiviral effect of interferon lambda against West Nile virus. Antiviral Res. 2009;83:53–60. doi: 10.1016/j.antiviral.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlakõiv T., Ritz D., Mordstein M., DeDiego M.L., Enjuanes L., Müller M.A., Drosten C., Staeheli P. Combined action of type I and type III interferon restricts initial replication of severe acute respiratory syndrome coronavirus in the lung but fails to inhibit systemic virus spread. J. Gen. Virol. 2012;93:2601–2605. doi: 10.1099/vir.0.046284-0. [DOI] [PubMed] [Google Scholar]
- Mahlakõiv T., Hernandez P., Gronke K., Diefenbach A., Staeheli P. Leukocyte-derived IFN-α/β and epithelial IFN-λ constitute a compartmentalized mucosal defense system that restricts enteric virus infections. PLoS Pathog. 2015;11:e1004782. doi: 10.1371/journal.ppat.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra S., Morcillo-Suárez C., Brassat D., Goertsches R., Lechner-Scott J., Urcelay E., Fernández O., Drulovic J., García-Merino A., Martinelli Boneschi F. IL28B polymorphisms are not associated with the response to interferon-β in multiple sclerosis. J. Neuroimmunol. 2011;239:101–104. doi: 10.1016/j.jneuroim.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Manuel O., Wójtowicz A., Bibert S., Mueller N.J., van Delden C., Hirsch H.H., Steiger J., Stern M., Egli A., Garzoni C., Swiss Transplant Cohort Study Influence of IFNL3/4 polymorphisms on the incidence of cytomegalovirus infection after solid-organ transplantation. J. Infect. Dis. 2015;211:906–914. doi: 10.1093/infdis/jiu557. [DOI] [PubMed] [Google Scholar]
- Marcello T., Grakoui A., Barba-Spaeth G., Machlin E.S., Kotenko S.V., MacDonald M.R., Rice C.M. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- Markowitz C.E. Interferon-beta: mechanism of action and dosing issues. Neurology. 2007;68(24, Suppl 4):S8–S11. doi: 10.1212/01.wnl.0000277703.74115.d2. [DOI] [PubMed] [Google Scholar]
- Marukian S., Andrus L., Sheahan T.P., Jones C.T., Charles E.D., Ploss A., Rice C.M., Dustin L.B. Hepatitis C virus induces interferon-λ and interferon-stimulated genes in primary liver cultures. Hepatology. 2011;54:1913–1923. doi: 10.1002/hep.24580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland A.P., Horner S.M., Jarret A., Joslyn R.C., Bindewald E., Shapiro B.A., Delker D.A., Hagedorn C.H., Carrington M., Gale M., Jr., Savan R. The favorable IFNL3 genotype escapes mRNA decay mediated by AU-rich elements and hepatitis C virus-induced microRNAs. Nat. Immunol. 2014;15:72–79. doi: 10.1038/ni.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miknis Z.J., Magracheva E., Li W., Zdanov A., Kotenko S.V., Wlodawer A. Crystal structure of human interferon-λ1 in complex with its high-affinity receptor interferon-λR1. J. Mol. Biol. 2010;404:650–664. doi: 10.1016/j.jmb.2010.09.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misumi I., Whitmire J.K. IFN-λ exerts opposing effects on T cell responses depending on the chronicity of the virus infection. J. Immunol. 2014;192:3596–3606. doi: 10.4049/jimmunol.1301705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordstein M., Kochs G., Dumoutier L., Renauld J.C., Paludan S.R., Klucher K., Staeheli P. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 2008;4:e1000151. doi: 10.1371/journal.ppat.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordstein M., Michiels T., Staeheli P. What have we learned from the IL28 receptor knockout mouse? J. Interferon Cytokine Res. 2010;30:579–584. doi: 10.1089/jir.2010.0061. [DOI] [PubMed] [Google Scholar]
- Mordstein M., Neugebauer E., Ditt V., Jessen B., Rieger T., Falcone V., Sorgeloos F., Ehl S., Mayer D., Kochs G. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J. Virol. 2010;84:5670–5677. doi: 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow M.P., Pankhong P., Laddy D.J., Schoenly K.A., Yan J., Cisper N., Weiner D.B. Comparative ability of IL-12 and IL-28B to regulate Treg populations and enhance adaptive cellular immunity. Blood. 2009;113:5868–5877. doi: 10.1182/blood-2008-11-190520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir A.J., Arora S., Everson G., Flisiak R., George J., Ghalib R., Gordon S.C., Gray T., Greenbloom S., Hassanein T., EMERGE study group A randomized phase 2b study of peginterferon lambda-1a for the treatment of chronic HCV infection. J. Hepatol. 2014;61:1238–1246. doi: 10.1016/j.jhep.2014.07.022. [DOI] [PubMed] [Google Scholar]
- Nice T.J., Strong D.W., McCune B.T., Pohl C.S., Virgin H.W. A single-amino-acid change in murine norovirus NS1/2 is sufficient for colonic tropism and persistence. J. Virol. 2013;87:327–334. doi: 10.1128/JVI.01864-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice T.J., Baldridge M.T., McCune B.T., Norman J.M., Lazear H.M., Artyomov M., Diamond M.S., Virgin H.W. Interferon-λ cures persistent murine norovirus infection in the absence of adaptive immunity. Science. 2015;347:269–273. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numasaki M., Tagawa M., Iwata F., Suzuki T., Nakamura A., Okada M., Iwakura Y., Aiba S., Yamaya M. IL-28 elicits antitumor responses against murine fibrosarcoma. J. Immunol. 2007;178:5086–5098. doi: 10.4049/jimmunol.178.8.5086. [DOI] [PubMed] [Google Scholar]
- O’Brien T.R., Prokunina-Olsson L., Donnelly R.P. IFN-λ4: the paradoxical new member of the interferon lambda family. J. Interferon Cytokine Res. 2014;34:829–838. doi: 10.1089/jir.2013.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odendall C., Kagan J.C. The unique regulation and functions of type III interferons in antiviral immunity. Curr. Opin. Virol. 2015;12:47–52. doi: 10.1016/j.coviro.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odendall C., Dixit E., Stavru F., Bierne H., Franz K.M., Durbin A.F., Boulant S., Gehrke L., Cossart P., Kagan J.C. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 2014;15:717–726. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabayashi T., Kojima T., Masaki T., Yokota S., Imaizumi T., Tsutsumi H., Himi T., Fujii N., Sawada N. Type-III interferon, not type-I, is the predominant interferon induced by respiratory viruses in nasal epithelial cells. Virus Res. 2011;160:360–366. doi: 10.1016/j.virusres.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Onoguchi K., Yoneyama M., Takemura A., Akira S., Taniguchi T., Namiki H., Fujita T. Viral infections activate types I and III interferon genes through a common mechanism. J. Biol. Chem. 2007;282:7576–7581. doi: 10.1074/jbc.M608618200. [DOI] [PubMed] [Google Scholar]
- Osterlund P.I., Pietilä T.E., Veckman V., Kotenko S.V., Julkunen I. IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-lambda) genes. J. Immunol. 2007;179:3434–3442. doi: 10.4049/jimmunol.179.6.3434. [DOI] [PubMed] [Google Scholar]
- Pagliaccetti N.E., Robek M.D. Interferon-λ in HCV Infection and Therapy. Viruses. 2010;2:1589–1602. doi: 10.3390/v2081589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Serti E., Eke O., Muchmore B., Prokunina-Olsson L., Capone S., Folgori A., Rehermann B. IL-29 is the dominant type III interferon produced by hepatocytes during acute hepatitis C virus infection. Hepatology. 2012;56:2060–2070. doi: 10.1002/hep.25897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera G.K., Di Meglio P., Nestle F.O. Psoriasis. Annu. Rev. Pathol. 2012;7:385–422. doi: 10.1146/annurev-pathol-011811-132448. [DOI] [PubMed] [Google Scholar]
- Pestka S. The interferons: 50 years after their discovery, there is much more to learn. J. Biol. Chem. 2007;282:20047–20051. doi: 10.1074/jbc.R700004200. [DOI] [PubMed] [Google Scholar]
- Pietilä T.E., Latvala S., Osterlund P., Julkunen I. Inhibition of dynamin-dependent endocytosis interferes with type III IFN expression in bacteria-infected human monocyte-derived DCs. J. Leukoc. Biol. 2010;88:665–674. doi: 10.1189/jlb.1009651. [DOI] [PubMed] [Google Scholar]
- Pott J., Mahlakõiv T., Mordstein M., Duerr C.U., Michiels T., Stockinger S., Staeheli P., Hornef M.W. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc. Natl. Acad. Sci. USA. 2011;108:7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokunina-Olsson L., Muchmore B., Tang W., Pfeiffer R.M., Park H., Dickensheets H., Hergott D., Porter-Gill P., Mumy A., Kohaar I. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z., Nie P., Secombes C.J., Zou J. Intron-containing type I and type III IFN coexist in amphibians: refuting the concept that a retroposition event gave rise to type I IFNs. J. Immunol. 2010;184:5038–5046. doi: 10.4049/jimmunol.0903374. [DOI] [PubMed] [Google Scholar]
- Reuter A., Soubies S., Härtle S., Schusser B., Kaspers B., Staeheli P., Rubbenstroth D. Antiviral activity of lambda interferon in chickens. J. Virol. 2014;88:2835–2843. doi: 10.1128/JVI.02764-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby R.E., Rehwinkel J. RNA degradation in antiviral immunity and autoimmunity. Trends Immunol. 2015;36:179–188. doi: 10.1016/j.it.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C.D., Griffiths S.J., Haas J. Interferon lambda genetic polymorphisms and viral infection: the tip of the iceberg? DNA Cell Biol. 2014;33:60–63. doi: 10.1089/dna.2013.2261. [DOI] [PubMed] [Google Scholar]
- Sabat R. IL-10 family of cytokines. Cytokine Growth Factor Rev. 2010;21:315–324. doi: 10.1016/j.cytogfr.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Sato A., Ohtsuki M., Hata M., Kobayashi E., Murakami T. Antitumor activity of IFN-lambda in murine tumor models. J. Immunol. 2006;176:7686–7694. doi: 10.4049/jimmunol.176.12.7686. [DOI] [PubMed] [Google Scholar]
- Sato S., Li K., Kameyama T., Hayashi T., Ishida Y., Murakami S., Watanabe T., Iijima S., Sakurai Y., Watashi K. The RNA sensor RIG-I dually functions as an innate sensor and direct antiviral factor for hepatitis B virus. Immunity. 2015;42:123–132. doi: 10.1016/j.immuni.2014.12.016. [DOI] [PubMed] [Google Scholar]
- Sheahan T., Imanaka N., Marukian S., Dorner M., Liu P., Ploss A., Rice C.M. Interferon lambda alleles predict innate antiviral immune responses and hepatitis C virus permissiveness. Cell Host Microbe. 2014;15:190–202. doi: 10.1016/j.chom.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard P., Kindsvogel W., Xu W., Henderson K., Schlutsmeyer S., Whitmore T.E., Kuestner R., Garrigues U., Birks C., Roraback J. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- Shindo H., Maekawa S., Komase K., Miura M., Kadokura M., Sueki R., Komatsu N., Shindo K., Amemiya F., Nakayama Y. IL-28B (IFN-λ3) and IFN-α synergistically inhibit HCV replication. J. Viral Hepat. 2013;20:281–289. doi: 10.1111/j.1365-2893.2012.01649.x. [DOI] [PubMed] [Google Scholar]
- Siegel R., Eskdale J., Gallagher G. Regulation of IFN-λ1 promoter activity (IFN-λ1/IL-29) in human airway epithelial cells. J. Immunol. 2011;187:5636–5644. doi: 10.4049/jimmunol.1003988. [DOI] [PubMed] [Google Scholar]
- Sommereyns C., Paul S., Staeheli P., Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorgeloos F., Kreit M., Hermant P., Lardinois C., Michiels T. Antiviral type I and type III interferon responses in the central nervous system. Viruses. 2013;5:834–857. doi: 10.3390/v5030834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza-Fonseca-Guimaraes F., Young A., Mittal D., Martinet L., Bruedigam C., Takeda K., Andoniou C.E., Degli-Esposti M.A., Hill G.R., Smyth M.J. NK cells require IL-28R for optimal in vivo activity. Proc. Natl. Acad. Sci. USA. 2015;112:E2376–E2384. doi: 10.1073/pnas.1424241112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S., Dai J., Eskdale J., Gallagher G.E., Megjugorac N.J., Gallagher G. Interferon-lambda1 (interleukin-29) preferentially down-regulates interleukin-13 over other T helper type 2 cytokine responses in vitro. Immunology. 2008;125:492–502. doi: 10.1111/j.1365-2567.2008.02862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stifter S.A., Feng C.G. Interfering with immunity: detrimental role of type I IFNs during infection. J. Immunol. 2015;194:2455–2465. doi: 10.4049/jimmunol.1402794. [DOI] [PubMed] [Google Scholar]
- Strange A., Capon F., Spencer C.C., Knight J., Weale M.E., Allen M.H., Barton A., Band G., Bellenguez C., Bergboer J.G., Genetic Analysis of Psoriasis Consortium & the Wellcome Trust Case Control Consortium 2 A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat. Genet. 2010;42:985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suppiah V., Moldovan M., Ahlenstiel G., Berg T., Weltman M., Abate M.L., Bassendine M., Spengler U., Dore G.J., Powell E. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat. Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- Swider A., Siegel R., Eskdale J., Gallagher G. Regulation of interferon lambda-1 (IFNL1/IFN-λ1/IL-29) expression in human colon epithelial cells. Cytokine. 2014;65:17–23. doi: 10.1016/j.cyto.2013.09.020. [DOI] [PubMed] [Google Scholar]
- Swiecki M., Colonna M. Type I interferons: diversity of sources, production pathways and effects on immune responses. Curr. Opin. Virol. 2011;1:463–475. doi: 10.1016/j.coviro.2011.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Nishida N., Sugiyama M., Kurosaki M., Matsuura K., Sakamoto N., Nakagawa M., Korenaga M., Hino K., Hige S. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat. Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- Thomas D.L., Thio C.L., Martin M.P., Qi Y., Ge D., O’Huigin C., Kidd J., Kidd K., Khakoo S.I., Alexander G. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas E., Gonzalez V.D., Li Q., Modi A.A., Chen W., Noureddin M., Rotman Y., Liang T.J. HCV infection induces a unique hepatic innate immune response associated with robust production of type III interferons. Gastroenterology. 2012;142:978–988. doi: 10.1053/j.gastro.2011.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson S.J., Goh F.G., Banks H., Krausgruber T., Kotenko S.V., Foxwell B.M., Udalova I.A. The role of transposable elements in the regulation of IFN-lambda1 gene expression. Proc. Natl. Acad. Sci. USA. 2009;106:11564–11569. doi: 10.1073/pnas.0904477106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travar M., Vucic M., Petkovic M. Interferon lambda-2 levels in sputum of patients with pulmonary Mycobacterium tuberculosis infection. Scand. J. Immunol. 2014;80:43–49. doi: 10.1111/sji.12178. [DOI] [PubMed] [Google Scholar]
- Urban T.J., Thompson A.J., Bradrick S.S., Fellay J., Schuppan D., Cronin K.D., Hong L., McKenzie A., Patel K., Shianna K.V. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 2010;52:1888–1896. doi: 10.1002/hep.23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte K., Gruetz G., Volk H.D., Looman A.C., Asadullah K., Sterry W., Sabat R., Wolk K. Despite IFN-lambda receptor expression, blood immune cells, but not keratinocytes or melanocytes, have an impaired response to type III interferons: implications for therapeutic applications of these cytokines. Genes Immun. 2009;10:702–714. doi: 10.1038/gene.2009.72. [DOI] [PubMed] [Google Scholar]
- Wobus C.E., Karst S.M., Thackray L.B., Chang K.O., Sosnovtsev S.V., Belliot G., Krug A., Mackenzie J.M., Green K.Y., Virgin H.W. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2004;2:e432. doi: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk K., Kunz S., Witte E., Friedrich M., Asadullah K., Sabat R. IL-22 increases the innate immunity of tissues. Immunity. 2004;21:241–254. doi: 10.1016/j.immuni.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Wolk K., Witte K., Witte E., Raftery M., Kokolakis G., Philipp S., Schönrich G., Warszawska K., Kirsch S., Prösch S. IL-29 is produced by T(H)17 cells and mediates the cutaneous antiviral competence in psoriasis. Sci. Transl. Med. 2013;5:204ra129. doi: 10.1126/scitranslmed.3006245. [DOI] [PubMed] [Google Scholar]
- Wollenberg A., Wetzel S., Burgdorf W.H., Haas J. Viral infections in atopic dermatitis: pathogenic aspects and clinical management. J. Allergy Clin. Immunol. 2003;112:667–674. doi: 10.1016/j.jaci.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Wongthida P., Diaz R.M., Galivo F., Kottke T., Thompson J., Pulido J., Pavelko K., Pease L., Melcher A., Vile R. Type III IFN interleukin-28 mediates the antitumor efficacy of oncolytic virus VSV in immune-competent mouse models of cancer. Cancer Res. 2010;70:4539–4549. doi: 10.1158/0008-5472.CAN-09-4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Brann T.W., Zhou M., Yang J., Oguariri R.M., Lidie K.B., Imamichi H., Huang D.W., Lempicki R.A., Baseler M.W. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J. Immunol. 2011;186:4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Valdez P.A., Danilenko D.M., Hu Y., Sa S.M., Gong Q., Abbas A.R., Modrusan Z., Ghilardi N., de Sauvage F.J., Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Hamming O.J., Ank N., Paludan S.R., Nielsen A.L., Hartmann R. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J. Virol. 2007;81:7749–7758. doi: 10.1128/JVI.02438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Cowled C., Marsh G.A., Shi Z., Wang L.F., Baker M.L. Type III IFN receptor expression and functional characterisation in the pteropid bat, Pteropus alecto. PLoS ONE. 2011;6:e25385. doi: 10.1371/journal.pone.0025385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Cowled C., Todd S., Crameri G., Virtue E.R., Marsh G.A., Klein R., Shi Z., Wang L.F., Baker M.L. Type III IFNs in pteropid bats: differential expression patterns provide evidence for distinct roles in antiviral immunity. J. Immunol. 2011;186:3138–3147. doi: 10.4049/jimmunol.1003115. [DOI] [PMC free article] [PubMed] [Google Scholar]