Abstract
Mitochondrial respiratory chain (RC) disease therapies directed at intra-mitochondrial pathology are largely ineffective. Recognizing that RC dysfunction invokes pronounced extra-mitochondrial transcriptional adaptations, particularly involving dysregulated translation, we hypothesized that translational dysregulation is itself contributing to the pathophysiology of RC disease. Here, we investigated the activities, and effects from direct inhibition, of a central translational regulator (mTORC1) and its downstream biological processes in diverse genetic and pharmacological models of RC disease. Our data identify novel mechanisms underlying the cellular pathogenesis of RC dysfunction, including the combined induction of proteotoxic stress, the ER stress response and autophagy. mTORC1 inhibition with rapamycin partially ameliorated renal disease in B6.Pdss2kd/kd mice with complexes I–III/II–III deficiencies, improved viability and mitochondrial physiology in gas-1(fc21) nematodes with complex I deficiency, and rescued viability across a variety of RC-inhibited human cells. Even more effective was probucol, a PPAR-activating anti-lipid drug that we show also inhibits mTORC1. However, directly inhibiting mTORC1-regulated downstream activities yielded the most pronounced and sustained benefit. Partial inhibition of translation by cycloheximide, or of autophagy by lithium chloride, rescued viability, preserved cellular respiratory capacity and induced mitochondrial translation and biogenesis. Cycloheximide also ameliorated proteotoxic stress via a uniquely selective reduction of cytosolic protein translation. RNAseq-based transcriptome profiling of treatment effects in gas-1(fc21) mutants provide further evidence that these therapies effectively restored altered translation and autophagy pathways toward that of wild-type animals. Overall, partially inhibiting cytosolic translation and autophagy offer novel treatment strategies to improve health across the diverse array of human diseases whose pathogenesis involves RC dysfunction.
Introduction
The mitochondrial respiratory chain (RC) consists of five multimeric protein complexes that collectively oxidize nutrient-derived substrates in an integrated process that transfers reducing equivalents and generates an electrochemical gradient to drive energy production in the chemical form of adenosine triphosphate (ATP) (1). A wide spectrum of seemingly unrelated complex diseases encompassing such variable symptoms as neurodegeneration, myopathy, cardiac disease, nephropathy, liver dysfunction, blindness, deafness and diabetes mellitus, shares a common pathophysiology of RC dysfunction. Indeed, primary mitochondrial RC diseases can impair nearly any body system, at any time, due to causative mutations in hundreds of distinct nuclear or mitochondrial DNA (mtDNA) genes (2). Further, diverse environmental and genetic factors commonly increase mitochondrial reactive oxygen species (ROS) generation, with a resultant induction of progressive mtDNA and membrane damage that eventually leads to secondary RC dysfunction and energy deficiency (3). Whether primary or secondary, the end result of impairment in RC electrochemical flux is reduced ATP production, increased NADH:NAD+ redox ratio with absolute cellular deficiencies of both reduced and oxidized nicotinamide adenine dinucleotide species (4) and increased oxidative stress (5). Yet, therapies aimed solely at targeting mitochondria-specific alterations, such as antioxidants, vitamins or cofactors intended to enhance residual RC enzyme function or quench ‘toxic’ metabolites, have proved to be generally ineffective in ameliorating disease manifestations of either primary or secondary mitochondrial dysfunction (6).
RC dysfunction disrupts global cellular function through mechanisms that are incompletely understood. Protein translation has proved to be one of the most consistently dysregulated basic cellular functions in RC disease (4,7). For example, transcriptome profiling of liver from B6.Pdss2kd/kd missense mutant mice that have RC complex I–III and II–III dysfunction due to coenzyme Q deficiency showed ‘ribosome’-related genes to be the most significantly upregulated biological pathway (8). Coenzyme Q deficiency results from its impaired biosynthesis in this model, since Pdss2 is one of two subunits of the prenyl diphosphate synthase required to isoprenylate benzoquinone to form coenzyme Q. The B6.Pdss2kd/kd missense mutant mice develop a focal-segmental glomerulosclerosis (FSGS)-like renal disease at ∼12 weeks of age (9), as well as metabolic alterations (8), neuromuscular dysfunction and a Parkinson's Disease-like phenotype (10). Differential transcriptional dysregulation of cytosolic and mitochondrial ribosomal genes has also been observed in skeletal muscle and fibroblasts from human patients with diverse RC diseases (4), a phenomenon that has proved to be highly consistent across nearly all species, tissues and RC disease subtypes (7). Protein translation in the cytosol and endoplasmic reticulum (ER) is a major energy-consuming process (11), where stimulation of messenger RNA (mRNA) translation initiation and elongation is directly regulated by the mTORC1 signaling pathway (12). mTORC1 activation also increases cell-cycle progression, selectively enhances ribosomal gene transcription and ribosome biogenesis to increase cellular proliferation and size (13) and inhibits autophagy (14,15). Recognizing that RC dysfunction invokes pronounced transcriptional dysregulation of translation-related processes, we hypothesized that translational dysregulation is itself contributing to the underlying pathophysiology of RC disease. Here, we investigated the effects of targeting mTORC1 and downstream mTORC1-regulated processes in murine, Caenorhabditis elegans, and human cell models of RC disease, generated using both genetic and pharmacological strategies. A major goal of these investigations was to distinguish the potential therapeutic benefits of inhibiting cytosolic translation relative to those of autophagy inhibition.
Cytosolic translation can be inhibited to variable degrees by a range of pharmacological approaches. Rapamycin is a known macrocyclic triene that forms a complex with FKBP12 to directly bind and inhibit mTOR, thereby partially and reversibly blocking protein synthesis (16). Interestingly, mTORC1 inhibition with rapamycin was recently shown to attenuate neurological disease progression, extend lifespan from a median of 50 days to 114 days in males and 111 days in females, and increase amino acid catabolism in an RC complex I deficient NDUFS4 Leigh syndrome murine model (17). We show that feeding B6.Pdss2kd/kd mice with rapamycin upon weaning (at 4 weeks of life) significantly ameliorates their renal glomerular disease to a similar extent as we previously showed occurs with coenzyme Q10 supplementation (8). However, no further synergy was gained by combining rapamycin with coenzyme Q10 treatments in this model. Rapamycin also rescued the short lifespan and improved the reduced mitochondrial content of complex I deficient C. elegans gas-1(fc21) worms that have a homozygous mutation in the complex I NDUFS2 subunit homolog (18). Finally, rapamycin partially protected cultured human podocytes with rotenone-induced RC complex I inhibition from autophagic death. Thus, we show that rapamycin has consistent, albeit modest, beneficial effects in diverse models of RC disease. However, significantly more effective and sustained beneficial effects in RC disease were achieved by treatment with probucol, which in addition to activating PPAR signaling pathways (8), is a drug that we show here also effectively inhibits mTORC1 activity. Probucol improved animal viability in gas-1(fc21) worms as well as the viability of diverse human cell types subjected to RC inhibition at complexes I, III or V.
The most dramatic beneficial effects in these RC disease models occurred upon treatment with a direct translation inhibitor, cycloheximide. Cycloheximide is a potent, reversible and specific inhibitor of eukaryotic cytosolic translation that works via direct binding to the ribosome, inhibiting eEF2-mediated translocation of the nascent protein, and halting elongation after one complete translocation cycle (19). Remarkably, cycloheximide treatment preserved cellular oxidative capacity despite direct RC inhibition, a surprising effect that we show results from upregulation of mitochondrial biogenesis and mitochondrial translation, with restoration of cellular levels of ATP. Beneficial effects of cytosolic translation inhibition in RC deficient models were strongly nutrient (glucose) dependent. Given the recent recognition that RC dysfunction activates proteotoxic stress (20), we also interrogated the ability of cycloheximide treatment to rebalance relative cellular protein production to degradative capacity. Indeed, proteotoxic stress in RC disease was significantly reduced with cycloheximide treatment via the selective inhibition of cellular protein translation, an important therapeutic effect of this drug that has not previously been recognized. Cycloheximide further prevented activation of the ER stress response and autophagy that occurred upon prolonged RC inhibition. Interestingly, targeted autophagy inhibition with either 3-methyladenine or lithium chloride substantially, and synergistically, could also rescue viability and total cellular respiratory capacity in cell models of RC inhibition. Transcriptome profiling of cycloheximide, probucol and rapamycin treatment effects in gas-1(fc21) mutant RC disease worms provided further evidence that these therapies effectively restored their altered translation and autophagy pathways toward that of wild-type animals. Collectively, these data identify novel mechanisms underlying the cellular pathogenesis of mitochondrial RC dysfunction, including the combined induction of proteotoxic stress, the ER stress response and autophagy. Further, these data strongly suggest that direct pharmacological inhibition of either cytosolic translation and/or autophagy offer novel therapeutic strategies to improve cell viability, health and mitochondrial physiology across diverse etiologies and presentations of mitochondrial RC disease.
Results
mTORC1 activity at the level of S6 phosphorylation is upregulated by RC disease and decreased by probucol treatment in both animal and cellular models of RC disease
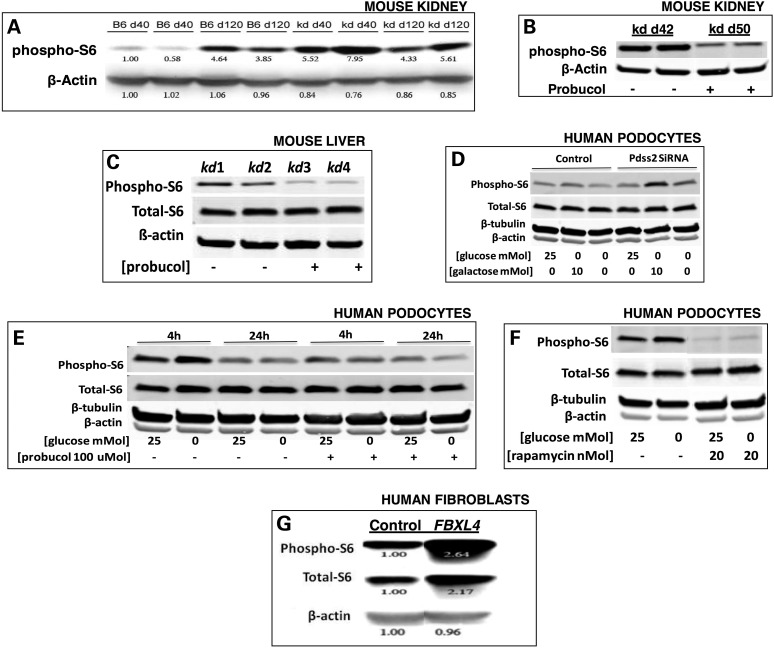
mTORC1 activity controls cell growth and proliferation by regulating protein synthesis. Through phosphorylation of S6 kinase, mTORC1 post-translationally activates the S6 ribosomal protein (S6) to transcriptionally upregulate the expression of proteins involved in cell-cycle progression and translation (12). We studied relative pS6 activity in kidney and liver from the B6.Pdss2kd/kd (‘kd/kd’) mutant mice that manifest an FSGS-like renal disease onset between 90 to 120 days (9), which is preceded by ultrastructural mitochondrial defects that are evident in renal tubular epithelial cells by the time endogenous coenzyme Q tissue content normally begins to rise on Day 40 (9). While pS6 expression levels rise with age in wild-type mice, the kidney of pre-symptomatic kd/kd mutant mice already exhibited significantly increased S6 phosphorylation relative to age-matched wild-type (B6) controls (40 days), with no significant difference in pS6 activity in symptomatic kd/kd animals relative to age-matched wild-type controls (120 days) (Fig. 1A). These data suggested that early mTORC1 activation may represent a physiological response to RC dysfunction and contribute to disease pathogenesis. Probucol is an oral anti-hyperlipidemic agent that we previously demonstrated to both prevent and reverse the FSGS-like renal disease in kd/kd mutant mice (8). Interestingly, probucol treatment of kd/kd mutant mice from weaning prevented not only their renal disease but also their early increase in pS6 expression in the kidney (Fig. 1B), with sustained pS6 reduction evident in the liver (Fig. 1C) of 120-day-old kd/kd animals.
Figure 1.
Increased mTORC1 activity occurs in B6.Pdss2kd/kd mouse kidney and in human cell models of genetic or pharmacological-based RC disease, and is reversed by probucol or rapamycin treatment. pS6 expression was measured by western blot analysis, with relative expression changes between groups quantified in ImageJ. (A) Kidney was studied by pS6 western blot analysis at day of life 40 or 120 from wild-type (B6) and B6.Pdss2kd/kd (‘kd’) mice. Relative quantitation, as calculated in ImageJ, is shown below each band. (B) Kidney was studied by pS6 western blot analysis from B6.Pdss2kd/kd mice treated from weaning to day of life 50 with probucol relative to untreated B6.Pdss2kd/kd littermates. (C) Liver from 120-day-old B6.Pdss2kd/kd mutant mice fed either normal chow or probucol from weaning. (D) Pdss2 knockdown by siRNA in human podocytes grown in variable nutrient conditions. Pdss2 siRNA and control human podocytes were cultured at 33°C for 24 h in 10% FBS, RPMI-1640 medium containing either high glucose (25 mm), galactose (10 mm) or no sugar. (E) Probucol effects on healthy human podocytes in variable nutrient conditions. Human podocytes were cultured with or without 100 µm probucol overnight in RPMI media containing either 25 mm glucose or no glucose at 33°C for either 4 or 24 h. (F) Rapamycin-treated healthy human podocytes in variable nutrient conditions. Human podocytes were cultured with or without 20 nm rapamycin at 33°C for 24 h in RPMI media with either 25 mm glucose or glucose-free media. (G) Human fibroblasts from patients with FBXL4-based primary RC disease. Human fibroblasts were cultured at 37°C for 24 h in 10% FBS, DMEM with 5 mm glucose medium from a healthy control subject or mitochondrial disease patient with primary (genetic-based) RC complex I and III deficiencies due to compound heterozygote mutations in FBXL4 (21). Relative quantitation of pS6 and total S6 protein is shown below each band, as calculated in ImageJ.
To assess the specificity of the effects of Pdss2-based mitochondrial dysfunction on mTORC1 activity, we depleted Pdss2 using siRNA in human podocytes. This was performed while controlling for the relative influence of cellular nutrient status, which is known to be a key factor in determining whether mTORC1 is activated to enable cells to grow and proliferate (2). Furthermore, RC-deficient cells rely upon ATP generation through upregulated glycolysis, a process that cannot use galactose. Therefore, changing the fuel source from glucose to galactose can reveal mitochondrial dysfunction because it forces cells to utilize oxidative phosphorylation (OXPHOS) for energy production. Similar to our findings in pre-symptomatic kd/kd mice, S6 phosphorylation was increased in the Pdss2 siRNA cells relative to controls when grown overnight at 33°C in glucose-free media either with or without 10 mm galactose (Fig. 1D). However, no increase in S6 phosphorylation occurred with Pdss2 depletion when cells were grown in abundant glucose (25 mm) media (Fig. 1D). These data support the hypothesis that Pdss2 activity normally attenuates mTORC1. They further demonstrate a significant influence of cellular nutrient status on modifying the physiological effects of RC dysfunction. We also observed that S6 phosphorylation in control podocytes is both transient and can be reduced by probucol treatment, with the most pronounced effect occurring in glucose-free conditions (Fig. 1E). Finally, probucol showed similar efficacy in terms of inhibiting S6 phosphorylation, regardless of glucose conditions (Fig. 1E), as did the well-established selective mTORC1 inhibitor, rapamycin (Fig. 1F).
Human cell lines models of primary RC dysfunction generated by direct pharmacological inhibition of OXPHOS capacity also show dysregulated mTORC1 activity (4). Fibroblast cell derived from a human patient with RC complex I–III dysfunction caused by pathogenic autosomal recessive mutations in the nuclear-encoded gene for the mitochondria-localized FBXL4 protein (21) had increased S6 phosphorylation (Fig. 1G). Encoding a member of the F-box protein family that can be involved in phosphorylation-dependent ubiquitination and/or G protein receptor coupling, FBXL4 has uniquely been shown to encode a mitochondrial intermembrane space protein of unknown function (21). Autosomal recessive mutations in FBXL4 have recently been recognized to cause a severe mitochondrial RC disorder characterized by early-onset encephalopathy, variable multi-systemic dysfunction, lactic acidemia and mitochondrial DNA depletion (21–23). Similarly, we previously showed that pS6 expression was increased in a complex I disease fibroblast line from a patient with Leigh syndrome whose disease results from pathogenic mtDNA mutations in both ND4 and ND6 (4). We conclude that primary RC dysfunction across diverse species and tissues is consistently associated with increased mTORC1 activity at the level of S6 phosphorylation.
AMPK is a direct sensor of mitochondrial function that negatively regulates mTORC1 by way of two key proteins, tuberous sclerosis 1 (TSC1) and Ras homolog enriched in brain (RHEB) (4). We therefore studied whether the observed increase in S6 phosphorylation correlated with a possible reduction of AMPK phosphorylation within the same models of RC dysfunction. However, liver from symptomatic kd/kd mutant mice showed increased AMPK phosphorylation (Supplementary Material, Fig. S1a). Interestingly, oral probucol treatment increased AMPK phosphorylation in liver from pre-symptomatic kd/kd mice, as well as in liver from wild-type B6 mice (Supplementary Material, Fig. S1b). Similarly, AMPK phosphorylation was increased in Pdss2 siRNA-treated podocytes relative to controls when grown overnight at 33°C in glucose-free media either alone or with 10 mm galactose, but not when abundant glucose (25 mm) was provided (Supplementary Material, Fig. S1c). Probucol (100 µm) treatment of healthy human podocytes also increased AMPK phosphorylation (Supplementary Material, Fig. S1d), a phenomenon that could be prevented by provision of abundant glucose (Supplementary Material, Fig. S1e). Direct RC complex I inhibition with rotenone increased AMPK phosphorylation in a dose-dependent fashion (Supplementary Material, Fig. S1f), but which could again be prevented by provision of abundant glucose (Supplementary Material, Fig. S1g). The degree of increased AMPK phosphorylation in human podocytes varied with the duration of RC inhibition at complexes I, III or V, respectively, by rotenone, antimycin A or oligomycin, with more pronounced activation at 5 h and a comparative decrease by 24 h (Supplementary Material, Fig. S1h). Finally, human fibroblasts with direct RC complex V inhibition from oligomycin showed the same early increase in AMPK phosphorylation that was relatively diminished by 24 h (data not shown), with a concentration-dependent activation of AMPK phosphorylation at 24 h that could be prevented through provision of ample (25 mm) glucose (Supplementary Material, Fig. S1i). We conclude that AMPK activity is consistently increased in both genetic and pharmacological inhibitor-based models of RC dysfunction, is further increased by probucol treatment both in vivo and in vitro, and normalizes both upon provision of ample glucose and with time. Thus, RC dysfunction activates both mTORC1 and AMPK activities. However, the substantial increase in AMPK activation caused by probucol treatment is consistent with its observed reduction in mTORC1 activity, and may potentially contribute to the therapeutic effect of probucol in the setting of RC disease.
Probucol and rapamycin, which both inhibit mTORC1 activity, improve health and viability in animal and cell models of RC disease
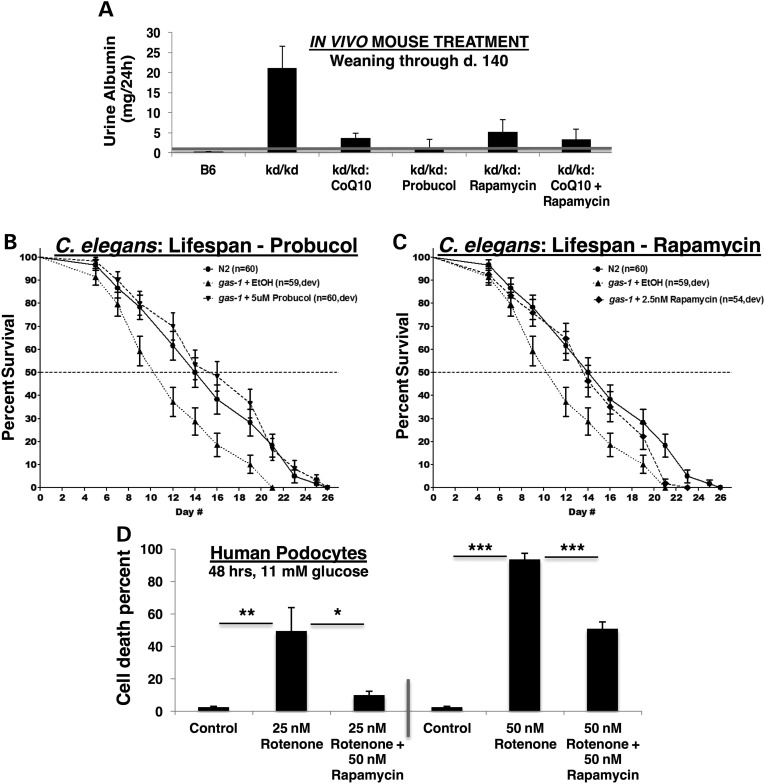
A major disease phenotype of kd/kd mice is albuminuria, with albumin wasting by the kidney increasing dramatically after age 90–120 days (9). We previously reported that oral treatment from weaning with coenzyme Q10 (CoQ10) in this genetic RC disease model can delay their onset of kidney disease by one month and reduce the severity of their renal disease, whereas oral probucol treatment both prevents and reverses active renal disease in kd/kd mice (8). Thus, probucol has several major physiological effects in kd/kd mice, some of which we previously reported to include restoration of PPAR signaling and reduced CoQ content in endogenous tissue (8), in addition to our current findings that suggest probucol effectively activates AMPK activity (Supplementary Material, Fig. S1) and inhibits mTORC1 activity (Fig. 1). To determine whether mTORC1 inhibition would be sufficient to prevent kidney disease in this model, we treated kd/kd animals with oral rapamycin. We found that treating mice with oral rapamycin in their chow from weaning led to a similar reduction of albuminuria (5.12 mg/24 h in rapamycin-treated versus 21.15 mg/24 h in untreated kd/kd mice) as is achieved with CoQ10 supplementation (Fig. 2A). Marginally improved efficacy that failed to achieve statistical significance was seen when combining oral rapamycin with CoQ10 treatments, although the animals remained frankly albuminuric (defined as >1 mg urine albumin in 24 h). These data are suggestive that upregulated mTORC1 activity and its downstream consequences only partially contribute to renal disease pathogenesis in the kd/kd mouse model of RC disease. Indeed, while mTORC1 activity was effectively inhibited by both probucol and rapamycin treatments, animal health was not fully restored by rapamycin alone.
Figure 2.
Pharmacological reversal of dysregulated mTORC1 activity is associated with significant health benefit across both animal and cellular models of RC disease. (A) Probucol or rapamycin treatments mitigate renal glomerular disease in B6.Pdss2kd/kd mutant mice. Twenty-four hour urine albumin levels (mg) from wild-type B6 (n = 26), untreated B6.Pdss2kd/kd mutants (n = 44) and either CoQ10 (n = 10, 1 mg/ml in drinking water), probucol (n = 25, 1% w/w chow diet), rapamycin (n = 7, 225 PPM chow diet) or CoQ10 (1 mg/ml in drinking water) plus rapamycin (n = 11) fed B6.Pdss2kd/kd mice from weaning to sacrifice at ∼140 days old. (B and C) Shortened lifespan of gas-1(fc21) complex I mutant C. elegans nematodes was rescued by feeding animals from early development at 20°C with (B) probucol (best effect at 5 µm) or (C) rapamycin (best effect at 2.5 nm (shown) and 25 nm on median and maximal lifespan, respectively). n, animal number studied per condition. Ethanol was used as buffer control for both drugs. Full statistical analyses and additional treatment dose trials for both drugs are provided in Supplementary Material, Table S1. (D) Rapamycin maintains cell viability in human podocytes despite direct RC inhibition by rotenone. Human podocytes were cultured at 33°C for 48 h in RPMI-1640 with 11 mm glucose, 10% FBS and the RC complex I inhibitor rotenone (25 nm or 50 nm) alone or with 50 nm rapamycin. n = 3 per condition. *P < 0.05; **P < 0.01; and ***P < 0.001 by Student's t-test.
To further dissect the relative efficacy of probucol versus rapamycin in RC disease, we studied the in vivo effects of exposing animals to log-order variation in concentration of both agents in a C. elegans nematode (worm) model of RC complex I deficiency. gas-1(fc21) mutant animals harbor an autosomal recessive missense mutation (p.R290K) in the nuclear-encoded complex I subunit NDUFS2 homolog and display significantly reduced median and maximal lifespan at 20°C (18). Treating mutant animals from early development (L1 stage) with low-dose probucol fully rescued both median and maximal lifespan (Fig. 2B), with greatest effects achieved at 5 µm (Supplementary Material, Table S1 and Fig. S2). Significant, although mildly less substantial, benefit was seen when probucol treatment was begun in adult-stage animals (Supplementary Material, Table S1 and Fig. S2). In comparison, only low-dose (2.5 nM) rapamycin treatment fully restored median lifespan (and 25 nm rapamycin improved maximal lifespan) in gas-1(fc21) mutant worms (Fig. 2C), although its benefit was not consistent even at 2.5 nm in all trials (Supplementary Material, Table S1 and Fig. S3). Collectively, these data suggest that the health benefit of either probucol or rapamycin treatments in RC disease likely relates to their partial inhibition of mTORC1 activity, as higher concentrations of either agent further diminished the already short lifespan of complex I deficient gas-1(fc21) worms (Supplementary Material, Table S1). Given that mTORC1 inhibits autophagy, which can variably function at the cellular level as either a pro-survival or pro-death signal, it is possible that the observed dose-dependent toxicity for these two mTORC1-inhibiting drugs relates to their moving the set-point on balance from pro-survival to pro-death in the setting of RC disease. Further, probucol's beneficial effect in restoring the overall physiology of RC complex I deficient worms appears to exceed that of rapamycin, as only probucol significantly and consistently rescued both their median and maximal lifespan to that typical of wild-type control animals. Thus, additional probucol effects beyond solely mTORC1 inhibition likely contribute to its health benefits in the setting of primary RC disease.
Rotenone is a potent RC complex I inhibitor that inhibits electron transport at the mitochondrial NADH:ubiquinone oxidoreductase. Similar to reduced animal viability as seen in RC deficient mice and nematodes, we found cell viability to be severely compromised by rotenone-based RC inhibition (Fig. 2D). Consistent with treatment effects in both mouse and C. elegans animal models, we discovered that cell viability upon exposure to an otherwise lethal level of rotenone-based RC inhibition can be significantly improved with 50 nm rapamycin treatment (Fig. 2D). These human cell studies confirm that upregulated mTORC1 activity consistently contributes to the pathogenesis of RC dysfunction, as directly inhibiting mTORC1 activity in RC-deficient cells significantly improves cellular health.
Cell death from RC dysfunction is effectively prevented by inhibiting cytosolic translation
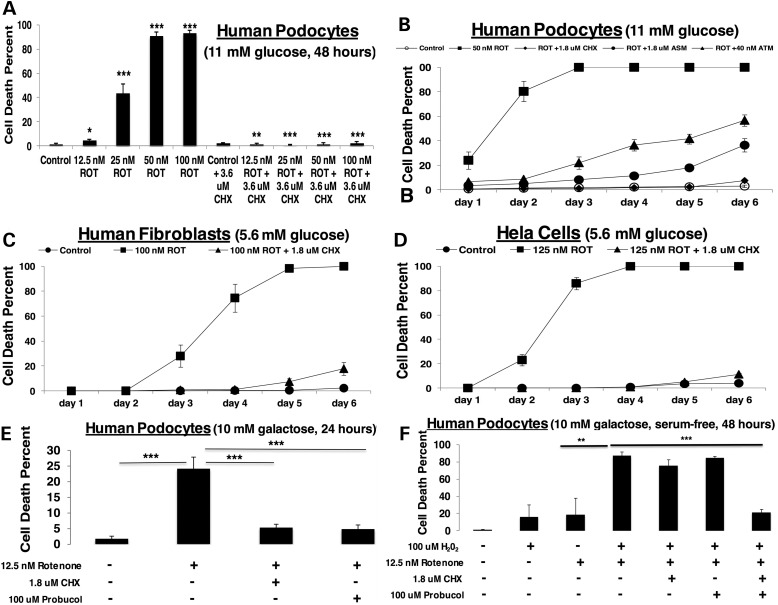
Given that mTORC1 has several major activities, we sought to determine whether its role in activating cytosolic translation was central to the pathology of RC disease. Remarkably, direct inhibition of cytosolic translation with cycloheximide dramatically rescued cell viability from rotenone-based RC dysfunction, even at otherwise completely lethal rotenone doses (Fig. 3A). The protective cellular effect of cycloheximide on viability was sustained through 6 days of rotenone treatment, with significant but somewhat less pronounced rescue also achieved with drugs that inhibit translation at either the level of protein synthesis (anisomycin, which inhibits peptidyl transferase) or its transcriptional regulation (actinomycin, which blocks RNA polymerase) (Fig. 3B). The protective effects of translation inhibition were not limited to human podocytes, as similar results were consistently obtained with other cell types, including human fibroblasts (Fig. 3C) and HeLa cells (Fig. 3D). However, no significant effect on the short lifespan of complex I deficient nematodes was achieved with cycloheximide at comparable micromolar, or even nanomolar, concentrations, regardless of whether treatment was begun at early development or only upon nematodes reaching the adult stage (Supplementary Material, Table S1 and Fig. S4). While cells grown in galactose media to stress their reliance on mitochondrial OXPHOS capacity can survive only very low levels of rotenone-based complex I inhibition, significantly enhanced cell viability in these stressed conditions could be achieved by treating cells with either cycloheximide (to directly inhibit translation) or probucol (to indirectly inhibit translation at the level of mTORC1, as detailed earlier) (Fig. 3E). While neither cycloheximide nor probucol rescued cell survival when direct RC inhibition in galactose-only media was compounded by oxidative stress in the form of hydrogen peroxide, their co-administration was able to significantly improve cell viability under these extreme conditions (Fig. 3F). This synergistic effect of both cycloheximide and probucol was suggestive that a greater degree of translation inhibition is required to survive oxidative stress in the setting of RC dysfunction. An alternative explanation could be that other physiological effects of probucol besides mTORC1-mediated translation inhibition, such as its antioxidant effect and/or its ability to stimulate PPAR signaling (8), contribute to health benefits from probucol in the setting of primary RC disease.
Figure 3.
Direct inhibition of cytosolic translation by cycloheximide preserves viability in RC deficient cells. (A) Rotenone dose range in human podocytes. Healthy human podocytes cultured for 48 h at 33°C in RPMI-1640, 11 mm glucose and 10% FBS media were treated with increasing concentrations of the potent RC complex I inhibitor, rotenone (12.5, 25, 50, 100 nm) alone or with 3.6 µm cycloheximide (CHX). n = 3 per condition. Bars and error bars convey mean and standard deviation, respectively. *P < 0.05; **P < 0.01; and ***P < 0.001 by Student's t-test. (B) Progressively potent inhibition of cytosolic translation preserves cell viability over time despite direct RC inhibition. Healthy human podocytes cultured for 6 days at 33°C in RPMI-1640, 11 mm glucose and 10% FBS media were treated with 50 nm rotenone (ROT) alone or with either 1.8 µm cycloheximide (CHX), 1.8 µm anisomycin (ASM) or 40 nm actinomycin (ATM). n = 3 per condition. Shapes and error bars convey mean and standard deviation, respectively. Statistical significance was determined by Student's t-test, where P < 0.01 at Day 1 and P < 0.001 at all other days for rotenone-treated relative to control. Viability for cycloheximide plus rotenone was not significantly different than viability of untreated control on any day. For anisomycin plus rotenone relative to control, P < 0.01 on Days 1 and 2 and P < 0.001 on all other days; and for actinomycin plus rotenone relative to control, P < 0.01 on Day 3 and P < 0.001 on all other days. Relative to rotenone-treated cells, drug treatment significantly restored viability with cycloheximide (P < 0.01 on Day 1 and P < 0.001 on all other days), anisomycin (P < 0.01 on Day 1 and P < 0.001 on all other days) or actinomycin (P < 0.05 on Day 1 and P < 0.001 on all other days). (C) Cycloheximide maintains viability in rotenone-treated human fibroblasts. Healthy human fibroblasts (FCL-F35) cultured for 6 days at 37°C in DMEM, 5.6 mm glucose and 10% FBS media were treated with 100 nm rotenone (ROT) alone or with 1.8 µm cycloheximide (CHX). n = 3 per condition. Shapes and error bars convey mean and standard deviation, respectively. Statistical significance was determined by Student's t-test. For rotenone relative to control, P < 0.01 on Days 3 and 4 and P < 0.001 on Days 5 and 6; for rotenone plus cycloheximide relative to control, P < 0.01 on Days 5 and 6; and for rotenone plus cycloheximide relative to rotenone alone, P < 0.01 on Day 3 and P < 0.001 on Days 4, 5 and 6. (D) Cycloheximide maintains viability in rotenone-treated HeLa cells. HeLa cells cultured for 6 days at 37°C in DMEM, 5.6 mm glucose and 10% FBS media were treated with 125 nm rotenone (ROT) alone or with 1.8 µm cycloheximide (CHX). n = 3 per condition. Shapes and error bars convey mean and standard deviation, respectively. Statistical significance was determined by Student's t-test, where for rotenone relative to control, P < 0.001 on all Days 2 through 6; for rotenone plus cycloheximide relative to control, P < 0.05 on Day 6 and for rotenone plus cycloheximide relative to rotenone alone, P < 0.001 on all Days 2 through 6. (E) Cycloheximide or probucol treatment prevents rotenone-induced cell death in galactose media. Healthy human podocytes were exposed to 12.5 nm rotenone (ROT) for 24 h in 10 mm galactose (no glucose) media with or without 1.8 µm cycloheximide (CHX) or 100 µm probucol. n = 3 per condition. Bars and error bars convey mean and standard deviation, respectively. ***P < 0.001 by Student's t-test. (F) Cycloheximide and probucol co-treatment is required to prevent cell death in galactose media from the additive effects of rotenone and oxidative stress. Healthy human podocytes were exposed to 12.5 nm rotenone (ROT) and/or 100 µm hydrogen peroxide (H2O2) for 48 h in 10 mm galactose (no glucose) serum-free media with or without 1.8 µm cycloheximide (CHX) and/or 100 µm probucol. n = 3 per condition. Bars and error bars convey mean and standard deviation, respectively. **P < 0.01 and ***P < 0.001 by Student's t-test.
Cycloheximide causes partial and selective inhibition of cytosolic translation
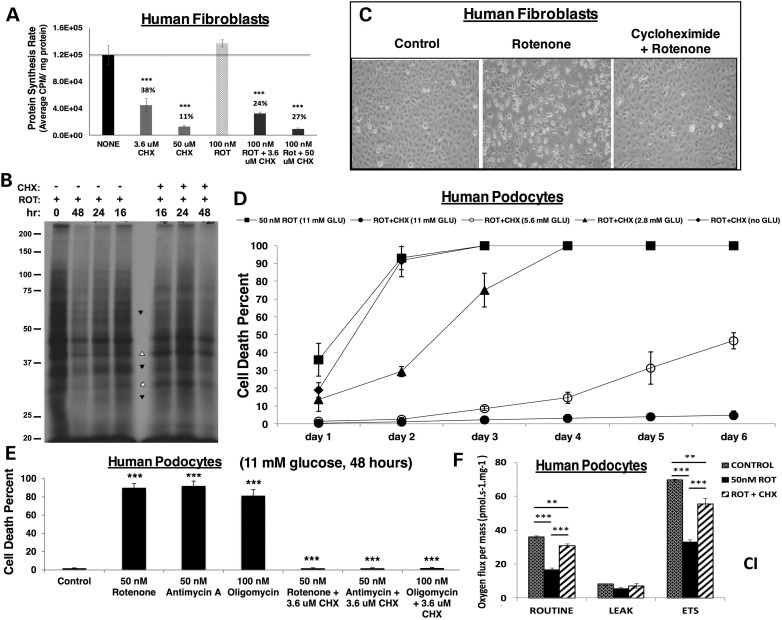
Given the established role of cycloheximide to specifically inhibit cytosolic translation, we sought to test the degree of translation inhibition achieved with cycloheximide at the low concentrations that proved sufficient to rescue cell viability in the setting of RC dysfunction. Cycloheximide caused a concentration-dependent inhibition of cytosolic translation in both human fibroblasts (Fig. 4A) and podocytes (Supplementary Material, Fig. S5a). Interestingly, low micromolar-range cycloheximide led to only partial (62% decrease) inhibition of cell translation in healthy fibroblasts, with a somewhat more substantial but still partial (76% decrease) effect in rotenone-treated fibroblasts (Fig. 4A). Rotenone itself had no significant effect on cellular protein synthesis rate (Fig. 4A). Most notably, micromolar-range cycloheximide treatment displayed selectivity in the specific cellular proteins whose synthesis was inhibited (Fig. 4B). In comparison, only marginal reduction of translation activity (Supplementary Material, Fig. S5a), with no evidence for similarly selective changes in the abundance of specific protein bands (Supplementary Material, Fig. S5b), was evident in cells treated with either rapamycin or probucol at the doses found to significantly rescue rotenone-mediated cell death. Collectively, these data are suggestive that proteotoxicity, which we define as impaired cellular function caused by an accumulation of misfolded, misplaced or otherwise dysfunctional proteins, may play a role in RC disease pathogenesis. Further, while rapamycin and probucol improve cell viability upon RC inhibition, their full mechanisms of action are likely distinct from that of cycloheximide. Indeed, only micromolar-range cycloheximide treatment leads to a full rescue from cell lethality in RC dysfunction and inhibits the translation of specific proteins. Future delineation of which proteins are selectively inhibited upon partial translation inhibition with micromolar-range cycloheximide may identify those proteins that underlie proteotoxicity in RC disease.
Figure 4.
Cycloheximide partially and selectively inhibits translation to rescue cell viability and total cellular respiratory capacity in direct RC inhibition, in a nutrient-dependent but RC complex site independent fashion. (A) The rate of cytosolic/ER protein synthesis is unchanged with rotenone but partially reduced by cycloheximide. Solid black line indicates control protein synthesis rate. Human fibroblasts were treated for 24 h with cycloheximide (CHX) at either 3.6 or 50 µm concentration alone or with 100 nm rotenone (ROT). ***P < 0.001 by Student's t-test relative to no treatment control. (B) Cycloheximide has a selective effect on cytosolic translation. Human podocytes were treated with either 50 nm rotenone alone or in combination with 3.6 µm CHX for the indicated time periods and then labeled for 40 min with 35S-Cys/Met. TCA precipitable counts were determined for each cell lysate. Portions of lysates containing approximately equal amounts of incorporated radioactivity were analyzed by SDS–PAGE. Protein bands were detected by fluorography. Arrowheads, protein bands whose intensity either increases (open arrowheads) or decreases (filled arrowheads) with the addition of CHX relative to rotenone treatment alone. (C) Cycloheximide preserves cell morphology despite RC inhibition. Healthy human podocytes were cultured for 48 h in RPMI-1640, 10% FBS and 11 mm glucose medium alone (left panel), with 50 nm rotenone (center panel) or with 50 nm rotenone and 1.8 µm cycloheximide (right panel). (D) Maintenance of cellular viability by cycloheximide with prolonged RC inhibition is nutrient-dependent. Human podocytes were treated at 33°C for 6 days with 50 nm rotenone (ROT) to inhibit RC complex I in DMEM, 10% FBS and 1.8 µm cycloheximide (CHX) with serial concentrations of glucose (11 mm, 5.6 mm, 2.8 mm, no glucose). n = 3 per condition. Shapes and error bars convey mean and standard deviation, respectively. Statistical significance was determined at each glucose concentration of rotenone plus cycloheximide relative to rotenone alone by Student's t-test. For both 11 mm glucose and 5.6 mm glucose P < 0.01 on Day 1 and P < 0.001 on Days 2 through 6; for 2.8 mm glucose P < 0.05 on Days 1 and 3 and P < 0.001 on Day 2; and for no glucose condition P < 0.05 on Day 1 only. (E) Maintenance of cellular viability by cycloheximide is independent of the RC complex site inhibited. Human podocytes were cultured at 33°C for 48 h in RPMI-1640, 11 mm glucose, 10% FBS medium alone or treated with 50 nm rotenone (complex I inhibitor), 50 nm antimycin A (complex III inhibitor) or 100 nm oligomycin (complex V inhibitor), with or without 3.6 µm cycloheximide (CHX). n = 3 per condition. Bars and error bars convey mean and standard deviation, respectively. ***P < 0.001 for each inhibitor either relative to control or relative to the same inhibitor plus cycloheximide, by Student's t-test. (F) Cellular respiratory capacity is preserved by cycloheximide despite direct RC complex I inhibition. High-resolution respirometry (Oxygraph 2K, Oroboros) was performed in healthy human podocytes in 11 mm glucose media alone or with 24 h exposure to 50 nm rotenone (ROT) and/or 1.8 µm cycloheximide (CHX). Respiratory states measured included routine (basal), leak (non-mitochondrial) and maximal (ETS, electron transport system). n = 3 per condition. Bars and error bars convey mean and standard deviation, respectively. **P < 0.01 and ***P < 0.001 by Student's t-test.
Cycloheximide rescues RC disease cell viability, morphology, respiratory capacity and mitochondrial content in a nutrient-dependent fashion
Cycloheximide (micromolar-range) treatment sufficient to rescue human fibroblast viability at 48 h from rotenone-based RC complex I inhibition was associated with maintenance of normal cell morphology (Fig. 4C). When testing cycloheximide effects in a genetic model of human RC disease, we achieved complete rescue of cell viability for 7 days despite exposure to an otherwise lethal RC complex III inhibitor, antimycin A, both in healthy fibroblasts and in FBXL4-mutant complex I–III deficient patient fibroblasts (Supplementary Material, Fig. S6a). Thus, cycloheximide benefits both pharmacologically inhibited and genetic disease-based human cell models of primary RC disease.
The ability of cycloheximide treatment to sustain cellular viability with prolonged RC inhibition was strongly glucose-dependent (Fig. 4D), where progressive loss of cell viability was achieved as glucose concentration was progressively halved and no benefit was obtained in glucose-free media. This result was not unexpected, as RC inhibited cells require glucose-dependent glycolytic activity to maintain cellular energy production. Protective effects of micromolar-range cycloheximide in normal glucose conditions (11 mm) were consistent regardless of the site of RC inhibition being at complex I (rotenone), complex III (antimycin A) or complex V (oligomycin) (Fig. 4E). As expected, RC inhibition at each of these sites inhibited total cellular respiratory capacity under both basal (routine, measuring integrated OXPHOS capacity of complexes I through V) and maximal (electron transport system, ETS, measuring integrated OXPHOS capacity of complexes I through IV) conditions (Fig. 4F; Supplementary Material, Fig. S6b and c). Surprisingly, cycloheximide treatment significantly rescued total cellular respiratory capacity in cells treated with each of these potent RC complex inhibitors (Fig. 4F; Supplementary Material, Fig. S6b and c). Thus, the ability of cycloheximide to maintain cell viability in the setting of RC dysfunction appeared likely to involve an upregulation in mitochondrial mass that was sufficient to maintain total cellular respiratory capacity despite a targeted reduction in the OXPHOS capacity of each mitochondrion.
To directly test whether cycloheximide treatment improved mitochondrial mass or physiology, rotenone-treated human podocytes were evaluated for relative mitochondrial content (Supplementary Material, Fig. S7a) and function at the level of mitochondrial membrane potential (Supplementary Material, Fig. S7b), which requires intact OXPHOS activity. Indeed, both mitochondrial mass and membrane potential were significantly reduced with rotenone treatment, and rescued by micromolar-range cycloheximide treatment. Further, cycloheximide treatment alone significantly improved mitochondrial membrane potential in uninhibited control cells (Supplementary Material, Fig. S7b). Similar effects were achieved in human fibroblasts obtained from a patient with FBXL4-based complex I–III deficiency (Supplementary Material, Fig. S7c), with a corresponding significant reduction of mitochondrial superoxide burden upon cycloheximide treatment (Supplementary Material, Fig. S6d). Quantitation of in vivo mitochondrial physiology following 24 h of cycloheximide treatment in RC-deficient gas-1(fc21) worms revealed similar findings of significantly improved mitochondrial mass and membrane potential, as were, respectively, assessed by relative fluorescence quantitation of terminal pharyngeal bulb mitotracker green and TMRE levels (Supplementary Material, Fig. S7d). Superoxide levels appeared to increase with cycloheximide treatment in this mutant worm model, although this result is most likely attributable to their increased mitochondrial mass, considering that the relative degree of oxidant increase was less than the increase observed in their mitochondrial content (Supplementary Material, Fig. S7d). This is an important caveat since matrix oxidant burden is the product of mitochondrial mass and oxidant levels per mitochondrion. In comparison, we studied effects on nematode mitochondrial physiology of the other drug treatments that we found to improve RC disease cell and animal viability. Rapamycin treatment in the mitochondrial deficient complex I mutant nematode model led to a significant but modest improvement in mitochondrial mass, whereas probucol treatment significantly reduced to a similar extent their mitochondrial mass, membrane potential and matrix superoxide burden (Supplementary Material, File S7d). Thus, the beneficial effect of either probucol or rapamycin treatment on animal lifespan could not be satisfactorily explained by the restoration of their mitochondrial physiology to that of wild-type worms. In contrast, while micromolar-range cycloheximide did not rescue complex I deficient gas-1(fc21) lifespan when administered throughout the full lifespan of the animal (Supplementary Material, Table S1 and Fig. S4), cycloheximide treatment for 24 h did improve toward wild-type levels all aspects of their mitochondrial physiology. Consistent with the quantifiable increases in mitochondrial mass observed in both cell and nematode RC disease models, cycloheximide treatment rescued levels of total cellular ATP, which were significantly reduced by rotenone-based RC inhibition (Supplementary Material, Fig. S7e). Overall, only direct inhibition of cytosolic translation with micromolar-range cycloheximide treatment consistently and significantly benefited both human cell and nematode RC disease models across the multi-dimensional aspects of mitochondrial physiology that were examined.
Cell death in RC disease occurs by autophagy, which is effectively inhibited by cycloheximide or lithium chloride
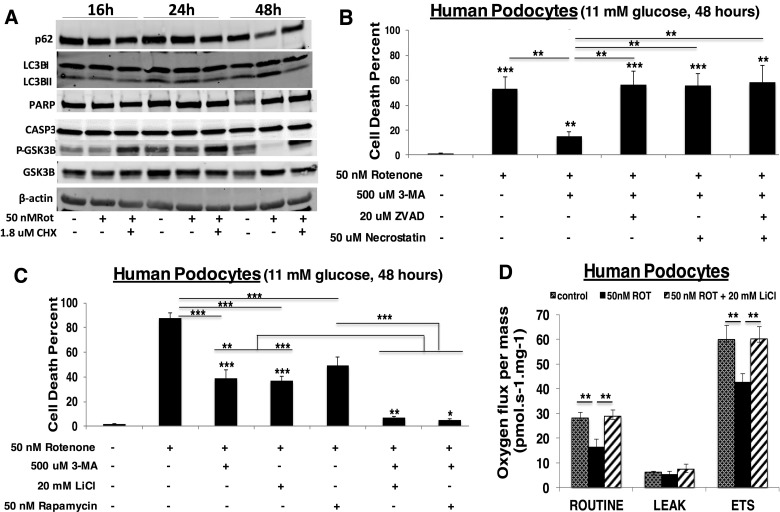
Given the dramatic ability of cycloheximide to rescue cell death from RC inhibition, we sought to determine the specific mechanism(s) involved (24). No activation of apoptosis (PARP or CASP3 cleavage) was evident upon RC inhibition by 50 nm rotenone for 48 h (Fig. 5A). Indeed, induction of apoptosis was only detectable at the level of CASP9 or PARP cleavage in human podocytes when very high (500 nm) concentrations of rotenone were used for short (6 h) duration (Supplementary Material, Fig. S8). However, cycloheximide effectively prevented the activation of autophagy that did occur by 48 h in podocytes (25) exposed to 50 nm rotenone treatment, as evidenced by p62 reduction, LC3B-II conversion and phospho-GSK3β reduction (Fig. 5A). When treated with pharmacological inhibitors of cell death that work at the level of autophagy (3-methyladenine) (26), apoptosis (ZVAD) (27) or necrosis (necrostatin) (28), only the autophagy inhibitor could rescue cell survival in rotenone (Fig. 5B). Given the clear loss of phospho-GSK3β (indicating its activation) that we detected with rotenone treatment (Fig. 5A), we tested whether a GSK3β inhibitor (29), lithium chloride, might also rescue cell viability from rotenone-induced RC dysfunction toxicity. Indeed, lithium chloride proved as effective as 3-methyladenine to preserve cell viability at 48 h, and had an even more pronounced extent than did 50 nm rapamycin (Fig. 5C). Most interesting was the dramatic synergistic effect on rescuing rotenone-induced cell death by combining autophagy inhibition from 3-methyladenine either with lithium chloride or rapamycin (Fig. 5C). Lithium chloride (GSK3β inhibitor) inhibits autophagy via an independent signaling pathway, whereas rapamycin (mTORC1 inhibitor) leads to partial inhibition of translation via inhibition of S6 phosphorylation with activation of autophagy via inhibition of 4E-BP1 (26). The profound protective effect on cell viability from RC inhibition that was achieved by combining inhibitors of autophagy and translation may relate to their off-target effects. However, we believe more likely explanations underlying these treatments' observed synergy are that autophagy can be more effectively inhibited by targeting it through independent signaling pathways, and/or that the cellular set-point at which autophagy changes from being pro-survival to pro-death is being reset. Indeed, lithium chloride (a known autophagy inhibitor) partially inhibited cellular translation activity, with a more pronounced effect in RC-inhibited than in control cells (Supplementary Material, Fig. S5A). Unlike cycloheximide (Fig. 4B), however, lithium chloride treatment did not selectivity modulate the translation of specific protein bands (Supplementary Material, Fig. S5B). Interestingly, lithium chloride treatment rescued not only cell viability (Fig. 5C) but also cellular respiratory capacity (Fig. 5D), a phenomenon that strongly suggests autophagy may play a contributory role in the pathogenesis of primary RC dysfunction. In other words, RC dysfunction appears to increase autophagy, which may reasonably include mitochondria-specific autophagy, or mitophagy. This conclusion is further supported by our detection of increased expression in FBXL4-mutant patient fibroblasts of the mitophagy marker, PINK1 (Supplementary Material, Fig. S9). Overall, these data suggest that inhibiting autophagy, either indirectly by cycloheximide treatment or with more direct autophagy inhibitors such as 3-methyladenine or lithium chloride, may have beneficial effects on both cellular viability and mitochondrial respiratory capacity in the setting of RC disease.
Figure 5.
Mechanistic investigation of cell death in mitochondrial dysfunction. (A) Cycloheximide prevents rotenone-induced autophagy. Western blot analysis of autophagy (p62, LC3BI/II, pGSK3β) and apoptosis (PARP, CASP3) activation markers in healthy human podocytes cultured in 10% FBS, RPMI, 11 mm glucose media with 50 nm rotenone and/or 1.8 µm cycloheximide (CHX) at 33°C for 16, 24 or 48 h. A representative blot is shown from three biological replicate experiments having identical results. (B) Inhibition of autophagy-mediated mitochondrial degradation in direct RC inhibition preserves cell viability. Human podocytes were exposed to 50 nm rotenone with 500 µm 3-methyladenine (3-MA) alone or combined with 20 µm ZVAD and/or 50 µm necrostatin at 33°C for 16, 24 and 48 h. n = 3 per condition. Bars and error bars convey mean and standard deviation, respectively. **P < 0.01 and ***P < 0.001 by Student's t-test. Asterices directly above bars convey significance relative to control, whereas asterices above horizontal lines convey significance of specified group comparisons. (C) Downregulation of both cell translation and mitochondrial degradation in direct RC inhibition maximally preserves cell viability. Human podocytes were cultured in 10% FBS, RPMI, 11 mm glucose and exposed for 48 h to 50 nm rotenone alone or with 500 µm 3-MA, 20 mm lithium chloride (LiCl) and/or 50 nm rapamycin. n = 3 per condition. Bars and error bars convey mean and standard deviation, respectively. *P < 0.05; **P < 0.01; and ***P < 0.001 by Student's t-test. Asterices directly above bars convey significance relative to control, whereas asterices above horizontal lines convey significance of specified group comparisons. (D) Inhibiting autophagy by lithium chloride treatment in direct complex I inhibition maintains total cellular total respiratory capacity. High-resolution respirometry (Oxygraph 2K, Oroboros) was performed in healthy human podocytes in 11 mm glucose media exposed for 24 h to 50 nm rotenone (ROT) alone or with 20 mm LiCl. Respiratory states measured included routine (basal), leak (non-mitochondrial) and maximal (ETS, electron transport system). n = 3 per condition. Bars and error bars convey mean and standard deviation, respectively. **P < 0.01 by Student's t-test.
Cycloheximide effects in RC dysfunction are mediated by induction of mitochondrial translation and reduction of both proteotoxic and ER stress
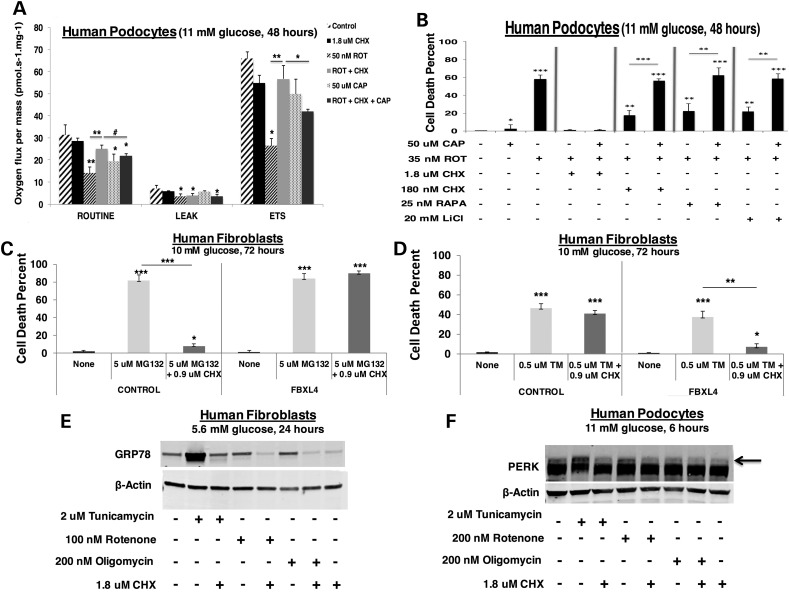
Given the surprising ability of cycloheximide-based cytosolic translation inhibition to rescue total cellular OXPHOS capacity despite direct RC inhibition, we investigated whether cycloheximide invoked a selective upregulation of mitochondrial translation. While specifically inhibiting mitochondrial translation with chloramphenicol significantly reduced total cellular OXPHOS capacity, chloramphenicol treatment only partially reversed cycloheximide's protective effect on total cellular respiratory capacity in rotenone-mediated RC inhibition (Fig. 6A). Further, the reduction of total cellular respiratory capacity that was seen with chloramphenicol treatment did not translate to increased cell death, as occurred with rotenone-based RC inhibition (Fig. 6B). Nor did chloramphenicol reverse the cytoprotective effects of cycloheximide (1.8 µm) on rotenone-induced cell death (Fig. 6B). Chloramphenicol treatment, however, did partially reverse the cell viability effects of cycloheximide with RC inhibition at either complex I (rotenone) or V (oligomycin) from a log-order lower dose of cycloheximide (180 nm) (Fig. 6B; Supplementary Material, Fig. S10a). An exception was that chloramphenicol did not significantly reverse low-dose cycloheximide effect with RC inhibition at complex III (antimycin A) (Supplementary Material, Fig. S10b). In contrast to the other RC inhibitors, antimycin A not only inhibits RC flux at complex III but also directly induces significant oxidative stress (30). Similar to effects seen when combined with cycloheximide treatment, chloramphenicol only reversed the partial cytoprotective effects of rapamycin and lithium chloride with RC inhibition at complex I (rotenone) (Fig. 6B) but not III (antimycin A) (Supplementary Material, Fig. S10b). Overall, these data suggest that cycloheximide-mediated inhibition of cytosolic translation in the setting of RC dysfunction upregulates mitochondrial translation, but this effect alone is insufficient to account for the dramatic health benefit of cycloheximide treatment.
Figure 6.
Cycloheximide treatment in RC-inhibited cells induces mitochondrial translation, rescues cellular ATP levels, and requires intact proteasome and glycosylation capacity. (A) Chloramphenicol partially reverses the protective effect of cycloheximide on total cellular respiratory capacity in RC complex I inhibited podocytes. High-resolution respirometry (Oxygraph 2K, Oroboros) was performed in healthy human podocytes in 11 mm glucose media exposed for 24 h to 50 nm rotenone (ROT) and/or 1.8 µm cycloheximide (CHX) with or without 50 µm chloramphenicol (CAP). Respiratory states measured included routine (basal), leak (non-mitochondrial) and maximal (ETS, electron transport system). n = 3 per condition. Bars and error bars convey mean and standard deviation, respectively. #P < 0.1; *P < 0.05; **P < 0.01; and ***P < 0.001 by Student's t-test. Asterices directly above bars convey significance relative to control, whereas asterices above horizontal lines convey significance of specified group comparisons. (B) Cycloheximide rescues cell viability in rotenone-treated human podocytes. Human podocytes were exposed alone or in combination to 50 µm chloramphenicol (CAP), 35 nm rotenone (ROT), 1.8 µm CHX, 25 nm rapamycin (RAPA) or 20 mm lithium chloride (LiCl) for 48 h in 11 mm glucose. n = 3 per condition. *P < 0.05; **P < 0.01; and ***P < 0.001 by Student's t-test. Asterices directly above bars convey significance relative to control, whereas asterices above horizontal lines convey significance of specified group comparisons. (C) Cycloheximide protection of cell viability in genetic-based RC disease requires intact proteasome activity. Control or FBXL4-mutant human fibroblasts were grown in 10 mm glucose for 72 h alone or with 5 µM MG-132 (to inhibit proteasome activity) and/or 0.9 µM CHX. n = 3 per condition. Bars and error bars convey mean and standard deviation, respectively. *P < 0.05 and ***P < 0.001 by Student's t-test. Asterices directly above bars convey significance relative to respective control conditions, whereas asterices above horizontal lines convey significance of specified group comparisons. (D) Cycloheximide protects cells from ER stress in genetic-based RC disease. Control or FBXL4-mutant human fibroblasts were grown in 10 mm glucose for 72 h alone or with 0.5 µm tunicamycin (TM, to induce ER stress) and/or 0.9 µm CHX. n = 3 per condition. Bars and error bars convey mean and standard deviation, respectively. *P < 0.05; **P < 0.01; and ***P < 0.001 by Student's t-test. Asterices directly above bars convey significance relative to respective control conditions, whereas asterices above horizontal lines convey significance of specific group comparisons. (E) Cycloheximide prevents activation of ER stress by RC inhibition at either complex I or V for 24 h. GRP78 protein expression was quantified by western blot analysis in healthy human fibroblasts cultured in DMEM containing 10% FBS with 5.6 mm glucose for 24 h and treated with either 2 µm tunicamycin, 200 nm rotenone or 100 nm oligomycin alone or with 1.8 µm CHX. (F) Cycloheximide prevents activation of ER stress by RC inhibition at either complex I or V for 6 h. PERK protein expression was quantified by western blot analysis in healthy human podocytes cultured in RPMI with 10% FBS and 11 mm glucose for 6 h and treated with either 2 µm tunicamycin, 200 nm rotenone or 200 nm oligomycin alone or with 1.8 µm CHX.
Therefore, we sought to investigate other possible mechanisms contributing to the beneficial health effect of cycloheximide. We specifically interrogated effects of cycloheximide treatment on key components of proteotoxic stress, since this has been recently recognized to be activated in RC dysfunction (20). Furthermore, it is plausible that inhibiting cell translation can resolve proteotoxic stress by rebalancing relative protein production to degradation capacity, as would be consistent with our discovery of the selective nature of cycloheximide's inhibition of cytosolic translation (Fig. 4B). Indeed, whereas cycloheximide treatment can protect from cell death upon proteasome inhibition (with MG-132) in otherwise healthy cells, no protection was attained in RC deficient FBXL4-mutant cells (Fig. 6C). This result demonstrates that cycloheximide protection of cell viability in the setting of RC disease requires intact proteasome activity. Interestingly, cycloheximide could only rescue cell death in FBXL4-mutant RC disease cells, but not in healthy cells, upon induction of ER stress by using tunicamycin to inhibit protein glycosylation (Fig. 6D). This finding suggests that the beneficial effect of cycloheximide in RC deficient cells involves mitigation of their increased ER stress.
Under ER stress, eIF2α becomes phosphorylated by PKR-like ER resident kinase (PERK), causing a repression of global translation but upregulated translation of specific gene transcripts such as activating transcription factor 4 (ATF4) (31–33). Indeed, ATF4 activation has previously been reported to occur with RC inhibition at complex I (34) or complex V (35). Similarly, we saw a 48% average upregulation of ATF4 in human fibroblasts treated with 50 nm rotenone for 48 h, which was significantly reduced by cycloheximide treatment to 36% of untreated control values (P = 0.017, n = 3, qRT–PCR gene expression data not shown). Western blot analysis of glucose-regulated protein 78 (GRP78), a major ER stress sensor that interacts with unfolded protein response activators (36), similarly suggested that ER stress at the level of GRP78 induction is progressively induced in human podocytes by 24 h of RC inhibition at either complex I or V (Fig. 6E), with a similar degree of PERK activation evident by this time (Supplementary Material, Fig. S11a). Interestingly, GRP78 activation became more pronounced with an increasing duration and degree of complex I inhibition (Supplementary Material, Fig. S11b), and also occurred with inhibition of the proteasome by MG-132 or of protein glycosylation by tunicamycin (Supplementary Material, Fig. S11c). In contrast, shorter-term RC inhibition for 6 h at either complex I (rotenone) or V (oligomycin) resulted in more pronounced PERK activation with rotenone treatment, to a similar degree as occurred with tunicamycin exposure (Fig. 6F; Supplementary Material, Fig. S12a). eIF2α phosphorylation was also increased by rotenone exposure for 6 h (Supplementary Material, Fig. S12). Thus, multiple read-outs of the ER stress pathway consistently showed that direct RC inhibition activates ER stress. Cycloheximide at micromolar concentrations in both human fibroblasts (Fig. 6E) and in human podocytes (Fig. 6F; Supplementary Material, Fig. S11a), regardless of whether exposed for either 6 or 24 h to direct RC inhibitors, substantially decreased both GRP78 and PERK expression. Cycloheximide also prevented GRP78 activation in response to either proteasome inhibition or tunicamycin-induced ER stress (Supplementary Material, Fig. S11c). These data strongly suggest that low-dose (micromolar-range) cycloheximide effectively inhibits the activation of ER stress that is induced by primary RC dysfunction.
RNAseq transcriptome profiling confirms that rapamycin, probucol and cycloheximide modulate translation and autophagy in mitochondrial complex I-deficient nematodes
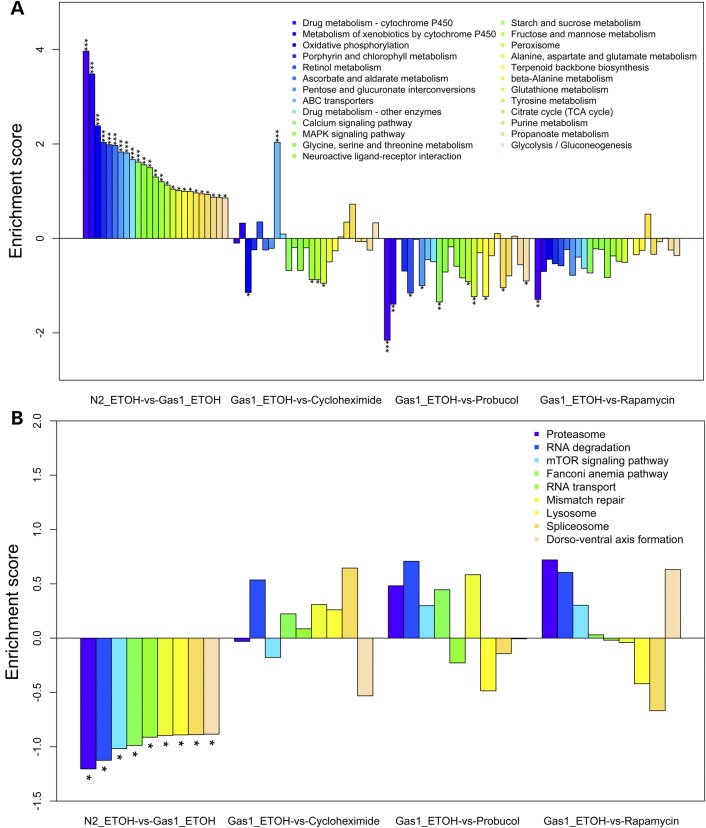
To interrogate the broad cellular mechanisms of these drug treatments in the setting of complex I deficiency, RNAseq analysis was performed in the C. elegans gas-1(fc21) model. Synchronized young adult worms were treated for 24 h with either 2.5 µm cycloheximide, 5 µm probucol, or 2.5 nm rapamycin and compared with gas-1(fc21) and N2 Bristol (wild-type) control worms exposed only to ethanol buffer. Similar to what we previously observed by microarray-based transcriptome profiling (37), pathway-level profiling confirmed that many intermediary metabolic and cell defense pathways, most notably oxidative phosphorylation and P450 metabolism, were upregulated in gas-1(fc21) mutants relative to wild-type worms (Fig. 7A). Pronounced and widespread normalization of these pathways occurred with probucol treatment, and to a lesser extent with rapamycin and cycloheximide (Fig. 7A). Validation by quantitative RT–PCR analysis confirmed that upregulation in gas-1(fc21) of the major mitochondrial antioxidant-defense enzyme, manganese superoxide dismutase (sod-3), was significantly decreased by either probucol or cycloheximide treatment (Supplementary Material, Fig. S13A and B). Cycloheximide treatment also normalized the upregulated expression in these animals of the C. elegans homolog of nrf (skn-1) that induces mitochondrial biogenesis (Supplementary Material, Fig. S13B). The most downregulated pathways in gas-1(fc21) worms relative to wild-type N2 controls were ‘proteasome’, ‘RNA degradation’ and ‘mTOR signaling pathway’, all three of which were largely reversed by treatment with either probucol or rapamycin (Fig. 7B). However, cycloheximide treatment reversed only the transcriptional downregulation seen in the ‘RNA degradation pathway’, without having any impact on the other two most downregulated KEGG pathways (Fig. 7B).
Figure 7.
Pathway-level RNAseq transcriptome effects of cycloheximide, probucol or rapamycin in mitochondrial complex I mutant adult worms. Effects of drug treatments on (A) significantly upregulated and (B) significantly downregulated KEGG pathways in gas-1(fc21) relative to N2 (wild-type) worms. The enrichment score is shown for KEGG pathways with significant dysregulation in gas-1(fc21) worms relative to N2 (P < 0.05), where ethanol was used as the buffer control. The relative effects on each of these pathways in gas-1(fc21) worm following 24 h treatments with either cycloheximide (2.5 µm), probucol (5 µm) or rapamycin (2.5 nm) are also shown, enabling assessment of the degree to which each treatment ameliorated dysregulation in individual KEGG pathways. Full results are provided in Supplementary Material, Fig. S1 ,Table S1, Fig. S13 and S14.
Transcriptome profiles were compared to assess common effects in this model of all three drugs. Each treatment significantly upregulated in gas-1(fc21) the KEGG pathways that were significantly downregulated in gas-1(fc21) relative to wild-type worms: ‘phagosome’, ‘DNA replication’, ‘pantothenate and CoA biosynthesis' and ‘RNA degradation’ (Supplementary Material, Fig. S1 and Fig. S14A–C). ‘Arachidonic acid metabolism’ was also commonly upregulated in gas-1(fc21) by each of the drug treatments, although it was not significantly changed in gas-1(fc21) relative to wild-type worms. Conversely, each treatment significantly downregulated KEGG pathways that were significantly upregulated in gas-1(fc21) relative to wild-type worms: ‘fatty acid degradation’, ‘fatty acid biosynthesis', ‘fructose and mannose metabolism’, ‘calcium signaling pathway’ and ‘peroxisome’ (Supplementary Material, Fig. S1 and Fig. S14A–C). Overall, the autophagy-related ‘phagosome’ pathway was the most significantly upregulated KEGG-defined basic cellular pathway in gas-1(fc21) young adult animals treated for 24 h with either probucol or rapamycin (Supplementary Material, Fig. S14B and C), with its marginal upregulation also seen from cycloheximide treatment (Supplementary Material, Fig. S14A). Further, ‘regulation of autophagy’ was significantly downregulated by each of the drug treatments in gas-1(fc21) worms, although this KEGG pathway was not significantly altered in gas-1(21) relative to N2 wild-type controls. The most pronounced impact on both the ‘phagosome’ and ‘regulation of autophagy’ pathways occurred with probucol treatment, for which gene-level expression changes within each pathway are shown upon comparison of drug-treated versus buffer-only treated control gas-1(fc21) worms (Supplementary Material, Fig. S14).
As expected, given its established role as a direct inhibitor of cytosolic translation, cycloheximide treatment in gas-1(fc21) animals also upregulated the expression of multiple pathways involved in translation and transcription. This included a significant upregulation of ‘ribosome’ (P < 0.001) and marginally significant upregulation of ‘protein processing in endoplasmic reticulum’, ‘splicesome’, ‘protein export’, ‘ribosome biogenesis in eukaryotes', ‘RNA degradation’ and ‘aminoacyl-tRNA biosynthesis'. Most interesting was the finding that the most upregulated pathway in gas-1(fc21) animals upon cycloheximide treatment was ‘ABC transporters’ (Supplementary Material, Table S1 and Fig. S14A), which are ATP-dependent membrane transporters having several members of subfamilies B and C that show mitochondrial localization (38). In particular, cycloheximide resulted in the significant upregulation of several members of subfamily B (ABCB1, ABCB5, ABCB9 and to a lesser degree, ABCB8) and subfamily C (ABCC5), as well as the modest downregulation of ABCB7 and ABCD3 (Supplementary Material, Fig. S13C). It is intriguing to postulate these transporters may play a role in the selective changes in cellular translation that we observed in RC mutant cells treated with cycloheximide (Fig. 4) (39).
Discussion
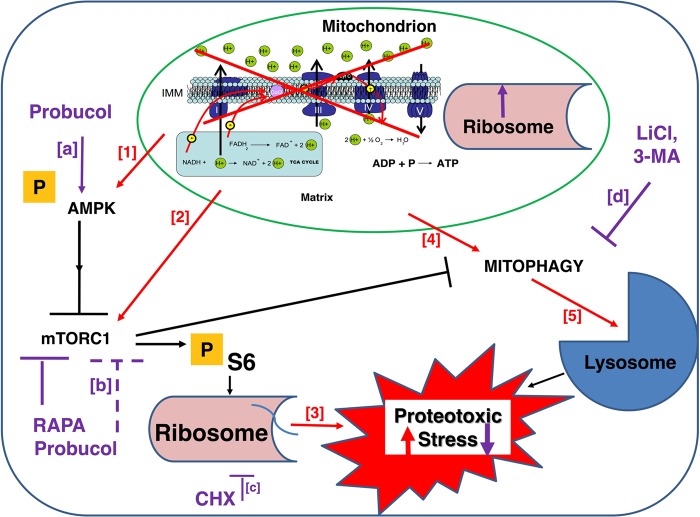
Diverse murine, C. elegans, and human cell models of primary RC disease have upregulated activities of mTORC1 (pS6) (Fig. 1) and AMPK (Supplementary Material, Fig. S1). Improved viability and health in these RC disease models can be effectively achieved, to variable degrees, by direct pharmacological inhibition of mTORC1 or its major downstream targets, cytosolic translation and autophagy. A schematic model is provided (Fig. 8) to depict the general cellular response that our data demonstrate occurs consistently in mitochondrial RC dysfunction, both within key nodes of the nutrient-sensing signaling network (AMPK, mTORC1) as well as in interacting organelles (ribosome, lysosome) and processes (translation, autophagy/mitophagy) that modulate cellular proteotoxic stress. This model further highlights novel therapeutic opportunities that exist to restore cellular health in mitochondrial RC disease by modulating these central determinants of proteotoxic stress (Fig. 8). Indeed, treatment with an mTORC1 inhibitor, rapamycin, significantly ameliorated renal disease in B6.Pdss2kd/kd mice with deficient coenzyme Q biosynthesis leading to RC complexes I–III and II–III deficiencies; improved animal viability and mitochondrial physiology in gas-1(fc21) nematodes with RC complex I deficiency; and prevented autophagy-mediated death in rotenone-treated human cells with RC complex I inhibition (Fig. 2). Even greater beneficial effect in each of these RC disease models was achieved by treatment with probucol (Fig. 2), which we previously showed activates PPAR pathways and here demonstrated also effectively activates AMPK (Supplementary Material, Fig. S1) and inhibits mTORC1 (Fig. 1) activities. Yet, the most pronounced and sustained health benefits in cellular RC disease models were obtained by partial, direct translation inhibition. Low-dose (1–3 µm range) cycloheximide treatment significantly rescued viability (Figs 3 and 4E; Supplementary Material, Fig. S6A) by inhibiting RC-disease induced autophagy (Fig. 5A) and preserving total cellular respiratory capacity (Fig. 4F; Supplementary Material, Fig. S6B and C) and ATP levels (Supplementary Material, Fig. S7E) across diverse human cell models of genetic and/or pharmacological RC inhibition at complexes I, III or V. The surprising health benefits of low-dose (micromolar-range) cycloheximide in RC disease further appears attributable to its reduction of proteotoxic and ER stress, as well as its secondary induction of mitochondrial translation and biogenesis (Fig. 6), likely via a uniquely selective reduction of potentially toxic proteins (Fig. 4B). Direct inhibition of autophagy by either 3-methyladenine or lithium chloride in these same RC disease models also led to a significant, and synergistic, rescue of cell viability and total cellular respiratory capacity (Fig. 5). Ultimately, our data demonstrate that cycloheximide and lithium chloride each leads to the significant inhibition of both cytosolic translation and autophagy in RC disease models. RNAseq-based transcriptome profiling of drug treatment effects in RC complex I deficient gas-1(fc21) mutant worms provided further evidence that these therapies effectively restore their altered expression of translation and autophagy pathways toward that of wild-type animals (Fig. 7). Collectively, these data identify novel mechanisms underlying the pathogenesis of mitochondrial RC dysfunction, including the combined induction of proteotoxic stress, the ER stress response and autophagy.
Figure 8.
Schematic model of general cellular response, and corresponding therapeutic approaches to restore cellular health, in mitochondrial RC dysfunction. Blue square and green oval depict a generic cell and mitochondrion, respectively. Arrows and bars convey activating and inhibiting effects, respectively. Orange ‘P’ indicates molecules whose activity is regulated by phosphorylation. Red font and arrows convey the direct effects on major components of cellular signaling and individual organelle responses to mitochondrial dysfunction, which culminate in increased proteotoxic stress. Bracketed red numbers highlight RC disease effects, which is supported by our experimental evidence as follows: (1) Supplementary Material, Figure S1A, C, F, G, H and I; (2) Supplementary Material, Figure S1A, D, E, F and G; (3) Figure 6E and F; (4) Figure 5A–C; and (5) Figure 7B. Purple font, arrows, and bars convey specific sites and consistent effects (solid lines, direct effect; dashed lines, likely indirect effect), respectively, of pharmacological agents that restore cellular physiological balance and cellular viability in the setting of RC dysfunction. Bracketed purple letters highlight individual drug effects, for which our supporting experimental evidence includes: (a) Supplementary Material, Figure S1B, D and E; (b) Figure 1B, C, E; (c) Figures 4B, 7 and Supplementary Material, Figure S5A; and (d) Fig. 5B. RAPA, rapamycin. CHX, cycloheximide. LiCl, lithium chloride. 3-MA, 3-methyladenine.
Most importantly, these findings suggest that partial inhibition of cytosolic translation and/or autophagy may offer novel treatment strategies for the wide range of human diseases whose pathogenesis involves mitochondrial RC dysfunction. For example, cycloheximide was shown to suppress mitochondrial degeneration in a yeast model of a human mitochondrial disease caused by a mutation in the adenine nucleotide translocase (ANT1) (40). Already available as an FDA-approved therapeutic agent for bipolar disorder, lithium chloride may be among the first agents in the class of ‘combined translation and autophagy inhibition therapy’ to warrant consideration for pursuit of clinical research trials in the setting of mitochondrial disease. Intriguingly, while mood disorders, including depression, are a common clinical problem in diverse mitochondrial RC diseases (41), lithium chloride has not been previously considered, or tested for multi-systemic benefit, in patients with mitochondrial diseases. Before pursuing empiric therapy in mitochondrial disease patients, however, it will be essential to conduct additional research investigations in mitochondrial disease patient cell lines, as well as in appropriate animal models and/or human patients, to identify specific molecular or biochemical subgroups of patients who might most substantially benefit from this treatment approach. It will also be important to determine whether additional therapeutic agents can be identified that achieve similar functional benefit at the level of translation and autophagy inhibition as seen with cycloheximide but have a more acceptable safety profile in human patients. The goal of such therapy would be to safely and effectively modulate the rheostat of cytosolic translation activity sufficiently to mitigate proteotoxic stress and potentially restore overall cellular health in RC disease. Pharmacological agents that target multiple aspects of the disrupted cellular signaling network that occurs in RC disease also hold promise to treat the wide spectrum of clinical manifestations that may occur. For example, probucol simultaneously activates PPAR and AMPK while inhibiting mTORC1 in the mouse and cellular RC disease models we tested. Probucol also offers an effective means to treat dyslipidemia, which is itself a known complication of mitochondrial RC disease (42). However, probucol is not a currently available medication in the USA and must be systematically studied to demonstrate safety and efficacy in rigorous clinical trials within the mitochondrial disease population (43) before its therapeutic use could be clinically contemplated. Despite such practical challenges, improved understanding of the broader pathogenesis of mitochondrial disease that we have gained offers novel insights into promising therapeutic targets that may lead to improved health for individuals living with the currently untreatable manifestations of RC disease.
Materials and Methods
Fibroblast cell line culture
Fibroblast cell lines (FCLs) were obtained from prior skin biopsies when available and/or established in the Clinical CytoGenomics Laboratory from skin biopsies performed in Mitochondrial-Genetics Diagnostic Clinic at The Children's Hospital of Philadelphia (M.J.F.). Informed consent was obtained per The Children's Hospital of Philadelphia Institutional Review Board approved study #08-6177 (M.J.F., PI). FCLs were grown in Dulbecco's modified Eagle's medium (DMEM, Gibco) containing 1 g/l glucose and supplemented with 20% FBS (Gibco), 1 mm sodium pyruvate (CellGro), 2 mm l-glutamine and 50 µg/ml uridine (Calbiochem). FCLs for western analysis were grown to 80–100% confluence in T75 flasks at 37°C before treatment with varying concentrations and duration of drug inhibitors or therapies.
Human podocyte culture
Conditionally immortalized human podocytes were a gift from Dr Duncan Johnstone (University of Pennsylvania, Philadelphia, PA, USA). The human podocyte cell line was cultured in RPMI 1640-based medium (25 mm glucose) supplemented with 10% fetal bovine serum (FBS) (Invitrogen), 2 g/l of sodium bicarbonate (NaHCO3), insulin–transferrin–selenium (ITS) supplement (Sigma-Aldrich) and 200 units/ml penicillin and streptomycin (Roche Applied Science), as described previously (44). Podocytes were grown in collagen-coated culture dishes at 33°C and 5% CO2.
Pdss2 gene siRNA knockdown
All materials were purchased from Santa Cruz, Inc. In six-well, 60 mm tissue culture dishes, human podocytes were grown to 50–70% confluence in antibiotic-free RPMI-1640, 10% FBS normal growth medium. Solution A: for each transfection, 15 μl of Pdss2 siRNA (sc-76100) were diluted into 135 μl shRNA Plasmid Transfection Medium (sc-108062). Solution B: for each transfection, 1–6 μl of shRNA Plasmid Transfection Reagent (sc-108061) were diluted with sufficient shRNA Plasmid Transfection Medium to bring the final volume to 150 μl. shRNA Plasmid DNA solution (Solution A) was added directly using a pipette to the dilute shRNA Plasmid Transfection Reagent (Solution B) and mixed gently by pipetting up and down, then incubated for 30 min at room temperature. Cells were washed twice with 2 ml of shRNA Transfection Medium and media was then aspirated. For each transfection, 1.5 ml shRNA Plasmid Transfection Medium was added to each well. Two hundred microliters shRNA Plasmid DNA/shRNA Plasmid Transfection Reagent Complex (Solution A + Solution B) was added drop-wise to each well to cover the entire layer and gently mixed by swirling. Cells were incubated for 6 h at 33°C in a CO2 incubator. Following incubation, 1.5 ml of antibiotic-free RPMI-1640 in 20% FBS normal growth medium was added and cells were incubated for an additional 24 h. Media was then replaced with fresh antibiotic-free RPMI-1640 in 10% FBS normal growth medium. Assays were performed in cells after an additional 48 h incubation in this media.
B6.Pdss2kd/kd missense mutant mice treatment groups
B6.Pdss2kd/kd mice were derived by backcrossing a recombinant chromosome derived by positional cloning onto the B6 background (45). B6.Pdss2kd/kd mice studied in this report were in the 13th backcross generation. Untreated mice were fed a standard LabDiet rodent chow obtained from PMI Nutrition International (Brentwood, MO, USA). Probucol-treated mice were given the same diet to which 1% w/w probucol was added (Animal Specialties and Provisions, Quakertown, PA, USA), which we estimate resulted in a final daily dosage of approximately 95 mg per kg body weight. CoQ10-treated mice received a standard chow with CoQ10 (LiQsorb; Tishcon Corp., Westbury, NY, USA) added to their drinking water from weaning (at 4 weeks of life). The amount of CoQ10 added was 1 mg/ml, which we estimate resulted in a final daily dosage of ∼400 mg CoQ10 per kg body weight. Rapamycin containing chow (225 PPM) was obtained from Dr Elizabeth Fernandez at The University of Texas. All studies were carried out in accordance with NIH guidelines and approved by the Institutional Animal Care and Use Committees of The University of Pennsylvania and The Children's Hospital of Philadelphia.
Mouse urine collection and albumin assay
B6.Pdss2kd/kd mice were placed in metabolic cages without food for 24 h with 0.45% NaCl and 2.5% sucrose in the drinking water. Total urine volumes were measured and aliquots of urine were tested for albumin concentration by enzyme-linked immunosorbent assay (ELISA). A mouse albumin ELISA quantitative kit was obtained from Bethyl Laboratories, Inc. High-binding capacity ELISA plates (Nunc, No: 442404) were coated with 100 μl of purified goat anti-mouse albumin antibody (10 μl/ml) in 50 mm carbonate-bicarbonate buffer, pH 9.6, by incubating at 4°C overnight. After several washes with distilled water, the wells were blocked with 200 μl of phosphate buffered saline (PBS)-1% bovine serum albumin (BSA) for 2 h at room temperature followed by washing three times with PBS-0.05% Tween 20. To each well was added 100 μl of serial diluted samples or standards, followed by incubation at room temperature for 1 h. After incubation, the plates were washed four times with PBS-0.05% Tween 20, and then incubated with 100 μl of goat anti-mouse albumin antibody conjugated with HRP at room temperature for 1 h. After washing four times with PBS-0.05% Tween 20, the reaction was incubated with 100 μl ABTS solution at room temperature for 20 min, and then with 100 μl 2 M H2SO4 stopping solution. Each plate was read with an ELISA reader at 405 nm.
Trypan blue exclusion assay
Cell death was determined by Trypan blue dye exclusion test (46). The assay is based on the fact that the dye is negatively charged and does not interact with the cell unless the membrane is damaged. At the described time points, cells from each group were detached by trypsinization and subsequently collected by centrifugation. After resuspension in PBS, equal volumes of cell suspension and 0.4% Trypan blue (Invitrogen) were mixed and incubated for 10 min at room temperature. The number of dead cells (blue stained cells) was counted using a hemocytometer from five random fields using a counting grid. Data are expressed as percent of total cells/field ± standard deviation.
Cellular respiratory capacity quantitation in intact cells by Oxygraph-2K (Oroboros) analysis
Cells were cultured in tissue culture flasks until reaching 80% confluence and then prepared for respiratory capacity analysis by high-resolution polarography using an Oxygraph-2K (Oroboros, Austria), as previously described (47). Cells were removed from culture flasks with trypsinization and collected in fresh culture medium. Control (normal) and mitochondrial disease cell lines were simultaneously analyzed in two separate chambers in 2 ml volume containing 1 million cells per chamber. Experiments were performed as previously described (48). Respiration was measured at 37°C in culture medium (DMEM), where inhibitors for the different mitochondrial respiratory chain complexes were added to the cells in the following order: oligomycin (2 µg/ml) (Sigma) to inhibit complex V (to assess non-mitochondrial respiratory capacity or leak rate), carbonyl cyanide 4- (trifluoromethoxy) phenylhydrazone (FCCP) (Sigma) uncoupler with step-wise titration in 2.5–1.5 µm increments (to assess maximal electron transport system respiratory capacity rate), rotenone (Sigma) in 0.5 µm final concentration to inhibit complex I, and antimycin A (Sigma) in 2.5 µm final concentration to inhibit complex III. Data were analyzed using DatLab4 (Oroboros, Austria) software. Statistical analyses were performed using ANOVA and paired t-tests to compare group means.
Cellular ATP content analysis
Cultured cells were treated in 10 mm dishes with 50 nm rotenone with or without 1.8 µm cycloheximide for 24 h. Cells were then washed with cold PBS and lysed in 100 µl of ATP Assay Buffer (Abcam ATP assay kit, ab83355). Cells were centrifuged at 15 000g for 2 min at 4°C to pellet insoluble materials, after which the supernatant was collected and 50 µl was added to 96-well plates with ATP standard. Optical density was measured at 570 nm by colorimetric assay.
Fluorescence-activated cell sorting (FACS) analysis of mitochondrial physiological parameters in human cells
Cells were collected with trypsin, and resuspended in Hank's buffered salt solution (HBSS) to obtain 1–5 × 105 cells per sample, as previously reported (21,49). Samples were loaded with either 50 nm MitoTracker Green (MTG) or 40 nm TetraMethylRhodamine, Ethyl ester (TMRE) at 37°C for 30 min. Duplicate measurements were performed and unloaded control samples were run in parallel. Fluorescence-activated cell sorting (FACS) was performed with an Accuri C6 flow cytometer (BD Biosciences) equipped with a 488 nm laser with 530/30 nm emission for MTG and 585/42 nm emission for TMRE. A total of 10 000 events were recorded per condition. Background subtracted geometric means of MTG or TMRE were calculated as percent change relative to same day controls.
Preparation of human cell and mouse tissues for western blot
(i) Cells were washed with HBSS, trypsinized, flash frozen in 15 ml conical tubes and cell pellets were lysed for 30 min on ice in 200 μl cold cell lysis buffer containing 20 mm HEPES pH 7.9, 1.5 mm MgCl2, 10 mm KCl, 0.25% v/v nonidet P-40, 0.2 mm EDTA, 0.5 mm dithiothreitol (DTT), protease inhibitor and phosphatase inhibitor cocktail (1:100) and 0.5 mm PMSF (Sigma-Aldrich). Samples were centrifuged at 10 000g for 5 min at 4°C and supernatants were then transferred to fresh tubes. Protein concentration was determined by DC Protein Assay (Bio-Rad). Protein extract samples were treated with Laemmli Sample Buffer (Bio-Rad) with β-mercaptoethanol at 95°C for 4 min for immunoblot analyses. (ii) Lobes of livers were dissected from euthanized mice and crushed with a metal clamp pre-frozen in liquid nitrogen, and stored at −80°C until homogenization. For a 50 mg piece of tissue, cold 200 µl cell lysis buffer was added, the sample was completely homogenized, cold 800 µl cell lysis buffer was added, briefly vortexed, kept on ice for 30 min, briefly vortexed and then centrifuged for 10 min at 10 000g at 4°C in a microcentrifuge. The supernatant was aspirated and then the same steps were followed as detailed earlier for cell culture.
Phosphokinase immunoblot analyses
Hundred micrograms of protein was separated by SDS–PAGE on a 4–15% Tris–Glycine gradient gel (Bio-Rad) at 30 mA ×1 h, transferred to nitrocellulose membranes at 350 mA for 1.5 h and incubated in Odyssey blocking buffer for 1 h. Phospho-proteins were labeled with rabbit anti-phospho antibodies at 1:2000 dilutions at 4°C for 1 h or overnight. Membranes were incubated with Odyssey IRDye goat anti-rabbit or anti-mouse secondary antibody at 1:10 000 dilutions for 30 min. Protein bands were visualized by the Odyssey Infrared Imaging System (LI-COR Biosciences). Specific antibodies used for immunoblot analyses were phospho-S6 ribosomal protein (2215), S6 ribosomal protein (2217), phospho-AMPK, AMPK, cleaved PARP, cleaved CASP9, P62, LC3B-II, phospho-GSK3β, PERK, eIF2α, phospho-eIF2α (Cell Signaling Technology), PINK1 (BC100-494, Novus Biologicals), GRIM19 (ab110240), SDHA (ab14715), UQCRC2 (ab14745), MTCO1 (ab14705) and ATP5A (ab14748) (MitoSciences). β-tubulin (Cell Signaling) and/or β-actin (Cell Signaling and GenScript) were used as loading controls.
Protein synthesis rate and western analysis
Human podocytes cultured at 33°C were treated with drugs as described subsequently for 16, 24 or 48 h. They were then starved for amino acids for 20 min and labeled with media containing 40 µCi/ml 35S-Cys/Met for 40–60 min at 33°C. Labeled cells were washed, trypsinized and lysed at 4°C with Tris-buffered saline containing 1% NP-40 and protease inhibitor cocktail. Lysates were clarified by centrifugation at 10 000g and aliquots were either counted to determine TCA-precipitable counts in each sample, or analyzed by SDS–PAGE and autoradiography to resolve protein bands.
Caenorhabditis elegans lifespan analyses
Animals were maintained at 20°C throughout the experiment. Synchronized nematode cultures were initiated by bleaching young adults to obtain eggs. Collected eggs were allowed to hatch overnight on 10 cm, unspread, Nematode Growth Media (NGM) plates, after which L1-arrested larvae were transferred to 10 cm NGM plates spread with OP50 E. coli. Drug treatment was applied by dissolving probucol, rapamycin and cycloheximide in ethanol, and then added to plates to the desired concentrations and allowed to dry. Worms were then added to 10 cm NGM plates spread with drug either beginning at the early development stage with synchronized (L1) larvae and/or upon reaching the first day of egg laying when synchronous young adults were moved to fresh 3.5 cm NGM drug plates seeded with bacteria (Lifespan experiment ‘Day 0’). The same volume of ethanol was added to control NGM plates. 60 nematodes were studied per condition, divided on three 3.5 cm NGM plates. Mortality was confirmed by stimulating nematodes lightly with a platinum wire; if the nematode did not move after stimulations it was scored as dead and removed from the plate. Worms that died of protruding/bursting vulva, bagging or crawling off the agar were censored. Statistical analysis was performed in GraphPad prism 5.0.
Caenorhabditis elegans fluorescence analyses of in vivo mitochondrial physiological parameters
Oxidant burden (MitoSOX Red), membrane potential (TMRE) and mitochondrial content (MitoTracker Green FM, MTG) were performed using terminal pharyngeal bulb fluorescence relative quantitation, as described previously, with minor modifications (50). Synchronous populations of Day 0 young adults were moved to 35 mm NGM plates spread with OP50 E. coli, with either drugs or appropriate buffer control, and with either 10 µm MitoSOX Red (matrix oxidant burden), 100 nm TMRE (mitochondrial membrane potential) or 10 µm MitoTracker Green FM (mitochondrial content) for 24 h. After 24 h, 50 worms per condition were transferred by picking onto 35 mm NGM plates spread with OP50 E. coli for 1 h to allow clearing of residual dye from the gut. Worms were then paralyzed in situ with 4 mg/ml levamisole. Photographs were taken in a darkened room at ×160 magnification with a Cool Snap cf2 camera (Nikon, Melville, NY, USA). A CY3 fluorescence cube set (MZFLIII, Leica, Bannockburn, IL, USA) was used for MitoSOX and TMRE. A GFP2 filter set (Leica) was used for MitoTracker Green FM. Exposure times were 2 s, 320 ms and 300 ms, respectively, for MitoSOX, TMRE and MitoTracker Green FM. The resulting images were background subtracted, and the nematode terminal pharyngeal bulb was manually circled to obtain mean intensity of the region by using Fiji Is Just ImageJ (51). Fluorescence data for each strain were normalized to its same day control to account for day-to-day variation. A minimum of three independent experiments of approximately 50 animals per replicate were studied per strain per dye. The significance of the difference in the mean fluorescence intensity between strains under different experimental conditions was assessed by mixed-effect ANOVA, which takes into account potential batch effect due to samples being experimentally prepared, processed, and analyzed on different days by including a batch random effect in the model. A statistical significance threshold was set at P < 0.05. All statistical analyses were performed in SAS 9.3.
Caenorhabditis elegans transcriptome profiling by RNAseq analysis
Wild-type (N2 Bristol) and mitochondrial RC complex I deficient gas-1(fc21) animals were maintained at 20°C by established protocol (52). Synchronous young adult populations of approximately 1000–2000 nematodes were treated beginning on the first day of egg laying for 24 h on NGM plates spread with 2.5 nm rapamycin, 5 µm probucol, 2.5 µm cycloheximide or ethanol buffer control, per established protocol (52). After drug treatment, total RNA was isolated and prepared for transcriptome profiling of four biological replicates per condition, as previously described (52). Briefly, total RNA was isolated using the Trizol method and RNA concentration was measured using the NanoDrop-1000. RNA quality was determined by using the Agilent Bioanalyzer in the Nucleic Acid Core Facility at The Children's Hospital of Philadelphia Research Institute. RIN number between 8 and 10 was required for further sample analysis. Library preparation was performed using the Illumina Truseq Stranded Total RNA Sample Preparation Kit (San Diego, CA, USA), with indexing to enable eight samples to be run per flow cell lane. Samples were submitted to the BGI @ CHOP Sequencing Core Facility at The Children's Hospital of Philadelphia Research Institute CHOP for next generation sequencing (RNAseq) analysis. Library quality was assessed by Bio A analysis to check concentration, library size and contamination, as well as by gel analysis to assess degradation. Quantitative PCR was then performed to determine optimal sample concentrations using the Applied Biosystems Step One Plus Real Time PCR machine to enable proper sample pooling. Samples were run on Illumina HiSeq 2000 instruments in High Throughput Mode of 100 bp, paired-end reads with eight samples per lane to generate an estimated 20 million reads per sample. FASTQ files were provided for downstream processing and bioinformatics analysis. KEGG pathway-level analyses were performed as previously described (4). RNAseq data for all 20 data sets were submitted to the public Sequence Read Archive (SRA Bioproject ID PRJNA284422).
Gene expression analysis by qRT–PCR
Human cells were collected in cold PBS, total RNA was isolated and reverse transcribed and qRT–PCR was performed on cDNA for relative gene expression studies, as previously described (26). Taqman Gene Expression Assays used in human cells were ATF4 (Hs00909569_gl, Applied Biosystems) as target gene and ACTB (Hs01060665_g1, Applied Biosystems) as endogenous gene. Taqman Gene Expression Assay for cdc-42 as endogenous gene (Ce02435136_g1, Applied Biosystems, Foster City, CA, USA) and for the target genes daf-16, skn-1 and sod-3 (Ce02422838_m1, Ce02407446_m1 and Ce02404515_g1, respectively, Applied Biosystems, Foster City, CA, USA) was used for quantitative analysis of changes in their expression levels in C. elegans. Quantitative analysis of expression changes in expression level starting with 40 ng cDNA per well, with triplicate analyses per experiment, was performed by qPCR reaction on the 7500 Fast Real Time PCR machine from Applied Biosystems (Applied Biosystems, Foster City, CA, USA). At least three biological replicates were performed per condition. Sequence Detection Software v2.0.6 was used for relative quantitative analysis of gene expression.
Supplementary Material
Author Contributions
M.P. and M.J.F. conceived the study design; M.P. performed all cell culture, cell viability and high-resolution respirometry experiments; M.P. and D.L.G. conducted mouse experiments including breeding, genotyping and urine albumin assays; M.P. and M.T. performed immunoblot analyses; E.P. performed RNAseq library preparation; Z.Z. performed RNAseq bioinformatics analyses; M.P., E.P., J.L. and E.M. performed signaling profiling experiments; J.O. performed C. elegans lifespan analyses; Y.J.K. performed C. elegans fluorescence analyses of mitochondrial physiological parameters; C.F. provided guidance on cell death mechanistic analyses; Y.A. and J.T. performed translation activity and western analyses and provided guidance on ER stress analyses; R.X. assisted with statistical analysis; M.J.F., M.P. and Y.A. wrote the manuscript.
Funding
This work was supported in part by grants from the National Institutes of Health (R01-HD065858-01A1 to M.J.F.; R01-AG18001 to Y.A.), The Kelsey Wright Foundation, The Juliet's Cure Mitochondrial Disease Research Fund, and the Genes, Genomes and Pediatric Disease (GGPD) Research Affinity Group at The Children's Hospital of Philadelphia Research Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. C.A.F. is the Joshua Kahan Endowed Chair in Pediatric Leukemia Research.
Supplementary Material
Acknowledgements
We are grateful to Drs Douglas C. Wallace, Richard Youle and Karen Urtishak for helpful discussions, Dr Eric Rappaport for guidance on RNAseq sample preparation, Dr Juan Carlos Perin for assistance with RNAseq data transfer, as well as Dr Duncan Johnstone who derived and generously provided the human podocyte control line.
Conflict of Interest statement. None declared.
References
- 1.DiMauro S., Schon E.A. (2003) Mitochondrial respiratory-chain diseases. N. Engl. J. Med., 348, 2656–2668. [DOI] [PubMed] [Google Scholar]
- 2.Koopman W.J., Willems P.H., Smeitink J.A. (2012) Monogenic mitochondrial disorders. N. Engl. J. Med., 366, 1132–1141. [DOI] [PubMed] [Google Scholar]
- 3.Pieczenik S.R., Neustadt J. (2007) Mitochondrial dysfunction and molecular pathways of disease. Exp. Mol. Pathol., 83, 84–92. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z., Tsukikawa M., Peng M., Polyak E., Nakamaru-Ogiso E., Ostrovsky J., McCormack S., Place E., Clarke C., Reiner G. et al. (2013) Primary respiratory chain disease causes tissue-specific dysregulation of the global transcriptome and nutrient-sensing signaling network. PloS one, 8, e69282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atkuri K.R., Cowan T.M., Kwan T., Ng A., Herzenberg L.A., Herzenberg L.A., Enns G.M. (2009) Inherited disorders affecting mitochondrial function are associated with glutathione deficiency and hypocitrullinemia. Proc. Natl Acad. Sci. USA., 106, 3941–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parikh S., Saneto R., Falk M.J., Anselm I., Cohen B.H., Haas R., Medicine Society T.M. (2009) A modern approach to the treatment of mitochondrial disease. Curr. Treat. Options. Neurol., 11, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z., Falk M.J. (2014) Integrated transcriptome analysis across mitochondrial disease etiologies and tissues improves understanding of common cellular adaptations to respiratory chain dysfunction. Int. J. Biochem. Cell. Biol., 50, 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falk M.J., Polyak E., Zhang Z., Peng M., King R., Maltzman J.S., Okwuego E., Horyn O., Nakamaru-Ogiso E., Ostrovsky J. et al. (2011) Probucol ameliorates renal and metabolic sequelae of primary CoQ deficiency in Pdss2 mutant mice. EMBO Mol. Med., 3, 410–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng M., Falk M.J., Haase V.H., King R., Polyak E., Selak M., Yudkoff M., Hancock W.W., Meade R., Saiki R. et al. (2008) Primary coenzyme Q deficiency in Pdss2 mutant mice causes isolated renal disease. PLoS Genet., 4, e1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ziegler C.G., Peng M., Falk M.J., Polyak E., Tsika E., Ischiropoulos H., Bakalar D., Blendy J.A., Gasser D.L. (2012) Parkinson's disease-like neuromuscular defects occur in prenyl diphosphate synthase subunit 2 (Pdss2) mutant mice. Mitochondrion, 12, 248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan K., Ito N., Nakajo A., Kurayama R., Fukuhara D., Nishibori Y., Kudo A., Akimoto Y., Takenaka H. (2012) The struggle for energy in podocytes leads to nephrotic syndrome. Cell Cycle, 11, 1504–1511. [DOI] [PubMed] [Google Scholar]
- 12.Abraham R.T. (2005) TOR signaling: an odyssey from cellular stress to the cell growth machinery. Curr. Biol., 15, R139–e69141. [DOI] [PubMed] [Google Scholar]
- 13.Fingar D.C., Blenis J. (2004) Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene, 23, 3151–3171. [DOI] [PubMed] [Google Scholar]
- 14.Castets P., Ruegg M.A. (2013) MTORC1 determines autophagy through ULK1 regulation in skeletal muscle. Autophagy, 9, 1435–1437. [DOI] [PubMed] [Google Scholar]
- 15.Gilkerson R.W., De Vries R.L., Lebot P., Wikstrom J.D., Torgyekes E., Shirihai O.S., Przedborski S., Schon E.A. (2012) Mitochondrial autophagy in cells with mtDNA mutations results from synergistic loss of transmembrane potential and mTORC1 inhibition. Hum. Mol. Genet., 21, 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider-Poetsch T., Ju J., Eyler D.E., Dang Y., Bhat S., Merrick W.C., Green R., Shen B., Liu J.O. (2010) Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol., 6, 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson S.C., Yanos M.E., Kayser E.B., Quintana A., Sangesland M., Castanza A., Uhde L., Hui J., Wall V.Z., Gagnidze A. et al. (2013) mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science, 342, 1524–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kayser E.B., Morgan P.G., Sedensky M.M. (1999) GAS-1: a mitochondrial protein controls sensitivity to volatile anesthetics in the nematode Caenorhabditis elegans. Anesthesiology, 90, 545–554. [DOI] [PubMed] [Google Scholar]
- 19.Poehlsgaard J., Douthwaite S. (2005) The bacterial ribosome as a target for antibiotics. Nat. Rev. Microbiol., 3, 870–881. [DOI] [PubMed] [Google Scholar]
- 20.Livnat-Levanon N., Kevei E., Kleifeld O., Krutauz D., Segref A., Rinaldi T., Erpapazoglou Z., Cohen M., Reis N., Hoppe T. et al. (2014) Reversible 26S proteasome disassembly upon mitochondrial stress. Cell Rep., 7, 1371–1380. [DOI] [PubMed] [Google Scholar]
- 21.Gai X., Ghezzi D., Johnson M.A., Biagosch C.A., Shamseldin H.E., Haack T.B., Reyes A., Tsukikawa M., Sheldon C.A., Srinivasan S. et al. (2013) Mutations in FBXL4, encoding a mitochondrial protein, cause early-onset mitochondrial encephalomyopathy. Am. J. Hum. Genet., 93, 482–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonnen P.E., Yarham J.W., Besse A., Wu P., Faqeih E.A., Al-Asmari A.M., Saleh M.A., Eyaid W., Hadeel A., He L. et al. (2013) Mutations in FBXL4 cause mitochondrial encephalopathy and a disorder of mitochondrial DNA maintenance. Am. J. Hum. Genet., 93, 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huemer M., Karall D., Schossig A., Abdenur J.E., Al Jasmi F., Biagosch C., Distelmaier F., Freisinger P., Graham B.H., Haack T.B. et al. (2015) Clinical, morphological, biochemical, imaging and outcome parameters in 21 individuals with mitochondrial maintenance defect related to FBXL4 mutations. J. Inherit. Metab. Dis., [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayir H., Kagan V.E. (2008) Bench-to-bedside review: Mitochondrial injury, oxidative stress and apoptosis--there is nothing more practical than a good theory. Crit. Care, 12, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng Y.C., Chang J.M., Chen C.A., Chen H.C. (2014) Autophagy modulates endoplasmic reticulum stress-induced cell death in podocytes: A protective role. Exp. Biol. Med., 240, 467–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Y., Wang X., Guo H., Zhang B., Zhang X.B., Shi Z.J., Yu L. (2013) Synthesis and screening of 3-MA derivatives for autophagy inhibitors. Autophagy, 9, 595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Misaghi S., Korbel G.A., Kessler B., Spooner E., Ploegh H.L. (2006) z-VAD-fmk inhibits peptide:N-glycanase and may result in ER stress. Cell Death Differ., 13, 163–165. [DOI] [PubMed] [Google Scholar]
- 28.Cho Y.S. (2014) Perspectives on the therapeutic modulation of an alternative cell death, programmed necrosis (review). Int. J. Mol. Med., 33, 1401–1406. [DOI] [PubMed] [Google Scholar]
- 29.O'Brien W.T., Harper A.D., Jove F., Woodgett J.R., Maretto S., Piccolo S., Klein P.S. (2004) Glycogen synthase kinase-3beta haploinsufficiency mimics the behavioral and molecular effects of lithium. J. Neurosci., 24, 6791–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Q., Vazquez E.J., Moghaddas S., Hoppel C.L., Lesnefsky E.J. (2003) Production of reactive oxygen species by mitochondria: central role of complex III. J. Biol. Chem., 278, 36027–36031. [DOI] [PubMed] [Google Scholar]
- 31.Tsukumo Y., Tsukahara S., Furuno A., Iemura S., Natsume T., Tomida A. (2014) TBL2 is a novel PERK-binding protein that modulates stress-signaling and cell survival during endoplasmic reticulum stress. PloS one, 9, e112761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruggiano A., Foresti O., Carvalho P. (2014) Quality control: ER-associated degradation: protein quality control and beyond. J. Cell Biol., 204, 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Endres K., Reinhardt S. (2013) ER-stress in Alzheimer's disease: turning the scale? Am. J. Neurodegener. Dis., 2, 247–265. [PMC free article] [PubMed] [Google Scholar]
- 34.Silva J.M., Wong A., Carelli V., Cortopassi G.A. (2009) Inhibition of mitochondrial function induces an integrated stress response in oligodendroglia. Neurobiol. Dis., 34, 357–365. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Reyes I., Sanchez-Arago M., Cuezva J.M. (2012) AMPK and GCN2-ATF4 signal the repression of mitochondria in colon cancer cells. Biochem. J., 444, 249–259. [DOI] [PubMed] [Google Scholar]
- 36.Martin S., Lovat P.E., Redfern C.P. (2014) Cell-type variation in stress responses as a consequence of manipulating GRP78 expression in neuroectodermal cells. J. Cell Biochem., 116, 438–449. [DOI] [PubMed] [Google Scholar]
- 37.Falk M.J., Zhang Z., Rosenjack J.R., Nissim I., Daikhin E., Nissim I., Sedensky M.M., Yudkoff M., Morgan P.G. (2008) Metabolic pathway profiling of mitochondrial respiratory chain mutants in C. elegans. Mol. Genet. Metab., 93, 388–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flanagan J.U., Huber T. (2007) Structural evolution of the ABC transporter subfamily B. Evol. Bioinform. Online, 3, 309–316. [PMC free article] [PubMed] [Google Scholar]
- 39.Arnold I., Wagner-Ecker M., Ansorge W., Langer T. (2006) Evidence for a novel mitochondria-to-nucleus signalling pathway in respiring cells lacking i-AAA protease and the ABC-transporter Mdl1. Gene, 367, 74–88. [DOI] [PubMed] [Google Scholar]
- 40.Wang X., Zuo X., Kucejova B., Chen X.J. (2008) Reduced cytosolic protein synthesis suppresses mitochondrial degeneration. Nat. Cell. Biol., 10, 1090–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scaglia F. (2010) The role of mitochondrial dysfunction in psychiatric disease. Dev. Disab. Res. Rev., 16, 136–143. [DOI] [PubMed] [Google Scholar]
- 42.Clarke C., Xiao R., Place E., Zhang Z., Sondheimer N., Bennett M., Yudkoff M., Falk M.J. (2013) Mitochondrial respiratory chain disease discrimination by retrospective cohort analysis of blood metabolites. Mol. Genet. Metab., 110, 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kerr D.S. (2013) Review of clinical trials for mitochondrial disorders: 1997–2012. Neurotherapeutics, 10, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saleem M.A., O'Hare M.J., Reiser J., Coward R.J., Inward C.D., Farren T., Xing C.Y., Ni L., Mathieson P.W., Mundel P. (2002) A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J. Am. Soc. Nephrol., 13, 630–638. [DOI] [PubMed] [Google Scholar]
- 45.Dell K.M., Li Y.X., Peng M., Neilson E.G., Gasser D.L. (2000) Localization of the mouse kidney disease (kd) gene to a YAC/BAC contig on Chromosome 10. Mamm. Genome, 11, 967–971. [DOI] [PubMed] [Google Scholar]
- 46.Stoddart M.J. (2011) Cell viability assays: introduction. Methods. Mol. Biol., 740, 1–6. [DOI] [PubMed] [Google Scholar]
- 47.Garedew A., Haffner B., Hutter E., Gnaiger E. (2008) In Mitochondrial Pathways and Respiratory Control, 2nd edn Oroboros MiPNet Publications, Innsbruck, pp. 55–61. [Google Scholar]
- 48.Hutter E., Renner K., Jansen-Durr P., Gnaiger E. (2002) Biphasic oxygen kinetics of cellular respiration and linear oxygen dependence of antimycin A inhibited oxygen consumption. Mol. Biol. Rep., 29, 83–87. [DOI] [PubMed] [Google Scholar]
- 49.Dingley S., Chapman K.A., Falk M.J. (2012) Fluorescence-activated cell sorting analysis of mitochondrial content, membrane potential, and matrix oxidant burden in human lymphoblastoid cell lines. Methods Mol. Biol., 837, 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dingley S., Polyak E., Lightfoot R., Ostrovsky J., Rao M., Greco T., Ischiropoulos H., Falk M.J. (2010) Mitochondrial respiratory chain dysfunction variably increases oxidant stress in Caenorhabditis elegans. Mitochondrion, 10, 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Polyak E., Zhang Z., Falk M.J. (2012) Molecular profiling of mitochondrial dysfunction in Caenorhabditis elegans. Methods Mol. Biol., 837, 241–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.