Abstract
Spinocerebellar ataxia type 6 (SCA6) is dominantly inherited neurodegenerative disease, caused by an expansion of CAG repeat encoding a polyglutamine (PolyQ) tract in the Cav2.1 voltage-gated calcium channel. Its key pathological features include selective degeneration of the cerebellar Purkinje cells (PCs), a common target for PolyQ-induced toxicity in various SCAs. Mutant Cav2.1 confers toxicity primarily through a toxic gain-of-function mechanism; however, its molecular basis remains elusive. Here, we studied the cerebellar gene expression patterns of young Sca6-MPI118Q/118Q knockin (KI) mice, which expressed mutant Cav2.1 from an endogenous locus and recapitulated many phenotypic features of human SCA6. Transcriptional signatures in the MPI118Q/118Q mice were distinct from those in the Sca1154Q/2Q mice, a faithful SCA1 KI mouse model. Temporal expression profiles of the candidate genes revealed that the up-regulation of genes associated with microglial activation was initiated before PC degeneration and was augmented as the disease progressed. Histological analysis of the MPI118Q/118Q cerebellum showed the predominance of M1-like pro-inflammatory microglia and it was concomitant with elevated expression levels of tumor necrosis factor, interleukin-6, Toll-like receptor (TLR) 2 and 7. Genetic ablation of MyD88, a major adaptor protein conveying TLR signaling, altered expression patterns of M1/M2 microglial phenotypic markers in the MPI118Q/118Q cerebellum. More importantly, it ameliorated PC loss and partially rescued motor impairments in the early disease phase. These results suggest that early neuroinflammatory response may play an important role in the pathogenesis of SCA6 and its modulation could pave the way for slowing the disease progression during the early stage of the disease.
Introduction
Spinocerebellar ataxias (SCAs) are a group of dominantly inherited disorders characterized by the loss of balance, progressive motor dysfunction and degeneration of the cerebellar Purkinje cells (PCs). Among these diseases, SCA1, 2, 3, 6, 7 and 17 are caused by an expansion of a CAG repeat encoding a polyglutamine (PolyQ) tract in their respective disease protein (1). Outside the PolyQ tract, the implicated proteins do not share homology, and each protein appears to be functionally distinct. In all disorders, the length of the CAG repeat correlates directly with disease severity: the longer the repeat, the more severe the symptoms and the earlier the onset. Although the cellular and molecular pathogenesis underlying these SCAs is not well understood, the resemblance in their pathologies raised the possibility that they might share common molecular mechanisms. Transcriptional analysis using faithful knockin (KI) mouse models of SCA1 and SCA7 revealed common cerebellar gene expression changes at their early disease stages (2).
SCA6 is caused by an expansion of a CAG repeat in the CaV2.1 voltage-gated calcium channel (3,4). The expanded CAG repeat per se does not affect the functional properties of the channels in SCA6 KI mice carrying expanded CAG repeats in the Cacna1a locus (5,6). The PolyQ-expanded protein confers virulence primarily through the toxic gain-of-function mechanism (7).
The major subcellular site of SCA6 pathogenesis remains elusive. The pathogenic mechanism of SCA6 appears to involve the endolysosomal degradation pathway because mutant Cav2.1 channels were found to accumulate in the lysosomes of SCA6 PCs (6). Consistent with this notion, the lack of Cathepsin B, a major lysosomal cysteine proteinase in the cerebellum, exacerbated PC loss and motor impairment in a KI mouse model of SCA6 (6). On the other hand, Du et al. (8) recently proposed that the carboxyl-terminal fragment of Cav2.1 (α1ACT), which contains the PolyQ tract, might be produced independently from the full-length Cav2.1 transcript by means of a cellular internal ribosomal entry site and translocation to the nucleus to function as a transcription factor. An expanded PolyQ tract may cause neuronal dysfunction by interrupting the transcriptional enhancer function of α1ACT.
To develop a model of SCA6, we previously generated several KI mouse models that expressed the mutant transcripts under the control of an endogenous promoter. Among them, Sca684Q/84Q mice expressed a PolyQ-expanded humanized Cav2.1 and developed slowly to progressive ataxia (5). Aggregation of mutant Cav2.1 channels was observed in the mutant PCs of these mice at an old age. Meanwhile, the MPI118Q/118Q mice expressed the mutant channels with longer PolyQ tracts at modestly enhanced levels because of the insertion of a splice acceptor site mutation at the beginning of exon 47 of the Cacna1a gene (6). Importantly, the MPI118Q/118Q mice recapitulated many key behavioral and neuropathological features of human SCA6, including progressive ataxia, selective PC loss, formation of non-ubiquitinated cytoplasmic inclusion and swollen PC axons.
We reasoned that these Sca6 KI models should replicate transcriptional alterations associated with SCA6 pathogenesis. Thus, using these models, we examined cerebellar gene expression to elucidate the molecular pathways associated with the pathogenesis of SCA6 and clarify whether common transcriptional alterations may be observed at the early symptomatic stages of several PolyQ-associated SCA mouse models.
Here, we showed that transcriptional signatures in the young MPI118Q/118Q mice were distinct from those in the young Sca1154Q/2Q mice, a faithful KI mouse model of SCA1. Our studies revealed that in the cerebellum of two Sca6 KI models, initiation of microglial activation preceded the onset of PC loss. Moreover, modulation of the neuroinflammtory response by genetically ablating myeloid differentiation factor 88 (MyD88), a major adaptor molecule essential for Toll-like receptor (TLR) signaling pathway, significantly delayed the progression of PC degeneration in the MPI118Q/118Q mice. Together, our data suggest a role of innate immune response in the pathogenesis of SCA6 as well as a novel mechanism that could be targeted to curb the disease progression.
Results
Cerebellar gene expression changes in early symptomatic MPI118Q/118Q mice
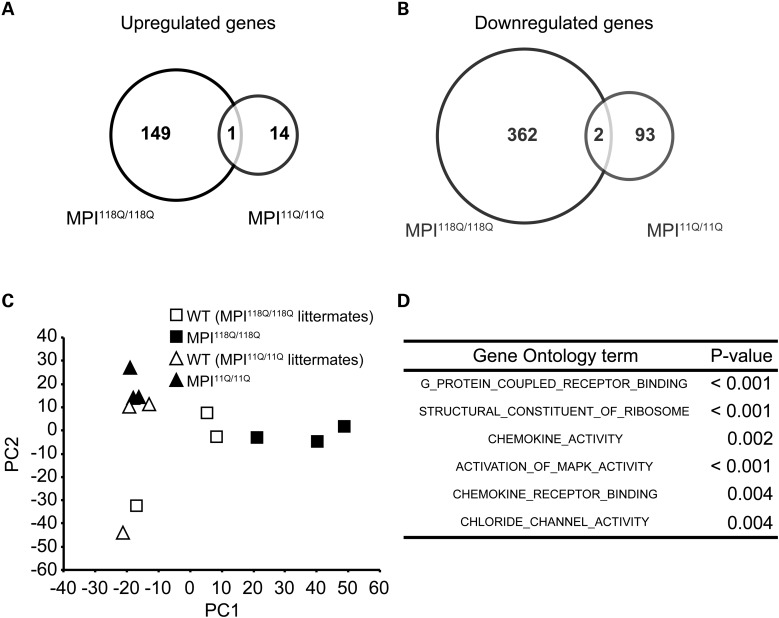
First, we surveyed transcriptional changes in the MPI118Q/118Q mouse cerebellum using the Affymetrix GeneChip Mouse Genome 430 2.0 Array. The experiments were conducted at an early symptomatic disease stage (6 weeks) when the mutant animals developed mild ataxia, and PC loss was yet to be observed (6). Comparison of the gene expression patterns between the MPI118Q/118Q mice (n = 3) and their wild-type (WT) littermates (n = 3) revealed that 150 genes were up-regulated and 364 genes were down-regulated in the MPI118Q/118Q mouse cerebellum (based on the cutoff values of P < 0.01 and fold change >1.5) (Fig. 1A and B and Supplementary Fig. S1).
Figure 1.
Gene expression profile of MPI118Q/118Q KI mice. (A) Venn diagram of genes that were significantly up-regulated in the MPI118Q/118Q mice but not in the MPI11Q/11Q mice. (B) Venn diagram of genes that were significantly down-regulated in the MPI118Q/118Q mice but not in the MPI11Q/11Q mice. (C) PCA of gene expression patterns in the MPI118Q/118Q and MPI11Q/11Q mice and their respective WT littermates. (D) GO enrichment analysis of the MPI118Q/118Q mice.
To exclude, the changes that were possibly caused by inserting a splice mutation and/or ‘humanization’ of exon 47 (6), we additionally compared the expression profiles of the 6-week-old MPI11Q/11Q KI mice (n = 3) with their WT littermates (n = 3). Both MPI11Q and MPI118Q alleles carry similar splice acceptor site mutations and humanized exon 47; however, the CAG repeat tract in the former has a normal (11 pure CAG repeats) size. In this analysis, we found that a total of 110 genes (15 up-regulated and 95 down-regulated genes) were differentially expressed (Fig. 1A and B and Supplementary Fig. S1). Of these, three genes (one up-regulated and two down-regulated genes) were commonly misregulated in both MPI11Q/11Q KI and MPI118Q/118Q mouse cerebella (Fig. 1A and B), suggesting that these changes were not specific to the MPI118Q/118Q mice. Therefore, we determined the up-/down-regulation of the remaining 149/362 genes as candidate changes caused by the expansion of a PolyQ tract in the channel.
To confirm the microarray data, quantitative real-time PCR (qPCR) analysis of these candidate genes was performed using the RNAs derived from another batch of 6-week-old MPI118Q/118Q mice and their WT littermates (n = 3, each). Of the 186 genes analyzed, the analysis successfully validated differential expression in the same direction for 150 genes (80.6%) (Supplementary Fig. S2A and B). To examine the overall difference among the gene expression profiles of the MPI118Q/118Q, MPI11Q/11Q and WT mice, we performed a principal component analysis (PCA). The gene expression profiles of the MPI118Q/118Q mice were clearly distinguished from those of the WT and MPI11Q/11Q mice by the first principal component (Fig. 1C). On the other hand, the MPI11Q/11Q and WT mice showed gene expression changes based on the second principal component (Fig. 1C). Therefore, the first principal component would reflect MPI118Q/118Q-specific gene expression patterns.
Comparison of cerebellar gene expression patterns with other SCA KI mouse models
Next, we compared the gene expression patterns of 6-week-old MPI118Q/118Q mice with those of Sca1154Q/2Q and Sca7266Q/5Q mice at their early symptomatic stages (2) (Supplementary Fig. S3). Both Sca1154Q/2Q and Sca7266Q/5Q mice carried expanded CAG repeats in the respective endogenous gene and faithfully recapitulated the phenotypes of SCA1 and SCA7, respectively. The mice used in these studies had a similar genetic background as those used in the present study, and the Affymetrix oligonucleotide arrays were used in all experiments. We confirmed significant concordance (Kendall's τ rank correlation coefficient 0.031, P < 0.001) between the Sca1154Q/2Q and Sca7266Q/5Q cerebellar expression patterns. We additionally found a weaker concordance in the cerebellar expression patterns between the MPI118Q/118Q and Sca7266Q/5Q mice, and unexpectedly, a pair of MPI118Q/118Q and Sca1154Q/2Q mice was found to be discordant (Table 1). We examined the gene expression levels of Igfbp5 in the MPI118Q/118Q cerebellum because the down-regulation of Igfbp5 has been implicated as a common pathogenic response in Sca1154Q/2Q and Sca7266Q/5Q mice (2). qPCR analysis, however, failed to detect any significant difference in the levels of Igfbp5 expression between the MPI118Q/118Q mice and their WT littermates between 4 and 8 weeks of age (Supplementary Fig. S4), suggesting that the insulin-like growth factor pathway may not be affected in SCA6. Overall, these results suggest that the transcriptional signatures in the MPI118Q/118Q cerebellum were distinct from those in the Sca1154Q/2Q cerebellum and that the molecular pathogenesis of SCA6 may be dissimilar from that in SCA1.
Table 1.
Concordance analysis in SCA model mice
| SCA model | Kendall’s τ rank correlation coefficient | P-Value |
|---|---|---|
| MPI118Q/118Q versus. Sca1154Q/2Q | −0.083 | <0.001 |
| MPI118Q/118Q versus. Sca7266Q/5Q | 0.020 | <0.001 |
| Sca1154Q/2Q versus. Sca7266Q/5Q | 0.031 | <0.001 |
Elevated expression of neuroinflammatory genes and microglial activation in the cerebellum of two Sca6 mouse models
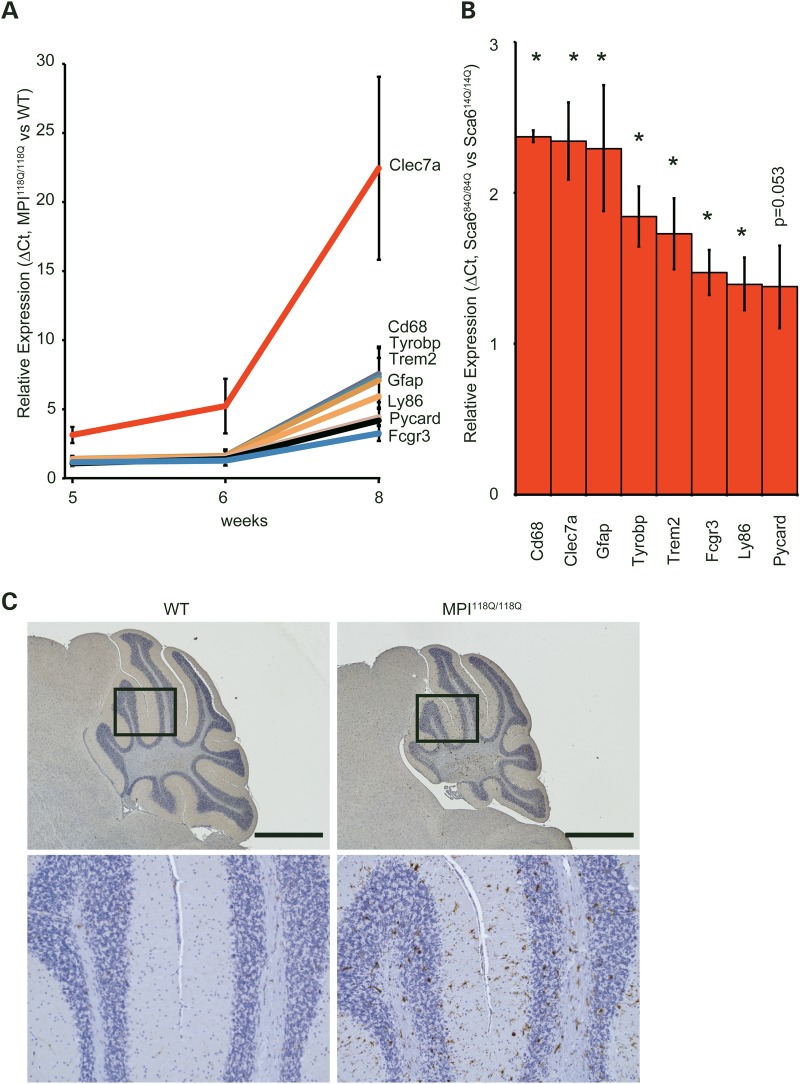
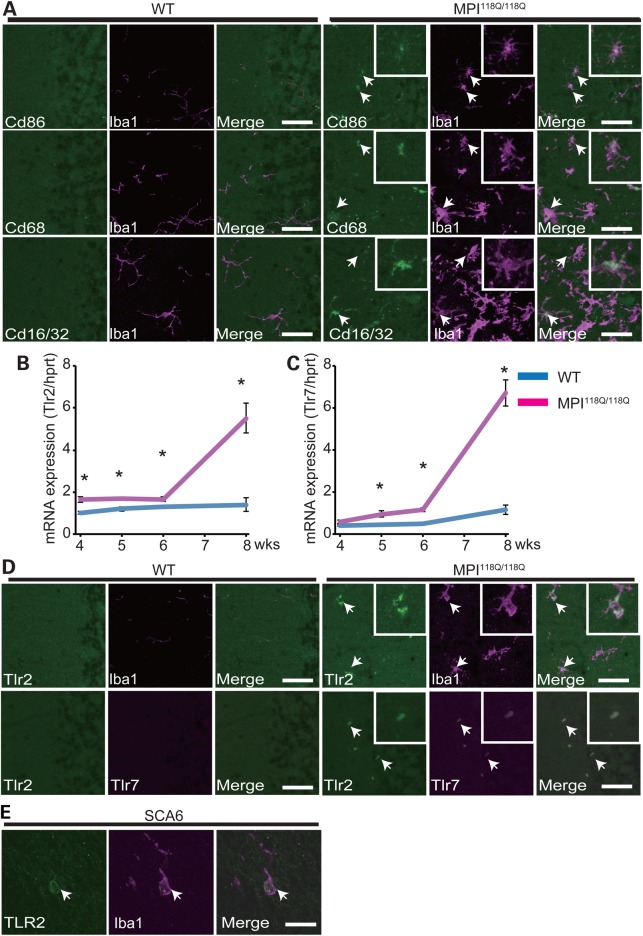
Gene ontology (GO) analysis revealed that the 6 GO terms were significantly over-represented for the MPI118Q/118Q cerebellum (Fig. 1D), whereas the similar analysis gave the over-representation of 36 terms for Sca1154Q/2Q cerebellum (Supplementary Fig. S5), further implying the difference in the gene expression patterns between these two models. As three terms ‘G-protein coupled receptor binding’, ‘chemokine activity’ and ‘chemokine receptor binding’ were significantly enriched, we hypothesized that the inflammatory response induced by the chemokine ligand binding to its receptor might be significantly enhanced in the MPI118Q/118Q mutant cerebellum. We therefore selected 8 neuroinflammation-related genes of 22 genes based on their relative expression levels at 6 weeks of age (Supplementary Fig. S2) and examined the temporal patterns of their expression in the MPI118Q/118Q cerebellum. It has been known that most of them are chiefly expressed in microglia and/or astroglia, and all of them showed more intense up-regulation at 8 weeks of age (9,10) (Fig. 2A). Several genes (Clec7a, Cd68, Gfap and Fcgr3) showed significantly enhanced expression even at 5 weeks of age when the neuritic degeneration of PC was not significant (6). These results suggest that a neuroinflammatory response involving microglial activation possibly occurs in the cerebellum of the MPI118Q/118Q mice at the early stage of the disease. To examine whether microglial activation occurred in the MPI118Q/118Q cerebellum, cerebellar sections with anti-Iba1 immunostaining were examined. As shown in Figs 2C and 3A, Iba1-positive hypertrophic microglial cells were detected in the cerebellum of 6-week-old MPI118Q/118Q mice. The majority (64.0 ± 2.6%) of these Iba1-positive cells showed co-immunofluorescence with Cd68, another microglial marker (Fig. 3A). Next, we examined whether a similar neuroinflammatory response might occur in the cerebellum of Sca684Q/84Q mice, which gradually developed progressive motor incoordination after 6 months of age but never showed a distinct neurodegenerative phenotype even at 24 months of age (5). qPCR analysis using the cerebellar RNAs obtained from 90-week-old animals showed that the expression levels of the 7 neuroinflammation-related genes were significantly increased in the Sca684Q/84Q mice, compared with their Sca614Q/14Q littermates (Fig. 2B). Immunohistochemical analysis using anti-Iba1 antibodies revealed that microglial activation was more intense in the cerebellum of the 90-week-old Sca684Q/84Q mice, compared with their Sca614Q/14Q littermates (Supplementary Fig. S6). We also examined whether similar microglial activation occurred in the cerebellum of aged heterozygous (MPI118Q/+) mice, as the majority of SCA6 patients are heterozygous for the mutant allele (11). Iba-1 immunohistochemistry elucidated the patchy distribution of activated microglia, which partially (87.8 ± 3.0%) showed Tlr2-coimmunofluorescence, in the 1-year-old MPI118Q/+ cerebellum but not in the cerebellum of WT littermates (Supplementary Fig. S8A and B). These results suggest that the neuroinflammatory response associated with microglial activation took place before the onset of PC degeneration in these two Sca6 mice models.
Figure 2.
(A) Temporal expression patterns of eight neuroinflammatory genes in the MPI118Q/118Q cerebellum. These eight genes (Clec7a, Cd68, Tyrobp, Trem2, Gfap, Ly86, Pycard and Fcgr3) were selected from the microarray data of the MPI118Q/118Q KI mice. The qPCR data were evaluated using the ΔCt method. The statistically significant differences were detected for Clec7a (5, 6 and 8 weeks, P < 0.02, <0.05 and <0.02, respectively), Cd68 (5, 6 and 8 weeks, P < 0.05, <0.05 and <0.02, respectively), Gfap (5, 6 and 8 weeks, P < 0.05, <0.05 and <0.02, respectively), Fcgr3 (5, 6 and 8 weeks, P < 0.05, <0.05 and <0.01, respectively), Ly86 (6 and 8 weeks, P < 0.05 and <0.01, respectively), Tyrobp (8 weeks, P < 0.05), Trem2 (8 weeks, P < 0.01) and Pycard (8 weeks, P < 0.02) by Student's t-test. (B) The expression of eight neuroinflammatory genes (Clec7a, Cd68, Tyrobp, Trem2, Gfap, Ly86, Pycard and Fcgr3) in the Sca684Q/84Q KI cerebellum at 90 weeks of age. The error bars represent standard errors. The asterisks indicate a statistically significant difference (P < 0.05). (C) Immunohistochemical detection of Iba1-positive microglia in the 6-week-old MPI118Q/118Q (right) and WT (left) mice. The cerebellar sections were immunostained with Iba1 antibodies and counter-stained with hematoxylin (scale bars indicate 1 mm.)
Figure 3.
Characterization of activated microglia in the cerebellum of the MPI118Q/118Q mice. (A) Double-immunofluorescence analysis of a microglial marker Iba1 and activated microglial marker (Cd86, Cd68 and Cd16/32) antibodies, stained cerebellar section of a 6-week-old MPI118Q/118Q mouse. (B–D) The infiltrating microglia express Tlr2 and Tlr7 in the Sca6 model. The scale bars indicate 50 μm. (B) qPCR analysis of Tlr2 in the MPI118Q/118Q cerebellum at 4, 5, 6 and 8 weeks of age. (C) qPCR analysis of Tlr7 in the MPI118Q/118Q cerebellum at 4, 5, 6 and 8 weeks of age. The asterisks indicate a statistically significant difference (P < 0.05). The error bars represent standard errors. (D) Co-immunofluorescence studies of the microglial marker Iba1 with Tlr2 (top) and Tlr2 with Tlr7 (bottom). The scale bars indicate 50 μm. The arrows indicate doubly labeled cells. (E) Co-immunofluorescence studies of the microglial marker Iba1 with TLR2 in SCA6 postmortem cerebellum. The scale bars indicate 25 μm. The arrows indicate doubly labeled cells.
Characterization of neuroinflammatory response in the cerebellum of Sca6 cerebella
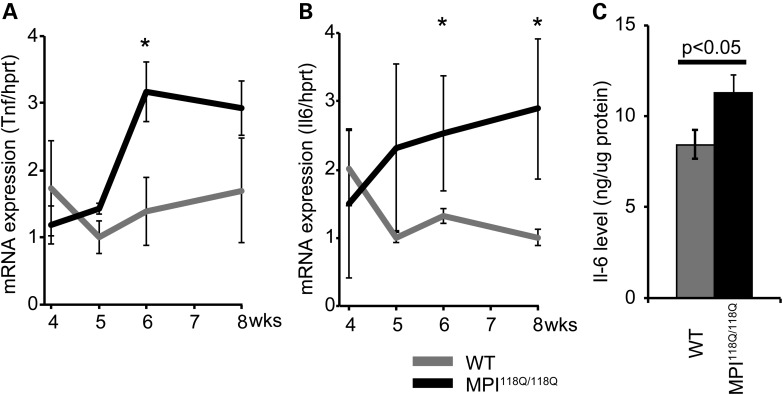
There is increasing evidence that activated microglia contribute to the pathogenesis of neurodegenerative diseases, such as Alzheimer's disease and Parkinson's disease (12,13); however, their role in SCA pathogenesis is unclear (14). Activated microglia can have heterogeneous phenotypes that range from ‘classically activated’ pro-inflammatory, cytotoxic M1 polarized cells to the alternatively activated anti-inflammatory, protective M2 polarized cells (15). To explore the role of early neuroinflammatory responses in Sca6 models, we assessed the expression of a variety of M1 and M2 markers in microglia in the MPI118Q/118Q cerebellum. Consistent with the increased transcriptional expression of Fcgr3 and Cd86 (Fig. 2A and Supplementary Fig. S9A), several Iba1-positive microglia revealed co-immunofluorescence with two M1 microglial markers, Cd16/32 (51.1 ± 3.3%) and Cd86 (73.4 ± 4.3%), in the cerebella of the 6-week-old MPI118Q/118Q mice (Fig. 3A). On the other hand, the mRNA expression levels of two M2 microglial markers, Arg1 and Sphk1, were not significantly changed in the MPI118Q/118Q cerebellum between 4 and 8 weeks of age (Supplementary Fig. S9B and C). We additionally examined the expression of tumor necrosis factor (Tnf) and interleukin-6 (Il-6) because M1 microglia produce these pro-inflammatory cytokines (16). qPCR analysis revealed that Tnf and Il-6 mRNAs were up-regulated in the cerebellum of the 6-week-old MPI118Q/118Q mice (Fig. 4A and B), and ELISA assay (Fig. 4C) showed that the Il-6 protein levels were higher in the cerebella of the 8-week-old MPI118Q/118Q mice, compared with their WT littermates. Altogether, these results suggest that M1-like pro-inflammatory microglia constituted a major part of the activated microglia in the cerebellum of the MPI118Q/118Q mice during the early stage of the disease.
Figure 4.
Expression analysis of two pro-inflammatory cytokines, Tnf and Il-6, in the MPI118Q/118Q cerebellum. The expression levels of the indicated pro-inflammatory cytokines, Tnf (A) and Il-6 (B) in the 4-, 5-, 6- and 8-week-MPI118Q/118Q cerebellum. The asterisks indicate P < 0.05. (C) The protein level of the cerebellum of the 8-week-MPI118Q/118Q mice measured using IL-6 ELISA. The error bars represent standard errors.
Next, we investigated the expression patterns of TLRs, major innate pattern recognition receptors expressed in microglia, because microglia initiate innate and adaptive immune responses through multiple TLRs, and TLRs have been associated with the pathogenesis of neurodegenerative diseases (12,13,17). Of nine known Tlr genes (18), which are commonly expressed in human and mice, the expression levels of Tlr1, Tlr2 and Tlr7 mRNAs, but not those of the other Tlr mRNAs, were significantly increased in the cerebella of the 8-week-old MPI118Q/118Q mice, compared with those in their age-matched WT littermates (n = 3, each) (Fig. 3B and C and Supplementary Fig. S10A). Meanwhile, qPCR analysis of the nine Tlr genes in the cerebella of the 90-week-old Sca684Q/84Q mice revealed that the expression levels of the Tlr2 and Tlr7 mRNAs were significantly enhanced compared with the levels in age-matched Sca614Q/14Q mice (Supplementary Fig. S10B), suggesting that the up-regulation of Tlr2 and Tlr7 mRNAs may reflect a common microglial response in these two Sca6 models. In the MPI118Q/118Q cerebellum, these changes were initiated before 5 weeks of age and augmented at 8 weeks of age (Fig. 3B and C). We additionally performed immunohistochemical analysis to examine the cellular localization of Tlr2 and Tlr7 in the MPI118Q/118Q cerebellum. Double-immunofluorescence studies with antibodies against Tlr2 and Iba1 revealed co-localization of the two markers in the microglial cells (Fig. 3D). In addition, double-immunofluorescence analysis with antibodies against Tlr2 and Tlr7 showed co-localization of these two Tlrs whereas Gfap-positive astroglia failed to show co-immunofluorescence with Tlr7 (Supplementary Fig. S7B), suggesting that both Tlr2 and Tlr7 were chiefly expressed in the activated microglia in the MPI118Q/118Q cerebellum (Fig. 3D). Similar co-immunofluorescence of Tlr2 and Iba1 or Tlr2 and Tlr7 were observed in the cerebellum of 2-year-old Sca684Q/84Q mice (Supplementary Fig. S10C) and MPI118Q/+ mice (Supplementary Fig. S8B). Altogether, these results suggest that Tlr2 and Tlr7 may play a role in activating microglia in the two Sca6 mouse models. We also conducted a double-immunofluorescence analysis on the postmortem cerebellar tissues of an SCA6 patient and found that Iba1-positive cells were scattered in the molecular layer and many of these cells showed co-IF with TLR2 (Fig. 3E).
Expression levels of Taf, Btg1, Pmca2 and Grn mRNAs in the MPI118Q/118Q cerebella
Using chromatin immunoprecipitation-based cloning, Du et al. (8) recently identified Taf, Btg1, Pmca2 and Grn as candidate genes for the transcriptional targets of α1ACT. They also reported that the mRNA expression levels of Btg1 were reduced in the cerebellar tissues of two SCA6 patients, compared with that in the normal control. To examine a similar phenomenon in the cerebellum of the MPI118Q/118Q mouse, we prepared mRNAs from the cerebella of the MPI118Q/118Q mice (from 4 to 8 weeks of age) and examined the expression levels of these genes using qPCR. We found that there was no significant difference in the relative amount of these RNAs, normalized to Hprt, between the MPI118Q/118Q mice and their WT littermates (Supplementary Fig. S11A) except for the expression of Grn in 8-week-old cerebellum. Similar results were obtained when we examined their expression in the cerebella of 90-week-old Sca684Q/84Q mice (Supplementary Fig. S11B). These results suggest that the reduced expression of these candidate genes may not be necessary for inducing PC degeneration in SCA6.
Deficiency of MyD88 alters the expression of microglial markers in the cerebellum of MPI-118Q KI mice
Upon ligand bindings, both Tlr2 and 7 have been shown to trigger inflammatory response in a MyD88-dependent manner (18). To test the possibility that Tlr2 and/or Tlr7-mediated microglial activation may play a role in the pathogenesis of SCA6, we next crossbred the MPI-118Q mice with well-characterized MyD88 deficient mice (MyD88−/−) (19). Doubly heterozygous matings of MPI118Q/+/MyD88+/− mice produced nine different genotypes with all of them showing frequencies closely matching the progeny expectation (data not shown). All the genotypes, including MPI118Q/118Q; MyD88−/− and MPI118Q/118Q; MyD88+/+, were indistinguishable in terms of cage behavior until P28.
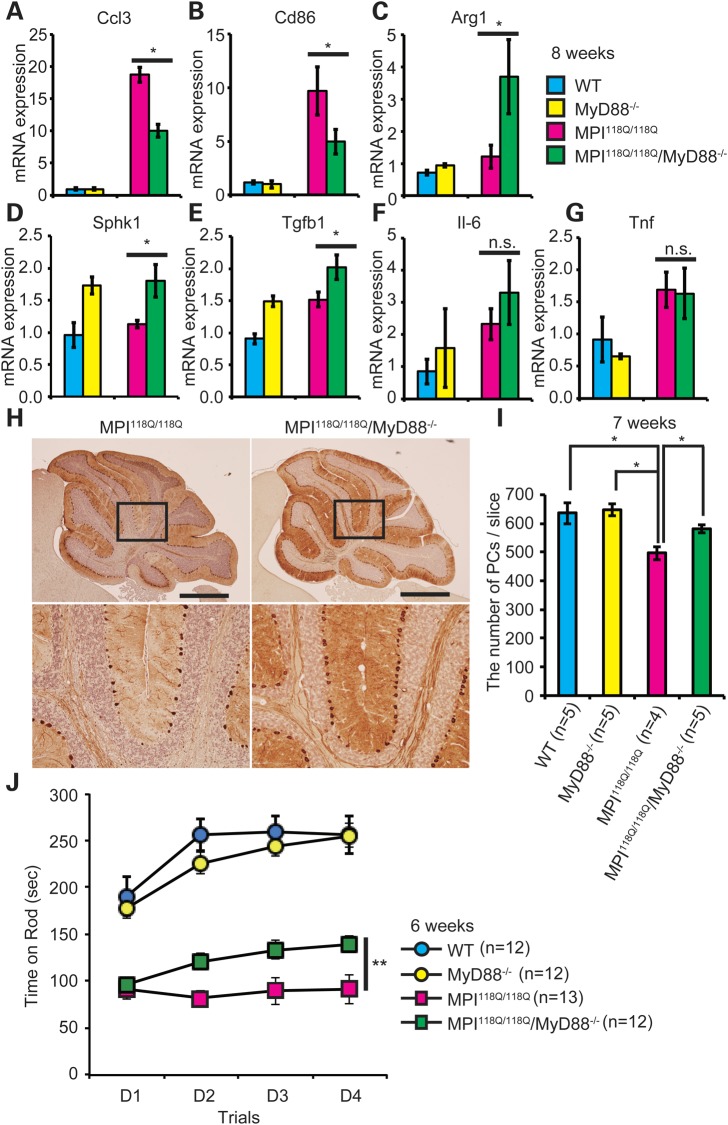
To examine whether the lack of MyD88 affected the neuroinflammatory response in the MPI-118Q cerebellum, we first evaluated the degree of microgliosis by quantifying Iba1 immunofluorescence in the cerebellum. As shown in Supplementary Fig. S12A and B, Iba1 immunofluorescence did not significantly differ in the cerebellum of MPI118Q/118Q; MyD88−/− (DM) in comparison with that of MPI118Q/118Q, suggesting that MyD88 deficiency did not affect the number of activated microglia in MPI118Q/118Q mice. Macrophage undergoes classical M1 activation upon stimulation by TLR ligands and IFN-γ (20), and recent studies have shown the evidence that MyD88-dependent signaling may influence macrophage polarization (21,22). We therefore next examined the expression levels of M1/M2 microglial phenotypic markers in the MPI118Q/118Q; MyD88−/− cerebellum by qPCR analysis. Interestingly, the expression levels of Cd86 were significantly decreased (Fig. 5B), whereas those of Arg1 and Sphk1 were significantly increased in the DM cerebellum (Fig. 5C and D) when compared with the MPI118Q/118Q cerebellum. We also examined the gene expression of several cytokines and chemokine, and found that the expression of Tgfb1, which is an anti-inflammatory cytokine known as an M2 marker, was significantly elevated in the DM cerebellum (Fig. 5E). In contrast, the expression levels of Ccl3, an M1 chemokine (23), were significantly suppressed (Fig. 5A) although those of Il-6 and Tnf were not significantly altered (Fig. 5F and G) in the DM cerebellum. Overall, these results suggest that MyD88 deficiency altered the neuroinflammatory response and perhaps shifted the balance of microglial activation toward M2 phenotype with a concomitant reduction of M1 response in the cerebellum of MPI-118Q KI mice.
Figure 5.
Deficiency of MyD88 ameliorated PC death and motor impairment in MPI118Q/118Q mice. (A–F) qPCR analyses of Ccl3 (A), Cd86 (B), Arg1 (C), Sphk1 (D), Tgfb1 (E), Il-6 (F) and Tnf (G) in the WT (n = 4), MyD88−/− (n = 3), MPI118Q/118Q (n = 5) and MPI118Q/118Q; MyD88−/− (n = 4) cerebellum at 8 weeks of age. Statistically significant difference P < 0.05 by Student's t-test. (H) Anti-calbindin Abs immunohistochemistry on cerebellar sections of 7-week-old WT, MyD88−/−, MPI118Q/118Q and MPI118Q/118Q; MyD88−/− mice (scale bars indicate 1 mm). (I) Quantitative analysis showed that a decrease in the number of PCs was more pronounced in the MPI118Q/118Q cerebellum compared with the MPI118Q/118Q; MyD88−/− cerebellum at 7-week-old age (*P < 0.05 by Student's t-test). Error bars indicate SEM. (J) Rota-rod analysis of the DM mice. Mice were trained for four trials per day for 4 days (D1–D4). Error bars indicate SEM. MPI118Q/118Q versus MPI118Q/118Q/MyD88−/− (**repeated measures ANOVA, group effect; F = 11.608, P = 0.006, time effect; F = 0.296, P = 0.693, group and time interaction; F = 4.052, P = 0.045).
Genetic ablation of MyD88 ameliorates PC loss and improves motor performance in MPI-118Q KI mice at the early stage of the disease
To evaluate the effects of MyD88 deficiency on PC loss in the MPI118Q/118Q mice, we quantified the number of remaining PCs on mid-sagittal cerebellar sections stained with anti-calbindin Abs. The number of remaining PCs was significantly larger in the DM cerebellum in comparison with that in the MPI118Q/118Q cerebellum (Fig. 5H and I) at 7 weeks of age, suggesting that the lack of MyD88 ameliorated PC loss in the MPI-118Q KI mice. To examine the effects of loss of MyD88 expression on the ataxic phenotype seen in the MPI118Q/118Q mice, we tested the animals on the accelerating Rota-rod at 6 weeks of age (Fig. 5J). MyD88−/− mice performed similarly to WT littermates, whereas the motor impairments of MPI118Q/118Q were reproduced in these litters. Importantly, DM mice performed significantly better than MPI118Q/118Q mice (Fig. 5J, **repeated measures ANOVA, group effect; F = 11.608, P = 0.0061), indicating that the lack of MyD88 expression improved motor coordination in the MPI118Q/118Q mice at 6 weeks. We further examined the effects of MyD88 deficiency at a later age. At 10 weeks of age, when more than 35% of total PCs were already lost in the MPI118Q/118Q cerebellum (6) (Supplementary Fig. S13B), the degree of PC loss was comparable between the two groups (Supplementary Fig. S13A and B). Overall, these results suggested that MyD88 deficiency ameliorated PC degeneration during the early stage of the disease in MPI118Q/118Q mice.
Discussion
This study showed that a set of genes involved in neuroinflammation and innate immune processes was up-regulated in the cerebella of both Sca684Q/84Q KI and MPI118Q/118Q mice before significant PC degeneration began. These changes were concomitant with microglial activation and became pronounced in the MPI118Q/118Q mice as they aged. Significance of such an early induction of neuroinflammatory responses in these Sca6 models was underscored by the observation that the loss of MyD88 gene, a key player in TLR signaling, not only altered the patterns of microglial activation but also ameliorated PC loss in the early phase of neurodegeneration in MPI118Q/118Q mice. Thus, we propose that the innate immune response could now emerge as a potential target in PC degeneration at least in the early phase of SCA6.
Comparison of microarray data between Sca1154Q/2Q and MPI118Q/118Q KI mice suggested that although cerebellar PCs are a major target for PolyQ-induced neurotoxicity in both SCA1 and SCA6, distinct pathogenic cascade(s) may be involved in the pathogenesis of each disease. This seems consistent with the notion that native biochemistry and function of the host protein are crucial for pathogenesis of PolyQ diseases (24). GO analysis showed that a couple of GO terms, such as ‘structural-constituent-of ribosome’ and ‘chloride-channel-function’, are specifically enriched in the MPI118Q/118Q cerebellum. Detailed analysis of these pathways would be useful for further dissecting the molecular pathogenesis of SCA6.
Innate immune response has been thought to play an important role in the progression of major neurodegenerative diseases, such as Alzheimer's disease and Parkinson's disease, but little is known about the role of microglial activation in the pathogenesis of SCAs (14). Our data indicated the predominance of M1-like microglia in the MPI118Q/118Q cerebellum. Although M1/M2 classification oversimplifies the complexity of microglial activation (15,25,26), these results suggest that the majority of activated microglia might mediate the pro-inflammatory cytotoxic response in the Sca6 models. Enhanced microglial activation and persistent skewing toward M1-like polarization in the MPI118Q/118Q cerebellum may be associated with PC loss because persistence of an inflammatory stimulus can result in neurodegenerative pathology (27). Partial recovery of PC loss in the DM cerebellum seemed to support the hypothesis, because it was concomitant with the elevated expression levels of three M2 markers (Arg1, Sphk1 and Tgfb1) as well as the decreased expression of Cd86 and Ccl3.
It is of interest that the expression levels of Tgfb1 were significantly increased in the DM cerebellum. It has been shown that astroglial overexpression of TGF-β1 protects adult mice against neurodegeneration (28), whereas the enhanced TGF-β signaling was shown to rescue PolyQ-induced cytotoxicity in a neuroblastoma cell line expressing the mutant androgen receptor (29). Thus, the elevated expression of Tgfb1 might play a protective role against neurotoxicity induced by the mutant Cav2.1 channel.
A characteristic neuropathological feature in SCA6 is the formation of a non-ubiquitinated inclusion in the cytoplasm (i.e. the cell body or cell processes) of the PCs. We previously revealed the evidence that the pathogenesis of SCA6 involves the endolysosomal degradation pathway. Lysosomal accumulation or ‘storage’ of mutant Cav2.1 appeared to correlate with disease progression in MPI118Q/118Q mice (6), leading to the hypothesis that the pathogenic mechanism underlying SCA6 might overlap, at least in part, with that in lysosomal storage diseases (LSDs). It has been shown that neuroinflammation is a common and universal feature in LSDs with CNS pathology (30), and in several mouse models of LSDs, such as GM1 gangliosidosis and globoid cell leukodystrophy, neuroinflammation was activated prior to the onset of clinical signs and neuronal loss (31). Thus, the early onset of the neuroinflammatory response in the two Sca6 models appeared to further support the hypothesis.
We found enhanced expression of the TLR family members Tlr2 and Tlr7 in the activated microglia in both the MPI118Q/118Q and Sca684Q/84Q cerebellum at the early stages of their conditions. The activation of Tlr2 on microglia has been linked with the pathogenesis of other neurodegenerative diseases, and it has been proposed that amyloid-β and mutant superoxide dismutase-1 can bind to Tlr2 on microglia and promote microglial activation (32). Up-regulation of Tlr2 mRNA was detected in the brains of other neurodegenerative disease mouse models, such as APP/PS1 KI mice and Thy1-A30P α-synuclein transgenic mice (33,34). Furthermore, one recent report identified enhanced Tlr2 expression on microglia as the earliest change in the cerebellum of twitcher mice, a murine model for globoid cell leukodystrophy (31). Therefore, although the cause of Tlr2 up-regulation and its ligands may differ in the respective disease, it appears that the neuroinflammatory response mediated through Tlr2 and its downstream signal transducer MyD88 (18) might be a common mechanism during the early phase of neurodegeneration.
Overexpression of the SCA6-expanded α1ACT fragment in mice was reported to cause mild ataxia at their late age without developing PC loss (8). It is hard to directly compare those α1ACT Tg mice with our MPI118Q/118Q mice because many parameters regarding mutant protein expression, such as protein length (fragment versus full-length), promotor (exogenous versus endogenous), repeat length (disease range versus hyper-expanded) and expression levels (highly enhanced versus modestly enhanced), were completely different between the two models. It should be noted, however, that we could not detect such a fragment in the cerebellum of the MPI118Q/118Q mice (6), suggesting that α1ACT fragment may not be essential for reproducing SCA6 in mice.
Recently, Cvetanovic et al. (35) reported that the initiation of microglial activation preceded the PC loss in the Sca1154Q/2Q cerebellum, but its significance is largely unknown. Future investigation on the role of neuroinflammatory response in the SCA1 pathogenesis would be expected to further advance our understanding on the role of neuroinflammation in the pathogenesis of SCAs.
In conclusion, our gene expression analysis of the two Sca6 KI mouse models revealed distinct patterns of cerebellar gene expression including an induction of innate immune response during the early disease stage. Modulation of innate immune response by genetically ablating MyD88 partially rescued the progression of PC loss and motor incoordination in MPI118Q/118Q mice in its early stage. These beneficial effects seemed to be disease stage-dependent as it was no longer apparent at 10 weeks. Further analysis is necessary to disclose the mechanism for such dependence but one possible reason is that the role of microglial response in the SCA6 pathogenesis may change as the disease progresses. In any case, our results suggest that neuroinflammation may play a pivotal role in the early SCA6 pathogenesis and that modulation of the innate immune response in the brain may pave the way for slowing disease progression during its early stage.
Materials and Methods
Mice
Sca6 KI models including, MPI118Q/118Q, MPI11Q/11Q, Sca684Q/84Q and Sca614Q/14Q mice were reported in the literature (5,6). MyD88 knockout mice (19) on a C57BL/6 background were obtained from Oriental Bio Service (Kyoto, Japan). All animal procedures were performed in accordance with the protocols approved by the Animal Experiment Committee of the Tokyo Medical and Dental University (0090078).
Human sample
Postmortem cerebellar tissues were obtained from an SCA6 patient who developed ataxia at 63 years of age and was 83 years old at the time of her death. Immunohistochemical analysis with antibodies raised against carboxyl-terminal part of Cav2.1 (36,37) revealed abundant cytoplasmic inclusion formation in the remaining PCs in this sample.
Gene expression analysis
Total RNA was extracted from the WT, MPI118Q/118Q and MPI11Q/11Q mice (n = 3 in each group) using the RNeasy mini kit (QIAGEN, Hilden, Germany). The quality of the RNA samples was assessed using Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA), and specimens with RNA integrity number values >7.5 were used. Sample labeling, hybridization and fluorescence detection were performed according to the manufacturer’'s instructions, using the GeneChip Mouse Genome 430 2.0 arrays (Affymetrix, Santa Clara, CA, USA). The gene expression data obtained were then normalized using the Robust Multi-array Average method (38). To avoid possible bias from differences in age, separate normalization was performed for 6-week-old mice. We used R statistical software version 2.1 (http://www.r-project.org/) and Bioconductor packages (http://www.bioconductor.org/) for analyzing the microarray data, unless otherwise noted. The gene expression data are available at the Gene Expression Omnibus database under the accession ID GSE61908. Statistical analysis of differently expressed genes (DEGs) was performed using the Linear Models for Microarray Data (limma) package (39). Genes with a P-value of <0.01 and with a 1.5-fold difference in the average expression levels were considered as DEGs. For biological interpretation of the gene expression changes, we used the Gene Set Enrichment Analysis version 2.0.14 with MSigDB 4.0 (40). The gene set category C5.all, which is based on the ontology terms defined in the GO database (http://www.geneontology.org/), was used.
Reverse transcription-quantitative PCR
The first strand of cDNA was synthesized using Superscript II Reverse Transcriptase (Life Technologies) and oligod(T)12–18 (Life Technologies). Five micrograms of total RNA was used in the RT reaction. The reaction of qPCR was quantified with Light cycler 480 SYBR Green I Master Mix (Roche Applied Science) and the Light cycler 480 instrument II system (Roche Applied Science).
Antibodies
Anti-Calbindin (McAB300), Iba1, Cd68 (clone ED-1), Tlr2 (clone T2.5) and Tlr7 antibodies were from Swant, Wako, Santa Cruz, Hycult biotech and Abcam, respectively. Anti-Cd86 (B7-2) and Cd16/32 (clone 93) were from eBiosciences.
Immunohistochemistry
Formalin-fixed, paraffin-embedded, 6-μm-thick sections were deparaffinized, exposed to microwave in a citrate buffer, and treated with primary antibodies. After treating with primary antibodies, the section was washed with phosphate-buffered saline (PBS) and incubated with biotin-conjugated secondary antibodies (Vector laboratories) for 1 h and colored with 3,3′-diaminobenzidine solution (simple stain DAB, Nichirei Bioscience). For double-immunofluorescence labeling, formalin-fixed, O.C.T. compound (Tissue-Tek)-embedded frozen sections were washed with PBS and incubated with primary antibodies in Dako REAL (Dako) overnight. After washing with PBS containing 0.01% Tween 20, the primary antibodies were detected using Alexa 488 or 555 conjugated secondary antibodies (Life Technologies) diluted at 1:500 for 3 h at room temperature. The sections were then washed, counter-stained with 4′,6-diamidine-2′-phenylindole dihydrochloride (Roche diagnostics) and examined under a confocal microscope (LSM 510META, Carl Zeiss).
Rota-rod test
Animals were placed on an accelerated rotating rod (UGO Basile) in four trials every day for a period of 4 days, as described in Ref. (41).
Statistical analysis
Student's t-test was used for quantifying qPCR, the biochemical analysis and immunohistochemical analysis. A one-way repeated measures ANOVA was used for quantifying the Rota-rod analysis.
Supplementary Material
Funding
This work was supported by the following: Grants-in-Aid for Scientific Research (grant number ‘25282241’) (to H.M. and K.W.), a Grant-in-Aid for Scientific Research on Innovative Areas (Foundation of Synapse and Neurocircuit Pathology, grant number ‘25110712’) (to K.W.) and a Grant-in-Aid for Young Scientists (grant number ‘26860661’) (to T.A.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan; and a grant from the Core Research for Evolutional Science and Technology (CREST), the Japan Science and Technology Agency (to H.T. and H.M.). Funding to pay the Open Access publication charges for this article was provided by the Japan Science and Technology Agency.
Supplementary Material
Acknowledgements
We acknowledge Drs Jennifer Gatchel, Chad Shaw and Huda Zoghbi for helping with the concordance analysis.
Conflict of Interest statement. None declared.
References
- 1.Orr H.T., Zoghbi H.Y. (2007) Trinucleotide repeat disorders. Annu. Rev. Neurosci., 30, 575–621.2. [DOI] [PubMed] [Google Scholar]
- 2.Gatchel J.R., Watase K., Thaller C., Carson J.P., Jafar-Nejad P., Shaw C., Zu T., Orr H.T., Zoghbi H.Y. (2008) The insulin-like growth factor pathway is altered in spinocerebellar ataxia type 1 and type 7. Proc. Natl Acad. Sci. U.S.A., 105, 1291–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhuchenko O., Bailey J., Bonnen P., Ashizawa T., Stockton D.W., Amos C., Dobyns W.B., Subramony S.H., Zoghbi H.Y., Lee C.C. (1997) Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the alpha(1A)-voltage-dependent calcium channel. Nat. Genet., 15, 62–69. [DOI] [PubMed] [Google Scholar]
- 4.Ishikawa K., Tanaka H., Saito M., Ohkoshi N., Fujita T., Yoshizawa K., Ikeuchi T., Watanabe M., Hayashi A., Takiyama Y. et al. (1997) Japanese families with autosomal dominant pure cerebellar ataxia map to chromosome 19p13.1-p13.2 and are strongly associated with mild CAG expansions in the spinocerebellar ataxia type 6 gene in chromosome 19p13.1. Am. J. Hum. Genet., 61, 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watase K., Barrett C.F., Miyazaki T., Ishiguro T., Ishikawa K., Hu Y., Unno T., Sun Y., Kasai S., Watanabe M. et al. (2008) Spinocerebellar ataxia type 6 knockin mice develop a progressive neuronal dysfunction with age-dependent accumulation of mutant Ca(v)2.1 channels. Proc. Natl Acad. Sci. U.S.A., 105, 11987–11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Unno T., Wakamori M., Koike M., Uchiyama Y., Ishikawa K., Kubota H., Yoshida T., Sasakawa H., Peters C., Mizusawa H. et al. (2012) Development of Purkinje cell degeneration in a knockin mouse model reveals lysosomal involvement in the pathogenesis of SCA6. Proc. Natl Acad. Sci. U.S.A., 109, 17693–17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kahle J.J., Gulbahce N., Shaw C.A., Lim J., Hill D.E., Barabasi A.L., Zoghbi H.Y. (2011) Comparison of an expanded ataxia interactome with patient medical records reveals a relationship between macular degeneration and ataxia. Hum. Mol. Genet., 20, 510–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du X.F., Wang J., Zhu H.P., Rinaldo L., Lamar K.M., Palmenberg A.C., Hansel C., Gomez C.M. (2013) Second cistron in CACNA1A gene encodes a transcription factor mediating cerebellar development and SCA6. Cell, 154, 118–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor P.R., Brown G.D., Reid D.M., Willment J.A., Martinez-Pomares L., Gordon S., Wong S.Y.C. (2002) The beta-glucan receptor, dectin-1, is predominantly expressed on the, surface of cells of the monocyte/macrophage and neutrophil lineages. J. Immunol., 169, 3876–3882. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K., Rochford C.D.P., Neumann H. (2005) Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J. Exp. Med., 201, 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi H., Ishikawa K., Tsutsumi T., Fujigasaki H., Kawata A., Okiyama R., Fujita T., Yoshizawa K., Yamaguchi S., Tomiyasu H. et al. (2004) A clinical and genetic study in a large cohort of patients with spinocerebellar ataxia type 6. J. Hum. Genet., 49, 256–264. [DOI] [PubMed] [Google Scholar]
- 12.Tahara K., Kim H.D., Jin J.J., Maxwell J.A., Li L., Fukuchi K. (2006) Role of toll-like receptor signalling in A beta uptake and clearance. Brain, 129, 3006–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panaro M.A., Lofrumento D.D., Saponaro C., De Nuccio F., Cianciulli A., Mitolo V., Nicolardi G. (2008) Expression of TLR4 and CD14 in the central nervous system (CNS) in a MPTP mouse model of Parkinson’s-like disease. Immunopharm. Immunot., 30, 729–740. [DOI] [PubMed] [Google Scholar]
- 14.Rüb U., Schöls L., Paulson H., Auburger G., Kermer P., Jen J.C., Seidel K., Korf H.W., Deller T. (2013) Clinical features, neurogenetics and neuropathology of the polyglutamine spinocerebellar ataxias type 1, 2, 3, 6 and 7. Prog. Neurobiol., 104, 38–66. [DOI] [PubMed] [Google Scholar]
- 15.David S., Kroner A. (2011) Repertoire of microglial and macrophage responses after spinal cord injury. Nat. Rev. Neurosci., 12, 388–399. [DOI] [PubMed] [Google Scholar]
- 16.Willment J.A., Lin H.H., Reid D.M., Taylor P.R., Williams D.L., Wong S.Y.C., Gordon S., Brown G.D. (2003) Dectin-1 expression and function are enhanced on alternatively activated and GM-CSF-treated macrophages and are negatively regulated by IL-10, dexamethasone, and lipopolysaccharide (vol 171, pg 4569, 2003). J. Immunol., 171, 6297. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen M.D., D’Aigle T., Gowing G.V., Julien J.P., Rivest S. (2004) Exacerbation of motor neuron disease by chronic stimulation of innate immunity in a mouse model of amyotrophic lateral sclerosis. J. Neurosci., 24, 1340–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akira S., Uematsu S., Takeuchi O. (2006) Pathogen recognition and innate immunity. Cell, 124, 783–801. [DOI] [PubMed] [Google Scholar]
- 19.Adachi O., Kawai T., Takeda K., Matsumoto M., Tsutsui H., Sakagami M., Nakanishi K., Akira S. (1998) Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity, 9, 143–150. [DOI] [PubMed] [Google Scholar]
- 20.Sica A., Mantovani A. (2012) Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest., 122, 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanke M.L., Angle A., Kielian T. (2012) MyD88-dependent signaling influences fibrosis and alternative macrophage activation during Staphylococcus aureus biofilm infection. PLoS ONE, 7, e42476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Halloran S., O'Leary A., Kuijper T., Downer E. (2014) MyD88 acts as an adaptor protein for inflammatory signalling induced by amyloid-β in macrophages. Immunol. Lett., 162, 109–118. [DOI] [PubMed] [Google Scholar]
- 23.Mantovani A., Sica A., Sozzani S., Allavena P., Vecchi A., Locati M. (2004) The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol., 25, 677–686. [DOI] [PubMed] [Google Scholar]
- 24.Orr H.T. (2012) Polyglutamine neurodegeneration: expanded glutamines enhance native functions. Curr. Opin. Genet. Dev., 22, 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heneka M.T., Kummer M.P., Stutz A., Delekate A., Schwartz S., Vieira-Saecker A., Griep A., Axt D., Remus A., Tzeng T.C. et al. (2013) NLRP3 is activated in Alzheimer's disease and contributes to pathology in APP/PS1 mice. Nature, 493, 674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kroner A., Greenhalgh A.D., Zarruk J.G., Passos dos Santos R., Gaestel M., David S. (2014) TNF and increased intracellular iron alter macrophage polarization to a detrimental M1 phenotype in the injured spinal cord. Neuron, 83, 1098–1116. [DOI] [PubMed] [Google Scholar]
- 27.Heneka M.T., Kummer M.P., Latz E. (2014) Innate immune activation in neurodegenerative disease. Nat. Rev. Immunol., 14, 463–477. [DOI] [PubMed] [Google Scholar]
- 28.Brionne T.C., Tesseur I., Masliah E., Wyss-Coray T. (2003) Loss of TGF-beta 1 leads to increased neuronal cell death and microgliosis in mouse brain. Neuron, 40, 1133–1145. [DOI] [PubMed] [Google Scholar]
- 29.Ehata S., Johansson E., Katayama R., Koike S., Watanabe A., Hoshino Y., Katsuno Y., Komuro A., Koinuma D., Kano M.R. et al. (2011) Transforming growth factor-beta decreases the cancer-initiating cell population within diffuse-type gastric carcinoma cells. Oncogene, 30, 1693–1705. [DOI] [PubMed] [Google Scholar]
- 30.Vitner E.B., Platt F.M., Futerman A.H. (2010) Common and uncommon pathogenic cascades in lysosomal storage diseases. J. Biol. Chem., 285, 20423–20427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snook E.R., Fisher-Perkins J.M., Sansing H.A., Lee K.M., Alvarez X., MacLean A.G., Peterson K.E., Lackner A.A., Bunnell B.A. (2014) Innate immune activation in the pathogenesis of a murine model of globoid cell leukodystrophy. Am. J. Pathol., 184, 382–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao W.H., Beers D.R., Henkel J.S., Zhang W., Urushitani M., Julien J.P., Appel S.H. (2010) Extracellular mutant SOD1 induces microglial-mediated motoneuron injury. Glia, 58, 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wirths O., Breyhan H., Marcello A., Cotel M.C., Bruck W., Bayer T.A. (2010) Inflammatory changes are tightly associated with neurodegeneration in the brain and spinal cord of the APP/PS1KI mouse model of Alzheimer's disease. Neurobiol. Aging, 31, 747–757. [DOI] [PubMed] [Google Scholar]
- 34.Letiembre M., Liu Y., Walter S., Hao W.L., Pfander T., Wrede A., Schulz-Schaeffer W., Fassbender K. (2009) Screening of innate immune receptors in neurodegenerative diseases: a similar pattern. Neurobiol. Aging, 30, 759–768. [DOI] [PubMed] [Google Scholar]
- 35.Cvetanovic M., Ingram M., Orr H., Opal P. (2015) Early activation of microglia and astrocytes in mouse models of spinocerebellar ataxia type 1. Neuroscience, 289, 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishiguro T., Ishikawa K., Takahashi M., Obayashi M., Amino T., Sato N., Sakamoto M., Fujigasaki H., Tsuruta F., Dolmetsch R. et al. (2010) The carboxy-terminal fragment of alpha(1A) calcium channel preferentially aggregates in the cytoplasm of human spinocerebellar ataxia type 6 Purkinje cells. Acta Neuropathol., 119, 447–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishikawa K., Fujigasaki H., Saegusa H., Ohkoshi N., Shoji S., Tanabe T., Mizusawa H. (1999) Abnormal expression of alpha 1A-voltage-dependent calcium channel protein in brains of spinocerebellar ataxia type 6 (SCA6). Am. J. Hum. Genet., 65, A276. [DOI] [PubMed] [Google Scholar]
- 38.Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. (2003) Summaries of affymetrix GeneChip probe level data. Nucleic Acids Res., 31, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smyth G.K. (2005) limma: Linear Models for Microarray Data. Springer, New York, NY. [Google Scholar]
- 40.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. U.S.A., 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watase K., Gatchel J.R., Sun Y.L., Emamian E., Atkinson R., Richman R., Mizusawa H., Orr H.T., Shaw C., Zoghbi H.Y. (2007) Lithium therapy improves neurological function and hippocampal dendritic arborization in a spinocerebellar ataxia type 1 mouse model. PLoS Med., 4, 836–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.