Abstract
Sirtuins (silent information regulator 2 [Sir2] proteins) belong to an ancient family of evolutionary conserved nicotinamide adenine dinucleotide (NAD)+-dependent enzymes with deacetylase and/or mono-ADP-ribosyltransferase activity. They regulate DNA repair and recombination, chromosomal stability, and gene transcription, and most importantly mediate the health-promoting effects of caloric restriction (CR), which includes the retardation of aging. At least seven Sir2 homologs, sirtuins (SIRT) 1 to 7 have been identified in mammals. Mammalian SIRT1, the most extensively studied family member, couples protein deacetylation with NAD+ hydrolysis and links cellular energy and redox state to multiple signaling and survival pathways. Cell-type and context-specific activation of sirtuins increases resistance to metabolic, oxidative, and hypoxic stress in different tissues. In particular, SIRT1 plays a central role in mediating the beneficial effects of CR, and its activation associates with longevity and the attenuation of metabolic disorders. SIRT1 in the kidney is cytoprotective and participates in the regulation of BP and sodium balance. Here, we review sirtuin biology and discuss how CR-triggered sirtuin-dependent pathways affect renal physiology and the pathogenesis of kidney diseases and related disorders.
In 1934, Clive M. McCay and colleagues observed that experimental caloric restriction (CR), without causing malnutrition, increases the life span of rats.1 This and other health-promoting effects of CR have been demonstrated in a wide range of organisms, including primates,2– 4 and positively affect a wide range of diseases, such as obesity and diabetes, atherosclerosis, neurodegenerative diseases, cancer, and age-associated renal injury.5–11
First clues about the molecular pathways triggered by CR that regulate aging come from observations in budding yeast, which serves as a useful model system for discovery of longevity genes. A major cause of aging in yeast is the accumulation of extra-chromosomal rDNA circles (ERCs), which are generated by homologous recombination between ribosomal DNA (rDNA) repeats.12 Using this model, Guarente and colleagues discovered that Sir2 (silent information regulator 2), the first sirtuin family member, is a suppressor of ERC formation and regulates life span. Increased copy numbers of the SIR2 gene not only increases life span in yeast but also delays aging in Caenorhabditis elegans and Drosophila melanogaster.13–15 These and other findings identify Sir2 and its orthologs as central modulators in the regulation of longevity in both lower and higher organisms. Furthermore, Sir2 and its homologs, Hst1 and Hst2, mediate CR-induced extension of yeast replicative life span,16,17 which may be different under nondividing conditions, and also appears to involve Sir2-independent pathways.18–20
At least seven homologs of Sir2, sirtuin (SIRT) 1 to 7, are present in mammals. SIRT1, the most extensively studied family member, couples protein deacetylation with NAD+ hydrolysis and integrates cellular energy and redox state with the regulation of multiple signaling and survival pathways. From an evolutionary point of view, SIRT1 has evolved as a universal regulator of a highly conserved cellular defense program that facilitates survival when nutrients are in short supply.21–23 Although its role in the regulation of longevity in mammals is still unclear, SIRT1 is required and sufficient for the induction of a CR phenotype in rodents.24–27
Although the molecular underpinnings of CR are complex and incompletely understood, the notion that pharmacologic targeting of sirtuins, SIRT1 in particular, could potentially revolutionize treatment of age-related disease conditions, such as diabetes, atherosclerosis, cancer, and certain forms of renal disease has been an incentive for major efforts in drug discovery.28 At least in experimental settings, the health benefits of caloric restriction such as its life-extending, anti-inflammatory, antioxidant, antidiabetic, and cancer-preventive effects are mimicked by the naturally occurring SIRT1 activator, resveratrol (trans-3,5,4′-trihydroxystilbene) and its analogs.29 Because resveratrol is found in foods, such as peanuts, blueberries, and grapes, and is a constituent of red wine, it has received much attention by the public media, although its beneficial effects for humans in naturally occurring amounts are unproven.
In the kidney, CR retards the normal aging process, which associates with structural and functional changes that include glomerulosclerosis and tubulo-interstitial fibrosis, a decrease in GFR and renal blood flow, and progressive loss of multiple tubular transport functions.30,7,8,31 Recent studies show that SIRT1 is expressed in the kidney and acts as a renal survival factor; however, its role in CR-mediated renoprotection is unclear. Here, we review sirtuin biology and discuss how sirtuin-dependent pathways affect renal physiology and the pathogenesis of kidney diseases and related disorders.
THE SIRTUIN FAMILY OF NAD+-DEPENDENT DEACETYLASES AND MONO-ADP-RIBOSYLTRANSFERASES: THE ESSENTIAL FACTS
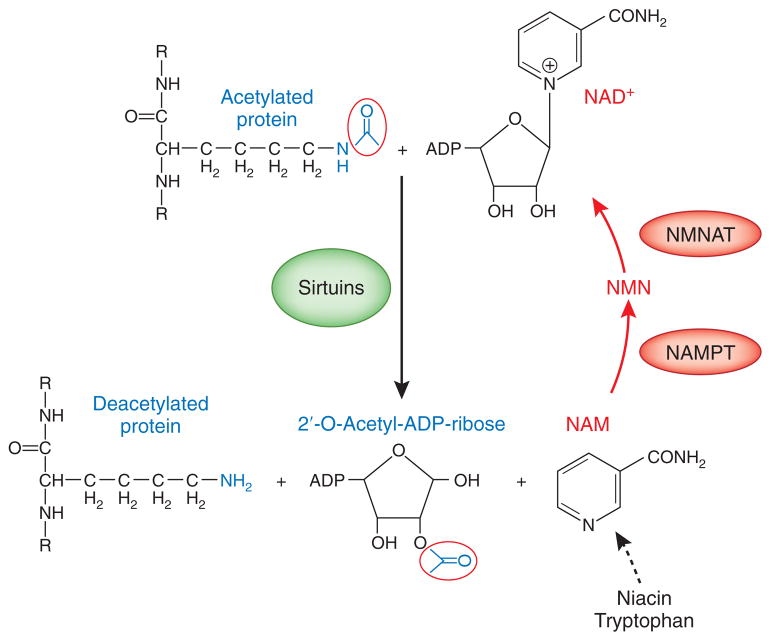
The first sirtuin family member, Sir2, originally known as mating-type regulator 1,32 was initially discovered as a silencer at mating-type loci in Saccharomyces cerevisiae.33 Later studies showed that Sir2 is also involved in the regulation of rDNA recombination34 and telomeric gene repression, which is associated with nucleosomal hypoacetylation.35,36 Detailed biochemical analyses demonstrate that Sir2 functions as a NAD+-dependent histone deacetylase,37,38 which couples deacetylation with hydrolysis of NAD+, generating nicotinamide (NAM) and 2′-O-acetyl-ADP-ribose (Figure 1).39,40
Figure 1.
Sirtuin-mediated protein deacetylation requries NAD+. Overview of the sirtuin deacetylation reaction and the NAD+ salvage pathway. Sirtuins deacetylate their target proteins at specific lysine residues. This reaction is dependent on the availability of NAD+, which serves as a cosubstrate, thereby integrating cellular energy and redox state with multiple signaling and survival pathways. The reaction results in the formation of a deacetylated protein, NAM, and 2′-O-acetyl-ADP-ribose, and is inhibited by NAM. The salvage pathway (red arrows) is used to regenerate NAD+ from NAM. NAMPT, the rate-limiting enzyme in this pathway, converts NAM to NMN, followed by conversion of NMN to NAD+ by NMNAT.
Sir2 also has mono-ADP-ribosyltransferase activity (transfer of ADP-ribose to other proteins), which is of minor importance for its biologic function. This is in contrast to two members of the mammalian sirtuins, SIRT4 and SIRT6, which primarily function as mono-ADP-ribosyltransferases. Unlike most other deacetylases, the catalytic activity of sirtuins is absolutely dependent on NAD+.41,42 Because the intracellular level of NAD+ (its ratio to its reduced form, NADH) is dependent on cellular energy and redox state, sirtuins function as molecular sensors of cellular energy balance.43,22
In mammals, seven homologs of yeast Sir2, SIRT1 through SIRT7, have been identified. Each sirtuin contains a conserved catalytic core domain of approximately 275 amino acids, which functions as a NAD+-dependent deacetylase and/or mono-ADP-ribosyltransferase. SIRT1 and SIRT5 primarily target proteins for deacetylation, which include histones, transcriptional regulators (p53, forkhead box O [FOXO] transcription factors, nuclear factor κB [NF-κB], and hypoxia-inducible factor [HIF]-2α), enzymes (acetyl-CoA synthase 1 [AceCS1]), and other signaling molecules such as androgen receptors, whereas SIRT2 and SIRT3 carry out both types of catalytic activity.23 SIRT4 and SIRT6, however, function as mono-ADP-ribosyltransferases and not as deacetylases.41,42 SIRT4, for example, mono-ADP-ribosylates glutamate dehydrogenase in pancreatic β cells and thereby inhibits amino acid–stimulated insulin secretion.42 Mammalian sirtuins differ not only with respect to their catalytic function but also in regard to their sub-cellular localization.44 – 46 SIRT1, SIRT6, and SIRT7 are primarily located in the nucleus, whereas SIRT2 is found in cytoplasm and SIRT3, SIRT4, and SIRT5 reside in the mitochondria.
REGULATION OF SIRTUIN ACTIVITY
Cellular sirtuin activity is controlled dynamically to meet metabolic and environmental challenges. It depends on protein expression levels, reversible posttranslational modifications, the availability of cosubstrate NAD+, and the presence or absence of interacting proteins that are capable of modulating sirtuin enzymatic activity. Expression of SIRT1 increases with CR, during starvation and nutrient deprivation, or when cells are acutely exposed to conditions that cause oxidative stress and DNA damage.47,48 Decreased expression of SIRT1 associates with a high fat diet, insulin resistance, high glucose, and senescence. CR increases the activity at the SIRT1 promoter through the interaction of FOXO3a with p53, whereby FOXO3a removes p53 from the SIRT1 promoter as a negative regulator.49
The increase in SIRT1 expression in response to acute stress, such as oxidative stress, is relatively short-lived, and controlled and counterbalanced on multiple regulatory levels. This involves complex regulation at the promoter, mRNA, and protein levels. Hypermethylated-in-cancer 1 and the adenoviral E1A protein-interacting C-terminal binding protein are part of a repressor complex that inhibits SIRT1 transcription and dissociates in response to changes in cellular redox state, which results in de-repression at the SIRT1 locus, increasing transcription in response to oxidative stress.50 Increased SIRT1 transcription after oxidative stress and following replicative senescence is counteracted by a reduction in the stability of mRNA encoding SIRT1. This results in part from decreased expression of a stabilizing mRNA binding protein, HuR, which maintains steady-state levels of the SIRT1 mRNA.51 p53 also induces micro RNA, miR34a, which targets SIRT1 mRNA and reduces level of SIRT1.52 An additional level of complexity in the regulation of SIRT1 biologic activity is added by reversible post-translational modifications, which include sumoylation53 and phosphorylation,54–56 and through biochemical interaction with inhibiting or activating factors, such as SIRT1 inhibitor DBC1 (deleted in breast cancer)57 or SIRT1 activator AROS (active regulator of SIRT1).58 Exposure to ultraviolet radiation or oxidative stress leads to desumoylation of SIRT1, and to increased formation of DBC1-SIRT complexes, both resulting in reduced enzymatic activity.53,57
SIRT1 enzymatic activity associates with the consumption of NAD+ and the production of NAM, which in turn, inhibits its catalytic activity. Recent studies indicate that the NAD+ salvage pathway plays a critical role in the regulation of SIRT1 enzymatic activity by lowering the concentration of inhibitory NAM and by increasing levels of sirtuin cosubstrate NAD+ (for a detailed review, see 22). The first reaction in the NAD+ salvage pathway is the generation of nicotinamide mononucleotide (NMN) from NAM by the activity of nicotinamide phosphoribosyltransferase (NAMPT), followed by conversion of NMN to NAD+ by NMN adenylyltransferase (NMNAT) (Figure 2). NAMPT is the rate-limiting enzyme in this pathway and therefore has a key role in regulating sirtuin activity. In cultured cells and in mouse tissues, its level of expression correlates with the cellular level of NAD+ and with sirtuin activity;59 that is, during caloric restriction NAMPT and SIRT activity both increase.21 NAMPT exists in intracellular and extracellular forms (iNAMPT and eNAMPT). eNAMPT, also known as PBEF (pre–B cell colony enhancing factor) or as visfatin (visceral fat–derived hormone),60,61 is present in plasma, where it synthesizes NMN, making it available for systemic distribution. As a systemic NAD+ biosynthetic enzyme, eNAMPT also regulates glucose homeostasis.22
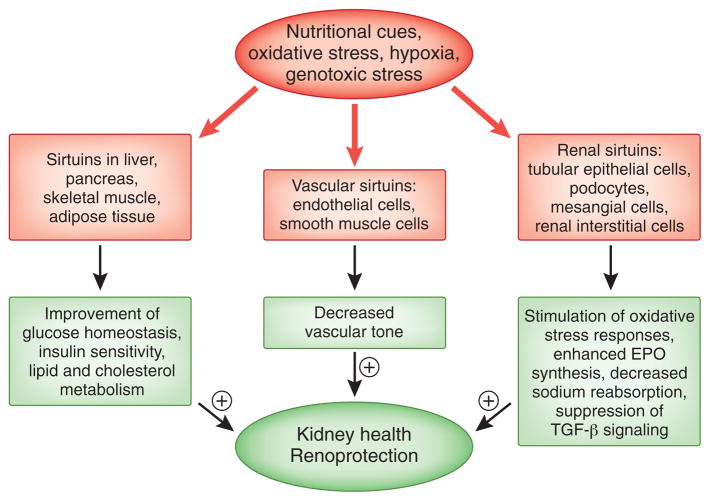
Figure 2.
Sirtuins protect kidney health. Shown is an overview of sirtuin-regulated processes with positive effect on kidney health. Nutritional cues and different forms of stress activate sirtuins in different tissues and cell types. This results in multiple local and systemic effects, which are likely to retard renal aging and result in renoprotection.
Interestingly, clinical studies show that serum levels of NAMPT increase in patients with chronic kidney disease.62,63 Its role in the pathogenesis and progression of chronic kidney disease, however, remains unclear at this point. A potential role for NAMPT in fibrogenesis has been suggested in mesangial cells, where NAMPT expression associates with increased synthesis of profibrotic molecules, such as TGF-β1, plasminogen activator inhibitor-1, and collagen type 1.64 This however may not involve SIRT1. Although increased NAMPT-mediated NAD+ biosynthesis enhances sirtuin activity, NAD+ is consumed by multiple enzymatic reactions that do not involve sirtuins, for example, covalent protein modifications by other ADP-ribosyltransferases.22
METABOLIC AND RENOPROTECTIVE EFFECTS OF SIRT1 ACTIVATION: RELEVANCE TO KIDNEY INJURY
Although the molecular underpinnings of CR are complex and understood incompletely, the health benefits of CR are well established. Among those is protection against many age-associated diseases such as diabetes, cancer, and inflammation. In rodents, caloric restriction or starvation increases SIRT1 levels in multiple tissues including brain, liver, and kidney.47,48 This response has also been observed in different human cell types, such as mononuclear cells, skeletal muscle, and adipose tissue.65–67 With regard to the kidney, caloric restriction associates with less injury in age-related and diabetic nephropathy models. This probably results from a combination of systemic and cell type-specific local effects, which include improved glucose and lipid metabolism, decreased accumulation of advanced glycosylation end-products, a reduction in oxidative stress, improved nitric oxide balance, and diminished angiotensin II signaling.7,8,68,31 Not surprisingly, intermittent fasting in a streptozotocin-induced diabetic nephropathy model is associated with increased renal SIRT1 expression.68 Although its role in the aging and diabetic kidney is only poorly understood, experimental studies in non-renal settings identify critical sirtuin-dependent signaling pathways that are triggered by CR, some of which are also activated in some renal cells. Without doubt, the metabolic benefits of systemic SIRT1 activation are likely to have a positive effect on the clinical outcome of age-associated or diabetic renal disease.
During fasting, SIRT1 stimulates hepatic glucose output by enhancing gluconeogenesis and by repressing glycolysis in the liver, thus contributing to the maintenance of plasma glucose homeostasis.48 This seems to occur through deacetylation and activation of peroxisome proliferator–activated receptor-γ coactivator 1α (PGC1-α) and FOXO1.48,69 Furthermore, SIRT1 promotes fatty acid mobilization in white adipose tissue during fasting by repressing peroxisome proliferator–activated receptor-γ (PPAR-γ) and by quenching its cofactors NCoR (nuclear receptor co-repressor) and SMRT (silencing mediator of retinoid and thyroid hormone receptors).27 Deletion of one SIRT1 allele in mice is sufficient to minimize this effect, and mice with complete SIRT1 deficiency fail to adapt to caloric restriction, a finding that underscores the central role of SIRT1 in executing the biologic effects of CR.25,24 In contrast to caloric restriction, high fat diet or the presence of obesity associates with reduced SIRT1 levels.70,67 Although it is unclear whether this reduction in SIRT1 is a primary event in the pathogenesis of obesity and its associated metabolic disorders, activation of SIRT1 in this context is beneficial, at least in the experimental setting.
Studies in rodents demonstrate that increasing SIRT1 activity by either pharmacologic or genetic means prolongs the life of obese animals and significantly improves their metabolic parameters, such as glucose tolerance, fasting blood glucose levels, and insulin resistance.71,72,10,73,26,74 Small-molecule sirtuin activators for the treatment of type 2 diabetes are currently in clinical trials and are likely to improve glucose and insulin homeostasis in humans, which certainly would benefit the kidney as well.
Aside from their positive effects on metabolism, sirtuins regulate pathways in renal cells that control cell viability, as has been shown in vitro and in vivo. SIRT1 promotes cell survival by modulating the cellular responses to different types of stress, including oxidative, genotoxic, and hypoxic stress.47,75–78 This survival-promoting role is triggered by CR and is important for the prevention of aging, as aging is associated with increased rates of stress-induced apoptosis.79,47 SIRT1 activation enhances resistance to apoptosis in human embryonic kidney cells47 and leads to cytoprotection in heart, brain, and pancreatic islets after ischemic/oxidative or cytokine-mediated injury.80 – 83 Deacetylation targets, which mediate these prosurvival effects, include FOXO transcription factors, p53, heat-shock protein-1, NF-κB, Ku70, Smad7, and uncoupling protein-2.75,84 – 86,76,87– 89
In the kidney, pretreatment with resveratrol reduces acute ischemia/reperfusion injury in rats.90,91 In mesangial cells, SIRT1 inhibits oxidative stress–induced apoptosis by deacetylation and inactivation of p53,92 in HK-2 cells through the activation of FOXO3a and the upregulation of catalase.93 SIRT1 also protects mesangial cells from TGF-β1–mediated apoptosis by deacetylating Smad7 at lysine residues 60 and 70, which accelerates its degradation by Smad ubiquitination regulatory factor-1.94
Recent studies from our laboratory demonstrate that SIRT1 is abundantly expressed in renal medullary interstitial cells, whereas only low levels of SIRT1 are found in the renal cortex by immunohistochemistry (C.M. Hao, unpublished data). In vitro, we find that SIRT1 protects primary renal medullary interstitial cells from oxidative stress–induced apoptosis after exposure to hydrogen peroxide. Deletion of one SIRT1 allele worsens apoptosis and fibrosis in the renal medulla after unilateral ureteral obstruction, whereas pharmacologic activation of SIRT1 is beneficial in this model (C.M. Hao, unpublished data). These findings suggest that SIRT1 has a robust cytoprotective role in renal medullary cells. SIRT1 activation could therefore be exploited therapeutically to stimulate potent antioxidant pathways, which promote the survival of cells that normally have to cope with high levels of oxidative stress generated by rapid changes in interstitial tonicity, relatively low blood flow, and low oxygen tension.95–97 SIRT1 is also expressed in podocytes (C.M. Hao, unpublished data). Whether it is cytoprotective in this cell type is unclear at the moment, and certainly warrants further investigation. For a summary of sirtuin-mediated effects on the kidney, see Figure 2.
SIRT1 IN THE REGULATION OF BP AND HANDLING OF RENAL SODIUM
Strong evidence supports a link between energy metabolism and the regulation of systemic BP,98,99 as hypertension is frequently clustered with obesity, hyperlipidemia, and hyperinsulinemia, collectively termed metabolic syndrome. CR, however, reduces BP in mildly hypertensive patients or reduces the number of antihypertensive drugs required to control hypertension.99,100 Several studies suggest that SIRT1 participates in BP regulation.101–103 Mechanistically, this seems to occur at least on two functional levels, the regulation of vascular tone and the regulation of renal sodium reabsorption in the collecting duct (Figure 2).
Miyazaki and colleagues report that overexpression of SIRT1 reduces angiotensin II AT1 receptor (AT1R) expression in vascular smooth muscle cells, whereas NAM, which inhibits SIRT1, increases vascular smooth muscle cell AT1R expression.101 Administration of resveratrol in mice not only suppresses AT1R expression in the aorta but also significantly blunts angiotensin II–induced hypertension.101 SIRT1, furthermore, promotes vasodilation by deacetylating endothelial nitric oxide synthase, thus increasing endothelial nitric oxide (NO) levels.103 Inhibition of endothelial SIRT1 inhibits vasodilation and decreases NO bioavailability.103
In the kidney SIRT1 participates in the regulation of sodium balance by repressing transcription of the α-subunit of the epithelial sodium channel, ENaC, in cultured inner medullary collecting duct cells.102 Interestingly, the inhibitory effect of SIRT1 on expression of α-ENaC is independent of its deacetylase activity, which is in contrast to most SIRT1 targets. SIRT1 interacts with disruptor of telomeric silencing-1 (Dot1), a methyl-transferase that methylates lysine residue 79 in the core domain of histone H3, which associates with the α-ENaC 5′-flanking region. The SIRT1/Dot1 interaction results in H3K79 hypermethylation in chromatin along the α-ENaC promoter and leads to transcriptional repression of α-ENaC.102 This effect on α-ENaC transcription is independent of mineralocorticoid receptor signaling. Interestingly, treatment with aldosterone decreases levels of mRNA encoding SIRT1. Because the effects of SIRT1 on α-ENaC transcription are independent of its enzymatic activity, changes in NAM levels or the use of SIRT1 activators are unlikely to modulate sodium reabsorption in the collecting duct.
SIRT1 MODULATION OF HYPOXIC RESPONSES: IMPLICATIONS FOR RENAL ERYTHROPOIETIN PRODUCTION
SIRT1 modulates cellular hypoxia responses by interaction with the α-subunit of HIF-2. HIF-2 is an oxygen-sensitive heterodimeric basic helix-loop-helix transcription factor that plays a central role in mediating cellular responses to hypoxic stress. It is responsible for the hypoxic induction of erythropoietin (EPO), vascular endothelial growth factor, and other oxygen-regulated genes.104 In the kidney, it is expressed in EPO-producing interstitial cells, endothelial, glomerular, and von Hippel-Lindau–deficient renal cancer cells. In nontransformed renal epithelial cells, HIF-2 is not active and hypoxic responses are mediated by HIF-1.104 SIRT1 stimulates the transcriptional activity of HIF-2, but not HIF-1, by selectively deacetylating HIF-2α under hypoxia.105 The ability of SIRT1 to modulate the transcriptional activity of HIF-2 under low oxygen conditions is reflected by a blunting of renal EPO responses when SIRT1 levels are reduced genetically.105 Conversely, SIRT1 activation enhanced renal EPO expression.105 Whether SIRT1 could become a therapeutic target in the treatment of renal anemia or can be used to stimulate other cytoprotective HIF-2 responses remains unclear.
SUMMARY AND OUTLOOK
In this brief review we provide an overview of sirtuin-modulated biologic processes and discuss how sirtuins could be exploited therapeutically to harness the health-promoting effects of CR. Although a large body of literature exists that deals with sirtuins in different model organisms, current understanding of sirtuin biology in the kidney is comparatively limited. Recent experimental findings, however, suggest that renal sirtuins are involved in the regulation of a wide range of physiologic processes in the kidney, which include oxidative stress responses, EPO production, and sodium homeostasis.
As sensors of redox and energy state, renal sirtuins are likely to integrate nutritional cues with cellular stress and survival responses in the kidney. In this role, renal sirtuins may be essential in protecting the kidney from aging and in mediating the beneficial effects of CR on renal health. Whether renal sirtuins take part in regulating the systemic aging process is unknown and warrants further investigation. Future studies using pharmacologic or genetic approaches are needed to better understand their role in renal physiology and pathology, and to determine whether they can serve as therapeutic targets to improve the clinical outcome of kidney diseases.
Footnotes
DISCLOSURES
V.H.H. is supported by the Krick-Brooks Chair in Nephrology and by research grants from the National Cancer Institute (NCI), and the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). C.M.H. is supported by grants from the NIDDK. The authors apologize to all those colleagues whose original work was not cited because of space limitations.
References
- 1.McCay CM, Crowell MF. Prolonging the life span. The Scientific Monthly. 1934;39:405–414. [Google Scholar]
- 2.Barger JL, Walford RL, Weindruch R. The retardation of aging by caloric restriction: Its significance in the transgenic era. Exp Gerontol. 2003;38:1343–1351. doi: 10.1016/j.exger.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 3.Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons HA, Kemnitz JW, Weindruch R. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop NA, Guarente L. Genetic links between diet and lifespan: Shared mechanisms from yeast to humans. Nat Rev Genet. 2007;8:835–844. doi: 10.1038/nrg2188. [DOI] [PubMed] [Google Scholar]
- 5.Piper MD, Bartke A. Diet and aging. Cell Metab. 2008;8:99–104. doi: 10.1016/j.cmet.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Campisi J, Yaswen P. Aging and cancer cell biology, 2009. Aging Cell. 2009;8:221–225. doi: 10.1111/j.1474-9726.2009.00475.x. [DOI] [PubMed] [Google Scholar]
- 7.Stern JS, Gades MD, Wheeldon CM, Borchers AT. Calorie restriction in obesity: Prevention of kidney disease in rodents. J Nutr. 2001;131:913S–917S. doi: 10.1093/jn/131.3.913S. [DOI] [PubMed] [Google Scholar]
- 8.Davis TA, Bales CW, Beauchene RE. Differential effects of dietary caloric and protein restriction in the aging rat. Exp Gerontol. 1983;18:427–435. doi: 10.1016/0531-5565(83)90021-9. [DOI] [PubMed] [Google Scholar]
- 9.Weindruch R. The retardation of aging by caloric restriction: Studies in rodents and primates. Toxicol Pathol. 1996;24:742–745. doi: 10.1177/019262339602400618. [DOI] [PubMed] [Google Scholar]
- 10.Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joseph J, Cole G, Head E, Ingram D. Nutrition, brain aging, and neurodegeneration. J Neurosci. 2009;29:12795–12801. doi: 10.1523/JNEUROSCI.3520-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinclair DA, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 13.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 15.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 17.Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- 18.Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 19.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, Longo VD. Sir2 blocks extreme life-span extension. Cell. 2005;123:655–667. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 21.Yang H, Lavu S, Sinclair DA. Nampt/PBEF/Visfatin: A regulator of mammalian health and longevity? Exp Gerontol. 2006;41:718–726. doi: 10.1016/j.exger.2006.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imai S. The NAD World: A new systemic regulatory network for metabolism and aging—Sirt1, systemic NAD biosynthesis, and their importance. Cell Biochem Biophys. 2009;53:65–74. doi: 10.1007/s12013-008-9041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michan S, Sinclair D. Sirtuins in mammals: Insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- 25.Boily G, Seifert EL, Bevilacqua L, He XH, Sabourin G, Estey C, Moffat C, Crawford S, Saliba S, Jardine K, Xuan J, Evans M, Harper ME, McBurney MW. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS ONE. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, Gu W, Guarente L. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 27.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milne JC, Denu JM. The Sirtuin family: Therapeutic targets to treat diseases of aging. Curr Opin Chem Biol. 2008;12:11–17. doi: 10.1016/j.cbpa.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 29.Yang H, Baur JA, Chen A, Miller C, Adams JK, Kisielewski A, Howitz KT, Zipkin RE, Sinclair DA. Design and synthesis of compounds that extend yeast replicative lifespan. Aging Cell. 2007;6:35–43. doi: 10.1111/j.1474-9726.2006.00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Epstein M. Aging and the kidney. J Am Soc Nephrol. 1996;7:1106–1122. doi: 10.1681/ASN.V781106. [DOI] [PubMed] [Google Scholar]
- 31.Nangaku M, Izuhara Y, Usuda N, Inagi R, Shibata T, Sugiyama S, Kurokawa K, van Ypersele de Strihou C, Miyata T. In a type 2 diabetic nephropathy rat model, the improvement of obesity by a low calorie diet reduces oxidative/carbonyl stress and prevents diabetic nephropathy. Nephrol Dial Transplant. 2005;20:2661–2669. doi: 10.1093/ndt/gfi096. [DOI] [PubMed] [Google Scholar]
- 32.Klar AJ, Fogel S, Macleod K. MAR1-a Regulator of the HMa and HMalpha Loci in SACCHAROMYCES CEREVISIAE. Genetics. 1979;93:37–50. doi: 10.1093/genetics/93.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 35.Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 36.Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 37.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 39.Landry J, Slama JT, Sternglanz R. Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem Biophys Res Commun. 2000;278:685–690. doi: 10.1006/bbrc.2000.3854. [DOI] [PubMed] [Google Scholar]
- 40.Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 42.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 43.Belenky P, Bogan KL, Brenner C. NAD+ metabolism in health and disease. Trends Biochem Sci. 2007;32:12–19. doi: 10.1016/j.tibs.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 44.Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 45.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 47.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 48.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 49.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Q, Wang SY, Fleuriel C, Leprince D, Rocheleau JV, Piston DW, Goodman RH. Metabolic regulation of SIRT1 transcription via a HIC1:CtBP corepressor complex. Proc Natl Acad Sci U S A. 2007;104:829–833. doi: 10.1073/pnas.0610590104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Abdelmohsen K, Pullmann R, Jr, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, Furneaux H, Gorospe M. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Y, Fu W, Chen J, Olashaw N, Zhang X, Nicosia SV, Bhalla K, Bai W. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007;9:1253–1262. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sasaki T, Maier B, Koclega KD, Chruszcz M, Gluba W, Stukenberg PT, Minor W, Scrable H. Phosphorylation regulates SIRT1 function. PLoS One. 2008;3:e4020. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ford J, Ahmed S, Allison S, Jiang M, Milner J. JNK2-dependent regulation of SIRT1 protein stability. Cell Cycle. 2008;7:3091–3097. doi: 10.4161/cc.7.19.6799. [DOI] [PubMed] [Google Scholar]
- 56.Zschoernig B, Mahlknecht U. Carboxy-terminal phosphorylation of SIRT1 by protein kinase CK2. Biochem Biophys Res Commun. 2009;381:372–377. doi: 10.1016/j.bbrc.2009.02.085. [DOI] [PubMed] [Google Scholar]
- 57.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 58.Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 59.Zhang T, Berrocal JG, Frizzell KM, Gamble MJ, DuMond ME, Krishnakumar R, Yang T, Sauve AA, Kraus WL. Enzymes in the NAD+ salvage pathway regulate SIRT1 activity at target gene promoters. J Biol Chem. 2009;284:20408–20417. doi: 10.1074/jbc.M109.016469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Samal B, Sun Y, Stearns G, Xie C, Suggs S, McNiece I. Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol Cell Biol. 1994;14:1431–1437. doi: 10.1128/mcb.14.2.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fukuhara A, Matsuda M, Nishizawa M, Segawa K, Tanaka M, Kishimoto K, Matsuki Y, Murakami M, Ichisaka T, Murakami H, Watanabe E, Takagi T, Akiyoshi M, Ohtsubo T, Kihara S, Yamashita S, Makishima M, Funahashi T, Yamanaka S, Hiramatsu R, Matsuzawa Y, Shimomura I. Visfatin: A protein secreted by visceral fat that mimics the effects of insulin. Science. 2005;307:426–430. doi: 10.1126/science.1097243. [DOI] [PubMed] [Google Scholar]
- 62.Axelsson J, Witasp A, Carrero JJ, Qureshi AR, Suliman ME, Heimburger O, Barany P, Lindholm B, Alvestrand A, Schalling M, Nordfors L, Stenvinkel P. Circulating levels of visfatin/pre-B-cell colony-enhancing factor 1 in relation to genotype GFR, body composition, and survival in patients with CKD. Am J Kidney Dis. 2007;49:237–244. doi: 10.1053/j.ajkd.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 63.Yilmaz MI, Saglam M, Carrero JJ, Qureshi AR, Caglar K, Eyileten T, Sonmez A, Cakir E, Yenicesu M, Lindholm B, Stenvinkel P, Axelsson J. Serum visfatin concentration and endothelial dysfunction in chronic kidney disease. Nephrol Dial Transplant. 2008;23:959–965. doi: 10.1093/ndt/gfm727. [DOI] [PubMed] [Google Scholar]
- 64.Song HK, Lee MH, Kim BK, Park YG, Ko GJ, Kang YS, Han JY, Han SY, Han KH, Kim HK, Cha DR. Visfatin: A new player in mesangial cell physiology and diabetic nephropathy. Am J Physiol Renal Physiol. 2008;295:F1485–F1494. doi: 10.1152/ajprenal.90231.2008. [DOI] [PubMed] [Google Scholar]
- 65.Crujeiras AB, Parra D, Goyenechea E, Martinez JA. Sirtuin gene expression in human mononuclear cells is modulated by caloric restriction. Eur J Clin Invest. 2008;38:672–678. doi: 10.1111/j.1365-2362.2008.01998.x. [DOI] [PubMed] [Google Scholar]
- 66.Civitarese AE, Carling S, Heilbronn LK, Hulver MH, Ukropcova B, Deutsch WA, Smith SR, Ravussin E. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4:e76. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pedersen SB, Olholm J, Paulsen SK, Bennetzen MF, Richelsen B. Low Sirt1 expression, which is upregulated by fasting, in human adipose tissue from obese women. Int J Obes (Lond) 2008;32:1250–1255. doi: 10.1038/ijo.2008.78. [DOI] [PubMed] [Google Scholar]
- 68.Tikoo K, Tripathi DN, Kabra DG, Sharma V, Gaikwad AB. Intermittent fasting prevents the progression of type I diabetic nephropathy in rats and changes the expression of Sir2 and p53. FEBS Lett. 2007;581:1071–1078. doi: 10.1016/j.febslet.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 69.Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem. 2005;280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 70.Qiao L, Shao J. SIRT1 regulates adiponectin gene expression through Foxo1-C/enhancer-binding protein alpha transcriptional complex. J Biol Chem. 2006;281:39915–39924. doi: 10.1074/jbc.M607215200. [DOI] [PubMed] [Google Scholar]
- 71.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Banks AS, Kon N, Knight C, Matsumoto M, Gutierrez-Juarez R, Rossetti L, Gu W, Accili D. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 75.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 76.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 77.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 78.Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 79.Higami Y, Shimokawa I. Apoptosis in the aging process. Cell Tissue Res. 2000;301:125–132. doi: 10.1007/s004419900156. [DOI] [PubMed] [Google Scholar]
- 80.Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 81.Hsu CP, Odewale I, Alcendor RR, Sadoshima J. Sirt1 protects the heart from aging and stress. Biol Chem. 2008;389:221–231. doi: 10.1515/BC.2008.032. [DOI] [PubMed] [Google Scholar]
- 82.Lee JH, Song MY, Song EK, Kim EK, Moon WS, Han MK, Park JW, Kwon KB, Park BH. Overexpression of SIRT1 protects pancreatic beta-cells against cytokine toxicity by suppressing the nuclear factor-kappaB signaling pathway. Diabetes. 2009;58:344–351. doi: 10.2337/db07-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Della-Morte D, Dave KR, DeFazio RA, Bao YC, Raval AP, Perez-Pinzon MA. Resveratrol pretreatment protects rat brain from cerebral ischemic damage via a sirtuin 1-uncoupling protein 2 pathway. Neuroscience. 2009;159:993–1002. doi: 10.1016/j.neuroscience.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 86.Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cohen HY, Lavu S, Bitterman KJ, Hekking B, Imahiyerobo TA, Miller C, Frye R, Ploegh H, Kessler BM, Sinclair DA. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 89.Jeong J, Juhn K, Lee H, Kim SH, Min BH, Lee KM, Cho MH, Park GH, Lee KH. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Exp Mol Med. 2007;39:8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- 90.Chander V, Chopra K. Protective effect of nitric oxide pathway in resveratrol renal ischemia-reperfusion injury in rats. Arch Med Res. 2006;37:19–26. doi: 10.1016/j.arcmed.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 91.Bertelli AA, Migliori M, Panichi V, Origlia N, Filippi C, Das DK, Giovannini L. Resveratrol, a component of wine and grapes, in the prevention of kidney disease. Ann N Y Acad Sci. 2002;957:230–238. doi: 10.1111/j.1749-6632.2002.tb02919.x. [DOI] [PubMed] [Google Scholar]
- 92.Kume S, Haneda M, Kanasaki K, Sugimoto T, Araki S, Isono M, Isshiki K, Uzu T, Kashiwagi A, Koya D. Silent information regulator 2 (SIRT1) attenuates oxidative stress-induced mesangial cell apoptosis via p53 deacetylation. Free Radic Biol Med. 2006;40:2175–2182. doi: 10.1016/j.freeradbiomed.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 93.Hasegawa K, Wakino S, Yoshioka K, Tatematsu S, Hara Y, Minakuchi H, Washida N, Tokuyama H, Hayashi K, Itoh H. Sirt1 protects against oxidative stress-induced renal tubular cell apoptosis by the bidirectional regulation of catalase expression. Biochem Biophys Res Commun. 2008;372:51–56. doi: 10.1016/j.bbrc.2008.04.176. [DOI] [PubMed] [Google Scholar]
- 94.Kume S, Haneda M, Kanasaki K, Sugimoto T, Araki S, Isshiki K, Isono M, Uzu T, Guarente L, Kashiwagi A, Koya D. SIRT1 inhibits transforming growth factor beta-induced apoptosis in glomerular mesangial cells via Smad7 deacetylation. J Biol Chem. 2007;282:151–158. doi: 10.1074/jbc.M605904200. [DOI] [PubMed] [Google Scholar]
- 95.Hao CM, Yull F, Blackwell T, Komhoff M, Davis LS, Breyer MD. Dehydration activates an NF-kappaB-driven, COX2-dependent survival mechanism in renal medullary interstitial cells. J Clin Invest. 2000;106:973–982. doi: 10.1172/JCI9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pallone TL. Is oxidative stress differentially regulated in the renal cortex and medulla? Nat Clin Pract Nephrol. 2006;2:118–119. doi: 10.1038/ncpneph0083. [DOI] [PubMed] [Google Scholar]
- 97.Burg MB, Ferraris JD, Dmitrieva NI. Cellular response to hyperosmotic stresses. Physiol Rev. 2007;87:1441–1474. doi: 10.1152/physrev.00056.2006. [DOI] [PubMed] [Google Scholar]
- 98.Garrison RJ, Kannel WB, Stokes J, 3rd, Castelli WP. Incidence and precursors of hypertension in young adults: The Framingham Offspring Study. Prev Med. 1987;16:235–251. doi: 10.1016/0091-7435(87)90087-9. [DOI] [PubMed] [Google Scholar]
- 99.Subcommittee on Nonpharmacological Therapy of the 1984 Joint National Committee on Detection, E, and Treatment of High Blood Pressure. Nonpharmacological approaches to the control of high blood pressure. Hypertension. 1986;8:444–467. [PubMed] [Google Scholar]
- 100.Group HPTR. The Hypertension Prevention Trial: Three-year effects of dietary changes on blood pressure. Arch Intern Med. 1990;150:153–162. [PubMed] [Google Scholar]
- 101.Miyazaki R, Ichiki T, Hashimoto T, Inanaga K, Imayama I, Sadoshima J, Sunagawa K. SIRT1, a longevity gene, downregulates angiotensin II type 1 receptor expression in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2008;28:1263–1269. doi: 10.1161/ATVBAHA.108.166991. [DOI] [PubMed] [Google Scholar]
- 102.Zhang D, Li S, Cruz P, Kone BC. Sirtuin 1 functionally and physically interacts with disruptor of telomeric silencing-1 to regulate alpha-ENaC transcription in collecting duct. J Biol Chem. 2009;284:20917–20926. doi: 10.1074/jbc.M109.020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mattagajasingh I, Kim CS, Naqvi A, Yamamori T, Hoffman TA, Jung SB, De-Ricco J, Kasuno K, Irani K. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc Natl Acad Sci U S A. 2007;104:14855–14860. doi: 10.1073/pnas.0704329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Haase VH. Hypoxia-inducible factors in the kidney. Am J Physiol Renal Physiol. 2006;291:F271–F281. doi: 10.1152/ajprenal.00071.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dioum EM, Chen R, Alexander MS, Zhang Q, Hogg RT, Gerard RD, Garcia JA. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science. 2009;324:1289–1293. doi: 10.1126/science.1169956. [DOI] [PubMed] [Google Scholar]