Abstract
Mutations affecting specific splicing regulatory elements offer suitable models to better understand their interplay and to devise therapeutic strategies. Here we characterize a meaningful splicing model in which numerous Hemophilia B-causing mutations, either missense or at the donor splice site (5′ss) of coagulation F9 exon 2, promote aberrant splicing by inducing the usage of a strong exonic cryptic 5′ss. Splicing assays with natural and artificial F9 variants indicated that the cryptic 5′ss is regulated, among a network of regulatory elements, by an exonic splicing silencer (ESS). This finding and the comparative analysis of the F9 sequence across species showing that the cryptic 5′ss is always paralleled by the conserved ESS support a compensatory mechanism aimed at minimizing unproductive splicing. To recover splicing we tested antisense oligoribonucleotides masking the cryptic 5′ss, which were effective on exonic changes but promoted exon 2 skipping in the presence of mutations at the authentic 5′ss. On the other hand, we observed a very poor correction effect by small nuclear RNA U1 (U1snRNA) variants with increased or perfect complementarity to the defective 5′ss, a strategy previously exploited to rescue splicing. Noticeably, the combination of the mutant-specific U1snRNAs with antisense oligonucleotides produced appreciable amounts of correctly spliced transcripts (from 0 to 20–40%) from several mutants of the exon 2 5′ss. Based on the evidence of an altered interplay among ESS, cryptic and the authentic 5′ss as a disease-causing mechanism, we provide novel experimental insights into the combinatorial correction activity of antisense molecules and compensatory U1snRNAs.
Introduction
Mutations affecting pre-mRNA splicing account for a significant proportion of human genetic disorders (1). The number of these mutations is largely underestimated because the impact on this finely orchestrated process (2–4) is hardly predictable, particularly when considering nucleotide variations within exons (5,6). As a matter of fact, the splicing process output depends on the type of regulatory elements affected by these mutations and on the network of interactions defining an exon in the specific gene sequence context. Taking into account the frequency of each class of splicing regulatory elements (7–9), it is predictable that their interplay, in the presence of inherited mutations, will produce an extremely variegate series of additive, negative or compensatory effects. The intriguing role of Exonic Splicing Enhancer and Silencer (ESE and ESS, respectively), sequences that substantially contribute to alternative splicing by regulating the splice site selection, is still largely unexplored, particularly when the proper exon inclusion is mandatory to encode a functional protein. The ESSs, which are estimated to be significantly less frequent in ‘real’ exons as compared with ‘pseudoexons’ (8), appear to evolve with a strength that correlates with that of the donor splice site (5′ss) (10). On the other hand, the influence of ESSs on cryptic 5′ss and their interplay with mutations could produce aberrant mRNA patterns (5,11–13) through poorly explored combinations of mechanisms, the definition of which would in turn help implementing our knowledge of ESS physiological functions.
In this study we provide an intriguing model for positive and negative interactions among regulatory elements leading to severe Hemophilia B (HB; OMIM 306900) forms.
First, we demonstrate that several mutations at the exon 2 5′ss of coagulation F9 gene produce aberrant splicing by inducing the usage of a strong exonic cryptic 5′ss, which is regulated by adjacent exonic splicing regulatory elements, both ESE and ESS. Intriguingly, numerous HB-causing missense changes at the ESS (14) increase the usage of the cryptic 5′ss. This provides the rationale for the use of antisense molecules (15–20) and variants of the small nuclear RNA U1 (U1snRNA) (21–31) for a tailored correction approach, exploiting the well-known advantages of RNA-based strategies (24). Antisense molecules, such as chemically modified oligonucleotides or engineered U7snRNA, or modified U1snRNA have been reported to counteract mutations and restore exon inclusion by masking either cryptic sites or improving exon definition in several models of human diseases.
For the first time we demonstrate that splicing correction, in the presence of a strong cryptic 5′ss and of mutations at the authentic one, can be achieved only by the combined effect of modified U1snRNA and antisense oligonucleotides, which also contribute to better understand the mechanisms underlying proper exon definition through the involvement of positive and negative splicing regulatory elements.
Results
We investigated a panel of mutations occurring at the positions +3 (c.252+3G>C), +5 (c.252 + 5G>A, c.252 + 5G>C, c.252 + 5G>T) and +6 (c.252 + 6T>C) of the 5′ss of exon 2 of F9 gene (Fig. 1A) that have been reported in the HB mutation database (http://www.factorix.org). All mutations cause severe HB forms characterized by factor IX (FIX) levels in plasma below 1%.
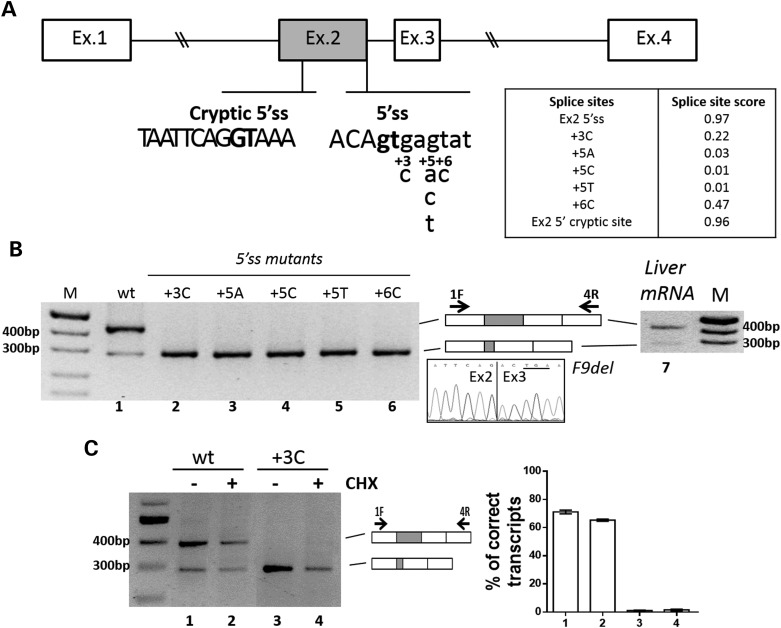
Figure 1.
Mutations at the authentic 5′ss induce the usage of a cryptic 5′ss in exon 2. (A) Schematic representation of the F9 genomic sequence cloned as minigene in the pCDNA3 vector. Exonic and intronic sequences are represented by boxes and lines, respectively. The sequences, with exonic and intronic nucleotides in upper and lower cases, respectively, report (i) the authentic 5′ss with the positions of the investigated changes detailed below, and (ii) the cryptic 5′ss. The highly conserved dinucleotide GT of authentic or cryptic 5′ss is in bold. The table reports the scores of the authentic and mutated 5′ss, and of the cryptic 5′ss. (B) Evaluation of F9 alternative splicing patterns in BHK cells transiently transfected with minigenes (left panel, lanes 1–6) or in human liver (right panel, lane 7). The schematic representation of the transcripts (with exons not in scale), and of primers used for the RT-PCR (arrows), is reported in the middle panel. Amplified products were separated on 2% agarose gel. M, 100 bp molecular weight marker. The chromatogram reports the sequence of the shorter transcript (F9del), which demonstrates the usage of the cryptic 5′ss in exon 2. The newly created nonsense triplet in exon 3 is underlined. (C) Evaluation of F9 alternative splicing patterns in BHK cells transiently transfected with pF9wt (lanes 1–2) and pF9+3C (lanes 3–4) alone (−) or upon treatment with 100 µm CHX added 4 h post-transfection. Amplified products were separated on 2% agarose gel. M, 100 bp molecular weight marker. The schematic representation of the transcripts, and of primers used for the RT-PCR (arrows), is reported in the central panel. Histograms report the relative percentage of correct transcripts, which is expressed as mean ± standard deviation (SD).
The F9 exon 2 5′ss mutations induce usage of an exonic cryptic 5′ss regulated by cis-elements, including an ESS
To investigate the effect of F9 exon 2 5′ss mutations on pre-mRNA splicing, we created a F9 minigene including the genomic region spanning exon 1 through 4 (Fig. 1A). Expression of the wild-type minigene in mammalian Baby Hamster Kidney (BHK) cells indicated that exon 2 was not completely included into the mature mRNA (Fig. 1B, lane 1). RT-PCR analysis and sequencing revealed a smaller amount (21 ± 6%) of an alternative transcript that originates from the usage of a cryptic 5′ss located 104 bp upstream of the authentic 5′ss (Fig. 1B). This gives rise to an aberrant F9 mRNA form (F9del) including only the 5′ portion (60 bp) of exon 2 that accounts for a deleted and frame-shifted mRNA harboring a premature nonsense triplet at position c.151 (underlined). To rule out the possibility that this result depends on minigene artifacts, we performed RT-PCR on human liver mRNA and demonstrated a comparable amount (∼20%) of the F9del transcript (Fig. 1B, lane 7). These findings are consistent with the computational analysis that predicts the cryptic 5′ss in exon 2 with a score (0.96) comparable to the authentic one (0.97) (Fig. 1A).
The mutations at the authentic F9 exon 2 5′ss were then inserted into the validated F9wt minigene. Expression studies demonstrated that all mutations, reducing the 5′ss score, lead to the virtually exclusive usage of the exonic cryptic 5′ss and synthesis of F9del transcripts (Fig. 1B, lanes 2–6).
However, the nonsense-mediated RNA decay (NMD) might lead to underestimate the levels of the F9del form, or prevent the evaluation of that potentially arising from exon 2 skipping, both frame-shifted and introducing a premature nonsense triplet in exon 3. To rule out this possibility, we performed splicing assays in the presence of 100 µm cycloheximide (CHX), a well-established NMD inhibitor. As shown in Figure 1C, the splicing patterns in cells expressing the F9wt (lanes 1–2) or the +3C (lanes 3–4) constructs were virtually unaffected upon CHX treatment. This finding points toward a negligible impact of NMD on splicing patterns of the F9 minigenes under investigation and is consistent with the fact that the premature nonsense triplet in the F9del form occurs in a very short exon (26 nucleotides), a condition that would prevent NMD (32).
The bioinformatics analysis of F9 exon 2 predicted several ESEs and also a Silencer element (AAAGAGGT) upstream of the cryptic 5′ss (Fig. 2A) and revealed that this region is highly conserved among mammals (Supplementary Material, Fig. S1). Intrigued by the hypothesis that a silencer favors the usage of the authentic 5′ss by down-regulating the cryptic one, we investigated its presence by exploiting natural models consisting of all missense mutations (c.135G>C, c.135G>T, c.137G>A, c.137G>C, c.138G>T, c.138G>C) (Fig. 2A) at the putative ESS sequence so far reported in moderate/severe HB patients (http://www.factorix.org). All of them, in addition to introduce amino acid substitutions (p.Lys45Asn, p.Lys45Asn, p.Arg46Lys, p.Arg46Lys, p.Arg46Ser, p.Arg46Ser), are predicted to slightly reduce the score of the putative ESS (from 0.92 to 0.75–0.80). Investigation of splicing patterns revealed that, with the exception of the c.135G>T, these mutations decreased the levels of normally spliced transcripts from 80 to ∼40% (Fig. 2B, lanes 1–6).
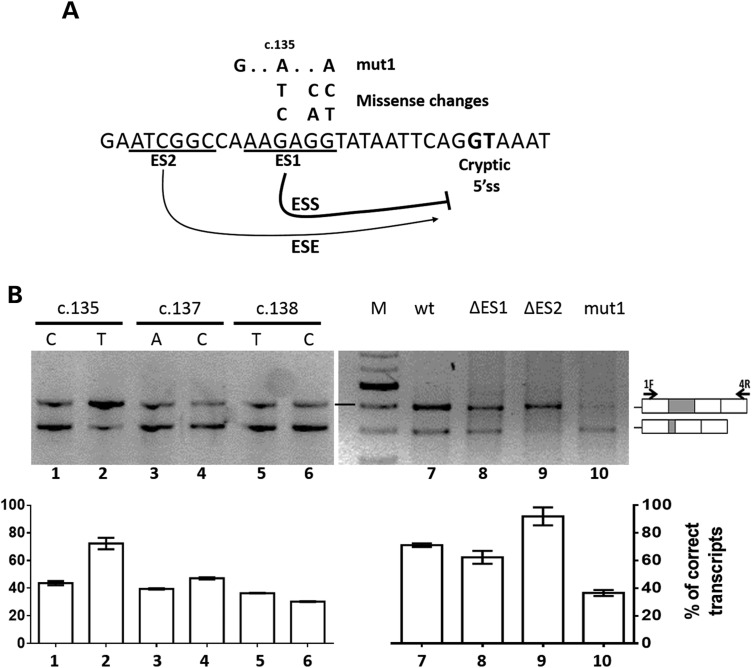
Figure 2.
The F9 exon 2 5′ss mutations induce usage of an exonic cryptic 5′ss regulated by cis-elements, including an ESS. (A) Sequence of the F9 exon 2 region upstream of the cryptic 5′ss (bold). The underlined sequences indicate the examers (ES1, ES2) deleted to identify functional regulatory elements, which appear to act as silencer and enhancer (curved arrows) of the cryptic 5′ss. The nucleotide changes introduced in the ES1 element, either missense or artificial (mut1), are reported above together with a reference position in the FIX cDNA. (B) Evaluation of F9 splicing patterns in BHK cells transiently expressing the F9wt (lane 7), missense variants (lane 1–6), mut1 (lane 10) or the deleted mutants ΔES1 (lane 8) and ΔES2 (lane 9). Histograms report the relative percentage of correct transcripts and results are expressed as mean ± SD from three independent experiments. The schematic representation of the transcripts, and of primers used for the RT-PCR (arrows), is reported in the right panel.
The function of this region was further explored by well-established approaches such as (i) multiple mutagenesis (three nucleotide changes, F9mut1) or (ii) deletion (six nucleotides, F9ΔES1) (Fig. 2A). The triple mutant F9mut1 resulted in the preferential usage of the cryptic 5′ss and, conversely, in the decreased use of the authentic 5′ss (Fig. 2B, lane 10). Coherently, albeit to a minor extent, the deletion in the F9ΔES1 minigene decreased the usage of the authentic 5′ss (lane 8). RNA secondary structure might influence these splicing outputs (33). Our prediction analysis (RNAfold) suggests that the accessibility of the cryptic 5′ss is maintained in the presence of nucleotide changes but appears to be reduced upon E1 deletion (data not shown). Therefore, it is tempting to speculate that the modest impact on splicing of F9ΔES1 minigene would arise from a compensatory effect in which the usage of the cryptic 5′ss is favored by the removal of the silencer element and concurrently disfavored by a reduced accessibility of the U1 snRNP.
Intriguingly, a 6-nucleotide deletion upstream of the putative ESS (F9ΔES2, Fig. 2A), made as control, decreased usage of the cryptic 5′ss (Fig. 2B, lane 9), consistent with the bioinformatics prediction of ESEs in this region.
Taken together data from these multiple and complementary mutants point toward the presence of a network of positive and negative regulatory elements, with the natural missense changes mapping an ESS sequence down-regulating the adjacent cryptic 5′ss.
Antisense oligonucleotides masking the cryptic 5′ss did not recover the selection of the defective F9 exon 2 5′ss but rescued missense mutations within the ESS
We designed and synthesized two synthetic 2′-Omethylphosphothioate antisense oligoribonucleotides (AON360, AON353) to mask and inhibit the cryptic 5′ss (Fig. 3A). To set up experimental conditions, we tested the AONs on the splicing pattern of the F9wt construct. At 10 nm, the AON360 had its maximal effect and remarkably reduced the relative amount of the F9del form as compared with the correctly spliced one (Fig. 3B; Supplementary Material, Fig. S2A, lanes 1–2), thus indicating that it effectively inhibited the usage of the cryptic 5′ss. On the other hand, the AON360, designed on the F9wt sequence, and hence not perfectly matching the mutated ESS, was also able to partially recover correct splicing of the F9 missense variants (Fig 3B; Supplementary Material, Fig. S2A, lanes 3–14), sharing with the F9wt a functional authentic 5′ss.
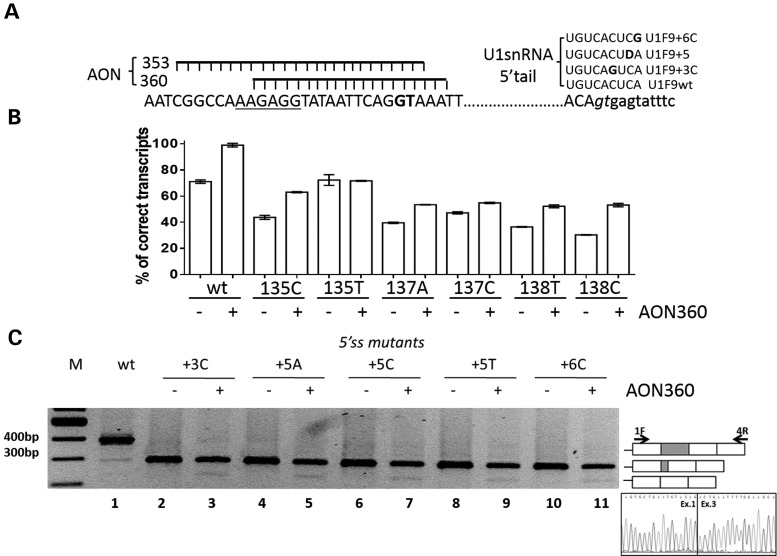
Figure 3.
Antisense oligonucleotides masking the cryptic 5′ss did not correct defective 5′ss mutants. (A) Sequence of the F9 gene region containing the ESS (underlined), the cryptic (bold) and the authentic (italics) 5′ss together with the schematic representation of the two antisense oligoribonucleotides AON353 and AON360 (left panel) and the sequence of the 5′ tail of the compensatory U1snRNAs (right panel, where D = U,G or A). (B) Evaluation of F9 alternative splicing patterns in BHK cells transiently expressing the F9 missense minigene variants alone (−) or with 10 nm AON360 (+). RT-PCR was conducted as in Figure 1, and a representative gel is provided in the Supplementary Material, Figure S2. Histograms report the relative percentage of correct transcripts and results are expressed as mean ± SD from three independent experiments. (C) Evaluation of F9 alternative splicing patterns in BHK cells expressing minigenes variants of the authentic 5′ss alone (−) or with 10 nm AON360 (+). RT-PCR products obtained with primers 1F and 4R (arrows in the right panel) were separated on 2% agarose gel. M, 100 bp molecular weight marker. The schematic representation of the transcripts is reported on the right. The chromatogram reports the sequence of the transcript lacking exon 2.
Conversely, the antisense approach failed to correct the F9 exon 2 5′ss mutations, and in addition induced complete skipping of exon 2 (Fig. 3C, lanes 2–11). Comparable results were obtained with the AON353 whereas a scrambled AON was ineffective (Supplementary Material, Fig. S2B, lanes 1–17).
U1snRNAs with increased complementarity to the defective F9 exon 2 5′ss have poor effects on the correct 5′ss selection
We designed a modified U1snRNA with perfect complementarity to the authentic F9 exon 2 5′ss (U1F9wt) (Fig. 3A), which therefore has a single mismatch for each mutant. Co-expression of the U1F9wt with each 5′ss mutant did not result in any appreciable rescue of splicing (Fig. 4A, lanes 2–11). On the other hand, the U1F9wt had a negligible effect on exon 2 definition even in the F9wt context, characterized by a strong authentic 5′ss (Supplementary Material, Fig. S3A).
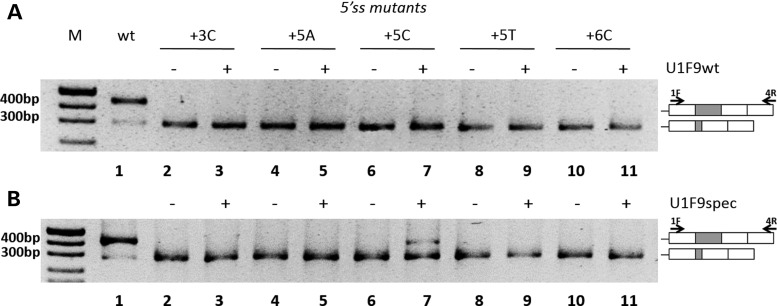
Figure 4.
U1snRNAs with improved (U1F9wt) or perfect (U1F9spec) complementarity to the mutated F9 exon 2 5′ss have poor correction effects. Evaluation of F9 alternative splicing patterns in BHK cells transiently transfected with minigenes without (−) or with (+) the pU1F9wt (A) or the pU1F9spec (B). The schematic representation of the transcripts and of primers used for the RT-PCR (arrows) are reported on the right. Amplified fragments were separated on 2% agarose gel. M, 100 bp molecular weight marker.
To further increase complementarity to mutant 5′ss, we created U1snRNAs specific for each mutation (U1F9spec; Fig. 3A). Complementation assays revealed that only the mutation +5C was partially corrected by the corresponding U1F9 + 5C variant, as indicated by the appreciable fraction of correct transcripts (26 ± 3% of total transcripts) (Fig. 4B, lane 6 versus 7). Conversely, the mutant-specific U1snRNA variants had no significant effects on aberrant splicing patterns generated by the expression of all the other 5′ss variants.
AONs and modified U1snRNAs have combinatorial correction effects
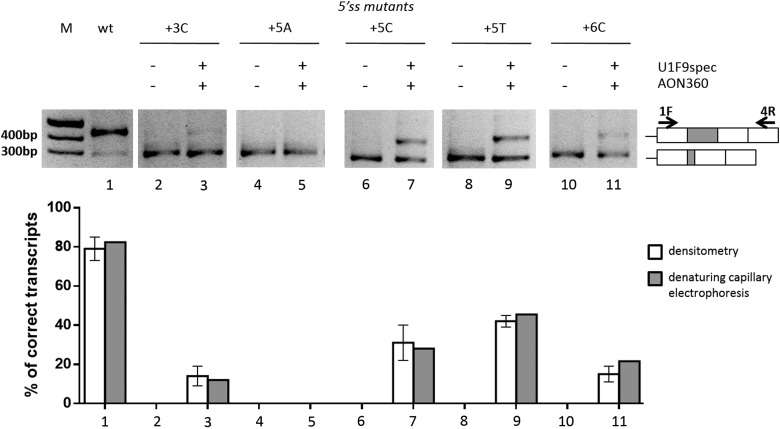
With the exception of the +5C mutant, neither the AON360, masking the cryptic 5′ss, nor the modified U1snRNAs, restoring complementarity to the 5′ss, were able to rescue exon 2 inclusion. We therefore explored the combination of these approaches to test their possible concerted effects. Strikingly, co-transfection of the AON360 with the mutant-specific U1snRNAs resulted in appreciable rescue of correctly spliced transcripts for the +3C (14 ± 5% of total transcripts), +5T (42 ± 3%) and +6C (15 ± 4%) mutations (Fig. 5, lanes 3, 9, 11) but not the +5A mutation (lane 5). The combined approach did not significantly improved the correction of the +5C mutant (31 ± 9%, lane 7) as compared with the U1F9spec alone (26 ± 3%). To rule out the potential confounding effect of heteroduplex forms, the RT-PCR fragments were fluorescently labeled and separated by denaturing capillary electrophoreses (Supplementary Material, Fig. S4), which provided comparable results (Fig. 5, gray histograms).
Figure 5.
Combinatorial correction effects of AON360 and mutation-specific U1snRNA variants. Evaluation of F9 alternative splicing patterns in BHK cells transiently transfected with minigenes alone (−) or with the combination of AON360 (10 nm) and the U1F9spec variants (+). RT-PCR and electrophoresis were conducted as in Figure 4. The schematic representation of the transcripts and of primers used (arrows) are reported on the right. The white histograms report the percentage of correct transcripts expressed as means ± SD from three independent experiments. The RT-PCR fragments were also fluorescently labeled and separated by denaturing capillary electrophoreses (Supplementary Material, Fig. S4). The gray histograms report the percentage of correct transcripts evaluated by analysis of peaks.
To provide insights into the specificity, we also tested the U1F9spec together a scrambled AON on the +5T mutant, which failed to rescue splicing (Supplementary Material, Fig. S3B). The U1-F9wt in combination with AON360 was ineffective for all mutants (data not shown).
Regarding the missense variants of the ESS, the additive contribution of the compensatory U1snRNA to the productive AON splicing rescue was reproducibly marginal (data not shown), a finding consistent with the presence of a normal authentic 5′ss.
Discussion
A crucial step in exon definition is represented by the selection of the proper 5′ss, which occurs in the presence of numerous sequences that resemble 5′ss but are not or inefficiently used in normal conditions (21). Among the signature of regulatory elements governing this choice and the definition of a given exon, the ESSs, which are less represented in constitutive exons (8), possess an intriguing and still poorly defined role.
Here we provide a paradigmatic example of the role of an unconventional ESS both in normal and pathological conditions, either in the presence of missense changes or of mutations at the authentic 5′ss. In particular, by exploiting the F9 exon 2 context as study model, we demonstrated that both mutation types, through distinct mechanisms, shift the balance toward the usage of a strong exonic cryptic 5′ss.
It has been suggested that the strength of ESS correlates with that of the 5′ss (10). Intriguingly, the comparative analysis of the F9 exon 2 across species (Supplementary Material, Fig. S1) (34) showed that the cryptic 5′ss is always paralleled by the conserved ESS. These observations support that the reported correlation between ESS and authentic 5′ss should be extended to and is particularly relevant for cryptic 5′ss leading to unproductive splicing, as in the model of F9 exon 2. Since the exon 2 encodes the γ-carboxyl glutamic domain, which is absolutely required for the interaction of the serine protease with membranes, we infer that the ESS plays an essential role for the FIX protein function. By reducing the efficiency of unproductive splicing, the ESS would limit the impact of the cryptic 5′ss on the expression of functional FIX (35).
On the other hand, natural mutations affecting the ESS would favor the cryptic 5′ss usage and reduce FIX expression. To support this mechanistic hypothesis we investigated all known missense mutations introducing changes within the ESS and potentially weakening it, and demonstrated that almost all of them favor the cryptic 5′ss recognition and significantly decrease the levels of correct transcripts. As such, they have an additional, and probably significant, detrimental impact on FIX expression through combination of reduced amounts of correct FIX mRNA (∼40%) with the alterations produced by amino acid substitutions. Interestingly, FIX plasma levels below 1%, which define a severe bleeding phenotype, were measured in the HB patients carrying these missense mutations.
Although we have not detailed the entire splicing regulatory network, these coherent results indicate a functional role for the ESS and offer an example of aberrant splicing caused by missense mutations that might represent a key determinant of the disease phenotype variability.
The splicing patterns in the F9 exon 2 context indicate a competition between 5′ss with comparable strength, and a modulatory role of the ESS that disfavors the selection of the cryptic 5′ss. The nucleotide changes at positions +3, +5 and +6 remarkably reduce the strength of the authentic 5′ss and its complementarity to the U1snRNA 5′ tail, thus shifting the balance to the usage of the cryptic one.
In the attempt to rescue splicing, we created antisense oligonucleotides designed to mask the cryptic 5′ss, a strategy that has been successfully used in several human disease models (18). This is a straightforward correction approach in the presence of the authentic 5′ss, as demonstrated by the remarkable inhibition of the cryptic 5′ss and rescue of correct splicing obtained with the AON360 in the context of the wild-type and of missense mutants. However, in the presence of mutations impairing the authentic 5′ss, and thus its recognition by the U1snRNP, this antisense strategy was ineffective and also led to exon skipping.
We therefore focused on improving complementarity of the 5′ tail of the U1snRNA to the defective 5′ss, which has repeatedly been shown to rescue splicing in many models of human disease characterized by different type of mutations (at 5′ss, at 3′ss, within exons) impairing exon definition (22–31). However, with the exception of the +5C mutant, the compensatory U1snRNA variants failed to elicit appreciable correction effects, a finding that might be attributable to the strong competition by the cryptic 5′ss.
We hypothesized that the concurrent increase in the efficiency of correct 5′ss selection by engineered U1snRNA and inhibition of the exonic cryptic 5′ss by antisense molecules would overcome the splicing defect. Our data demonstrated that the combination of mutant-specific U1snRNAs and AON360 was able to significantly increase the selection of the authentic 5′ss and rescue splicing (from 0 up to 40% of correct transcripts) in the presence of all, but one, mutations. Noticeably, the U1F9wt that differs for one mismatch only from the mutant-specific U1snRNAs was ineffective on F9 exon 2 5′ss mutants, thus strengthening the notion that the complementarity requirements between the U1snRNAs 5′ tail and the 5′ss for a functional splicing outcome are far from being known (21).
In conclusion, by molecular characterization of a series of severe HB mutations affecting the F9 exon 2 and its 5′ss, we propose, within a network of splicing regulatory elements, a function for ESS in the down-regulation of cryptic 5′ss in exons that are essential for protein function, which better defines the ESS relevance in normal and pathological conditions.
Based on the evidence for the altered interplay among the ESS, cryptic and authentic 5′ss as a disease-causing mechanism, we produced novel experimental insights into the combinatorial activity of antisense oligonucleotides and compensatory U1snRNA in inducing splicing correction.
Material and Methods
Creation of expression vectors
To create the pF9wt vector, the genomic regions of human F9 gene (NG_007994.1) spanning (i) the first codon of exon 1 to nt. +173 in intron 1 (fragment 1), (ii) nt.−264 of intron 1 to nt. +2254 of intron 3 (fragment 2) and (iii) nt. −1435 bp of intron 3 to the last codon of exon 4 (fragment 3) were amplified from genomic DNA of a normal subject with primers 1F-1R, 2F-3R and 3F-4R using high-fidelity PfuI DNA-polymerase (Transgenomic, Glasgow, UK). The F9 regions were sequentially cloned into the expression vector pCDNA3 (Life Technologies, Carlsbad, CA, USA) by exploiting the KpnI-BamHI (fragment 1), BamHI–NotI (fragment 2) and NotI-XhoI (fragment 3) restriction sites inserted within primers.
To create the pF9 + 3C, pF9 + 5A, pF9 + 5C, pF9 + 5T, pF9 + 6C, pF9c135C, pF9c135T, pF9c137A, pF9c137C, pF9c138T, pF9c138C, pF9mut1, pF9ΔES1 and pF9ΔES2 vectors, the nucleotide changes/deletion were introduced into the pF9wt minigene by the QuickChange II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA).
The pU1F9wt, pU1F9 + 3C, pU1F9 + 5C, pU1F9 + 5A, pU1F9 + 5T and pU1F9 + 6C expression vectors for the modified U1snRNAs were created by replacing the sequence between the sites BclI and BglII with oligonucleotides as previously reported (22).
Sequences of oligonucleotides are provided in Supplementary Material, Table S1. All vectors have been validated by sequencing.
Antisense oligonucleotides
AON353 and AON360 against the F9 exon 2 cryptic 5′ss, and the scrambled AON, contain 2′-O-methyl modified ribonucloetides and full-length phosphorothioate backbone (36).
Expression in mammalian cells and mRNA studies
BHK cells were cultured as previously described (13). Cells were seeded on 12-well plates and transfected with Lipofectamine 2000 reagents (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's protocol.
One microgram of pF9 minigene variants was transfected alone, with the AON (10 nm) and/or a molar excess (1.5×) of the pU1 plasmids. Total RNA was isolated 24 h post-transfection with Trizol (Life Technologies, Carlsbad, CA, USA), reverse-transcribed and amplified using the SuperScript III One-Step RT-PCR System (Life Technologies, Carlsbad, CA, USA) with primers 1F and 4R. A similar approach was used to evaluate F9 splicing patterns in human liver.
The correct and F9del fragments were also cloned into the pGEM vector and used as templates at known concentrations to verify the amplification efficiency, which appeared to be comparable (data not shown).
Densitometric analysis for the quantification of correct and aberrant transcripts was performed using the ImageJ software. For denaturing capillary electrophoresis analysis, the RT-PCR amplified fragments were fluorescently labeled by using primers 1F and the 4R labeled with FAM and run on a ABI-3100 instrument.
Computational analysis
Computational prediction of splice sites and/or splicing regulatory elements was conducted by using the http://www.fruitfly.org/seq_tools/splice.html, the Human Splicing Finder (http://www.umd.be/HSF/) and Rescue-ESE (http://genes.mit.edu/burgelab/rescue-ese/) online softwares. Computational prediction of RNA secondary structure was performed using the RNAfold software (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi).
Supplementary Material
Funding
This work was supported by Telethon-Italy (GGP14190 to D.B., E.B., D.S. and M.P.) and Ministero della Salute—Agenzia Italiana del Farmaco (AIFA 2008—Bando per le malattie rare—Progetto RF-null-2008–1235892 to F.B. and M.P.). Funding to pay the Open Access publication charges for this article was provided by Telethon Italy Foundation.
Supplementary Material
Acknowledgments
Conflict of Interest statement: M.P. and F.B. are founders of the start-up company Raresplice. The remaining authors declare no competing financial interests.
References
- 1.Sterne-Weiler T., Howard J., Mort M., Cooper D.N., Sanford J.R. (2011) Loss of exon identity is a common mechanism of human inherited disease. Genome Res., 21, 1563–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen M., Manley J.L. (2009) Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell. Biol., 10, 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wahl M.C., Will C.L., Luhrmann R. (2009) The spliceosome: design principles of a dynamic RNP machine. Cell, 136, 701–718. [DOI] [PubMed] [Google Scholar]
- 4.Nilsen T.W., Graveley B.R. (2010) Expansion of the eukaryotic proteome by alternative splicing. Nature, 463, 457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teraoka S.N., Telatar M., Becker-Catania S., Liang T., Onengut S., Tolun A., Chessa L., Sanal O., Bernatowska E., Gatti R.A. et al. (1999) Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am. J. Hum. Genet., 64, 1617–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ars E., Serra E., Garcia J., Kruyer H., Gaona A., Lazaro C., Estivill X. (2000) Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum. Mol. Genet., 9, 237–247. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X.H., Chasin L.A. (2004) Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev., 18, 1241–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kralovicova J., Vorechovsky I. (2007) Global control of aberrant splice-site activation by auxiliary splicing sequences: evidence for a gradient in exon and intron definition. Nucleic Acids Res., 35, 6399–6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stadler M.B., Shomron N., Yeo G.W., Schneider A., Xiao X., Burge C.B. (2006) Inference of splicing regulatory activities by sequence neighborhood analysis. Plos Genet., 2, e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao X., Wang Z., Jang M., Burge C.B. (2007) Coevolutionary networks of splicing cis-regulatory elements. Proc. Natl. Acad. Sci. USA, 104, 18583–18588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cartegni L., Krainer A.R. (2002) Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet., 30, 377–384. [DOI] [PubMed] [Google Scholar]
- 12.Pagani F., Baralle F.E. (2004) Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet., 5, 389–396. [DOI] [PubMed] [Google Scholar]
- 13.Cavallari N., Balestra D., Branchini A., Maestri I., Chuamsunrit A., Sasanakul W., Mariani G., Pagani F., Bernardi F., Pinotti M. (2012) Activation of a cryptic splice site in a potentially lethal coagulation defect accounts for a functional protein variant. Biochim. Biophys. Acta, 1822, 1109–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green P.M., Bentley D.R., Mibashan R.S., Nilsson I.M., Giannelli F. (1989) Molecular pathology of haemophilia B. EMBO J., 8, 1067–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruno I.G., Jin W., Cote G.J. (2004) Correction of aberrant FGFR1 alternative RNA splicing through targeting of intronic regulatory elements. Hum. Mol. Genet., 13, 2409–2420. [DOI] [PubMed] [Google Scholar]
- 16.Meyer K., Marquis J., Trub J., Nlend Nlend R., Verp S., Ruepp M.D., Imboden H., Barde I., Trono D., Schumperli D. (2009) Rescue of a severe mouse model for spinal muscular atrophy by U7 snRNA-mediated splicing modulation. Hum. Mol. Genet., 18, 546–555. [DOI] [PubMed] [Google Scholar]
- 17.Nlend Nlend R., Meyer K., Schumperli D. (2010) Repair of pre-mRNA splicing: prospects for a therapy for spinal muscular atrophy. RNA Biol., 7, 430–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spitali P., Aartsma-Rus A. (2012) Splice modulating therapies for human disease. Cell, 148, 1085–1088. [DOI] [PubMed] [Google Scholar]
- 19.Porensky P.N., Mitrpant C., McGovern V.L., Bevan A.K., Foust K.D., Kaspar B.K., Wilton S.D., Burghes A.H. (2012) A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum. Mol. Genet., 21, 1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Havens M.A., Duelli D.M., Hastings M.L. (2013) Targeting RNA splicing for disease therapy. Wiley interdisciplinary reviews. RNA, 4, 247–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roca X., Krainer A.R., Eperon I.C. (2013) Pick one, but be quick: 5′ splice sites and the problems of too many choices. Genes Dev., 27, 129–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinotti M., Rizzotto L., Balestra D., Lewandowska M.A., Cavallari N., Marchetti G., Bernardi F., Pagani F. (2008) U1-snRNA-mediated rescue of mRNA processing in severe factor VII deficiency. Blood, 111, 2681–2684. [DOI] [PubMed] [Google Scholar]
- 23.Pinotti M., Balestra D., Rizzotto L., Maestri I., Pagani F., Bernardi F. (2009) Rescue of coagulation factor VII function by the U1+5A snRNA. Blood, 113, 6461–6464. [DOI] [PubMed] [Google Scholar]
- 24.Pinotti M., Bernardi F., Dal Mas A., Pagani F. (2011) RNA-based therapeutic approaches for coagulation factor deficiencies. J. Thromb. Haemost., 9, 2143–2152. [DOI] [PubMed] [Google Scholar]
- 25.Fernandez Alanis E., Pinotti M., Dal Mas A., Balestra D., Cavallari N., Rogalska M.E., Bernardi F., Pagani F. (2012) An exon-specific U1 small nuclear RNA (snRNA) strategy to correct splicing defects. Hum. Mol. Genet., 21, 2389–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baralle M., Baralle D., De Conti L., Mattocks C., Whittaker J., Knezevich A., Ffrench-Constant C., Baralle F.E. (2003) Identification of a mutation that perturbs NF1 gene splicing using genomic DNA samples and a minigene assay. J. Med. Genet., 40, 220–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Susani L., Pangrazio A., Sobacchi C., Taranta A., Mortier G., Savarirayan R., Villa A., Orchard P., Vezzoni P., Albertini A. et al. (2004) TCIRG1-dependent recessive osteopetrosis: mutation analysis, functional identification of the splicing defects, and in vitro rescue by U1 snRNA. Hum. Mutat., 24, 225–235. [DOI] [PubMed] [Google Scholar]
- 28.Tanner G., Glaus E., Barthelmes D., Ader M., Fleischhauer J., Pagani F., Berger W., Neidhardt J. (2009) Therapeutic strategy to rescue mutation-induced exon skipping in rhodopsin by adaptation of U1 snRNA. Hum. Mutat., 30, 255–263. [DOI] [PubMed] [Google Scholar]
- 29.Schmid F., Glaus E., Barthelmes D., Fliegauf M., Gaspar H., Nurnberg G., Nurnberg P., Omran H., Berger W., Neidhardt J. (2011) U1 snRNA-mediated gene therapeutic correction of splice defects caused by an exceptionally mild BBS mutation. Hum. Mutat., 32, 815–824. [DOI] [PubMed] [Google Scholar]
- 30.Glaus E., Schmid F., Da Costa R., Berger W., Neidhardt J. (2011) Gene therapeutic approach using mutation-adapted U1 snRNA to correct a RPGR splice defect in patient-derived cells. Mol. Ther., 19, 936–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balestra D., Faella A., Margaritis P., Cavallari N., Pagani F., Bernardi F., Arruda V.R., Pinotti M. (2014) An engineered U1 small nuclear RNA rescues splicing-defective coagulation F7 gene expression in mice. J. Thromb. Haemost., 12, 177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kervestin S., Jacobson A. (2012) NMD: a multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol., 13, 700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qian W., Liu F. (2014) Regulation of alternative splicing of tau exon 10. Neurosci. Bull., 30, 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davidson C.J., Tuddenham E.G., McVey J.H. (2003) 450 million years of hemostasis. J. Thromb. Haemost., 1, 1487–1494. [DOI] [PubMed] [Google Scholar]
- 35.Butenas S., Orfeo T., Gissel M.T., Brummel K.E., Mann K.G. (2004) The significance of circulating factor IXa in blood. J. Biol. Chem., 279, 22875–22882. [DOI] [PubMed] [Google Scholar]
- 36.Rimessi P., Fabris M., Bovolenta M., Bassi E., Falzarano S., Gualandi F., Rapezzi C., Coccolo F., Perrone D., Medici A. et al. (2010) Antisense modulation of both exonic and intronic splicing motifs induces skipping of a DMD pseudo-exon responsible for x-linked dilated cardiomyopathy. Hum. Gene Ther., 21, 1137–1146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.