Abstract
Considerable variation in antibody response (AR) was observed among recipients of an injectable typhoid vaccine and an oral cholera vaccine. We sought to find whether polymorphisms in genes of the immune system, both innate and adaptive, were associated with the observed variation in response. For both vaccines, we were able to discover and validate several polymorphisms that were significantly associated with immune response. For the typhoid vaccines, these polymorphisms were on genes that belonged to pathways of polysaccharide recognition, signal transduction, inhibition of T-cell proliferation, pro-inflammatory signalling and eventual production of antimicrobial peptides. For the cholera vaccine, the pathways included epithelial barrier integrity, intestinal homeostasis and leucocyte recruitment. Even though traditional wisdom indicates that both vaccines should act as T-cell-independent antigens, our findings reveal that the vaccines induce AR using different pathways.
Keywords: immunoglobulin, genetic polymorphism, polysaccharide antigen, lipopolysaccharide, association
1. Introduction
The impact of vaccines on the control and eradication of many infectious diseases cannot be overstated. Yet, vaccines do not confer equal protection to all vaccine recipients (table 1). A recent dramatic example is that among the 1521 cases of a mumps outbreak in a 2009 summer camp in New York, only 9.1% had not previously received a mumps vaccination [1]. Because vaccines primarily work through the immune system of the recipient, it is important to quantify variability in immune response among vaccinees and to understand the causes of variability. As a matter of fact, a new branch of omics—called vaccinomics—has evolved that aims to understand the mechanisms of heterogeneity in immune responses to vaccines [2]. Many twin and family studies [3–6] have shown that vaccine immune response is highly heritable. Vaccinomics, therefore, posits that heterogeneity in host genetic markers results in variations in vaccine-induced immune responses and aims to identify these factors for predicting and minimizing vaccine failures or adverse events. While there are multiple parameters by which to measure immune response in a vaccinee [7–9], levels of various immunoglobulins have traditionally been used. This is because immune response usually involves the production of antibodies or immunoglobulins by B lymphocytes. It is now well established that for various vaccines, genetic effects play a dominant role in immune response [10–12]. Epigenomic effects are also becoming evident [13]. Quantification of immune response at various stages after vaccination is also important, because the level of early response may predict later response, which is known to be important is determining the extent of protection offered by the vaccine. In a study on a yellow fever vaccine [14], gene expression signatures in the blood a few days after vaccination—early innate response-related molecular signature—have been found that could predict later antibody titres with about 90% accuracy. Furthermore, these investigators also identified that a calcium/calmodulin-dependent protein kinase type IV largely controls antigen-specific antibody production, thus providing insights into the mechanism of immune response to the vaccine. With respect to a hepatitis B vaccine [15], an extended HLA signature has been found to be associated with non-response, indicating that individuals who possess this signature may require multiple doses or vaccines containing increased amounts of antigen. Studies on immune response to vaccines and on other aspects of vaccine research have spawned the development of an online network called vaccine investigation and online information network [16]. Vaccine immunogenicity is determined in part not only by the chemical and physical nature of microbial antigens and adjuvants, but also by the genetic make-up of vaccine recipients [17]. The impact of genetic variation in vaccinees on AR has also been obtained in some detail [18] for some vaccines, but not for vaccines against enteric diseases such as typhoid and cholera.
Table 1.
Impact of vaccines in the twentieth and twenty-first centuries. Data source: http://www.cdc.gov/vaccines/pubs/pinkbook/downloads/appendices/G/impact-of-vaccines.pdf.
| disease | % decrease in annual morbidity | disease | % decrease in annual morbidity |
|---|---|---|---|
| smallpoxa | 100 | Haemophilus influenzaea (<5 years of age) | 99 |
| diphtheriaa | 100 | hepatitis Ab | 91 |
| pertussisa | 89 | hepatitis Bb (acute) | 83 |
| tetanusa | 99 | pneumococcusb (invasive) | |
| polioa (paralytic) | 100 | all ages | 30 |
| measlesa | >99 | <5 years of age | 74 |
| mumpsa | 98 | rotavirusb (hospitalizations <5 years of age) | 88 |
| rubellaa | >99 | varicellab | 89 |
| CRSa | 100 | ||
aBased on a comparison of twentieth century annual morbidity and current morbidity.
bBased on a comparison of pre-vaccine era estimated annual morbidity and current morbidity.
2. The typhoid vaccine study
Primary failure, as assessed by post-vaccination antibody levels, occurs in 23–40% of recipients of typhoid vaccine [19,20]. The available typhoid vaccine is a polysaccharide (PS) vaccine, an inactivated subunit vaccine composed of long chains of sugar molecules that make up the surface capsule of Salmonella enterica serotype typhi (Salmonella typhi). The essential feature of immune response to a pure PS vaccine is that it is typically T-cell-independent. A PS vaccine stimulates B cells and non-cognate T cells without the assistance of cognate T cells [21,22]. Because data on the role of genetic variation in vaccinees on AR to PS vaccines are scanty, we undertook a study using an available typhoid vaccine. The objective of this study was to quantify the extent of variation in AR to a widely used Vi PS vaccine for typhoid and to discover associations of polymorphisms in candidate genes of vaccinees with AR.
(a). Methods of vaccinee recruitment, biospecimen collection, assays and statistical analyses
Potential vaccinees were recruited with written informed consent, provided that (i) she/he had not had fever lasting for more than three consecutive days in the past 1 year, and (ii) she was not pregnant or lactating. Individuals (n = 1000), unrelated at least to the first-cousin level, aged 12 years or older, inhabiting a socioeconomically depressed locality of Kolkata (formerly Calcutta), India, were thus recruited.
Each individual recruited into the study was provided, by intramuscular injection (marketed in India as Typherix® by GlaxoSmithKline), 0.5 ml vaccine containing 25 μg of the cell surface Vi PS extracted from S. typhi Ty2 strain with the excipients sodium chloride, sodium dihydrogen phosphate dihydrate, disodium phosphate dihydrate, phenol and water. From each study participant, a blood sample was collected immediately prior to vaccination and 28 days post-vaccination. Serum and DNA were isolated from these samples, using standard protocols. The use of Vi PS as a coating antigen in enzyme-linked immunosorbent assay (ELISA) to measure vaccine-induced anti-Vi ARs has been reported to be problematic. The use of PSs (lipopolysaccharide (LPS), Haemophilus influenzae type b capsular PS, Vi PS) as coating antigens for immunoassays is plagued by problems such as a poor binding of PSs to ELISA plates and inconsistent results [23 and references therein]. We therefore developed and standardized the Bio-Plex assay that provides much more robust and consistent results than ELISA [24]. The bead assay was performed using the Bio-Plex (Bio-Rad, Hercules, CA) platform. The Vi PS antigen (supplied by Fina Biosolutions, Rockville, MD) was conjugated to Bio-Rad beads (COOH (028)) at a concentration 8.33 × 106 beads ml−1 in a total volume of 1.50 ml. Aliquots (50 µl) of human serum were prepared at various dilutions in human serum diluent (PBS, 1% w/v, 5% v/v/goat serum, 0.05% Tween 20 v/v and 0.1% Kathon v/v). Day-0 serum was diluted 1 : 50, 1 : 400 and 1 : 3200; day-28 serum was diluted 1 : 200, 1 : 1600 and 1 : 12 800 in human serum diluent before incubation with the Vi beads for 3 h. After washing the beads, the bound human IgG was detected by goat anti-human IgG-RPE (1 : 5000 in human serum diluent) by incubation for 30 min. Following the final wash steps, the beads were analysed using the Bio-Plex instrument. A pooled serum with a high anti-Vi IgG response was used to create a standard curve that was assigned an arbitrary value of 200 ELISA units (EU) per ml. The responses in the individual subjects were compared with the standard curve to calculate anti-Vi IgG EU ml−1 in the day-0 and day-28 serum.
Single nucleotide polymorphisms (SNPs) in and around 283 autosomal genes from immunological pathways were chosen, using a statistical protocol that took into account differences in interpopulation variation of linkage disequilibrium and allele frequencies, from the HapMap database (http://www.hapmap.org). Genotyping was done using Sequenom (55.7% of assayed SNPs) and Illumina (remaining 44.3% of SNPs) platforms.
AR to vaccination, measured as the difference between 28-day post-vaccination and pre-vaccination antibody levels (EU ml−1), was log10-transformed (logAR) to induce normality to the frequency distribution. The genotype data were curated by removing (i) loci with less than 90% call rate (8.9% of loci), (ii) non-polymorphic loci with minor allele frequency (MAF) < 0.05 (24.7% of loci with call rate > 90%), (iii) significantly deviant from Hardy–Weinberg equilibrium (4.2% of polymorphic loci), (iv) individuals with less than 75% of loci with valid genotype calls, (v) individuals on whom AR could not be properly assayed (8 of 1000 individuals) and (vi) individuals showing cryptic relationships (more than 80% of loci with identical genotypes: six pairs of individuals satisfied this criterion; one individual from each pair was randomly chosen for final analysis). After data curation, the final dataset comprised genotypes of 984 individuals at 2040 SNPs in 283 genes. Analysis of variance (ANOVA) was performed to test equality of mean values of logAR among subgroups of vaccinees.
For association analysis, we sorted the AR of the vaccinees and grouped the vaccinees into five pentile (20 percentile) groups. We then computed the correlation coefficient between the MAF at the locus and mean AR of vaccinees belonging to the pentile groups, and tested the significance of each correlation coefficient non-parametrically using a resampling technique. Genotypes of individual vaccinees at a locus were randomly permuted, and the correlation coefficient was computed in this permuted sample. This procedure was repeated 1000 times to produce a histogram of correlation coefficients under the null hypothesis r = 0. This histogram was then used to compute the p-value associated with the observed r at this locus. After generating the p-values for all loci, the false discovery rate procedure [25] was used to adjust p-values for multiple testing and to identify statistically significant values of r at the 5% level of significance. All significant allelic associations with logAR detected using the above-described procedures were also validated using genotype data using PLINK v. 1.06 [26] (http://pngu.mgh.harvard.edu/~purcell/plink/). We have used the quantitative trait association module of PLINK, and have used empirical significance levels. We have also tested the equality of mean values of the response among individuals stratified by genotype at each locus using PLINK.
Because association studies are known to result in many false-positive inferences, we generated an internal cross-validation sample by randomly splitting the 984 vaccinees into two half-samples of approximately 500 vaccinees each (with approx. 100 vaccinees in each of the five pentile groups based on AR within each half-sample). The first half-sample was used for association discovery and the second half-sample was used for validation of discovered associations. Haplotypes were inferred using PHASE [27] (http://www.stat.washington.edu/stephens/phase.html). Haplotype association analysis was performed using the above-described permutation algorithm for single SNP markers.
(b). Enormous variation in antibody titres among vaccine recipients
The scatter diagram of Vi-PS pre- (day 0) and post- (day 28) vaccination antibody titres among the 984 vaccinees are depicted in figure 1. The extent of variation was enormous. The correlation between pre- and post-vaccination antibody levels was low (0.188) but statistically significant (p < 0.0005). The contributions of variation in age, gender or religious backgrounds to the variation in difference between post- and pre-vaccination antibody titres were statistically non-significant (p > 0.05).
Figure 1.
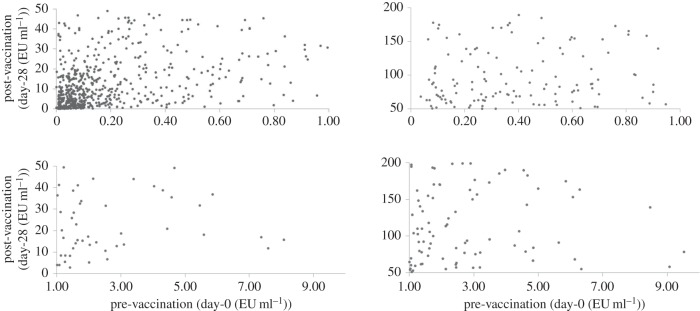
Scatter diagram depicting variation in pre- and post-vaccination Vi-polysaccharide antibody titres among typhoid vaccine recipients. (The enormity of the extent of variation necessitated splitting the single scatter diagram into four quadrants for display.)
(c). Genomic association discovery and validation
No statistically significant genetic stratification or substructuring among the vaccines could be detected [28]. The demographic characteristics of the vaccinees in the discovery and validation half-samples were similar and statistically non-significant. In the discovery half-sample, 54 SNPs in and around 43 genes showed significant correlation (p-values were estimated by data permutation) at the 5% level (using multiple-testing correction by FDR method) with AR. The discovered associations were cross-validated using the data on the validation half-sample. Of these 54 SNPs, significant association was found only for eight SNPs—all intronic—in seven genes (table 2). These genes were saturated with additional SNPs, genotype data were generated for the additional SNPs and haplotype associations with AR were calculated; the correlations were all statistically significant [28].
Table 2.
Details of the eight SNPs in seven genes that showed significant cross-validated association with antibody response.
| gene | chromo-some no. | rs no. | nt position | SNP location within gene (mRNA transcript no. in NCBI database) | ra in discovery half-sample (n = 492) | r in validation half-sample (n = 492) | p-value using PLINK in the total sample (n = 984) |
|---|---|---|---|---|---|---|---|
| IL1RL1 | 2 | 10208293 | 102332742 | Intron 10 (NM_016232.4) | 0.95 | 0.91 | 0.002 |
| CTLA4 | 2 | 231779 | 204442732 | Intron 1 (NM_005214.3) | −0.99 | −0.93 | 0.028 |
| CD86 | 3 | 17203439 | 123278196 | Intron 1 (NM_175862.3) | 0.95 | 0.97 | 0.016 |
| TLR1 | 4 | 5743572 | 38481786 | Intron 2 (NM_003263.3) | 0.98 | 0.93 | 0.05 |
| DEFB1 | 8 | 2978873 | 6717373 | Intron 1 (NM_005218.3) | 0.98 | 0.98 | 0.016 |
| 2977772 | 6720606 | Intron 1 (NM_005218.3) | 0.91 | 0.94 | 0.013 | ||
| MAPK8 | 10 | 10857564 | 49282578 | Intron 1 (NM_002750.2) | −0.95 | −0.96 | 0.03 |
| IL17D | 13 | 1888001 | 20183906 | Intron 2 (NM_138284.1) | 0.94 | 0.91 | 0.018 |
ar is the correlation between minor allele frequency and antibody response in EU ml−1.
3. The cholera vaccine study
Cholera is caused by toxicogenic strains of the Gram-negative bacterium Vibrio cholerae. Ingestion of contaminated food or water causes cholera. Infected individuals suffer from moderate-to-severe watery diarrhoea that leads to dehydration. Cholera can be fatal if left untreated. Lack of safe water is the major cause of endemicity of cholera in many countries. In recent times, there have been two major outbreaks: in Zimbabwe in 2009 and in Haiti in 2010. Several thousand people died of cholera. These outbreaks prompted the World Health Organization to encourage administration of oral cholera vaccine. V. cholerae, has two major serogroups—O1 (serotypes Inaba and Ogawa) and O139—that are responsible for most cholera cases. Members of the serogroups differ in the LPS structures of the outer membrane. Each dose of the oral cholera vaccine contains: formalin-killed V. cholerae Inaba, El Tor biotype (strain Phil 6973; 600 EU), heat-killed V. cholerae Ogawa classical biotype (Cairo 50; 300 EU), formalin-killed V. cholerae Ogawa classical biotype (Cairo 50; 300 EU), heat-killed V. cholerae Inaba, classical biotype (Cairo 48; 300 EU) and formalin-killed V. cholerae O139 (4260B; 600 EU). Two doses are orally administered 14 days apart.
(a). Methods
The methodology of recruitment of 1000 individuals into the cholera vaccine study was similar to that of the typhoid vaccine study. Serum anti-V. cholerae (Inaba 569) LPS—that is, total anti-LPS antibody—was assayed using the Bio-Plex (Bio-Rad) platform. Anti-LPS Ig EU ml−1 was estimated using methods similar to those described for the typhoid vaccine study. A vibriocidal assay was performed for each individual with V. cholerae O1 Inaba (OS-418), Ogawa (MAK757) and uncapsulated O139 (MO-10T4) strains using sera collected during pre- and post-vaccine trials. Commercially available guinea pig serum (Rockland, Gilbertsville, PA) was used as a source of complement. The sera (100 ml) were added to 100 ml of PBS in the first well to give twofold dilution and subsequent dilutions were made reciprocally up to 4800. Reference rabbit antisera against V. cholera O1 Inaba, Ogawa and O139 were included in each set of assay as controls. In every batch of assay, serum obtained from a healthy volunteer and a high-titre antiserum obtained from one of the volunteers in this study were included as negative and positive controls, respectively.
(b). Genomic association discovery and validation
After data curation, data on 948 vaccine recipients were available for statistical analyses. Although the vaccine is administered two times, 14 days apart, we considered the antibody level 28 days after the first administration of the vaccine as the post-vaccination response. Similar to the typhoid study, there was considerable variation in immune response to the cholera vaccine (table 3) as reflected by the large standard deviations of pre-vaccination antibody level, as also of 28-days post-vaccination level and fold-change (defined as the ratio of post-vaccination to pre-vaccination antibody levels). The effects of age, gender and religion on variation in fold-change were all statistically non-significant (p > 0.05). We tested for existence of cryptic relatedness and population stratification using Eigenstrat [29]; no evidence was detected as all individuals formed a single cluster in the scatterplot of the first three principal components [30] (figure 1).
Table 3.
Mean and s.d. of serum anti-V. cholerae LPS antibody in 948 cholera vaccine recipients pre- and 28 days post-vaccination, and fold change.
| mean | s.d. | |
|---|---|---|
| pre-vaccination (EU ml−1) | 22.54 | 37.95 |
| post-vaccination (EU ml−1) | 76.89 | 114.88 |
| fold change (pre/post) | 5.42 | 12.41 |
In the discovery subset of vaccinees, statistically significant correlations with AR, as measured by fold-change, were detected for 159 SNP loci on 93 genes. These correlations were then validated in the validation subset. Most discovered correlations could not be validated, except seven SNPs that are located in and around five genes (rs17180481 and rs17180600 in the flanking region and intron, respectively, of MARCO (2q14.2); rs598493 in an intron of TNFAIP3 (6q23.3); rs3012694 and rs266087 in introns of RFX3 (9p24.2); rs266087 in an intron of CXCL12 (10q11.21) and rs6567272 in an intron of TNFRSF11A (18q22.1)). Mean AR for vaccinees belonging to the three genotypes for each of these seven SNP loci differed significantly for only the four SNPs in and around MARCO, TNFAIP3 and CXCL12 (rs17180481: p = 0.023, effect size = 1.8 ± 0.3; rs17180600: p = 0.026, effect size = 1.6 ± 0.3; rs598493: p = 0.001, effect size = 3.9 ± 0.8; rs266087: p = 0.014, effect size = 2.4 ± 0.6), but not for the remaining three.
4. Discussion
Many non-protein antigens, such as PSs and lipids, are not processed and presented along with MHC proteins and hence are not recognized by helper T cells. Therefore, these stimulate antibody production in the absence of helper T cells and are T-independent antigens. Being composed of multiple identical epitopes, most T-independent antigens are polyvalent. Polyvalent antigens can induce cross-linking of surface Ig molecules on B cells, leading to activation of B cells without the requirement of helper T cells.
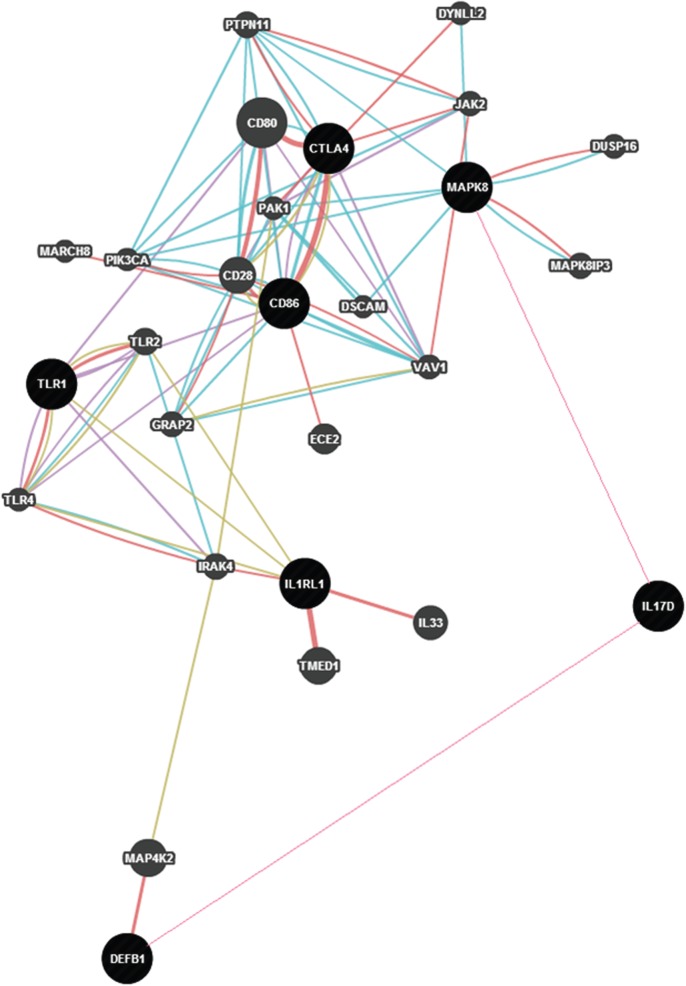
SNPs in seven genes—CD80, CTLA4, TLR1, IL1RL1, MAPK8, IL17D and DEFB1—were found to be associated with variation in AR to the typhoid vaccine. The network relationship among these genes based on their co-expression, co-localization, etc. is presented in figure 2. It has been noted [28] that the B7 family of co-stimulatory molecules, CD86 along with CD80, present on the antigen-presenting cells, interact with CD28/CTLA4 receptors on T cells to provide major non-cognate co-stimulatory signals [31]. The non-cognate T-cell dependence is indicated by the finding of SNPs in CD86 and CTLA4 being associated with AR to the PS vaccine. A critical quantitative role for CTLA-4 in governing T- and B-cell homeostasis has also been recently discovered [32]. TLR1 showed association with the host response to the vaccine. An important immune evasion mechanism in some bacterial pathogens is by downregulation of TLR-mediated immune responses through diminished TLR-mediated cell signalling or expression [33,34]. TLR1 coexpresses with CD86 (figure 2). All Toll-like receptors have a Toll-interleukin 1 domain that is responsible for signal transduction. IL1RL1 participates in this signalling. We have found SNPs in this gene to be significantly associated with AR induced by the Vi-PS vaccine. MAP kinases are involved in a wide variety of cellular processes such as proliferation, differentiation, transcription regulation and development. Therefore, the association of an SNP in MAPK8 with AR is not unexpected. The involvement of a MAP kinase and IL17D, which stimulates the production of other cytokines, can result in pro-inflammatory signals that can induce upregulation of DEFB1 [35]. We have found multiple SNPs in DEFB1 to be significantly associated with Vi-PS vaccine response. Thus, the overall picture that emerges from our study is that polymorphisms in genes involved in PS recognition, signal transduction, inhibition of T-cell proliferation, pro-inflammatory signalling and eventual production of antimicrobial peptides are associated with AR to the Vi-PS vaccine for typhoid.
Figure 2.
Network of genes associated with antibody response to the typhoid vaccine constructed using GeneMania and depicts physical interaction (red), co-expression (purple), physical interaction (turquoise) and shared protein domains (grey) among the genes. (Online version in colour.)
The cholera vaccine is an LPS vaccine. This vaccine, like the Vi-PS typhoid vaccine, is expected to induce antibody production without support of T-helper cells. However, we have found no commonality among the genes that modulate AR to the typhoid and cholera vaccines. One reason for this may be that the routes of administration of these vaccines are different.
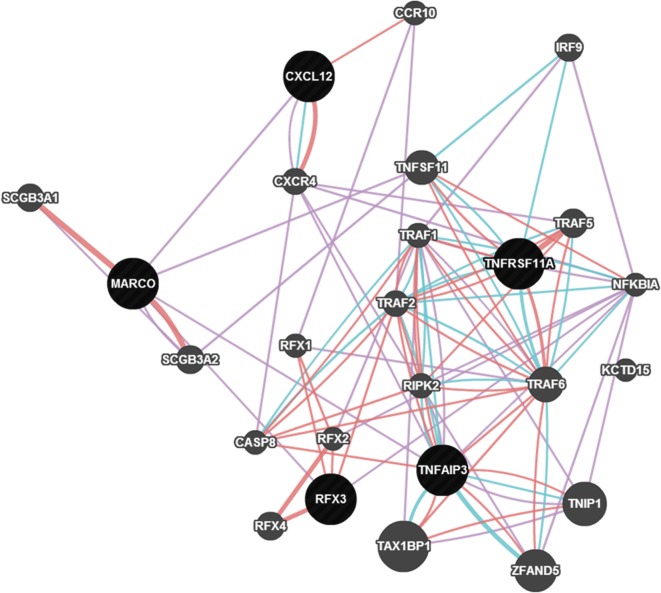
Three genes—MARCO, TNFAIP3 and CXCL12—were found to have significant roles in modulating AR to the oral cholera vaccine. A single cell layer of the mucosal epithelium essentially regulates intestinal homeostasis [30]. This physical barrier is a prominent part of the innate immune system with damage-healing and infection-limiting properties. CXCL12 is a homeostatic or constitutive chemokine. The product of this gene plays a key role in damage-healing by stimulating the activation of laminin-specific integrins [36]. V. cholerae infection produces strong inflammatory manifestations [37]. CXCL12 is a neutrophil and lymphocyte chemoattractant. It is upregulated in response to V. cholerae infection, which results in increased recruitment of polymorphonuclear leucocytes to the site of infection [38]. The LPS in the cholera vaccine possibly provides signals that mimic those of the live bacterium, without actually causing disease. MARCO is a type II transmembrane protein of the class A scavenger-receptor family [39]. The major component of the cholera vaccine is LPS which is a very potent activator of innate immune responses. Surface expression of MARCO is upregulated in a dose- and time-dependent manner by LPS stimulation [40]. MARCO is upregulated in bacterial infections in macrophages of most tissues; variants in this gene have crucial roles in bacterial binding [41]. MARCO and CXCL12 are co-expressed (figure 3). Epithelial barrier integrity is a possible requirement of adequate response to the whole-cell-killed cholera vaccine. Tight junctions between intestinal epithelial cells mediate the permeability of the intestinal barrier. TNFAIP3 has a role in promoting intestinal epithelial barrier integrity and in enabling maintenance of intestinal homeostasis through tight junction protein regulation [42].
Figure 3.
Network of genes associated with antibody response to the cholera vaccine. (Online version in colour.)
One of the greatest successes of modern medicine has been the discovery of vaccines. Vaccines have eradicated many infectious diseases. Vaccines have saved millions, possibly billions, of lives. Yet many vaccine recipients do not get adequate protection from infection. Unfortunately, our understanding of the causes of interindividual variation in immune response resulting from vaccination is woefully inadequate. While various factors, such as age at vaccination, number of doses, amount per dose, nutrition, presence of other pathogens and commensals, surely contribute to the variability in immune response, it has also been recognized that the genetic background of a vaccine recipient plays a major role in modulating AR to a vaccine. In the past decade or so, a large number of studies have been carried out to associate variations in genetic backgrounds of vaccine recipients to their ARs to the vaccine. These studies have not only resulted in many association discoveries, but have also yielded insights into mechanisms of response. We conducted this study using an injectable vaccine for typhoid that is known to have a high level of primary vaccine failure, and an oral vaccine for cholera for which hardly anything is known regarding immune response induced by the vaccine. We have found significant evidence of genes and pathways, belonging to both innate and adaptive systems, which are associated with the modulation of AR to these vaccines. Even though traditional wisdom indicates that both vaccines should act as T-cell-independent antigens, our findings reveal that the vaccines induce AR using different pathways. However, we note that these two vaccines have other differences, notably (i) their routes of administration are different and (ii) the typhoid vaccine is a single-dose vaccine, whereas the cholera vaccine is a two-dose vaccine. The latter difference may be critical in activating different pathways for mounting immune response.
It is clear from our studies that host genomics plays a significant role in modulating immune response to vaccines. Understanding these genomic correlates will help identify individuals who may require larger doses of vaccines to mount an adequate protective response, or even to formulate better vaccines by possibly using small synthetic peptides homologous to sequences of relevant genes as adjuvants.
Acknowledgements
I am grateful to Dr Diane Wagener, Dr Herman Staats, Dr Carol Whisnant, Dr James Stephenson, Neeta Sarkar-Roy, Binuja Verma, K. Narayanasamy, Bijan Bairagya, Biplab Dey, Uposoma Dey, Ardhendu Endow, Souvik Mukherjee, Sonia Poddar, Priya Sengupta and Debabrata Sutradhar, Shaun M. Kirwan and investigators, phlebotomists and clinicians who helped in participant recruitment, vaccination, biospecimen collection and monitoring of adverse events.
Ethical statement
This study was approved by the Institutional Ethics Committee of the TCG-ISI Centre for Population Genomics, Kolkata; Health Ministry Screening Committee, Government of India, New Delhi and the Drug Controller General of India, New Delhi. Recruitment, vaccination and biospecimen collection were done with individual written informed consent. The study was approved by the institutional ethics committee.
Funding statement
This work was supported by NIAID, National Institutes of Health, USA (contract no. HHSN200400067C). As a condition of the contract, the raw data have been submitted in ImmPORT of DAIT, NIAID, NIH.
Competing interests
I have no competing interests.
References
- 1.Centers for Disease Contol and Prevention 2010. Update: mumps outbreak—New York and New Jersey, June 2009–January 2010. MMWR 59, 125–129. [PubMed] [Google Scholar]
- 2.Poland GA, Ovsyannikova IG, Kennedy RB, Haralambieva IH, Jacobson RM. 2011. Vaccinomics and a new paradigm for the development of preventive vaccines against viral infections. OMICS 15, 625–636. ( 10.1089/omi.2011.0032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacobson RM, Ovsyannikova IG, Targonski PV, Poland GA. 2007. Studies of twins in vaccinology. Vaccine 25, 3160–3164. ( 10.1016/j.vaccine.2007.01.048) [DOI] [PubMed] [Google Scholar]
- 4.Klein NP, Fireman B, Enright A, Ray P, Black S, Dekker CL. 2007. A role for genetics in the immune response to the varicella vaccine. Pediatr. Infect. Dis. J. 26, 300–305. ( 10.1097/01.inf.0000257454.74513.07) [DOI] [PubMed] [Google Scholar]
- 5.Newport MJ, Goetghebuer T, Weiss HA, Whittle H, Siegrist C-A, Marchant A. 2004. Genetic regulation of immune responses to vaccines in early life. Genes Immun. 5, 122–129. ( 10.1038/sj.gene.6364051) [DOI] [PubMed] [Google Scholar]
- 6.Tan PL, Jacobson RM, Poland GA, Jacobsen SJ, Pankratz SV. 2001. Twin studies of immunogenicity-determining the genetic contribution to vaccine failure. Vaccine 19, 2434–2439. ( 10.1016/S0264-410X(00)00468-0) [DOI] [PubMed] [Google Scholar]
- 7.Gaucher D, et al. 2008. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J. Exp. Med. 205, 3119–3131. ( 10.1084/jem.20082292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plotkin SA. 2010. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 17, 1055–1065. ( 10.1128/CVI.00131-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shrimpton A, Duddridge M, Ziegler-Heitbrock L. 2006. Vaccination with polysaccharide-conjugate-vaccines in adult patients with specific antibody deficiency. Vaccine 24, 3574–3580. ( 10.1016/j.vaccine.2006.01.063) [DOI] [PubMed] [Google Scholar]
- 10.Filali-Mouhim A, et al. 2014. Integrated systems biology analysis reveals contrasting role for innate immune response genes in conferring risk of infection in RV144 trial. AIDS Res. Hum. Retroviruses 30 (Suppl. 1), A15–A16. ( 10.1089/aid.2014.5017a) [DOI] [Google Scholar]
- 11.Lambert ND, Haralambieva IH, Kennedy RB, Ovsyannikova IG, Pankratz VS, Poland GA. 2014. Polymorphisms in HLA-DPB1 are associated with differences in rubella virus-specific humoral immunity after vaccination. J. Infect. Dis. 211 (Suppl. 1), 898–905. ( 10.1093/infdis/jiu553) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan K, Cai W, Cao F, Sun H, Chen S, Xu R, Wei X, Shi X, Yan W. 2013. Genetic effects have a dominant role on poor responses to infant vaccination to hepatitis B virus. J. Hum. Genet. 58, 293–297. ( 10.1038/jhg.2013.18) [DOI] [PubMed] [Google Scholar]
- 13.Lu Y, Cheng Y, Yan W, Nardini C. 2014. Exploring the molecular causes of hepatitis B virus vaccination response: an approach with epigenomic and transcriptomic data. BMC Med. Genomics 7, 12 ( 10.1186/1755-8794-7-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Querec TD, et al. 2009. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 10, 116–125. ( 10.1038/ni.1688) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kruskall MS, Alper CA, Awdeh Z, Yunis EJ, Marcus-Bagley D. 1992. The immune response to hepatitis B vaccine in humans: inheritance patterns in families. J. Exp. Med. 175, 495–502. ( 10.1084/jem.175.2.495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He Y, et al. 2014. Updates on the web-based VIOLIN vaccine database and analysis system. Nucleic Acids Res. 42, D1124–D1132. () [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pulendran B, Kumar P, Cutler CW, Mohamadzadeh M, Van Dyke T, Banchereau J. 2001. Lipopolysaccharides from distinct pathogens induce different classes of immune responses in vivo. J. Immunol. 167, 5067–5076. ( 10.4049/jimmunol.167.9.5067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimman TG, Vandebriel RJ, Hoebee B. 2007. Genetic variation in the response to vaccination. Community Genet. 10, 201–217. ( 10.1159/000106559) [DOI] [PubMed] [Google Scholar]
- 19.Gupta D, Faridi MM, Aggarwal A, Kaur I. 2008. Seroprevalence of anti-Vi antibodies and immunogenicity of Typhim Vi vaccine in children. Hum. Vaccin. 4, 305–308. ( 10.4161/hv.4.4.5824) [DOI] [PubMed] [Google Scholar]
- 20.Sur D, et al. 2009. A cluster-randomized effectiveness trial of Vi typhoid vaccine in India. N. Engl. J. Med. 361, 403–405. ( 10.1056/NEJMe0905519) [DOI] [PubMed] [Google Scholar]
- 21.Vos Q, Lees A, Wu ZQ, Snapper CM, Mond JJ. 2000. B-cell activation by T cell-independent type 2 antigens as an integral part of the humoral immune response to pathogenic microorganisms. Immunol. Rev. 176, 154–170. ( 10.1034/j.1600-065X.2000.00607.x) [DOI] [PubMed] [Google Scholar]
- 22.Snapper CM. 2006. Differential regulation of protein- and polysaccharide-specific Ig isotype production in vivo in response to intact Streptococcus pneumoniae. Curr. Protein Pept. Sci. 7, 295–305. ( 10.2174/138920306778017972 [DOI] [PubMed] [Google Scholar]
- 23.Peterfi Z, Kocsis B. 2000. Comparison of blocking agents for an ELISA for LPS. J. Immunoassay 21, 341–354. ( 10.1080/01971520009349541) [DOI] [PubMed] [Google Scholar]
- 24.Staats HF, Kirwan SM, Whisnant CC, Stephenson JL, Wagener D, Majumder PP. 2010. Development of a bead immunoassay to measure Vi polysaccharide-specific serum IgG after vaccination with the Salmonella enterica Serovar Typhi Vi polysaccharide. Clin. Vaccine Immunol. 17, 412–419. ( 10.1128/CVI.00354-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. B 57, 289–300. [Google Scholar]
- 26.Purcell S, et al. 2007. PLINK: a toolset for whole genome association and population-based linkage analysis. Am. J. Hum. Genet. 81, 559–575. ( 10.1086/519795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens M, Smith N, Donnelly P. 2001. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68, 978–989. ( 10.1086/319501) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majumder PP, et al. 2009. Genetic determinants of immune-response to a polysaccharide vaccine for typhoid. HUGO J. 3, 17–30. ( 10.1007/s11568-010-9134-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909. ( 10.1038/ng1847) [DOI] [PubMed] [Google Scholar]
- 30.Majumder PP, Sarkar-Roy N, Staats H, Ramamurthy T, Maiti S, Chowdhury G, Whisnant CC, Narayanasamy K, Wagener DK. 2013. Genomic correlates of variability in immune response to an oral cholera vaccine. Eur. J. Hum. Genet. 21, 1000–1006. ( 10.1038/ejhg.2012.278) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan N, Ghousunnissa S, Jegadeeswaran SKM, Thiagarajan D, Hasnain SE, Mukhopadhyay S. 2007. Anti-B7–1/B7–2 antibody elicits innate-effector responses in macrophages through NF-kB-dependent pathway. Int. Immunol. 19, 477–486. ( 10.1093/intimm/dxm012) [DOI] [PubMed] [Google Scholar]
- 32.Kuehn HS, et al. 2014. Immune dysregulation in human subjects with heterozygous germline mutations in CTLA4. Science 345, 1623–1627. ( 10.1126/science.1255904) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alvarez JI. 2005. Inhibition of Toll like receptor immune responses by microbial pathogens. Front. Biosci. 10, 582–587. ( 10.2741/1554) [DOI] [PubMed] [Google Scholar]
- 34.Babu S, et al. 2009. Attenuation of Toll-like receptor expression and function in latent tuberculosis by coexistent filarial infection with restoration following antifilarial chemotherapy. PLoS Negl. Trop. Dis. 3, e489 ( 10.1371/journal.pntd.0000489) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Neil DA, et al. 1999. Expression and regulation of the human b-defensins hBD1 and hBD2 in intestinal epithelium. J. Immunol. 163, 6718–6724. [PubMed] [Google Scholar]
- 36.Agle KA, Vongsa RA, Dwinell MB. 2011. Chemokine stimulation promotes enterocyte migration through laminin-specific integrins. Am. J. Physiol. Gastrointest. Liver Physiol. 301, G968–G980. ( 10.1152/ajpgi.00208.2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ou G, Rompikuntal PK, Bitar A, Lindmark B, Vaitkevicius K, Wai SN, Hammarström M-L. 2009. Vibrio cholerae cytolysin causes an inflammatory response in human intestinal epithelial cells that is modulated by the PrtV protease. PLoS ONE 4, e7806 ( 10.1371/journal.pone.0007806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poggi A, et al. 2003. Migration of Vd1 and Vd2T cells in response to CXCR3 and CXCR4 ligands in healthy donors and HIV-1-infected patients competition by HIV-1 Tat . Blood 103, 2205–2213. ( 10.1182/blood-2003-08-2928) [DOI] [PubMed] [Google Scholar]
- 39.Kraal G, van der Laan LJW, Elomaa O, Tryggvason K. 2000. The macrophage receptor MARCO. Microbes Infect. 2, 313–316. ( 10.1016/S1286-4579(00)00296-3) [DOI] [PubMed] [Google Scholar]
- 40.Van der Laan LJW, et al. 1999. Regulation and functional involvement of macrophage scavenger receptor MARCO in clearance of bacteria in vivo. J. Immunol. 162, 939–947. [PubMed] [Google Scholar]
- 41.Sankala M, et al. 2002. Characterization of recombinant soluble macrophage scavenger receptor MARCO. J. Biol. Chem. 277, 33 378–33 385. ( 10.1074/jbc.M204494200) [DOI] [PubMed] [Google Scholar]
- 42.Fung EY, Smyth DJ, Howson JM, Cooper JD, Walker NM, Stevens H, Wicker LS, Todd JA. 2009. Analysis of 17 autoimmune disease-associated variants in type 1 diabetes identifies 6q23/TNFAIP3 as a susceptibility locus. Genes Immunity 10, 188–191. ( 10.1038/gene.2008.99) [DOI] [PubMed] [Google Scholar]