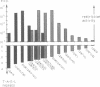Abstract
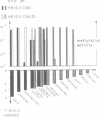
We have investigated the role of a four amino acid element that is repeated twice and three times, respectively, in the specificity polypeptides of the two allelic restriction-modification systems EcoR124 and EcoR124/3. We had earlier shown that this difference in amino acid sequence between the two systems is solely responsible for the different DNA sequence specificities of the two systems. The effect of single amino acid substitutions and small insertion and deletion mutations on restriction activity and modification specificity was determined in vivo by phage infection assays and in vitro by methylation of DNA with purified modification methylases. Mutant restriction-modification systems with changes in the number and the length of the central amino acid repeats exhibited decreased restriction activity and in some cases relaxed substrate specificity. Our data strongly support the idea that the repetitive amino acid motif in the specificity polypeptides forms part of a flexible interdomain linker. It may be responsible for positioning on the DNA the two major specificity polypeptide domains which are thought to contact independently the half sites of the split recognition sequences typical for all type I restriction-modification systems.
Full text
PDF




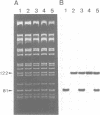
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler S. P., Nathans D. Studies of SV 40 DNA. V. Conversion of circular to linear SV 40 DNA by restriction endonuclease from Escherichia coli B. Biochim Biophys Acta. 1973 Mar 19;299(2):177–188. doi: 10.1016/0005-2787(73)90340-7. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Chamberlain J. P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979 Sep 15;98(1):132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of protein conformation. Biochemistry. 1974 Jan 15;13(2):222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- Cowan G. M., Gann A. A., Murray N. E. Conservation of complex DNA recognition domains between families of restriction enzymes. Cell. 1989 Jan 13;56(1):103–109. doi: 10.1016/0092-8674(89)90988-4. [DOI] [PubMed] [Google Scholar]
- Eskin B., Linn S. The deoxyribonucleic acid modification and restriction enzymes of Escherichia coli B. J Biol Chem. 1972 Oct 10;247(19):6192–6196. [PubMed] [Google Scholar]
- Evans R. M., Hollenberg S. M. Zinc fingers: gilt by association. Cell. 1988 Jan 15;52(1):1–3. doi: 10.1016/0092-8674(88)90522-3. [DOI] [PubMed] [Google Scholar]
- Firman K., Price C., Glover S. W. The EcoR124 and EcoR124/3 restriction and modification systems: cloning the genes. Plasmid. 1985 Nov;14(3):224–234. doi: 10.1016/0147-619x(85)90006-x. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace F. V., Cowan G. M., Murray N. E. EcoA and EcoE: alternatives to the EcoK family of type I restriction and modification systems of Escherichia coli. J Mol Biol. 1985 Nov 5;186(1):65–75. doi: 10.1016/0022-2836(85)90257-8. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace F. V., Murray N. E. Two DNA recognition domains of the specificity polypeptides of a family of type I restriction enzymes. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9368–9372. doi: 10.1073/pnas.83.24.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Horiuchi K., Zinder N. D. Cleavage of bacteriophage fl DNA by the restriction enzyme of Escherichia coli B. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3220–3224. doi: 10.1073/pnas.69.11.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JACOB F., WOLLMAN E. L. Etude génétique d'un bactériophage tempéré d'Escherichia coli. l. Le système genétique du bactériophage. Ann Inst Pasteur (Paris) 1954 Dec;87(6):653–673. [PubMed] [Google Scholar]
- Kannan P., Cowan G. M., Daniel A. S., Gann A. A., Murray N. E. Conservation of organization in the specificity polypeptides of two families of type I restriction enzymes. J Mol Biol. 1989 Oct 5;209(3):335–344. doi: 10.1016/0022-2836(89)90001-6. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Linder P., Doelz R., Gubler M., Bickle T. A. An anticodon nuclease gene inserted into a hsd region encoding a type I DNA restriction system. Nucleic Acids Res. 1990 Dec 11;18(23):7170–7170. doi: 10.1093/nar/18.23.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. E., Gough J. A., Suri B., Bickle T. A. Structural homologies among type I restriction-modification systems. EMBO J. 1982;1(5):535–539. doi: 10.1002/j.1460-2075.1982.tb01205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Nagaraja V., Shepherd J. C., Bickle T. A. A hybrid recognition sequence in a recombinant restriction enzyme and the evolution of DNA sequence specificity. Nature. 1985 Jul 25;316(6026):371–372. doi: 10.1038/316371a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Price C., Lingner J., Bickle T. A., Firman K., Glover S. W. Basis for changes in DNA recognition by the EcoR124 and EcoR124/3 type I DNA restriction and modification enzymes. J Mol Biol. 1989 Jan 5;205(1):115–125. doi: 10.1016/0022-2836(89)90369-0. [DOI] [PubMed] [Google Scholar]
- Price C., Shepherd J. C., Bickle T. A. DNA recognition by a new family of type I restriction enzymes: a unique relationship between two different DNA specificities. EMBO J. 1987 May;6(5):1493–1497. doi: 10.1002/j.1460-2075.1987.tb02391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sain B., Murray N. E. The hsd (host specificity) genes of E. coli K 12. Mol Gen Genet. 1980;180(1):35–46. doi: 10.1007/BF00267350. [DOI] [PubMed] [Google Scholar]
- Skrzypek E., Piekarowicz A. The EcoDXX1 restriction and modification system: cloning the genes and homology to type I restriction and modification systems. Plasmid. 1989 May;21(3):195–204. doi: 10.1016/0147-619x(89)90043-7. [DOI] [PubMed] [Google Scholar]
- Stalder R., Arber W. Characterization of in vitro constructed IS30-flanked transposons. Gene. 1989;76(2):187–193. doi: 10.1016/0378-1119(89)90159-5. [DOI] [PubMed] [Google Scholar]
- Suri B., Bickle T. A. EcoA: the first member of a new family of type I restriction modification systems. Gene organization and enzymatic activities. J Mol Biol. 1985 Nov 5;186(1):77–85. doi: 10.1016/0022-2836(85)90258-x. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeyar M. A., Weiner M. P., Hutton C. J., Batt C. A. A simple and rapid method for the selection of oligodeoxynucleotide-directed mutants. Gene. 1988 May 15;65(1):129–133. doi: 10.1016/0378-1119(88)90425-8. [DOI] [PubMed] [Google Scholar]
- Vinson C. R., Sigler P. B., McKnight S. L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989 Nov 17;246(4932):911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- Wood W. B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]