ABSTRACT
Communication between the mitochondrial and nuclear genomes is vital for cellular function. The assembly of mitochondrial enzyme complexes, which produce the majority of cellular energy, requires the coordinated expression and translation of both mitochondrially and nuclear-encoded proteins. The joint genetic architecture of this system complicates the basis of mitochondrial diseases, and mutations both in mitochondrial DNA (mtDNA)- and nuclear-encoded genes have been implicated in mitochondrial dysfunction. Previously, in a set of mitochondrial-nuclear introgression strains, we characterized a dual genome epistasis in which a naturally occurring mutation in the Drosophila simulans simw501 mtDNA-encoded transfer RNA (tRNA) for tyrosine (tRNATyr) interacts with a mutation in the nuclear-encoded mitochondrially localized tyrosyl-tRNA synthetase from Drosophila melanogaster. Here, we show that the incompatible mitochondrial-nuclear combination results in locomotor defects, reduced mitochondrial respiratory capacity, decreased oxidative phosphorylation (OXPHOS) enzyme activity and severe alterations in mitochondrial morphology. Transgenic rescue strains containing nuclear variants of the tyrosyl-tRNA synthetase are sufficient to rescue many of the deleterious phenotypes identified when paired with the simw501 mtDNA. However, the severity of this defective mito-nuclear interaction varies across traits and genetic backgrounds, suggesting that the impact of mitochondrial dysfunction might be tissue specific. Because mutations in mitochondrial tRNATyr are associated with exercise intolerance in humans, this mitochondrial-nuclear introgression model in Drosophila provides a means to dissect the molecular basis of these, and other, mitochondrial diseases that are a consequence of the joint genetic architecture of mitochondrial function.
KEY WORDS: Disease, Epistasis, Mitochondria
Summary: Interacting mutations in a mitochondrially encoded tRNA and nuclear-encoded tRNA synthetase result in a suite of pathologies, including altered locomotor capacity, mitochondrial function and morphology, and mitochondrial translation.
INTRODUCTION
The endosymbiotic origin of mitochondria permitted gene transfer from the evolving mitochondrial genome to the nuclear genome. The result of this 2-billion-year association is that ‘mito-nuclear’ cross-talk is now vital to cellular function (Rand et al., 2004; Woodson and Chory, 2008). Mitochondria are semi-autonomous organelles that contain their own genome and are maternally inherited in most species. The mitochondrial genome encodes 13 of the polypeptide subunits involved in oxidative phosphorylation (OXPHOS), in addition to two ribosomal RNAs (rRNAs) and 22 transfer RNAs (tRNAs) required for mitochondrial protein translation (Rotig, 2011). Additional machinery required for mitochondrial protein synthesis [including aminoacyl-tRNA synthetases; initiation, elongation and termination factors; and ribosomal protein subunits) are encoded in the nucleus, imported into the mitochondria and integrated into the system for proper function (Woodson and Chory, 2008; Smits et al., 2010). Owing to the joint genetic architecture of this system, mitochondrial disorders can result from either mutations in the mitochondrial DNA (mtDNA) (Greaves et al., 2012) or in nuclear-encoded genes (nDNA) that function in the mitochondria (Rotig, 2011).
In higher organisms, the link between mutations in the mtDNA or nDNA and mitochondrial disease pathologies is often unclear. Combinations of genetic variants might result in synergistic interactions that alter phenotypes, making disease manifestation a complex problem (Klose, 1999). Additionally, environmental factors have the ability to modify phenotypes, resulting in intricate gene-gene-environment (G×G×E) interactions (Arnqvist et al., 2010; Zhu et al., 2014). The contribution of mtDNA mutations to disease traits is influenced by mito-nuclear interactions, and the pathogenic properties of a mtDNA mutation might depend on the nuclear background in which it is expressed (Dowling et al., 2008). This makes assessing the contribution of individual factors to the expression of complex disease traits difficult and, thus, examination of both mitochondrial and nuclear interacting factors is required to fully understand the dynamics that underlie mitochondrial disease expression.
In humans, over 500 point mutations and rearrangements in the mtDNA are associated with disease pathologies [Jacobs, 2003; Wong, 2007; Mitomap (http://mitomap.org/MITOMAP)]. Mitochondrial tRNA (mt-tRNA) genes compose less than 10% of the mitochondrial genome but are responsible for more than half of the identified mutations, implicating them as important disease targets. Mitochondrial tRNAs play a crucial role in the translation of mtDNA-encoded proteins in the mitochondrial matrix, where they interact with a nuclear-encoded partner, a mitochondrial aminoacyl-tRNA synthetase (mtAATS). The mtAATS is responsible for ‘charging’ the appropriate tRNA with the correct amino acid (Konovalova and Tyynismaa, 2013). Once charged, the tRNA brings the amino acid to the ribosomal complex for integration into a growing polypeptide chain based on complementary anticodon base-pairing. The interacting tRNA and synthetase are encoded in separate genomes, providing the opportunity for dual-genome epistatic interactions. Mutations in any component involved might result in translation and protein synthesis defects. In humans, mutations in either mt-tRNAs or their associated synthetases can result in a suite of pathologies, including respiratory-chain deficiencies (Kemp et al., 2011), exercise intolerance (Pulkes et al., 2000), anemia, encephalopathy and cardiomyopathy (Konovalova and Tyynismaa, 2013), among others. A simple model would predict that mutations in any mtAATS would result in general disruption of mitochondrial translation and similar pathologies but, interestingly, mtAATS mutations result in distinct clinical pathologies that are specific to the synthetase affected (Konovalova and Tyynismaa, 2013).
TRANSLATIONAL IMPACT.
Clinical issue
Mitochondrial diseases often display patterns of incomplete penetrance, and the genotype-to-phenotype relationship is complex. The contribution of mitochondrial DNA (mtDNA) mutations to disease traits is influenced by both genetic and environmental (cellular and external) factors, and the pathogenic properties of a mtDNA mutation might depend on the nuclear background in which it is expressed. This makes it difficult to assess the contribution of individual factors to the expression of complex disease traits. Thus, the analysis of both mitochondrial and nuclear interacting factors is required to fully understand mitochondrial disease expression. Drosophila is an exceptionally useful animal model to investigate this because it allows the manipulation of the mitochondrial and nuclear genomes simultaneously.
Results
The mtDNA from Drosophila simulans (Dsim) was introduced into Drosophila melanogaster (introgression) to generate strains that simultaneously express a naturally occurring mutation in the Dsim mtDNA and a mutation in the Drosophila melanogaster (Dmel) nuclear DNA. Both mutations affect genes that encode mitochondrial-function-related enzymes. This approach allowed the characterization of a dual-genome mitochondrial-nuclear incompatibility on a range of traits, providing a spectrum of the pathological phenotypes that result from the interacting mutations. The model is able to recapitulate a number of pathologies observed in human mitochondrial disease, including disrupted mitochondrial function, abnormal mitochondrial morphology and decreased exercise ability. A transgenic approach is used to rescue these deleterious phenotypes, identifying genetic interactions that could be manipulated for therapeutic purposes.
Implications and future directions
This study provides insights into the mitochondrial and nuclear genetic architecture that regulates cellular energy metabolism and influences the expression of complex mitochondrial disease traits. The research focuses on a specific example of a mtDNA mutation that displays different phenotypic effects based on the nuclear background in which it is expressed, and offers a plausible mechanistic explanation for the variable penetrance observed in human mitochondrial diseases. The results provide strong evidence that the combined analysis of mitochondrial and nuclear genotypes might be a better predictor of the physiological and disease consequences of mtDNA mutations. Additionally, these results suggest a paradigm for further characterization of mitochondrial diseases mechanisms, and for identifying potential therapeutic targets and strategies.
In order to dissect the role of mitochondrial-nuclear interactions in mitochondrial diseases, our lab previously developed a Drosophila model in which both genomes can be jointly manipulated (Rand et al., 2006; Montooth et al., 2010). mtDNA from Drosophila simulans (Dsim) was introduced across species boundaries to generate a series of novel strains that contain Dsim mtDNA in controlled Drosophila melanogaster (Dmel) nuclear backgrounds. A specific mito-nuclear combination displayed a strong interaction effect. When simw501 (Dsim) mtDNA was paired with OreR (Dmel) nuclear chromosomes, a suite of abnormal phenotypes was observed, including: delayed development time, reduced fecundity and shortened scutellar bristles. These phenotypes appeared normal when other Dsim mtDNAs were placed on OreR chromosomes, or when the same simw501 mtDNA was placed on an Austria (Dmel) nuclear background. These data implicated a locus in the Austria (Aut) nuclear genome that is able to rescue the performance defects unique to the (simw501); OreR (mito; nuclear) combination. Genetic mapping of OreR identified a mutation in the tyrosyl-mtAATS [aminoacyl-tRNA synthetase for tyrosine in the mitochondria: Aats-tyr-m (Aatm)], and sequencing of the complete simw501 mtDNA revealed a potential interacting mutation in the mt-tRNA for tyrosine (tRNATyr). To confirm the source of this epistasis, a transgenic approach was used to generate rescue strains with genomic insertions of the alternative OreR and Aut alleles of Aatm (Meiklejohn et al., 2013). The OreR and Aut Aatm alleles differ by one nonsynonymous mutation in the OreR sequence that changes a highly conserved alanine to valine at amino acid position 275 in the Aatm peptide, and one synonymous site. An Aatm rescue allele was constructed using the complete OreR Aatm coding sequence but with a replacement of the single nonsynonymous single-nucleotide polymorphism (SNP) that restores the conserved alanine at position 275 of the Aatm protein. This rescue allele, referred to as AatmOre_V275A, allows us to test whether the A275V SNP is responsible for the incompatibility with the simw501 mtDNA. Here, we use both the mito-nuclear introgression strains and transgenic rescue strains to develop a model for mitochondrial translation diseases.
In humans, mitochondrial diseases often display patterns of incomplete penetrance, and the genotype-to-phenotype relationship is complex (Zeviani and Di Donato, 2004; Schon et al., 2012; Riley et al., 2013). Thresholds for mtDNA mutations differ by organ and tissue type, and tissues with high OXPHOS demands (brain, heart, muscle, etc.) might be more sensitive to mtDNA mutations (DiMauro and Schon, 2003). In many instances the same genetic mutation varies in phenotypic effect, implicating the importance of environmental and genetic interacting factors (Jacobs, 2003). Here, we test whether the interacting mutations will differentially affect a variety of traits in multiple nuclear genetic contexts. The traits characterized range from being tightly associated biochemically to the interacting mutations and mitochondrial function, such as mitochondrial translation and OXPHOS enzyme activity, to multifactorial behavioral traits that differ in their energy demands, such as flight and climbing. Using this system, we test the hypothesis that the mito-nuclear genetic interaction exhibits uniform phenotypes versus varied pathologies that are trait specific. The goal of this study is to examine a more complete spectrum of the pathological capacity of interacting mitochondrial and nuclear mutations.
RESULTS
Two sets of Drosophila strains were used in this study, referred to as ‘introgression strains’ and ‘rescue strains’. The first set contains four mito-nuclear introgression strains consisting of two different mtDNA genotypes, each of which is placed onto two different controlled, homozygous nuclear chromosomal backgrounds. The mito-nuclear introgression strains were generated by placing alternative D. melanogaster OreR mtDNA (ore) and D. simulans simw501 mtDNA on two inbred wild-type D. melanogaster nuclear backgrounds, OreR and AutW132 (supplementary material Table S1) (Montooth et al., 2010). The second set consists of transgenic Aatm rescue strains and matching sibling controls generated as described in Meiklejohn et al., 2013 (supplementary material Table S1 and Fig. S1). The rescue strains pair the simw501 mtDNA with three alternative nuclear Aatm alleles: the defective AatmOre, the wild-type AatmAut or the transgenic rescue AatmOre_V275A. The transgenic Aatm strains allow us to test the phenotypic causality of the interacting mtDNA tRNATyr and Aatm SNPs by isolating the nuclear interacting factor and placing the experimental alleles into a genetically controlled nuclear background to be tested in conjunction with the incompatible simw501 mtDNA.
Mito-nuclear incompatibility alters scutellar bristle length
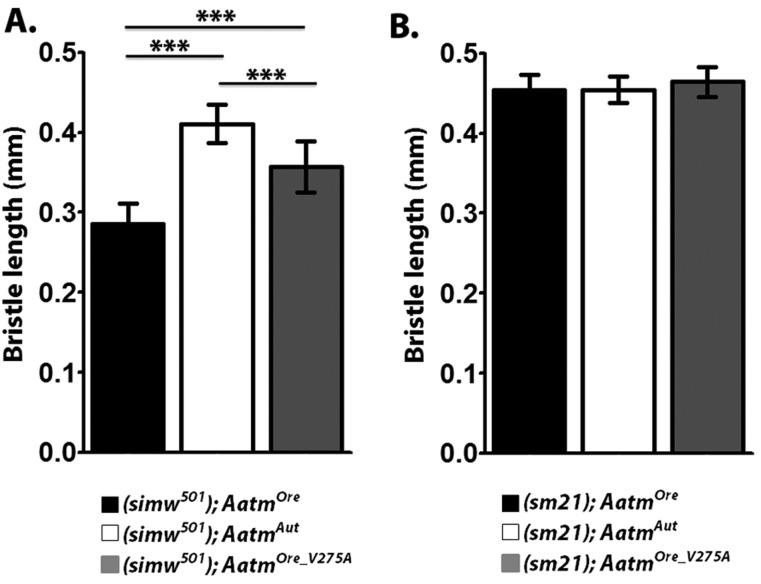
Previously, we have shown that the mito-nuclear introgression strain (simw501); OreR (mito; nuclear) has shortened bristle length (Meiklejohn et al., 2013). To test whether the interacting mito-nuclear SNPs were responsible for the bristle phenotype, scutellar bristle length was measured in the set of transgenic Aatm rescue strains and sibling controls. When simw501 mtDNA was paired with the AatmOre allele in a standardized and controlled nuclear background, bristle length was shorter compared with (simw501); AatmAut (PTukey<0.0001) (Fig. 1A). When the transgenic rescue allele of Aatm was paired with the simw501 mtDNA, this genotype, (simw501); AatmOre_V275A, displayed a partial rescue of bristle length, potentially due to additional modifiers in the nuclear background [AatmOre_V275A allele compared with both AatmOre and AatmAut (PTukey<1.0e-5)]. Additionally, when the closely related sm21 mtDNA from D. simulans was substituted in place of simw501 mtDNA on the same set of transgenic nuclear backgrounds, no difference in bristle length was observed across the transgenic genotypes (Fig. 1B, PANOVA>0.4). The sm21 and simw501 mtDNAs differ at six SNPs across their entire coding region: three synonymous sites (located in ATPase6, ND1 and ND5), two changes at non-conserved sites in the large-subunit ribosomal RNA (lrRNA), and an SNP in the anticodon stem of the tRNATyr gene; the latter is conserved across Drosophila species and is the putative functional defect in the simw501 mtDNA (Meiklejohn et al., 2013). This demonstrates the specificity of interaction between the simw501 mtDNA and AatmOre allele, and recapitulates our previous findings in the mito-nuclear introgression strains. The sibling control strains were generated from the same cross (supplementary material Fig. S1), and there were no differences in bristle length between the sibling controls in either the simw501 or sm21 transgenics (supplementary material Fig. S2A,B; PANOVA>0.05 for both A and B).
Fig. 1.
Effects of the epistatic interaction on scutellar bristle length. The transgenic rescue strains pair the simw501 mtDNA with three alternative nuclear Aatm alleles: AatmOre, AatmAut or AatmOre_V275A. The sibling control strains are generated from the same cross (supplementary material Fig. S1). (A) Scutellar bristle length of transgenic rescue strains (mean±s.d., n=15, F2,42=79.48, ***PANOVA<5e-15). The incompatible interaction shortens scutellar bristle length. (B) Bristle length measured in transgenic control sibling strains does not differ (mean±s.d., n=15, F2,42=0.285, PANOVA=0.75).
Flight ability is compromised in the epistatic genotype
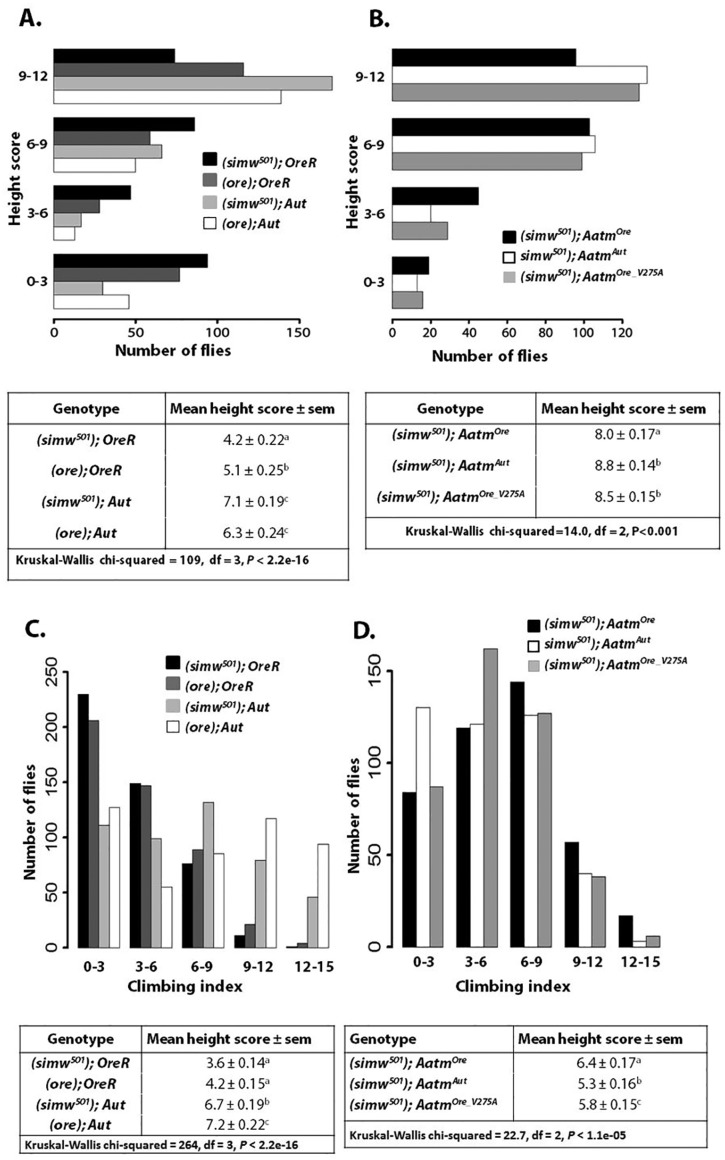
In humans, diseases that compromise mitochondrial translation often result in exercise defects and early fatigue (Pulkes et al., 2000; Riley et al., 2010). Drosophila flight muscles are highly metabolically active and predominantly aerobic (Sacktor, 1976; Deak, 1977; Beenakkers et al., 1984), whereas climbing muscles favor anaerobic metabolism (glycolysis) as their energy source (Demontis et al., 2013). We performed flight and climbing (negative geotaxis) assays to measure two different aspects of locomotor performance as proxies for muscle function. Custom-designed flight chambers (modified from Benzer, 1973) were used to test whether the mito-nuclear incompatibility results in performance defects. A cylindrical column with a sticky coating contained height marks used to score where individual flies landed in the column; strong flyers land near the top and generate higher scores, whereas poor flyers fall lower on the column. In Fig. 2, the bars indicate the number of flies recorded at each height. Mito-nuclear introgression strains with Aut nuclear backgrounds displayed stronger flight ability than those with an OreR nuclear background (Fig. 2A). The simw501 mtDNA further reduced flight capacity in the OreR nuclear background, but had no effect in the Aut background (supplementary material Table S4).
Fig. 2.
The mito-nuclear incompatibility alters flight and climbing ability. (A) Flight distributions of mito-nuclear introgression strains. The distribution of (simw501); OreR is downshifted and displays the lowest average flight score. (simw501); Aut and (ore); Aut distributions do not differ significantly (PWilcox=0.33). All other comparisons between genotypes are significantly different (pairwise contrasts in supplementary material Table S4). (B) Flight distributions of transgenic rescue strains. (simw501); AatmOre displays the lowest distribution of height scores. The distribution of (simw501); AatmOre differs significantly from (simw501); AatmAut and (simw501); AatmOre_V275A (pairwise contrasts in supplementary material Table S4). The distributions of (simw501); AatmAut and (simw501); AatmOre_V275A do not differ (PWilcox>0.7). (C) Climbing profile of mito-nuclear introgression strains. (simw501); OreR and (ore); OreR distributions do not differ significantly (PWilcox>0.1). All other comparisons between genotypes are significantly different (pairwise contrasts in supplementary material Table S4). (D) Climbing profile of transgenic rescue strains. The distribution of (simw501); AatmOre differs significantly from (simw501); AatmAut and (simw501); AatmOre_V275A (pairwise contrasts in supplementary material Table S4). The distributions of (simw501); AatmAut and (simw501); AatmOre_V275A are marginally different (PWilcox=0.04). All P-values associated with Wilcoxon tests are Bonferroni-corrected to account for multiple testing. Different letters signify significant differences in pairwise contrasts.
In the transgenic Aatm strains, flight ability displayed similar patterns to the mito-nuclear introgression strains. When the AatmAut or AatmOre_V275A alleles were paired with the simw501 mtDNA, both strains displayed strong flight performance, and the distribution of the height scores were very similar (Fig. 2B, PWilcox>0.7; supplementary material Table S4). (simw501); AatmOre displayed poor flight performance, with an overall lower distribution of height scores (supplementary material Table S4). Flight ability was not assessed in sibling control strains because the Curly wing marker is present on the second chromosome balancer and these flies do not fly (supplementary material Table S1).
Climbing and flight performance have distinct mito-nuclear genetic bases
Climbing ability was measured to determine whether a distinct, complex locomotor trait that is less reliant on mitochondrial function would be influenced by the interacting mutations. Flies were placed into a climbing apparatus, and the vertical height that flies reached at a set time point was recorded to generate a climbing index. Over 300 flies per genotype were tested and the climbing index distributions are displayed in Fig. 2C,D. Similar to flight capacity, mito-nuclear introgression strains with Aut nuclear backgrounds were stronger climbers than OreR flies (Fig. 2C). The simw501 mtDNA marginally reduced climbing ability in the OreR nuclear background, although the effect was not significant (PWilcox>0.1, supplementary material Table S4). Unexpectedly, the simw501 mtDNA significantly reduced climbing performance in the Aut nuclear background (Fig. 2C).
In the transgenic rescue strains, (simw501); AatmOre were the strongest climbers, whereas (simw501); AatmAut were the worst climbers (PWilcox<4e-6) (Fig. 2D, supplementary material Table S4). (simw501); AatmOre_V275A displayed an intermediate distribution. There was no difference observed in the climbing ability of the transgenic control siblings (PKruskal–Wallis=0.493) (supplementary material Fig. S3, Table S4). Interestingly, in the rescue strains, the flight and climbing performance of the genotypes was inverted. The strongest flyers were the poorest climbers, and the poorest flyers were the strongest climbers (Fig. 2B,D).
Altered mitochondrial morphology from mito-nuclear interaction
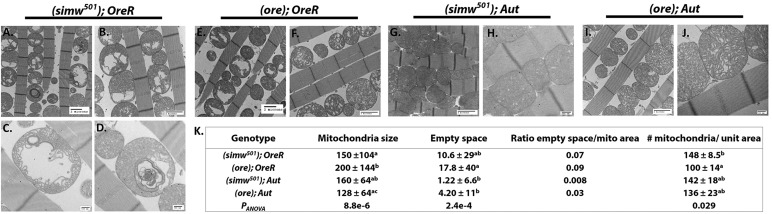
Transmission electron microscopy was used to assess changes in mitochondrial morphology and structure associated with the mitochondrial phenotypes observed. Preparations from indirect flight muscles from the four mito-nuclear introgression genotypes are shown in Fig. 3. Similar to several of the other phenotypes characterized, the difference between the OreR and Aut nuclear backgrounds was pronounced, with the simw501 mtDNA having a more significant impact in the OreR background. The cristae structure of (simw501); OreR mitochondria appeared loosely packed and displayed large gaps (Fig. 3A-D). Additionally, several instances of swirled cristae structure potentially due to cristae collapse were observed (Fig. 3C,D). (simw501); OreR contained the most mitochondria per unit area, potentially to compensate for mitochondrial defects (Fig. 3K). (ore); OreR also displayed large gaps in cristae structure, although no other instances of abnormal morphology were observed (Fig. 3E,F). The mitochondrial morphology of (ore); Aut and (simw501); Aut appeared healthier. The cristae were densely packed and fewer holes in cristae structure were observed. Quantification of mitochondrial ultrastructure measurements is displayed in the table in Fig. 3K (pairwise contrasts in supplementary material Table S5).
Fig. 3.
Characterization of mitochondrial morphology in the mito-nuclear introgression strains. TEMs of indirect flight muscle reveal structural abnormalities in (simw501); OreR. (A-D) (simw501); OreR display a loose cristae structure and matrix gaps. (simw501); OreR contained the most mitochondria per unit area, and several examples of swirled concentric cristae structures were observed (A,D). (E,F) (ore); OreR displayed some loose cristae structure and matrix gaps, and on average contained the largest mitochondria. (G-J) (simw501); Aut and (ore); Aut displayed tight cristae packing and healthy mitochondrial morphology. (K) Quantification of mitochondrial morphology parameters in mito-nuclear introgression strains. Approximately 100 mitochondria per genotype were analyzed to generate size data. Different superscript letters signify significant differences in pairwise comparisons (values for PTukey pairwise contrasts are displayed in supplementary material Table S5). Mitochondria were counted in 50 μm×50 μm sections to generate density data. The average number of mitochondria per unit area of >3 sections are displayed. Scale bars: 2 μm (A,B,E-G,I); 500 nm (C,D,H,J).
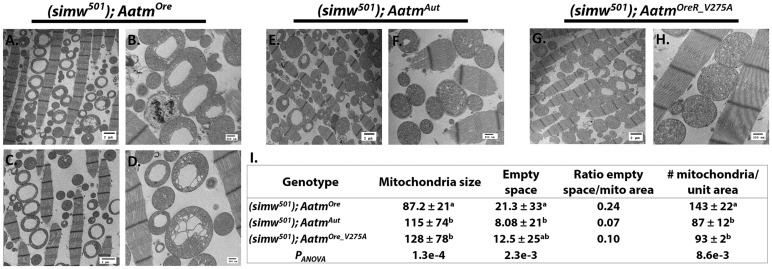
In the transgenic Aatm strains, the defective genotype (simw501); AatmOre contained the most mitochondria per unit area (Fig. 4I and supplementary material Table S5), and displayed large gaps in cristae structure similar to (simw501); OreR. Several examples of severely abnormal morphology were observed (Fig. 4A-D), although these did seem to be distinct from the morphological defects seen in (simw501); OreR (Fig. 3C,D). The (simw501); AatmAut and the rescue genotype (simw501); AatmOre_V275A strains displayed fewer gaps in cristae structure (Fig. 4E-H). Mitochondrial size was smaller in the (simw501); AatmOre strain (average area of 87.2 μm), as compared to the (simw501); AatmAut and (simw501); AatmOre_V275A strains (average of 115 and 128 μm, respectively). A ratio of empty matrix space/overall mitochondrial area was calculated to quantify cristae structure gaps. This ratio was more than double in the (simw501); AatmOre strain as compared to the AatmAut or AatmOre_V275 strains (ratio of 0.24 vs 0.07 and 0.10, respectively) (Fig. 4I).
Fig. 4.
Characterization of mitochondrial morphology in the transgenic rescue strains. The incompatible mito-nuclear pair results in mitochondrial morphological defects. (A-D) Large matrix holes and unidentified inclusions were observed in (simw501); AatmOre. On average (simw501); AatmOre contains the smallest mitochondria, but the most per unit area. (E,F) (simw501); AatmAut displays some matrix holes, but maintains a relatively low ratio of empty matrix space to mitochondrial area. (G,H) The transgenic rescue genotype (simw501); AatmOre_V275A displays a healthy cristae structure and normal mitochondrial morphology. (I) Quantification of mitochondrial morphology parameters in transgenic rescue strains. Approximately 100 mitochondria per genotype were analyzed to generate size data. Different letters signify significant differences in pairwise comparisons (values for PTukey pairwise contrasts are displayed in supplementary material Table S5). Mitochondria were counted in 50 μm×50 μm sections to generate density data. The average number of mitochondria per unit area of >3 sections are displayed. Scale bars: 2 μm (A,C,E,G); 500 nm (B,D,F,H).
Mitochondrial respiration capacity is reduced in the (simw501); OreR combination
To test whether the identified mito-nuclear incompatibility alters overall mitochondrial oxidative capacity, respiration was measured in vitro in isolated mitochondrial preparations. In the mito-nuclear introgression strains, a nuclear background effect on mitochondrial respiratory capacity was observed. Introgression strains generated with Aut nuclear backgrounds displayed higher state III (ADP-dependent) respiration rates. No difference was seen in the state IV (ADP-independent) basal respiration rates (Table 1). An uncoupled rate was induced by addition of the chemical uncoupler FCCP to evaluate maximum OXPHOS capacity that is independent of membrane potential and ATP synthesis. (simw501); OreR displayed reduced state III and uncoupled rates, indicative of a lower overall maximum respiratory capacity (pairwise contrasts in supplementary material Table S6).
Table 1.
Effects of the mito-nuclear interaction on mitochondrial respiration

In the transgenic rescue strains, the defective genotype (simw501); AatmOre had reduced uncoupled rates compared with (simw501); AatmAut(PTukey<0.05), indicating that the rate of maximal oxygen consumption is compromised, and the rescue genotype (simw501); AatmOre _V275A displayed an intermediate phenotype (Table 1, supplementary material Table S6). (simw501); AatmOre had comparable state III respiration rates to both (simw501); AatmAut and (simw501); AatmOre _V275A (PANOVA=0.24, for AatmOre, AatmAut and AatmOre_V275A alleles). The transgenic control siblings displayed no difference in any of the respiratory measurements (PANOVA>0.5, all parameters) (supplementary material Tables S2 and S6). The parameters measured are from functional and intact mitochondrial isolates, as all respiratory control ratio (RCR) values (state III O2 consumption rate/state IV O2 consumption rate) reported for all genotypes are >4 (Table 1), supporting the validity of the measured values (Schriner et al., 2009).
Mito-nuclear incompatibility alters OXPHOS enzyme abundance and activity
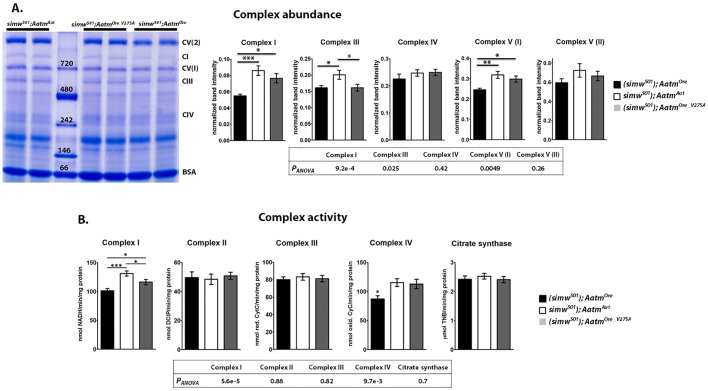
To determine how the interacting mutations influence the relative abundance of assembled OXPHOS enzyme complexes, blue native polyacrylamide gels were performed. Amongst the transgenic rescue strains, the abundance of all complexes was reduced in (simw501); AatmOre compared with (simw501); AatmAut (Fig. 5A), with some variation across complexes. This effect was significant for Complexes I, III and V(I) (PTukey<0.05, for all comparisons, supplementary material Table S7). The rescue genotype, (simw501); AatmOre_V275A, did not significantly differ from (simw501); AatmAut for Complex I, V(I) or V(II) abundance (PTukey>0.4, for all comparisons, supplementary material Table S7). Across all complexes the rank order of the genotypes was maintained (Fig. 5A). In the transgenic sibling controls there were no differences detected in the abundance of any of the mitochondrial complexes (supplementary material Fig. S4, PANOVA>0.6, for all comparisons in all complexes, supplementary material Table S7).
Fig. 5.
OXPHOS enzyme abundance and activity is reduced in mitochondrially translated complexes containing the most tyrosines. BN-PAGE analysis of mitochondrial protein isolates. (A) Example of BN-PAGE gel for transgenic rescue strains (note: full lanes moved from original gel locations for genotype consistency, no manipulations within lanes). Quantification of individual band intensity corresponding to Complex I, III and V relative to BSA. Across all complexes (simw501); AatmOre displayed reduced protein levels to varied degrees (individual contrasts in supplementary material Table S7). (B) Enzymatic activity of OXPHOS complexes. (simw501); AatmOre has significantly reduced activity of Complex I compared to (simw501); AatmAut and (simw501); AatmOre_V275A (PTukey=3.18e-5 and PTukey<0.05, respectively). The (simw501); AatmOre_V275A rescue strain has marginally lower Complex I activity compared with (simw501); AatmAut (PTukey<0.05). Complex II and Complex III activity does not differ between the rescue strains (PANOVA>0.8, each complex). Complex IV activity is reduced by the incompatibility in (simw501); AatmOre (PANOVA<0.01). There is no difference in citrate synthase among the rescue genotypes (PANOVA=0.7). The complexes with more mitochondrially encoded tyrosines display the most severe defect (Complexes I and IV). Enzymes that are completely nuclearly encoded (Complex II and citrate synthase) or those with few mitochondrially encoded tyrosines (Complex III) show little disruption of enzyme capacity. *P<0.05; **P<0.01; ***P<0.001.
To assess functionality of OXPHOS complexes, enzyme activity was measured in vitro. Previously in the introgression strains we found that complexes jointly encoded by both mtDNA and nDNA (Complex I, Complex III and Complex IV) were compromised in (simw501); OreR, whereas Complex II and citrate synthase, both of which are completely nuclearly encoded, showed no difference among the introgression genotypes in enzyme activity (Meiklejohn et al., 2013).
To determine whether the simw501–Aatm interaction was responsible for these phenotypes, complex activity was evaluated in the transgenic rescue strains. (simw501); AatmOre displayed compromised Complex I and IV activity (Complex I PANOVA<6e-5, and Complex IV PANOVA<0.01) (Fig. 5B, supplementary material Table S7). (simw501); AatmOre_V275A was able to partially or fully rescue these effects. The severity of the defect corresponds in rank order with the number of mitochondrially encoded tyrosines present in each complex (supplementary material Table S3). Complex I was more severely compromised than Complex IV; this is consistent with a model in which the number of tyrosines that are mitochondrially translated will determine the severity of the defect. In the transgenic sibling controls there was no difference in complex activity (PANOVA>0.3, each complex). Citrate synthase activity was marginally reduced in (simw501); AatmAut Sib (PANOVA>0.02) (supplementary material Fig. S4, Table S7).
Mito-nuclear incompatibility alters translation initiation rates in the mitochondria
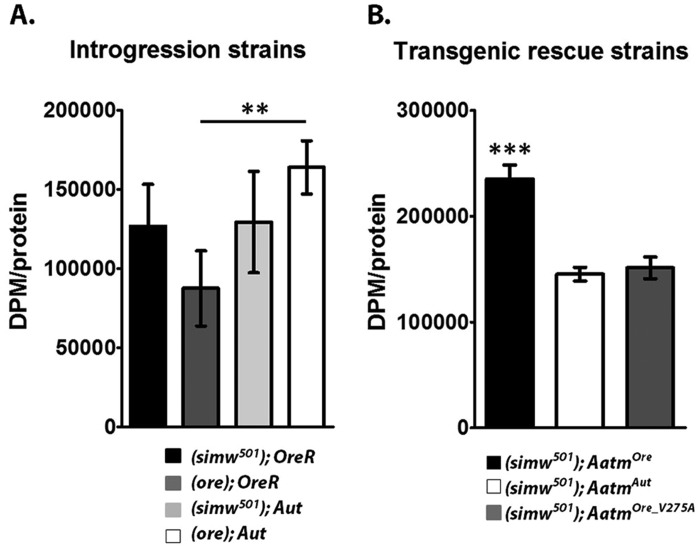
De novo translation initiation was measured via 35S-methionine incorporation to assess how the interacting mutations, which putatively influence tRNATyr charging, alter protein synthesis rates. Adult flies of each genotype were placed on standard media supplemented with 100 µCi/ml 35S-methionine for 48 h. Mitochondrial fractions were prepared, and 35S-methionine incorporation was found to differ amongst the mito-nuclear introgression strains (PANOVA<0.005), and strains with Aut nuclear backgrounds displayed higher rates of mitochondrial translation initiation (Fig. 6A). In the OreR nuclear background the simw501 mtDNA increased mitochondrial translation initiation, although this effect did not reach significance (supplementary material Table S8, PTukey=0.12). This result was confirmed in an independently performed experiment (Pt-test<0.02, supplementary material Fig. S6, data not combined as 35S-methionine batch differed and Aut introgression strains were not run). Translation initiation rates in (simw501); OreR did not differ significantly from either of the Aut introgression strains (Fig. 6A, supplementary material Table S8).
Fig. 6.
Mitochondrial translation is upregulated by the mito-nuclear incompatibility. Quantification of de novo translation as assessed by 35S-methionine incorporation into newly synthesized proteins, normalized to total mitochondrial protein. (A) 35S-methionine incorporation in introgression strains. (simw501); OreR displays a mild increase compared to (ore); OreR, and no significant difference is seen between (simw501); OreR compared to either (simw501); Aut or (ore); Aut. (B) 35S-methionine incorporation is significantly increased in (simw501); AatmOre compared to the other transgenic rescue strains (PTukey<4e-6 for both comparisons). DPM, disintegrations per minute. **P<0.01; ***P<0.001.
In the transgenic rescue strains a similar pattern was observed, and translation initiation rates were increased in (simw501); AatmOre compared to either (simw501); AatmAut or (simw501); AatmOre_V275A (Fig. 6B, PTukey<4e-6 for both comparisons). Additionally, no difference was observed between (simw501); AatmAut and (simw501); AatmOre_V275A (Fig. 6B, PTukey=0.9). In the transgenic sibling controls 35S-methionine incorporation was also marginally increased in (simw501); AatmOre Sib compared to (simw501); AatmAut Sib (supplementary material Fig. S5, PANOVA=0.013, supplementary material Table S8).
DISCUSSION
Here we characterize a broad range of phenotypes that are affected by a mitochondrial-nuclear incompatibility. In a previous study, we showed that a mutation in the mtDNA-encoded tRNATyr interacted negatively with a mutation in the nuclear-encoded aminoacyl-tRNA synthetase for tyrosine in the mitochondria, Aats-tyr-m (Aatm) gene. Transgenic rescue strains confirmed that the nuclear mutation was causally associated with a developmental delay in this system (Meiklejohn et al., 2013). The present study builds on this model and here we analyze the pathological outcomes of the interacting mutations starting with whole organismal phenotypes and working down to the level of mitochondrial function and translation. By comparing the specific mito-nuclear interaction in two sets of genetic backgrounds (‘introgression strains’ and ‘transgenic rescue strains’), we have shown that the incompatible mito-nuclear ‘disease’ genotype has distinct phenotypic consequences across different traits. The tissue-specific or variable penetrance of mitochondrial dysfunction is a common condition in human mitochondrial diseases, and this Drosophila model offers a promising system for dissecting this complexity.
The mito-nuclear incompatibility affects organismal performance and morphological phenotypes
A mutation identified in the human tyrosyl-tRNA synthetase gene, YARS2, primarily affects muscles and results in fatigue (Konovalova and Tyynismaa, 2013). We examined locomotor traits in our system and found that the simw501 and AatmOre mutations impact flight and climbing ability differentially, and these effects were nuclear background dependent. During insect flight a number of enzymes involved in aerobic metabolism have been found to function at near maximum capacity (Suarez et al., 1996), and flight muscles might account for >90% of organismal oxygen consumed (Suarez, 2000). The interacting mutations in (simw501); OreR likely contribute to subpar flight performance due to decreased aerobic metabolic capacity, supported by the reduced mitochondrial respiratory capacity and enzyme activity described below. Additionally, the flight defect observed in the defective genotype is consistent between the introgression strains and transgenic rescue strains.
Analysis of climbing ability did not reveal a consistent defect between the introgression strains and transgenic rescue strains when the simw501 and AatmOre mutations co-occur. Flight and climbing are separable behaviors that utilize morphologically distinct muscle groups in Drosophila (Peckham et al., 1990; Rhodenizer et al., 2008; Augustin and Partridge, 2009). Flight muscles contain abundant large mitochondria (30-40% of fiber volume) (Shafiq, 1963; figure 2 in O'Donnell et al., 1989), whereas leg muscles required for jumping and climbing [tergal depressor of the trochanter (TDT) and tergotrochanteral depressor muscle (TTM)] contain small and infrequently distributed mitochondria (figure 4 in O'Donnell et al., 1989). The metabolic profiles of these muscles also differ in insects; indirect flight muscles display a very low LDH/GAPDH ratio, and high TCA cycle activity (LDH/GAPDH, see figure 1 and table 2 in Beenakkers et al., 1984; SDH and IDH, see figure 1 in Deak, 1977), but TDT and TTM do not. The signature of enzymatic activity observed in indirect flight muscles is indicative of exclusive substrate processing in the mitochondria.
In mitochondrial disease states these muscle types are also differentially affected: knockdown of Drosophila proteins that play a role in mitochondria quality control (PINK1 and parkin) results in severe disruption of muscle fiber arrangement in the indirect flight muscles, whereas leg muscles are only mildly affected (Pesah et al., 2004; Yang et al., 2006). Additionally, Parkin mutants completely lack the capacity to fly, whereas jumping ability is partially maintained (Greene et al., 2003; Pesah et al., 2004). Thus, different energy demands of muscle types likely contribute to the contrast observed in the transgenic rescue strains, where (simw501); AatmOre has decreased flight performance but increased climbing ability. Based on muscle type differences described above it is predicted that flight but not climbing behavior will be more tightly linked to the interacting tRNA-synthase mutations. Additionally, we observed that flight capacity is higher across all transgenic rescue strains compared to the introgression strains, whereas climbing ability falls within a similar range across the groups. The introgression strains have inbred nuclear backgrounds whereas the transgenic rescue strains are F1 hybrids, potentially masking any deleterious recessive alleles in the nuclear genome and resulting in enhanced flight ability. This difference in genetic background between the introgression and the transgenic rescue strains might account for some of the differences in rescue effects in these two sets of strains.
A third organismal trait found to be influenced by the simw501–Aatm interaction was bristle length. We previously showed that (simw501); OreR results in a short bristle phenotype (Meiklejohn et al., 2013), and these results are recapitulated in the (simw501); AatmOre strain. We find that the AatmOre_V275A allele is only able to partially rescue this effect, indicating that the Ore A275V SNP is the main effect, but subtle background effects could be modifiers. Additionally, when a different mtDNA is placed on this background, (sm21); AatmOre, bristle length is completely restored, demonstrating that both mutations are required for the complete bristle defect. The bristle phenotype is similar to those previously identified in Drosophila Minute mutants in which cytosolic ribosomal components are disrupted. Translational defects result in reduced growth because protein synthesis cannot keep up with cellular demands (Lambertsson, 1998). A mutation in the mitochondrial import machinery, Tim50, results in a similar bristle defect, reduced cell proliferation and growth defects. Defective mitochondrial protein import likely results in suboptimal mitochondrial performance and the inability to produce sufficient energy, resulting in these growth defects (Sugiyama et al., 2007). Thus, defects in protein synthesis are a common component in other genetic models that result in short bristles, lending support for the model of defective mitochondrial translation presented here.
At the subcellular level, the simw501–Aatm interaction revealed dramatic effects on mitochondria morphology, including large gaps in the mitochondrial matrix and a collapsed cristae structure similar to those seen in human mitochondrial diseases (Carta et al., 2005; Cogliati et al., 2013). The defects observed in (simw501); OreR and (simw501); AatmOre appear morphologically distinct (Figs 3 and 4), and the hollow matrix is more severe and smooth in (simw501); AatmOre. Both of the defective genotypes in the introgression and transgenic rescue strains displayed an increase in mitochondrial number, which could be indicative of a compensatory mechanism to maintain energy production levels (discussed more below).
In Drosophila, disruption of proteins that play a role in mitochondria quality control, PINK1 and parkin, results in similar mitochondrial morphological defects, including swollen mitochondria, fragmented cristae structure and matrix degeneration (Park et al., 2006; Saini et al., 2011; Klein et al., 2014). In the case of the simw501 mtDNA and AatmOre interaction, it seems that mutations in both genomes have the ability to affect mitochondrial morphology. The defective mitochondrial complexes might be influencing structure formation, and previous studies have noted that specific complex dimerization can regulate membrane curvature (Dudkina et al., 2005; Daum et al., 2013). Additionally, a stoichiometric imbalance of mitochondrial and imported nuclear-encoded proteins might also influence structural properties (Houtkooper et al., 2013). Thus, our genetic model might provide the means to dissect mtDNA and nDNA contributors to mitochondrial morphology.
Alteration of mitochondrial function by the mito-nuclear incompatibility
A number of mitochondrial functional parameters were measured in live respiring mitochondria as well as frozen isolated samples. Integrated OXPHOS measurements of state III and uncoupled respiration rates revealed a strong nuclear background effect in the introgression strains (OreR nuclear background displayed lower rates than Aut), indicating that additional nuclear factors are influencing mitochondrial respiratory performance in conjunction with the identified tRNA-synthetase interaction. In the transgenic strains, uncoupled respiration was reduced in (simw501); AatmOre compared to (simw501); AatmAut, supporting the main effect of the tRNA-synthetase interaction, and suggesting that the mito-nuclear incompatibility results in decreased OXPHOS and O2 consumption, potentially contributing to some of the locomotor phenotypes characterized.
When analyzing individual OXPHOS enzyme complexes, we previously showed that all complexes jointly encoded by mtDNA and nDNA were compromised in (simw501); OreR (Meiklejohn et al., 2013). Upon controlled dissection of the interaction with the transgenic (simw501); AatmOre strain we observed a pattern of disruption that corresponds with the number of mitochondrially translated tyrosines present in each enzyme complex. The most severely affected complex, Complex I, contains the most tyrosines (98), followed by Complex IV with 39 tyrosines. The (simw501); AatmOre_V275A strain was able to partially rescue complex I activity and fully rescue complex IV activity. This implicates the tRNA-synthetase interaction as a main effect, but additional factors are likely to be involved. Complex III contains only one mtDNA-encoded subunit and 19 tyrosines. A defect in Complex III was not observed in the transgenic strains, potentially due to a lack of sensitivity in the assay, in addition to a predicted milder effect because fewer mitochondrially encoded tyrosines need to be incorporated into this complex. In the simw501–Aatm interaction we find that the absolute number of tyrosines in each complex is a better predictor of the severity of the defect than the proportion of tyrosine present (which is relatively consistent across complexes). This supports a model in which each tyrosine that must be incorporated into the protein complex can disrupt mitochondrial translation. In humans, similar defects in OXPHOS are observed when mitochondrial protein synthesis is compromised (Jacobs, 2003), and specific tRNA mutations result in varied biochemical defects, potentially due to the amino acid composition of each complex.
Compensatory mitochondrial biogenesis and retrograde signaling
Based on the physical location of the interacting mutations in our model, it is predicted that aminoacylation of the tRNATyr is compromised (Moreno-Loshuertos et al., 2011; Meiklejohn et al., 2013). In turn, this should impact translation rates and any functions that depend on mtDNA-encoded proteins. Our experiments on translation initiation reveal that genotypes harboring the incompatible mutations show elevated levels of 35S-methionine incorporation, whereas no such effect was observed when the AatmOre_V275A rescue allele was paired with the simw501 mtDNA. These results are consistent with the hypothesis that the defective mito-nuclear genotypes are mounting a compensatory response by stimulating additional translation. These results agree with additional evidence that mtDNA copy number is increased in (simw501); OreR compared to the other mito-nuclear genotypes (Zhu et al., 2014). Upregulation of mtDNA has been proposed as a compensatory mechanism of mt-tRNA mutations (Moreno-Loshuertos et al., 2011), and has been observed in humans with mitochondrial diseases (Yen et al., 2002). In further support of compensatory mitochondrial biogenesis, we observed increased mitochondrial numbers in both the introgression and transgenic rescue strains harboring the simw501 and AatmOre mutations (Figs 3 and 4).
The increase in mtDNA copy number, translation initiation and mitochondrial number indicate that the cell is able to sense disruption of mitochondrial activity and respond in an attempt to maintain function. Mitochondrial replication, transcription and translation require the import of nuclear proteins (Smits et al., 2010). The proposed functional consequences of the interacting mutations lies within the mitochondrion, suggesting that a mitochondrial retrograde signal mediates a nuclear transcriptional response to generate the necessary proteins required for increased mtDNA replication, translation and structural components. Retrograde signaling pathways result in the activation of nuclear transcription factors, and one relevant example might be the transcriptional coactivator PGC1α (peroxisome proliferator-activated receptor γ, coactivator 1 α), which is a master regulator of mitochondrial biogenesis in mammals and is induced by mitochondrial dysfunction (Butow and Avadhani, 2004; Chae et al., 2013). Here we observed decreased OXPHOS complex abundance and activity in the transgenic rescue strain carrying both the simw501 and AatmOre mutations, indicating that the compensatory upregulation of mitochondrial translation initiation cannot fully offset the effects of these mutations. Additionally, without this putative compensatory response, the pathological outcome of the interacting mutations might be more severe.
Nuclear modifiers of the epistatic interaction
In general, the ‘introgression strains’ and ‘transgenic rescue strains’ display similar phenotypic outcomes for the simw501–Aatm interaction. However, there are traits for which these strains show distinct phenotypes, suggesting that nuclear modifiers of the epistatic interaction exist. The mito-nuclear introgression strains were built on two separate sets of nuclear chromosomes, OreR and Aut. Nuclear chromosomal effects, in which Aut strains outperform OreR strains, were apparent in a number of phenotypes characterized, including mitochondrial respiration (Table 1: state III respiration), flight ability (Fig. 2A) and climbing ability (Fig. 2C). In these whole-genome introgression strains there are likely a large number of factors that differ between the OreR and Aut genomes that impact mitochondrial function, some of which might directly influence the effects of the simw501–Aatm interaction.
In order to control for these chromosomal effects and to test the simw501–Aatm interaction in a common nuclear background, transgenic strains were developed (Meiklejohn et al., 2013). We find that the simw501–Aatm interaction impacts a number of traits, and the phenotypes are influenced by nuclear background (Fig. 2A vs 2B: flight; Fig. 2C vs 2D: climbing; Table 1: mitochondrial state III respiration). Additionally, the transgenic rescue strains reveal that the Ore A275V SNP is the main effect, but subtle background effects could be modifiers that result in partial rescue of the primary defect. When characterizing the transgenic rescue strains in several of the phenotypes measured, the AatmOre_V275A allele displayed a partial rescue when compared to the AatmAut allele (Fig. 1: bristle length; Fig. 5: complex I activity). In other traits the AatmOre_V275A allele was able to fully rescue the defective phenotype and was indistinguishable from AatmAut (Fig. 2B: flight; Fig. 5: complex III and IV activity). These different traits seem to have additive or dominant effects in the simw501–Aatm interaction. Additionally, there was no clear dosage effect in the transgenic sibling controls. In most parameters measured the transgenic sibling control strains displayed similar performance (supplementary material Table S2, Figs S3 and S4), indicating that a single compatible Aatm allele is sufficient to rescue the deleterious phenotypes.
The mitochondrial defects displayed variable penetrance in the simw501–Aatm interaction, implying that specific traits might be differentially susceptible to the main effect of the interaction. Traits that are strongly influenced by mitochondrial translation and function (OXPHOS complex activity, Fig. 5) display distinct patterns of disruption corresponding to the simw501–Aatm interaction, whereas multifactorial traits with less direct biochemical dependence on mitochondria (climbing, Fig. 2) show mild or no disruption. This highlights how the pathogenic properties of mtDNA mutations can be altered by the nuclear background in which they are expressed, and these effects mirror the complexity observed in human mitochondrial diseases. Our model could provide an effective means to dissect this complexity and can be used to screen for additional nuclear factors that modify the simw501–Aatm interaction.
Human diseases of mitochondrial translation
In humans, separate pathogenic mutations have been identified in both the mitochondrial tRNATyr and nuclear tyrosyl-tRNA synthetase genes. The mutations in the mitochondrially localized tyrosyl-tRNA synthetase gene, YARS2, result in mitochondrial myopathy, lactic acidosis and sideroblastic anemia (MLASA syndrome) (Konovalova and Tyynismaa, 2013). Similar to the phenotypes seen in our flies, affected individuals display exercise intolerance and decreased electron transport chain (ETC) complex activity (Riley et al., 2013; Shahni et al., 2013). Interestingly, two patients homozygous for the same mutation (p.Phe52Leu) varied in the severity of the disease pathologies presented; one being mildly affected whereas the other was severely affected (Riley et al., 2013). These patients had different mtDNA haplogroups and nuclear genetic material, possibly contributing to the observed phenotypic variability. In a separate study a pathogenic mutation associated with exercise intolerance was identified in the mitochondrial tRNATyr gene (Pulkes et al., 2000). The specific phenotypes and similarities observed between humans with mitochondrial tRNA and synthetase mutations and the flies described here demonstrates the effectiveness of our Drosophila model, and exemplify the complexity of mitochondrial diseases.
Recent studies have shown that overexpression of specific mtAATSs is able to partially rescue molecular defects resulting from pathogenic mt-tRNA mutations in cell lines (Rorbach et al., 2008; Li and Guan, 2010; Perli et al., 2014; Tran Hornig-Do et al., 2014). In one example, the mt-tRNAVal mutation (m.1624C>T) resulted in destabilization of the uncharged tRNA, and overexpression of the nuclear synthetase, VARS2, was able to increase charging of the mutant tRNA and rescue molecular phenotypes (Rorbach et al., 2008). Our results demonstrate that expression of a compatible Aatm allele is able to partially or fully rescue a series of deleterious phenotypes when paired with simw501 mtDNA at the level of a whole organism. Several additional studies demonstrate the effectiveness of Drosophila models in dissecting mitochondrial diseases (Toivonen et al., 2001; Bayat et al., 2012; Vartiainen et al., 2014). Currently there are no effective treatments for mitochondrial tRNA diseases in humans. Our study and those described above suggest that expression and import of nuclear synthetases might provide a novel therapy for mitochondrial tRNA diseases. The recent approval of mitochondrial replacement therapies is one promising approach to treating these diseases, but the mito-nuclear interactions resulting from such ‘three-parent babies’ raise the possibility of unintended interactions between mtDNA and nDNA SNPs that could generate novel pathologies (Reinhardt et al., 2013). Additional studies examining the complexity of mito-nuclear genetic interactions are warranted as these treatments move into actual clinical practice.
MATERIALS AND METHODS
Drosophila stocks and maintenance
The four mito-nuclear genotypes used in this study are a subset of the strains described in Montooth et al., 2010. Alternative D. melanogaster OreR mtDNA (ore) and D. simulans simw501 mtDNA are placed on two inbred, wild-type D. melanogaster nuclear backgrounds, OreR and AutW132, using balancer chromosome extraction. The transgenic rescue strains are described in Meiklejohn et al., 2013. In brief, the AatmOre, AatmAut and AatmOre_V275A transgenic constructs containing the complete Aatm coding sequence were inserted into the same genomic location using ΦC31-mediated integration. The incompatible AatmOre allele differs from the AatmOre_V275A rescue allele by the A275V SNP in the synthetase coding sequence, and the sequence-verified AatmAut and AatmOre alleles differ by an additional single synonymous SNP that was not manipulated. Stocks homozygous for the transgenes were established, and carry a deficiency spanning the endogenous location of the Aatm allele on the second chromosome, balanced over Cyo. Cyo contains a wild-type copy of the Aatm allele matching the D.mel reference sequence. Transgenic stocks are crossed to virgin female (simw501); OreR to generate a series of experimental and transgenic control strains (supplementary material Fig. S1). The genetic notation used throughout is: (mito); nuclear, with the nuclear being the full homozygous chromosome sets for the introgression strains or the inserted Aatm allele for the transgenic strains. All fly stocks were kept in a 25°C temperature-controlled incubator with a 12 h on/off light cycle. Flies were density controlled and raised on standard media (2% yeast, 5.2% cornmeal, 11% sugar, 1% tegosept, 0.8% agar). Unless otherwise noted flies used in assays were mated females 15 days post-eclosion.
Bristle length
Posterior scutellar macrochaetae were measured using a Nikon dissecting microscope at 50× power (10× ocular, 5× zoom objective) with a micrometer scale built into the ocular lens. Flies were frozen at −20°C and bristles were measured from the tip to the base, with flies placed on a bed of cotton so that the shaft of the bristle could be aligned to be parallel with the focal plane of the microscope, allowing standardized measurements to be obtained. 15 flies for each genotype were measured in two blocks from separate genotype builds.
Flight assay
To test overall flight performance a cylindrical flight tester was used as originally described by Benzer (1973). A 500 ml graduated cylinder was coated on the inside with paraffin oil and flies were gently released through an affixed funnel at the top. Flies will fly immediately outwards and stick to the oil as they strike the walls of the cylinder. Flies with poor flight ability fall further before striking the wall. The cylinder was divided into 10 height bins, and the number of flies stuck at each height was quantified and plotted. 15-day-old female flies were used in this assay; n>300.
Climbing assay
Climbing ability (negative geotaxis) was measured in a custom-engineered lab apparatus. 10-ml plastic pipettes were used as cylindrical tubes (30 cm height×1 cm diameter) and were affixed to a stable backboard. Five flies were placed into a tube and the bottom was then stoppered with foam plugs. These specifications minimize crowding while discouraging flight. The backboard was tapped onto a hard surface applying universal force to knock the flies to the base of the tubes. The apparatus is stood upright and flies are given 30 s to climb, at which time digital photos were taken and the height climbed was measured for each individual fly. The apparatus is rested horizontally and flies are given 15 min of recovery time before being tested again. 15-day-old female flies were used in this assay; n>300.
Mitochondrial morphology and TEM
Indirect flight muscles were dissected from 15-day-old female flies into cold Schneider's Drosophila media, and fixed in a 2.5% Glutaraldehyde/2% Paraformaldehyde solution containing 0.1 M NaCaCo and 0.05 M sucrose. Samples were post-fixed in osmium tetroxide, washed and dehydrated in an ethanol series. Samples were embedded in low viscosity Spurr's resin. Samples were cut on a Reichert Ultracut E microtome in 90-nm sections. A Philips 410 transmission electron microscope was used for imaging.
Mitochondrial isolation
30 flies were sorted by sex on light CO2 and were given at least 48 h recovery time prior to homogenization. Flies were gently homogenized in 1 ml chilled isolation buffer (225 mM mannitol, 75 mM sucrose, 10 mM MOPS, 1 mM EGTA, 0.5% fatty-acid-free BSA, pH 7.2) using a glass-teflon dounce homogenizer. The extracts were centrifuged at 300 g for 5 min at 4°C. The obtained supernatant was then centrifuged at 6000 g for 10 min at 4°C to obtain a mitochondrially enriched pellet. The pellet was resuspended in 100 μl respiration buffer (225 mM mannitol, 75 mM sucrose, 10 mM KCl, 10 mM Tris-HCl, 5 mM KH2PO4, pH 7.2). Freshly prepared mitochondrial isolations were used for respiration or aliquoted and frozen at −80°C for the mitochondrial enzyme complex activity assays. Quantification of mitochondrial protein obtained from the isolation was determined using the BCA protein assay (Thermo Scientific, Rockford, IL, USA).
Mitochondrial respiration
Respiration rates were determined by oxygen consumption using a Clark-type electrode and metabolic chamber (Hansatech Instruments, Norfolk, UK). A combination of NADH-linked substrate, 5 µmol of pyruvate plus 5 µmol malate, was added to an isolated mitochondrial suspension in 1 ml or respiration buffer held in the respiration chamber at 30°C. 125 nmol ADP was added to generate state III respiration, representative of the maximal ability to generate ATP. State IV, or basal, respiration rates were measured. The ratio of state III to state IV was calculated to define the respiratory control ratio (RCR). Ratios of >3 were considered functionally acceptable for fresh mitochondria preparations (Schriner et al., 2009). Uncoupled respiration rates, indicative of maximal rate of O2 consumption, were generated by the addition of 0.5 nmol of FCCP [carbonyl cyanide 4-(trifluoro-methoxy) phenylhydrazone], a chemical uncoupler.
Blue-native polyacrylamide gel electrophoresis (BN-PAGE)
To obtain mitochondrial complexes in the native state, mitochondrial preps were carried out as described above and the mitochondrial pellets were resuspended in 100 µl 1× Native Sample Buffer (Invitrogen, NY, USA) containing 1% digitonin and 1× protease inhibitors (Roche, CA, USA). Samples were incubated for 15 min on ice, and were centrifuged at 16.1 g at 4°C for 40 min. Supernatant was aliquoted and frozen at −80°C. Protein was quantified using the BCA Protein Assay (Thermo Scientific, Rockford, IL, USA). 15 μg of mitochondrial protein were run on 3-12% Bis-Tris Native PAGE gels per manual specifications (Novex, Life Technologies, NY, USA). 1× Native PAGE Running buffer and 1× Dark Blue cathode buffer were used (Invitrogen) for electrophoresis. NativeMark Protein standard (Invitrogen) was used as the molecular weight marker. ImageJ software was used to quantify band intensity. Band intensity was normalized to internal BSA band present in samples (BSA added during mitochondrial isolation procedure). In images, full lanes were moved from original gel locations for genotype consistency, no manipulations were made within lanes.
Mitochondrial complex activity assays
Mitochondrial isolations were performed as described above. Enzyme activity assays were performed as previously described (Meiklejohn et al., 2013). In brief, protein quantification was used to standardize the amount of protein added to each reaction. The specific activity of Complex I was determined following the oxidation of NADH at 340 nm with the coenzyme Q analog decylubiquinone as the electron acceptor. The catalytic activity of Complex II was monitored by the reduction of DCPIP at 600 nm, and was inhibited with 400 mM malonate. Complex III activity was measured by monitoring the reduction of cytochrome c at 550 nm, and was inhibited with 5 µg/ml antimycin A. Complex IV (cytochrome c oxidase) activity was measured by determining the rate of oxidation of reduced cytochrome c at 550 nm, and was inhibited with 4 mM KCN. To measure citrate synthase activity, the rate-limiting reaction of citrate synthase was coupled to a chemical reaction in which DTNB reacts with CoA-SH and the absorbance of the product is measured at 412 nm. Specific reaction mixtures are described in Meiklejohn et al., 2013.
35S-methionine incorporation
De novo translation was measured by incorporation of 35S-methionine into synthesized proteins. Female flies were placed on standard media supplemented with 100 µCi/ml 35S-methionine for 48 h; n>4, 20 flies per n. Mitochondrial preparations were carried out as described above (using a BSA free isolation buffer: 210 mM Mannitol, 70 mM Sucrose, 5 mM HEPES, 1 mM EDTA, pH 7.4, 1× protease inhibitor), and the supernatant from the low speed spin was taken as the cytosolic fraction. 1% triton was added to fractions and proteins were precipitated in 10% trichloroacetic acid. A Beckman LS 6500 scintillation counter was used to measure radiolabeled methionine incorporation. Values were normalized to protein content in each fraction (BCA method).
Statistical analyses
Analysis of variance models were used in conjunction with Tukey's post-hoc contrasts to compare the phenotypic effects of the transgenic alleles. For non-normally distributed data sets (climbing and flight), Kruskal–Wallis one-way analysis of variance were used in conjunction with Wilcoxon rank sum tests. Bonferroni-corrected P-values are provided to account for multiple testing. Analyses were conducted using the statistical package R version 2.15.0.
Supplementary Material
Acknowledgements
Credit is given to Colin Meiklejohn and Kristi Montooth for developing and providing the mito-nuclear introgression strains and transgenic rescue strains. We would like to thank Geoff Williams for assistance with TEM imaging, and Brian C. Jones for assistance with protocol optimization.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
M.A.H. and D.M.R. designed the experiments and wrote the manuscript. M.A.H. and J.R.D. performed the experiments. E.V.-C. optimized methods and provided experimental feedback. M.A.H. analyzed the data prepared the figures.
Funding
This work was supported by grants 1F31AG040925-02 awarded to M.A.H., grant R15GM113156 from NIGMS awarded to E.V.-C., and grant R01GM067862 from NIGMS and grant R01AG027849 from NIA awarded to D.M.R.
Supplementary material
Supplementary material available online at http://dmm.biologists.org/lookup/suppl/doi:10.1242/dmm.019323/-/DC1
References
- Arnqvist G., Dowling D. K., Eady P., Gay L., Tregenza T., Tuda M. and Hosken D. J. (2010). Genetic architecture of metabolic rate: environment specific epistasis between mitochondrial and nuclear genes in an insect. Evolution 64, 3354-3363. 10.1111/j.1558-5646.2010.01135.x [DOI] [PubMed] [Google Scholar]
- Augustin H. and Partridge L. (2009). Invertebrate models of age-related muscle degeneration. Biochim. Biophys. Acta 1790, 1084-1094. 10.1016/j.bbagen.2009.06.011 [DOI] [PubMed] [Google Scholar]
- Bayat V., Thiffault I., Jaiswal M., Tétreault M., Donti T., Sasarman F., Bernard G., Demers-Lamarche J., Dicaire M.-J., Mathieu J. et al. (2012). Mutations in the mitochondrial methionyl-tRNA synthetase cause a neurodegenerative phenotype in flies and a recessive ataxia (ARSAL) in humans. PLoS Biol. 10, e1001288 10.1371/journal.pbio.1001288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenakkers A. M. T., Van der Horst D. J. and Van Marrewijk W. J. A. (1984). Insect flight muscle metabolism. Insect Biochem. 14, 243-260. 10.1016/0020-1790(84)90057-X [DOI] [Google Scholar]
- Benzer S. (1973). Genetic dissection of behavior. Sci. Am. 229, 24-37. 10.1038/scientificamerican1273-24 [DOI] [PubMed] [Google Scholar]
- Butow R. A. and Avadhani N. G. (2004). Mitochondrial signaling: the retrograde response. Mol. Cell 14, 1-15. 10.1016/S1097-2765(04)00179-0 [DOI] [PubMed] [Google Scholar]
- Carta A., Carelli V., D'Adda T., Ross-Cisneros F. N. and Sadun A. A. (2005). Human extraocular muscles in mitochondrial diseases: comparing chronic progressive external ophthalmoplegia with Leber's hereditary optic neuropathy. Br. J. Ophthalmol. 89, 825-827. 10.1136/bjo.2004.060590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae S., Ahn B. Y., Byun K., Cho Y. M., Yu M.-H., Lee B., Hwang D. and Park K. S. (2013). A systems approach for decoding mitochondrial retrograde signaling pathways. Sci. Signal. 6, rs4 10.1126/scisignal.2003266 [DOI] [PubMed] [Google Scholar]
- Cogliati S., Frezza C., Soriano M. E., Varanita T., Quintana-Cabrera R., Corrado M., Cipolat S., Costa V., Casarin A., Gomes L. C. et al. (2013). Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell 155, 160-171. 10.1016/j.cell.2013.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum B., Walter A., Horst A., Osiewacz H. D. and Kühlbrandt W. (2013). Age-dependent dissociation of ATP synthase dimers and loss of inner-membrane cristae in mitochondria. Proc. Natl. Acad. Sci. USA 110, 15301-15306. 10.1073/pnas.1305462110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak I. I. (1977). A histochemical study of the muscles of Drosophila melanogaster. J. Morphol. 153, 307-316. 10.1002/jmor.1051530209 [DOI] [PubMed] [Google Scholar]
- Demontis F., Piccirillo R., Goldberg A. L. and Perrimon N. (2013). Mechanisms of skeletal muscle aging: insights from Drosophila and mammalian models. Dis. Model. Mech. 6, 1339-1352. 10.1242/dmm.012559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro S. and Schon E. A. (2003). Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 348, 2656-2668. 10.1056/NEJMra022567 [DOI] [PubMed] [Google Scholar]
- Dowling D. K., Friberg U. and Lindell J. (2008). Evolutionary implications of non-neutral mitochondrial genetic variation. Trends Ecol. Evol. 23, 546-554. 10.1016/j.tree.2008.05.011 [DOI] [PubMed] [Google Scholar]
- Dudkina N. V., Heinemeyer J., Keegstra W., Boekema E. J. and Braun H.-P. (2005). Structure of dimeric ATP synthase from mitochondria: an angular association of monomers induces the strong curvature of the inner membrane. FEBS Lett. 579, 5769-5772. 10.1016/j.febslet.2005.09.065 [DOI] [PubMed] [Google Scholar]
- Greaves L. C., Reeve A. K., Taylor R. W. and Turnbull D. M. (2012). Mitochondrial DNA and disease. J. Pathol. 226, 274-286. 10.1002/path.3028 [DOI] [PubMed] [Google Scholar]
- Greene J. C., Whitworth A. J., Kuo I., Andrews L. A., Feany M. B. and Pallanck L. J. (2003). Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl. Acad. Sci. USA 100, 4078-4083. 10.1073/pnas.0737556100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper R. H., Mouchiroud L., Ryu D., Moullan N., Katsyuba E., Knott G., Williams R. W. and Auwerx J. (2013). Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 497, 451-457. 10.1038/nature12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs H. T. (2003). Disorders of mitochondrial protein synthesis. Hum. Mol. Genet. 12 Suppl. 2, R293-R301. 10.1093/hmg/ddg285 [DOI] [PubMed] [Google Scholar]
- Kemp J. P., Smith P. M., Pyle A., Neeve V. C. M., Tuppen H. A. L., Schara U., Talim B., Topaloglu H., Holinski-Feder E., Abicht A. et al. (2011). Nuclear factors involved in mitochondrial translation cause a subgroup of combined respiratory chain deficiency. Brain 134, 183-195. 10.1093/brain/awq320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein P., Müller-Rischart A. K., Motori E., Schönbauer C., Schnorrer F., Winklhofer K. F. and Klein R. (2014). Ret rescues mitochondrial morphology and muscle degeneration of Drosophila Pink1 mutants. EMBO J. 33, 341-355. 10.1002/embj.201470060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose J. (1999). Genotypes and phenotypes. Electrophoresis 20, 643-652. [DOI] [PubMed] [Google Scholar]
- Konovalova S. and Tyynismaa H. (2013). Mitochondrial aminoacyl-tRNA synthetases in human disease. Mol. Genet. Metab. 108, 206-211. 10.1016/j.ymgme.2013.01.010 [DOI] [PubMed] [Google Scholar]
- Lambertsson A. (1998). The minute genes in Drosophila and their molecular functions. Adv. Genet. 38, 69-134. 10.1016/S0065-2660(08)60142-X [DOI] [PubMed] [Google Scholar]
- Li R. and Guan M. X. (2010). Human mitochondrial leucyl-tRNA synthetase corrects mitochondrial dysfunctions due to the tRNALeu(UUR) A3243G mutation, associated with mitochondrial encephalomyopathy, lactic acidosis, and stroke-like symptoms and diabetes. Mol. Cell. Biol. 30, 2147-2154. 10.1128/MCB.01614-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn C. D., Holmbeck M. A., Siddiq M. A., Abt D. N., Rand D. M. and Montooth K. L. (2013). An incompatibility between a mitochondrial tRNA and its nuclear-encoded tRNA synthetase compromises development and fitness in Drosophila. PLoS Genet. 9, e1003238 10.1371/journal.pgen.1003238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montooth K. L., Meiklejohn C. D., Abt D. N. and Rand D. M. (2010). Mitochondrial-nuclear epistasis affects fitness within species but does not contribute to fixed incompatibilities between species of Drosophila. Evolution 64, 3364-3379. 10.1111/j.1558-5646.2010.01077.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Loshuertos R., Ferrín G., Acín-Pérez R., Gallardo M. E., Viscomi C., Pérez-Martos A., Zeviani M., Fernández-Silva P. and Enríquez J. A. (2011). Evolution meets disease: penetrance and functional epistasis of mitochondrial tRNA mutations. PLoS Genet. 7, e1001379 10.1371/journal.pgen.1001379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell P. T., Collier V. L., Mogami K. and Bernstein S. I. (1989). Ultrastructural and molecular analyses of homozygous-viable Drosophila melanogaster muscle mutants indicate there is a complex pattern of myosin heavy-chain isoform distribution. Genes Dev. 3, 1233-1246. 10.1101/gad.3.8.1233 [DOI] [PubMed] [Google Scholar]
- Park J., Lee S. B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J.-M. et al. (2006). Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157-1161. 10.1038/nature04788 [DOI] [PubMed] [Google Scholar]
- Peckham M., Molloy J. E., Sparrow J. C. and White D. C. S. (1990). Physiological properties of the dorsal longitudinal flight muscle and the tergal depressor of the trochanter muscle of Drosophila melanogaster. J. Muscle Res. Cell Motil. 11, 203-215. 10.1007/BF01843574 [DOI] [PubMed] [Google Scholar]
- Perli E., Giordano C., Pisano A., Montanari A., Campese A. F., Reyes A., Ghezzi D., Nasca A., Tuppen H. A., Orlandi M. et al. (2014). The isolated carboxy-terminal domain of human mitochondrial leucyl-tRNA synthetase rescues the pathological phenotype of mitochondrial tRNA mutations in human cells. EMBO Mol. Med. 6, 169-182. 10.1002/emmm.201303198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesah Y., Pham T., Burgess H., Middlebrooks B., Verstreken P., Zhou Y., Harding M., Bellen H. and Mardon G. (2004). Drosophila parkin mutants have decreased mass and cell size and increased sensitivity to oxygen radical stress. Development 131, 2183-2194. 10.1242/dev.01095 [DOI] [PubMed] [Google Scholar]
- Pulkes T., Siddiqui A., Morgan-Hughes J. A. and Hanna M. G. (2000). A novel mutation in the mitochondrial tRNA(TYr) gene associated with exercise intolerance. Neurology 55, 1210-1212. 10.1212/WNL.55.8.1210 [DOI] [PubMed] [Google Scholar]
- Rand D. M., Haney R. A. and Fry A. J. (2004). Cytonuclear coevolution: the genomics of cooperation. Trends Ecol. Evol. 19, 645-653. 10.1016/j.tree.2004.10.003 [DOI] [PubMed] [Google Scholar]
- Rand D. M., Fry A. and Sheldahl L. (2006). Nuclear-mitochondrial epistasis and drosophila aging: introgression of Drosophila simulans mtDNA modifies longevity in D. melanogaster nuclear backgrounds. Genetics 172, 329-341. 10.1534/genetics.105.046698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt K., Dowling D. K. and Morrow E. H. (2013). Medicine. Mitochondrial replacement, evolution, and the clinic. Science 341, 1345-1346. 10.1126/science.1237146 [DOI] [PubMed] [Google Scholar]
- Rhodenizer D., Martin I., Bhandari P., Pletcher S. D. and Grotewiel M. (2008). Genetic and environmental factors impact age-related impairment of negative geotaxis in Drosophila by altering age-dependent climbing speed. Exp. Gerontol. 43, 739-748. 10.1016/j.exger.2008.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley L. G., Cooper S., Hickey P., Rudinger-Thirion J., McKenzie M., Compton A., Lim S. C., Thorburn D., Ryan M. T., Giegé R. et al. (2010). Mutation of the mitochondrial tyrosyl-tRNA synthetase gene, YARS2, causes myopathy, lactic acidosis, and sideroblastic anemia--MLASA syndrome. Am. J. Hum. Genet. 87, 52-59. 10.1016/j.ajhg.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley L. G., Menezes M. J., Rudinger-Thirion J., Duff R., de Lonlay P., Rotig A., Tchan M. C., Davis M., Cooper S. T. and Christodoulou J. (2013). Phenotypic variability and identification of novel YARS2 mutations in YARS2 mitochondrial myopathy, lactic acidosis and sideroblastic anaemia. Orphanet. J. Rare Dis. 8, 193 10.1186/1750-1172-8-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorbach J., Yusoff A. A., Tuppen H., Abg-Kamaludin D. P., Chrzanowska-Lightowlers Z. M. A., Taylor R. W., Turnbull D. M., McFarland R. and Lightowlers R. N. (2008). Overexpression of human mitochondrial valyl tRNA synthetase can partially restore levels of cognate mt-tRNAVal carrying the pathogenic C25U mutation. Nucleic Acids Res. 36, 3065-3074. 10.1093/nar/gkn147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotig A. (2011). Human diseases with impaired mitochondrial protein synthesis. Biochim. Biophys. Acta 1807, 1198-1205. 10.1016/j.bbabio.2011.06.010 [DOI] [PubMed] [Google Scholar]
- Sacktor B. (1976). Biochemical adaptations for flight in the insect. Biochem. Soc. Symp. 111-131. [PubMed] [Google Scholar]
- Saini N., Georgiev O. and Schaffner W. (2011). The parkin mutant phenotype in the fly is largely rescued by metal-responsive transcription factor (MTF-1). Mol. Cell. Biol. 31, 2151-2161. 10.1128/MCB.05207-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon E. A., DiMauro S. and Hirano M. (2012). Human mitochondrial DNA: roles of inherited and somatic mutations. Nat. Rev. Genet. 13, 878-890. 10.1038/nrg3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schriner S. E., Abrahamyan A., Avanessian A., Bussel I., Maler S., Gazarian M., Holmbeck M. A. and Jafari M. (2009). Decreased mitochondrial superoxide levels and enhanced protection against paraquat in Drosophila melanogaster supplemented with Rhodiola rosea. Free Radic. Res. 43, 836-843. 10.1080/10715760903089724 [DOI] [PubMed] [Google Scholar]
- Shafiq S. A. (1963). Electron microscopic studies on the indirect flight muscles of Drosophila melanogaster: I. Structure of the myofibrils. J. Cell Biol. 17, 351-362. 10.1083/jcb.17.2.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahni R., Wedatilake Y., Cleary M. A., Lindley K. J., Sibson K. R. and Rahman S. (2013). A distinct mitochondrial myopathy, lactic acidosis and sideroblastic anemia (MLASA) phenotype associates with YARS2 mutations. Am. J. Med. Genet. A 161, 2334-2338. 10.1002/ajmg.a.36065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits P., Smeitink J. and van den Heuvel L. (2010). Mitochondrial translation and beyond: processes implicated in combined oxidative phosphorylation deficiencies. J. Biomed. Biotechnol. 2010, 737385 10.1155/2010/737385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez R. K. (2000). Energy metabolism during insect flight: biochemical design and physiological performance. Physiol. Biochem. Zool. 73, 765-771. 10.1086/318112 [DOI] [PubMed] [Google Scholar]
- Suarez R. K., Lighton J. R., Joos B., Roberts S. P. and Harrison J. F. (1996). Energy metabolism, enzymatic flux capacities, and metabolic flux rates in flying honeybees. Proc. Natl. Acad. Sci. USA 93, 12616-12620. 10.1073/pnas.93.22.12616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama S., Moritoh S., Furukawa Y., Mizuno T., Lim Y.-M., Tsuda L. and Nishida Y. (2007). Involvement of the mitochondrial protein translocator component tim50 in growth, cell proliferation and the modulation of respiration in Drosophila. Genetics 176, 927-936. 10.1534/genetics.107.072074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toivonen J. M., O'Dell K. M., Petit N., Irvine S. C., Knight G. K., Lehtonen M., Longmuir M., Luoto K., Touraille S., Wang Z. et al. (2001). Technical knockout, a Drosophila model of mitochondrial deafness. Genetics 159, 241-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran Hornig-Do H., Montanari A., Rozanska A., Tuppen H. A., Almalki A. A., Abg-Kamaludin D. P., Frontali L., Francisci S., Lightowlers R. N. and Chrzanowska-Lightowlers Z. M. (2014). Human mitochondrial leucyl tRNA synthetase can suppress non cognate pathogenic mt-tRNA mutations. EMBO Mol. Med. 6, 183-193. 10.1002/emmm.201303202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen S., Chen S., George J., Tuomela T., Luoto K. R., O'Dell K. M. C. and Jacobs H. T. (2014). Phenotypic rescue of a Drosophila model of mitochondrial ANT1 disease. Dis. Model. Mech. 7, 635-648. 10.1242/dmm.016527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L.-J. C. (2007). Pathogenic mitochondrial DNA mutations in protein-coding genes. Muscle Nerve 36, 279-293. 10.1002/mus.20807 [DOI] [PubMed] [Google Scholar]
- Woodson J. D. and Chory J. (2008). Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9, 383-395. 10.1038/nrg2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Gehrke S., Imai Y., Huang Z., Ouyang Y., Wang J.-W., Yang L., Beal M. F., Vogel H. and Lu B. (2006). Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl. Acad. Sci. USA 103, 10793-10798. 10.1073/pnas.0602493103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen M.-Y., Chen C.-S., Wang A.-G. and Wei Y.-H. (2002). Increase of mitochondrial DNA in blood cells of patients with Leber's hereditary optic neuropathy with 11778 mutation. Br. J. Ophthalmol. 86, 1027-1030. 10.1136/bjo.86.9.1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeviani M. and Di Donato S. (2004). Mitochondrial disorders. Brain 127, 2153-2172. 10.1093/brain/awh259 [DOI] [PubMed] [Google Scholar]
- Zhu C.-T., Ingelmo P. and Rand D. M. (2014). G×G×E for lifespan in Drosophila: mitochondrial, nuclear, and dietary interactions that modify longevity. PLoS Genet. 10, e1004354 10.1371/journal.pgen.1004354 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.