Abstract
Serine hydrolase inhibitors, which facilitate enzyme function assignment and are used to treat a range of human disorders, often act by an irreversible mechanism that involves covalent modification of the serine hydrolase catalytic nucleophile. The portion of mammalian serine hydrolases for which selective inhibitors have been developed, however, remains small. Here, we show that N-hydroxyhydantoin (NHH) carbamates are a versatile class of irreversible serine hydrolase inhibitors that can be modified on both the staying (carbamylating) and leaving (NHH) groups to optimize potency and selectivity. Synthesis and screening of a small library of NHH carbamates by competitive activity-based protein profiling furnished selective, in vivo-active inhibitors and tailored activity-based probes for multiple mammalian serine hydrolases, including palmitoyl protein thioesterease-1 (PPT1), mutations of which cause the human disease infantile neuronal ceroid lipofuscinosis.
Serine hydrolases (SHs) represent one of the largest and most diverse enzyme classes in Nature and perform myriad biochemical functions in physiology and disease (Long and Cravatt, 2011). SHs use a conserved mechanism involving a base-activated serine nucleophile to hydrolyze amide, ester, and thioester bonds in biomolecules; however, these enzymes also display markedly different structures and folds, distribute across virtually all subcellular compartments in the cell, and accept an expansive array of small-and macro-molecule substrates. In accordance with their diverse biological activities, SHs are targeted by drugs that are used to treat a wide range of diseases, including cognitive dementia (Racchi et al., 2004), obesity (Nelson and Miles, 2005), diabetes (Thornberry and Weber, 2007), and bacterial (Staub and Sieber, 2008) and viral (Marks and Jacobson, 2012) infections.
Many structurally diverse SH inhibitors have been developed that include compounds with reversible and irreversible mechanisms of action (Bachovchin and Cravatt, 2012). Irreversible inhibitors are largely electrophilic compounds that covalently modify the conserved catalytic serine nucleophile of SHs. Activated carbamates, for instance, irreversibly carbamylate the catalytic serine of SHs, and, in select cases, have been advanced to furnish selective and in vivo-active inhibitors and drugs for individual SHs (Bachovchin and Cravatt, 2012). Carbamate inhibitors can be operationally deconstructed into two primary elements – the 'staying group' (SG), which remains covalently bound to the SH catalytic serine, and the 'leaving group' (LG), which is displaced by the enzyme’s serine nucleophile. Even though the LG does not ultimately remain part of the covalent enzyme-inhibitor adduct, it still has substantial influence over both inhibitor recognition and reactivity due to its structure and electrophilic character, respectively. As new subtypes of carbamate inhibitors have been discovered, differences in the LG are often found to make large contributions to potency and selectivity (Chang et al., 2012; Chang et al., 2011). Nonetheless, many of the most effective LGs, such as the p-NO2-phenoxy (Long et al., 2009), hexafluoroisopropyloxy (Chang et al., 2012), and N-hydroxysuccinimide (NHS) (Chang et al., 2013) structures, are limited in their potential for structural diversification.
We hypothesized that efforts to expand the number of SHs targeted by selective inhibitors would benefit from carbamates that are more amenable to LG structural diversification. Indeed, in as much as the substrate selectivity of enzymes is predictive of features important for inhibitor binding, it is noteworthy that the catalytic efficiencies of SHs can be strongly influenced by the LG structures of substrates (Gross, 1983; Jenkins et al., 2004; Navia-Paldanius et al., 2012; Saghatelian et al., 2004). Here, we describe a class of N-hydroxyhydantoin (NHH) carbamates that incorporate the tempered electrophilicity of NHS-carbamates, but provide greater synthetic access to diverse and extended LG structures. We identify lead and optimized NHH carbamate inhibitors for several SHs using competitive activity-based protein profiling (ABPP) methods (Niphakis and Cravatt, 2014), including enzymes that heretofore have lacked selective chemical probes. Prominent among these enzymes was palmitoyl-protein thioesterase 1 (PPT1) (Bellizzi et al., 2000), mutations of which cause the human disease infantile neuronal ceroid lipofuscinosis (CLN1, INCL, or infantile Batten’s disease) (Kyttala et al., 2006). We show that PPT1, despite being inert toward traditional broad-spectrum SH activity-based probes containing fluorophosphonate (FP) electrophiles (Wang et al., 2013), reacts efficiently with NHH carbamates. We further describe NHH carbamates that act as selective and in vivo-active inhibitors, as well as tailored activity-based probes, for PPT1, thus providing valuable chemical probes to characterize the function of this enzyme.
RESULTS
Serine Hydrolase (SH) Inhibition by N-Hydroxyhydantoin (NHH) Carbamates
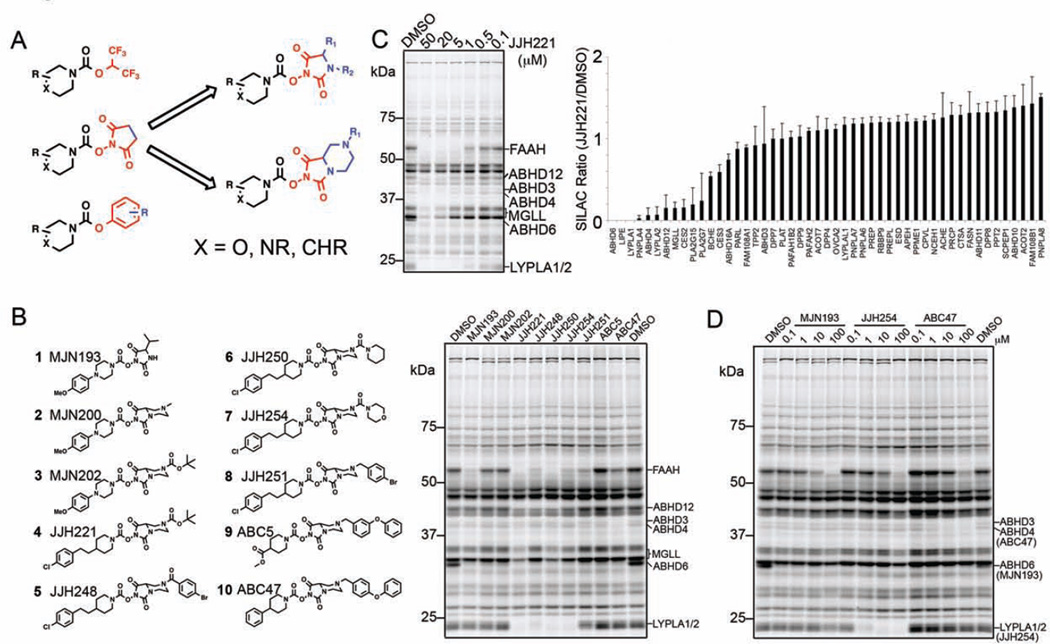
In considering ways to increase the structural diversity of established chemotypes for serine hydrolase inhibition, we turned our attention to N-hydroxysuccinimides (NHSs), a compound class that has furnished selective and in vivo-active inhibitors of monoacylglycerol lipase (MAGL, or MGLL) (Niphakis et al., 2013), as well as lead inhibitors for additional serine hydrolases, while showing minimal cross reactivity with other proteins in the proteome (Chang et al., 2013). In principle, the succinimidyl LG provides a good scaffold for structural elaboration, especially when compared to other established types of carbamate SH inhibitors that possess, for instance, p-NO2-phenoxy and hexafluoroisopropyloxy LGs (Figure 1A). Synthetic derivatization of the NHS group is not, however, straightforward, and we therefore wondered whether the structurally related N-hydroxyhydantoin (NHH), which was expected to exhibit similar LG potential while being more amenable to synthetic modification (Figure 1A), could serve as the basis for creating carbamates with broad SH inhibition potential.
Figure 1. Synthesis and initial characterization of NHH carbamates as inhibitors of SHs.
(A) Representative classes of carbamate inhibitors of SHs with variable leaving groups in red and potential sites for structural elaboration in blue. NHS carbamates (middle) were prioritized for further exploration over HFIP (top) or O-aryl (bottom) carbamates due to the synthetic tractability and structural diversification afforded by their elaboration (right). (B) Structures of representative NHH carbamates tested for SH inhibition by gelbased competitive ABPP and their in vitro inhibition profiles in a mouse brain membrane proteome. Brain proteome was pre-treated with NHH carbamates (20 µM, 30 min, 37 °C) followed by reaction with FP-Rh (1 µM, 30 min, room temperature) and analysis by SDSPAGE and in-gel fluorescence scanning. (C) Concentration-dependent competitive gelbased ABPP (left) and ABPP-SILAC (right) analyses of JJH221 (4). Gel-based ABPP corresponds to mouse brain membrane proteome. For the ABPP-SILAC analysis, heavy- and light-isotopically labeled PC3 cells were treated with JJH221 (20 µM, 4 h) or DMSO, respectively, prior to processing for ABPP-SILAC. SHs showing SILAC ratios (JJH221/DMSO) less than 0.33 were considered targets of JJH221. Data represent average values ± S.D. for two independent experiments. (D) Concentration-dependent inhibition of mouse brain membrane SHs by representative NHH carbamates as measured by gel-based competitive ABPP. SH targets of the tested NHH carbamates are marked on the right side of the gel. Assignment of SH enzyme activities in competitive ABPP gels are based on gel migration patterns consistent with past studies (e.g., (Blankman et al., 2007; Hoover et al., 2008)).
We synthesized a small library of NHH carbamates and screened these compounds for inhibitory activity against mouse brain membrane SHs by competitive ABPP using the SH-directed probe fluorophosphonate-rhodamine (FP-Rh) (Patricelli et al., 2001) (Figure 1B). Most of the NHH carbamates showed activity against one or more brain SHs, some of which could be assigned based on previous studies (Blankman et al., 2007; Hoover et al., 2008) (Figure 1B). NHH carbamates with relatively small extensions to the LG (e.g., the isopropyl and fused methyl piperazine found in MJN193 and MJN200, respectively) showed restricted reactivity in mouse brain membranes, with notable potency and selectivity for ABHD6. In contrast, compounds with more elaborated LGs (JJH221, JJH248, JJH250, JJH254, JJH251, ABC5, ABC47; Compounds 4–10, respectively) showed broader reactivity with brain SHs, suggesting that distal modifications to the LG substantially influenced these NHH carbamate-SH interactions (Figure 1B).
We selected one of the more promiscuous compounds – JJH221 (4) – for analysis by mass spectrometry (MS)-based ABPP methods to assess the range of SHs targeted by NHH carbamates. JJH221-sensitive SHs were identified using the quantitative MS method ABPP-SILAC (Stable isotope labeling by amino acids in cell culture; (Mann, 2006)) (Adibekian et al., 2011). In brief, proteomes from isotopically heavy- and light-amino acid-labeled human PC3 cells were treated with JJH221 (20 µM) or DMSO, respectively, for 4 h followed by the biotinylated FP probe FP-biotin (Liu et al., 1999) (2.5 µM, 1 h). FP-labeled proteomes were then combined and processed for LC-MS/MS analysis as described previously (Adibekian et al., 2011). JJH221 (4) was found to inhibit several human SHs (defined as proteins showing a three-fold or greater decrease in signal in the JJH221-treated proteome), including enzymes previously shown to be sensitive to NHS-carbamates (e.g., ABHD6, MGLL) and others for which selective inhibitors are lacking (e.g., ABHD4, ABHD12, PLA2G15, PNPLA4) (Figure 1C).
We next selected a subset of NHH carbamates with diverse target profiles in the mouse brain membrane proteome for concentration-dependent profiling, which identified compounds with good potency and selectivity for ABHD6 (MJN193; 1) and LYPLA1/2 (JJH254; 7), as well as promising lead inhibitors for ABHD3 and ABHD4 (ABC34 (13) and ABC47 (10)) (Figure 1D and Figure S1). Various structural classes of ABHD6 (Hsu et al., 2013; Janssen et al., 2014; Li et al., 2007; Marrs et al., 2011) and LYPLA1/2 (Adibekian et al., 2012; Biel et al., 2006; Dekker et al., 2010) inhibitors have been described and a probe report has been submitted for JJH254 (7) as part of the NIH Molecular Libraries Program Network (Hulce et al., 2014). We therefore focused our attention on the lead inhibitors for ABHD3 and ABHD4.
Optimization of NHH Carbamate Inhibitors for ABHD3 and ABHD4
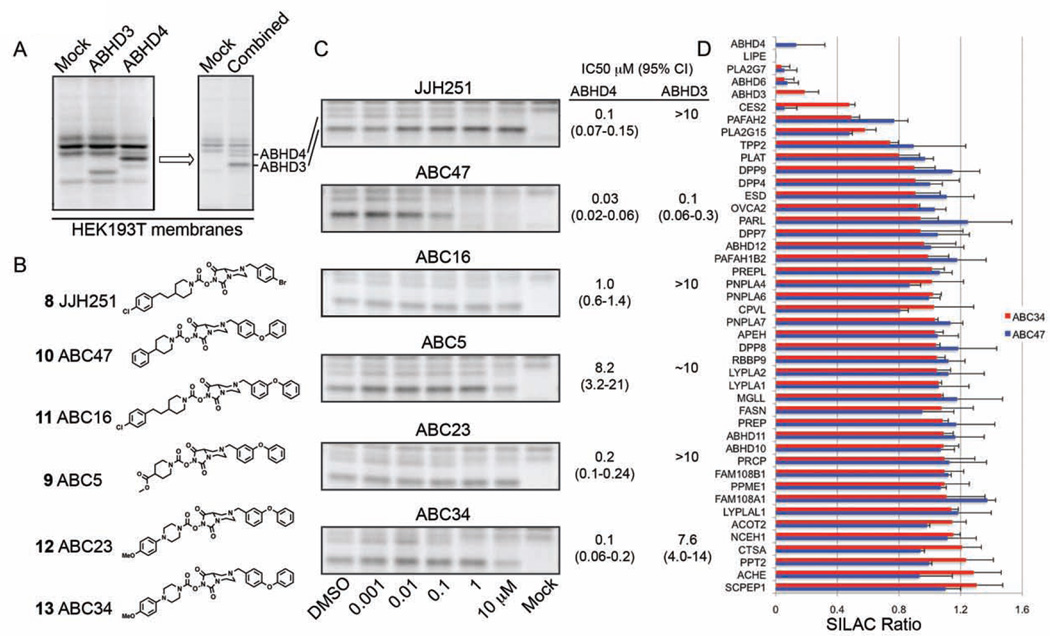
ABHD3 and ABHD4 hydrolyze distinct classes of phospholipids – medium chain (Long et al., 2011) and N-acyl (Lee et al., 2015; Simon and Cravatt, 2006) phospholipids, respectively – and selective and cell-active inhibitors are lacking for these enzymes. ABHD3 and ABHD4 were recombinantly expressed by transient transfection in HEK293T cells, and the cell lysates combined to provide a convenient assay for monitoring inhibition of both enzymes by gel-based ABPP (Figure 2A). A focused library of ABC47 (10) analogues was synthesized, where each compound retained the basic tertiary amine substituent on the NHH LG and was diversified off of this amine or on the staying group (SG) of the carbamate (Figure 2B). Gel-based ABPP confirmed that ABC47 (10) acts as a potent dual inhibitor of ABHD3 and ABHD4 (IC50 values of 0.13 and 0.03 µM, respectively) and identified additional compounds that preferentially inhibited ABHD4 over ABHD3 (e.g., ABC34 (13), ABC23 (12), and JJH251 (8)) (Figure 2C and Figure S2). Among the ABHD4-preferring inhibitors, JJH251 was deprioritized because we had already found that it inhibited additional serine hydrolases such as FAAH and LYPLA1/2 in the mouse brain membrane proteome (Figure 1B).
Figure 2. NHH carbamate inhibitors of ABHD3 and ABHD4.
(A) Recombinant expression of mouse ABHD3 and human ABHD4 by transient transfection in HEK293T cells and detection of both enzymes in a combined lysate of transfected cells by gelbased ABPP with the FP-Rh probe. (B) Additional NHH carbamates screened against recombinant ABHD3 and ABHD4. (C) Concentration-dependent inhibition of ABHD3 and ABHD4 by NHH carbamates using gel-based competitive ABPP. See Figure S2 for curves used to generated the reported IC50 values. Data represent average values and 95% confidence limits (CI) for three independent experiments. (D) ABPP-SILAC analysis using FP-biotin as a probe to enrich SHs, showing overlaid in situ SH inhibition profiles for ABC47 (10) (500 nM, 4 h) and ABC34 (13) (1 µM, 4 h) in PC3 cells. Data represent average values ± S.D. for two independent experiments.
We next assessed the activity and selectivity of dual ABHD3/4 and ABHD4-preferring inhibitors in PC3 cells by ABPP-SILAC. Isotopically heavy and light PC3 cells were treated in situ with inhibitor (ABC34 (13) (1 µM) or ABC47 (10) (0.5 µM)) or DMSO for 4 h and then lysed and processed for ABPP-SILAC analysis. ABC47 (10) was found to inhibit both ABHD3 and ABHD4 in PC3 cells with good selectivity (Figure 2D). Among the 44 serine hydrolases detected in this study, only four additional targets were observed for ABC47 – LIPE, PLA2G7, ABHD6, and CES2 (Figure 2D). ABC34 (13) completely inhibited ABHD4 and shared a similar off-target profile with ABC47 (10). Surprisingly, however, partial inhibition (~80%) of ABHD3 was also observed, which indicates that the selectivity window for ABHD4 over ABHD3 may be compressed in living cells compared to cell lysates.
These results, taken together indicate that NHH carbamates can inhibit both ABHD3 and ABHD4 with good potency and selectivity in human cells.
Identification of PPT1 as a Target of NHH Carbamates
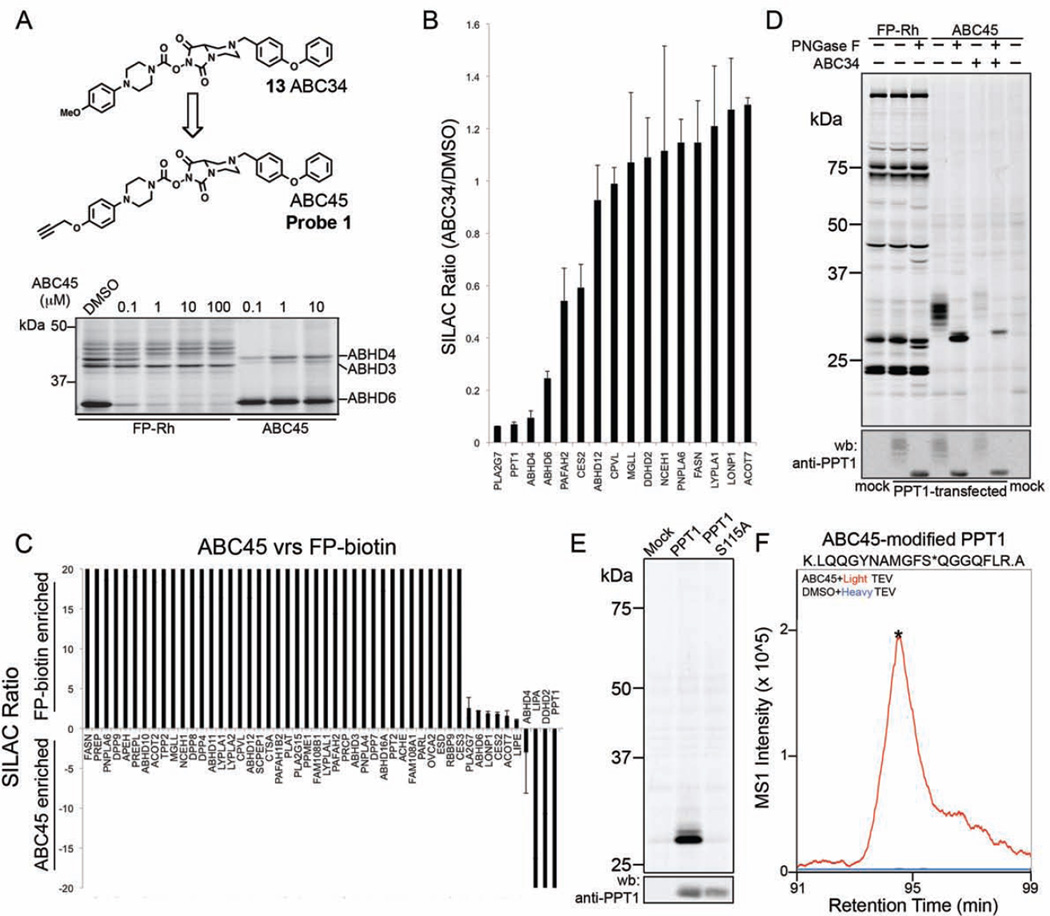
Designating ABC34 (13) as a promising lead ABHD4 inhibitor, we next generated an alkyne-derivatized analog of this compound for direct detection and identification of covalent protein targets by conjugation to azide reporter tags (Speers and Cravatt, 2004) using copper-catalyzed azide-alkyne cycloaddition (CuAAC or click) chemistry (Rostovtsev et al., 2002). We found it most synthetically feasible to replace the methoxy group of ABC34 (13) with a propargyloxy substituent to yield the ‘click’ probe ABC45 (probe 1) (Figure 3A). ABC45 inhibited ABHD4, but not ABHD3, in transfected HEK293T cell proteome in a concentration-dependent manner as detected by competitive ABPP with FP-Rh (Figure 3A), and the ABC45-ABHD4 adduct could, in a complementary manner, be directly visualized by CuAAC conjugation to a rhodamine-azide (Rh-N3) tag (Figure 3A). ABC45 also identified an ABC34-sensitive protein in mouse brain that matches the expected molecular mass (~40 kDa) of ABHD4 and was absent in brain tissue from ABHD4−/− mice (Figure S3A). These data indicate that ABC45 acts as a tailored activity-based probe for the convenient gel-based detection of ABHD4 in complex biological systems.
Figure 3. Discovery of PPT1 as a target of NHH carbamates.
(A) Structure of the click probe ABC45 and its reactivity with recombinant ABHD4 in transfected cell lysates. Left side of gel shows a competitive ABPP gel of ABHD4-transfected cell lysates treated with the indicated concentrations of ABC45 (30 min 37 °C) followed by FP-Rh. Right side of gel shows direct labeling of ABHD4 and other SHs by ABC45 as measured by CuAAC to an Rh-N3 tag. (B) ABPP-SILAC analysis showing the in situ SH inhibition profile for ABC34 (13) (1 µM, 4 h) in PC3 cells, where SH enrichment and inhibition were measured with the ABC45 probe. Data represent average values ± S.D. for two independent experiments. (C) ABPP-SILAC analysis comparing the reactivity and enrichment of SHs with FP-biotin (2.5 µM, 1 h) versus ABC45 (5 µM, 1 h) in PC3 cell proteomes. Data represent average values ± S.D. for two independent experiments. (D) Verification that recombinant human PPT1 expressed by transient transfection in HEK293T cells reacts with ABC45 (1 µM), but not FP-Rh (1 µM), and the ABC45 reactivity is blocked by ABC34 (5 µM, 4 h). PNGaseF treatment causes a shift in PPT1 signals by SDS-PAGE, consistent with the reported glycosylation state of this protein (Camp et al., 1994; Lyly et al., 2007). Upper, ABPP gel; lower, anti-PPT1 western blot. (E) ABC45 labels recombinant wild type PPT1, but not the catalytic serine mutant (S115A) of this enzyme. All samples were treated with PNGaseF prior to analysis by gelbased ABPP. (F) ABC45 labels the catalytic serine S115 of recombinant PPT1 as determined by isoTOP-ABPP analysis of PPT1-transfected cell lysates. Asterisk in the ABC45-modified tryptic peptide corresponds to the predicted site of ABC45 modification (S115) as determined by tandem MS analysis. Asterisk located on the MS1 peak for the ABC45-modified tryptic peptide marks the time point of MS2 acquisition.
We next asked whether ABC45 could be used as a probe in competitive ABPP-SILAC experiments to identify targets of ABC34 (13). Isotopically heavy- and light-labeled PC3 cells were treated with ABC34 (13) (1 uM) or DMSO for 4 h, respectively, lysed, and then treated with ABC45 (5 µM, 1 h). The heavy and light proteomic samples were then combined and analyzed by LC-MS/MS, which identified a small subset of SHs, including ABHD4, that were inhibited by ABC34 (13) (Figure 3B and Table S1). Among the handful of additional SH targets of ABC34 were ABHD6 and PLA2G7, which were expected based on our ABPP-SILAC studies with FP-biotin (see Figure 2D), and palmitoyl-protein thioesterase 1 (PPT1), which was inhibited by more than 90%. As we and others have reported previously, PPT1 shows poor reactivity with broad-spectrum FP probes (Martin et al., 2012; Wang et al., 2013). Accordingly, PPT1 was not detected in our previous ABPP-SILAC studies with FP-biotin (Figure 2D). We confirmed the selective enrichment of PPT1 by ABC45 by performing a probe-versus-probe ABPP-SILAC study, where heavy and light PC3 proteomes were treated with FP-biotin (2.5 µM, 1 h) and ABC45 (5 µM, 1 h), respectively. PPT1, along with two additional SHs, LIPA and DDHD2, were preferentially enriched by ABC45 over FP-biotin, while most of the other SHs were more strongly enriched by FP-biotin (Figure 3C and Table S1). A handful of SHs corresponding mostly to enzymes that were identified in our ABPP-SILAC studies as targets of ABC34 (13) (Figures 2D and 3B), were equivalently enriched by ABC45 and FP-biotin (Figure 3C).
The selective enrichment of PPT1 by ABC45 and the near-complete blockade of this enrichment by ABC34 (13) suggested that this enzyme was covalently modified and inhibited by NHH carbamates. We further explored this possibility by treating HEK293T cells transfected with human PPT1 (hPPT1) with ABC34 (13) (5 µM, 4 h pre-treatment) or DMSO followed by cell lysis and treatment with ABC45 (5 µM, 1 h). A strong, but diffuse ABC34-sensitive, ABC45-labeled protein band was detected by gel-based ABPP in hPPT1-transfected, but not mock-transfected cell lysates (Figure 3D), and this diffuse signal was compressed to a tight, faster-migrating band matching the predicted molecular weight of PPT1 following treatment with the glycosidase PNGaseF (Figure 3D). Western blotting with an anti-PPT1 antibody confirmed a similar migration pattern for hPPT1 protein in transfected cell lysates (Figure 3D, bottom panel). These data indicate that both ABC34 (13) and ABC45 react with PPT1 and are consistent with previous studies demonstrating that this enzyme is glycosylated (Camp et al., 1994; Lyly et al., 2007). In contrast, FP-Rh failed to detect hPPT1 in transfected cell lysates (Figure 3D).
We next aimed to confirm whether NHH carbamates react with the catalytic serine of PPT1 – S115 (Bellizzi et al., 2000). HEK293T cells were transfected with wild type and an S115A mutant of PPT1, lysed, and treated with ABC45 followed by CuAAC conjugation to an azide-Rh tag, PNGaseF treatment, and analysis by SDS-PAGE. This study revealed strong labeling of wild-type PPT1, but not the S115A mutant (Figure 3E). We also identified S115 as the site of ABC45 labeling on PPT1 using a quantitative MS method termed isoTOP-ABPP (Weerapana et al., 2010). In brief, PPT1-transfected cell lysates were treated with DMSO or the ABC45 probe (5 µM) for 1 h and then incubated with isotopically heavy or light azide-biotin tags, respectively, under CuAAC reaction conditions. Each azide biotin tag also contained an intervening TEV protease cleavage site (Weerapana et al., 2010). Heavy and light probe-labeled proteins were then combined, enriched by streptavidin chromatography, digested sequentially on-bead with trypsin (to remove non-probe-modified peptides) and TEV to release ABC45-modified peptide(s), which were analyzed by LC-MS/MS on an LTQ-Orbitrap instrument. A single ABC45-modified peptide, corresponding to amino acids 105–122, was detected in the isotopically light, but not heavy samples, and tandem MS analysis assigned the ABC45 modification site to S115 (Figure 3F and Figure S3B, C). Taken together, these data indicate that NHH carbamates react with PPT1 through carbamylation of the enzyme’s conserved serine nucleophile.
Selective, Cell-Active NHH Carbamate Inhibitors for ABHD4 and PPT1
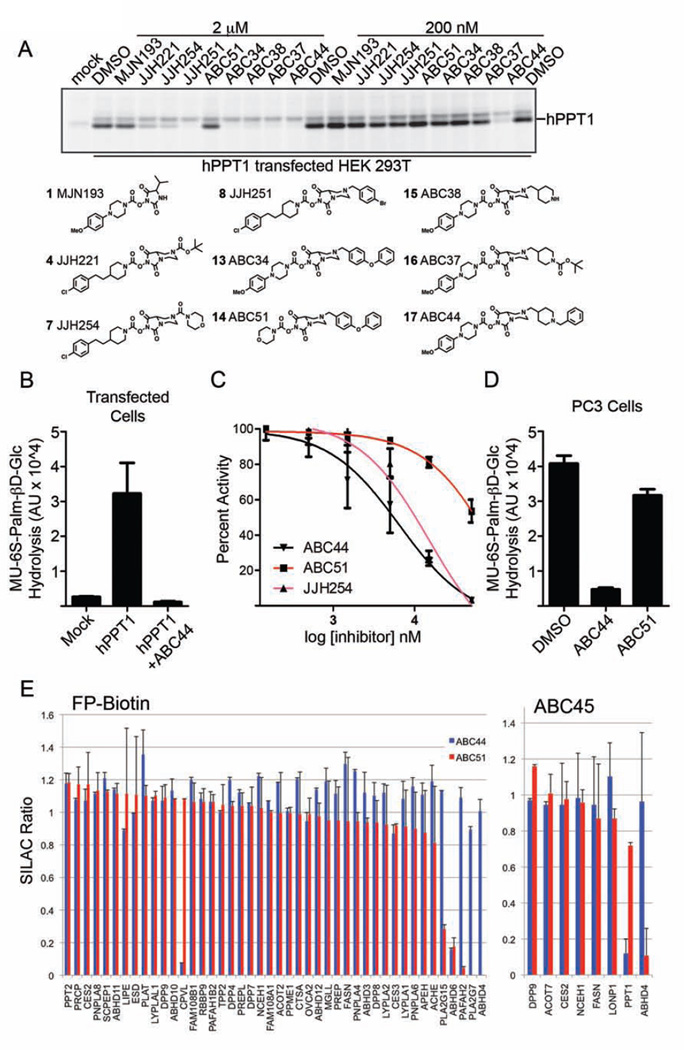
Excited that our analysis of NHH targets led to the serendipitous discovery of a lead inhibitor and tailored activity-based probe for PPT1, we sought to more deeply investigate the structure-activity relationship (SAR) of the NHH carbamate class in the hopes of identifying selective inhibitors for both PPT1 and ABHD4. We first assayed a structurally diverse panel of NHH carbamates at 2 or 0.2 µM for in situ inhibition of hPPT1 in transfected HEK293T cells (Figure 4A). While many compounds exhibited inhibitory activity toward PPT1 at 2 uM, only ABC44 (17) showed complete inhibition of hPPT1 at 200 nM (Figure 4A). ABC44 (17) displayed an in situ IC50 value for inhibiting recombinant PPT1 of 0.1 µM as measured by gel-based competitive ABPP (Table 1 and Figure S4A).
Figure 4. Discovery of cell-active NHH carbamate inhibitors of PPT1 and ABHD4.
(A) Screening of a subset of NHH carbamates in hPPT1-transfected HEK293T cells in situ, where PPT1 inhibition was measured by competitive ABPP using the ABC45 probe. Structures of tested compounds shown below the ABPP gel. (B) hPPT1-transfected cell lysates show much greater hydrolytic activity with the MU-6S-Palm-βD-Glc substrate compared to mock-transfected cell lysates or hPPT1-transfected cell lysates pre-treated with ABC44 (17) (50 µM, 30 min). Data represent average values ± S.E. for three independent experiments. (C) Concentration-dependent inhibition of MU-6S-Palm-βD-Glc hydrolytic activity of hPPT1-transfected cell lysates by ABC44 (17), ABC51 (14), and JJH254 (7). Data represent average values ± S.E. for three independent experiments. (D) PC3 cells treated in situ with ABC44, but not ABC51 (500 nM inhibitor; 4 h) showed substantial reductions in 4-MU-6S-Palm-βD-Glc hydrolysis activity compared to DMSO-treated control cells. Data represent average values ± S.E. for three independent experiments. (E) ABPP-SILAC analysis showing overlaid in situ SH inhibition profiles for ABC44 (0.1 µM, 4 h) and ABC51 (1 µM, 4 h) in PC3 cells where SH enrichment and inhibition were measured with the FP-biotin (left) or ABC45 (right) probes. Data represent average values ± S.D. for two independent experiments.
Table 1.
IC50 values (µM; with 95% confidence intervals in parentheses) for inhibition of human PPT1 by representative NHH carbamates measured in PPT1-transfected cells (in situ, competitive ABPP) or cell lysates (in vitro, pH 5.0, MU-6S-Palm-βD-Glc substrate).
| NHH carbamate | in situ (ABPP) | in vitro (substrate) |
|---|---|---|
| ABC44 (17) | 0.1 (0.04–0.2) | 6.5 (3.6–12) |
| ABC51 (14) | 3.9 (2.0–7.4) | > 50 |
| JJH254 (7) | 0.5 (0.3–1.0) | 15 (9.5–25) |
We next tested whether ABC44 inhibited PPT1 activity using a substrate assay. Endogenous substrates for PPT1 remain poorly characterized, but this enzyme can be assayed with a fluorogenic substrate 4-methylumbelliferyl- 6-thio-palmitate-βD-glucopyranoside (MU-6S-Palm-βD-Glc) (van Diggelen et al., 1999). In this assay, PPT1 hydrolyzes the substrate's palmitoyl thioester bond, and exogenous sweet almond β-glucosidase deglycosylates and frees the fluorophore, 4-methylumbelliferone (4-MU), which can be measured to assess PPT1 activity. PPT1-transfected cells displayed a strong increase in MU-6S-Palm-βD-Glc hydrolysis compared to mock-transfected cells (Figure 4B), and treatment of these transfected lysates at pH 5.0 with ABC44 (17) fully blocked PPT1 activity with an IC50 value of 6.5 µM (Figure 4C and Table 1). Additionally, in situ treatment of PC3 cells with ABC44 (17) (500 nM, 4 h) showed a 90% reduction in endogenous PPT1 activity based on the MU-6S-Palm-βD-Glc substrate assay (Figure 4D).
A second compound of interest was ABC51 (14), which showed limited activity against PPT1 in situ (Figure 4A, D and Table 1) or in vitro (Figure 4B and Table 1), but was a good inhibitor of ABHD4 in vitro (IC50 value of 0.5 µM) as measured with an N-acyl phosphatidylethanolamine (NAPE) substrate hydrolysis assay performed with lysates from ABHD4-transfected HEK293T cells (Figure S4B). In contrast, neither ABC44 (17) nor the LYPLA1/2 inhibitor JJH254 (7) altered the NAPE hydrolysis activity of ABHD4 (Figure S4B). JJH254 did, however, inhibit PPT1 activity both in vitro (Figure 4C and Table 1) and in situ (Figure 4A, Table 1, and Figure S4A), albeit with less potency than ABC44.
To evaluate the broader selectivity of ABC44 (17) and ABC51 (14) across the SH class, we treated heavy and light isotopically-labeled PC3 cells with inhibitor (ABC51 (1 µM); or ABC44 (1 or 0.1 µM); 4 h) and DMSO, respectively, followed by ABPP-SILAC analysis with either the FP-biotin or ABC45 probes. ABC44 (17), tested at 1 or 0.1 µM, near-completely inhibited PPT1 (~90%+; as measured by ABC45) and showed good selectivity, only cross-reacting with two of the >40 quantified SH activities – ABHD6 and CPVL (Figure 4E, Figure S4C, and Supplementary Table 1). Conversely, ABC51 (14) inhibited > 90% of ABHD4 activity, as measured with either the FP-biotin or ABC45 probes, and cross-reacted with four additional targets – ABHD6, PAFAH2, PLA2G7, and PLA2G15 (Figure 4C). We were initially surprised that ABC44 (17), when tested at 0.1 µM, produced near-complete blockade of endogenous PPT1 in ABPP-SILAC experiments, given that this concentration equals the inhibitor’s in situ IC50 value measured with recombinant PPT1 (see Table 1 and Figure S4A). We speculate that the potency of ABC44 (17) may be blunted in transfected cells due to the higher cellular concentration of PPT1 compared to the endogenous levels of this enzyme found in other cells. Finally, even when tested at concentrations substantially above those required to inhibit their primary targets, ABC44 (17) and ABC51 (14) did not cause cytotoxicity (cell toxicity IC50 values > 50 µM).
These results, taken together, demonstrate that ABC44 (17) and ABC51 (14) are cell-active inhibitors that display good potency and selectivity for PPT1 and ABHD4, respectively. While we do not yet understand the basis for the markedly enhanced potency displayed by ABC44 (and JJH254 (7)) for PPT1 in situ versus in vitro, it is possible that these compounds preferentially accumulate in the lysosomal compartment of cells to promote PPT1 inactivation.
In Vivo Activity of NHH Carbamates
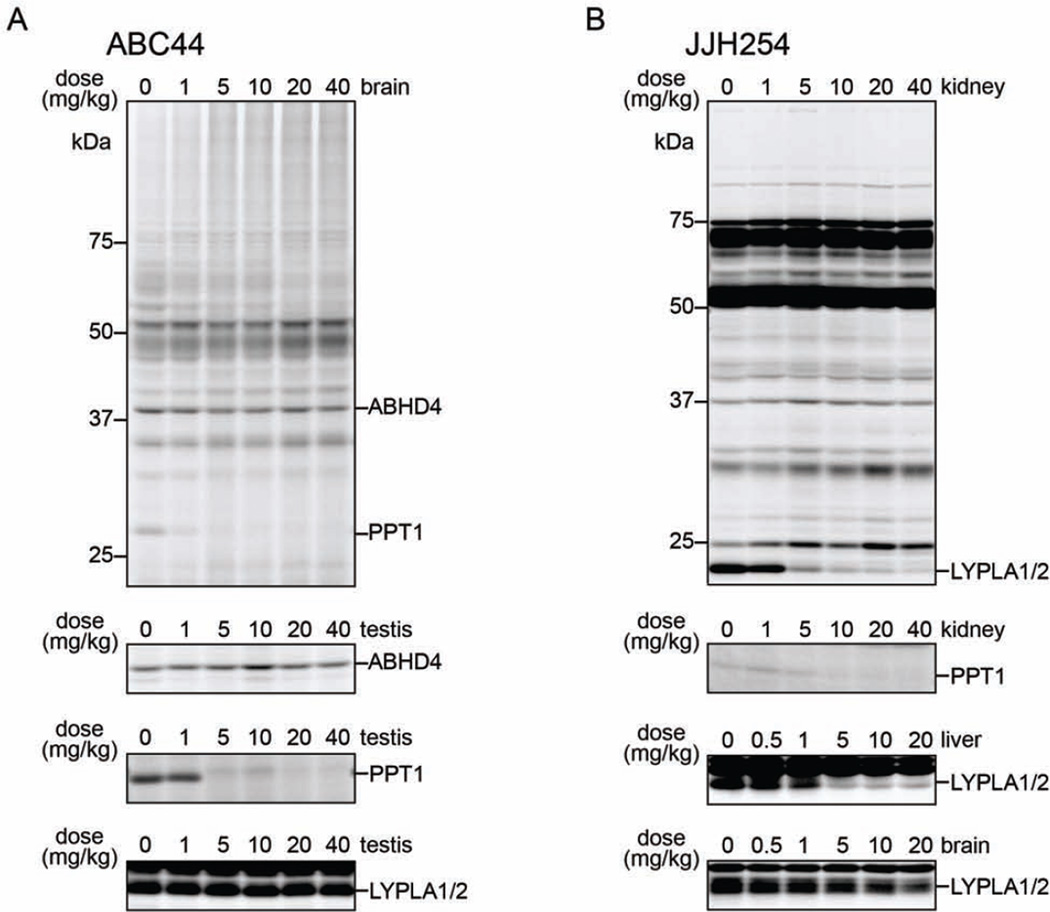
We next asked whether representative NHH carbamates could inhibit their SH targets in vivo. ABC44 (17), ABC51 (14), and JJH254 (7) were administered across a dose range of 1–40 mg/kg to C57BL6 mice by intraperitoneal injection, and, after 4 h, animals were sacrificed and tissues harvested for analysis by gel-based ABPP using the FP-Rh and ABC45 probes. ABC44 (17) showed potent, dose-dependent inhibition of an ABC45-reactive protein matching the molecular mass of deglycosylated PPT1 in PNGaseF-treated central and peripheral tissues, producing near-complete blockade of this enzyme activity at 1–5 mg/kg (Figure 5A and Figure S5A). ABC44 (17) showed excellent selectivity for PPT1 and did not inhibit other SHs detected in brain and testis tissues treated with the FP-Rh (Figure S5A) or ABC45 (Figure 5A and Figure S5A) probes. JJH254 (7) also showed good in vivo activity, producing near-complete blockade of LYPLA1 and LYPLA2 in peripheral (e.g., liver, kidney), but not central (brain) tissues at a dose of 5 mg/kg as measured with the FP-Rh probe (Figure 5B and Figure S5A). JJH254 (7) also showed limited cross-reactivity with other SHs, consistent with competitive ABPP-SILAC studies in human cells (Figure S5B; and (Hulce et al., 2014)), but did inhibit PPT1 at doses of 10 mg/kg or greater as measured with ABC45 (Probe 1) (Figure 5B). Contrasting with these results, ABC51 (14) did not block the activity of ABHD4 in any tissue examined (data not shown), possibly indicating that improvements in potency and/or drug-like properties are required to convert this compound into an in vivo-active probe
Figure 5. NHH carbamates inhibit PPT1 and other SHs in vivo.
(A) Gel-based competitive ABPP analysis of SH activities in tissues from ABC44 (17)- (A) or JJH254 (7)- (B) treated mice. The activities of ABHD4 and PPT1 were visualized using the ABC45 probe in tissue proteomes treated with PNGaseF (post-ABC45 labeling), while the activities of LYPLA1 and LYPLA2 were visualized using the FP-Rh probe.
DISCUSSION
In the present work, we have examined an underexplored class of carbamate inhibitors bearing extended N-hydroxyhydantoin (NHH) leaving groups against the serine hydrolase (SH) enzyme class using competitive ABPP. NHH carbamates inhibited SHs that have proven largely insensitive to other carbamate scaffolds, including ABHD3, ABHD4, and PPT1. That PPT1 is also poorly reactive with FPs (Martin et al., 2012; Wang et al., 2013) agents, which represent arguably the most generic SH-directed electrophile, underscores the remarkable selectivity individual SHs can display for specific reactive chemotypes. Such restricted reactivity relationships, when uncovered, can form the basis for the design of selective inhibitors and tailored ABPP probes, as we have shown herein for PPT1 and ABHD4.
Our most advanced PPT1 inhibitor ABC44 (17) contains an NHH leaving group that is nearly isosteric with that of ABC34 (13), but features two trialkylated basic amines. This dibasic property could contribute to the enhanced potency of ABC44 in cells compared to cell lysates, since positively charged amine molecules have been shown to accumulate in the lysosome (Goldman et al., 2009), the subcellular compartment where PPT1 resides. However, the explanation for enhanced in situ potency of NHH carbamates for PPT1 is likely more complex, as JJH254 (7), which lacks basic nitrogens, was also much more active against PPT1 in cells. It is possible that other features shared between ABC44 and JJ254 promote their accumulation in the lysosome to facilitate PPT1 inhibition. That several other lysosomal hydrolases were unaffected by ABC44 (17), including CTSA, PRCP, SCPEP1, and PPT2, indicates, regardless of mechanism, the increased potency displayed by this NHH carbamate for PPT1 in cells compared to lysates does not unduly compromise selectivity. Conversely, ABC44 (17) inhibited ABHD6, which is not considered a lysosomal protein, indicating that this NHH carbamate can access additional compartments in the cell.
In recombinant expression systems, PPT1 exhibited a minor, upper molecular weight (MW) form (Figures 3D and 3E) that was less sensitive to ABC34 (13) (Figure 3D). We speculate that this upper MW species could represent a transitional form of PPT1 as the protein migrates from the endoplasmic reticulum to the lysosome and undergoes maturation through post-translational modification. Whether such inhibitor-resistant forms of PPT1 reflect an artifact of overexpression or might also exist endogenously is not yet clear, but our ABPP-SILAC studies, where 90%+ of PPT1 activity was blocked by ABC44 (17) (0.1–1 µM) in PC3 cells (Figure 4D, E and Figure S4C), would argue that most, if not all of endogenous PPT1 is sensitive to ABC44 inhibition. From a methodological perspective, these data underscore the importance of methods, like competitive ABPP, that can determine target engagement for inhibitors in living systems (Simon et al., 2013), as this parameter may differ substantially from inhibitor potencies measured in vitro due to compound and/or target localization, or other parameters that affect protein function in cells.
Deleterious mutations that impair PPT1 expression cause a rare, Mendelian distributed genetic disorder termed infantile neuronal ceroid lipofuscinosis (INCL) or infantile Batten’s disease (Kyttala et al., 2006). Peptide-based reversible inhibitors of PPT1 have been described (Dawson et al., 2010; Meng et al., 1998) with the potential to serve as pharmacological chaperones that could facilitate the transport of mutant PPT1 variants to the lysosome (Dawson et al., 2010), where improved enzyme stability may be observed in the lower pH environment of this organelle. Whether these peptide-based inhibitors can affect PPT1 transport and/or function in vivo remains unclear. Assuming that restoration of PPT1 activity (and not merely expression) is required to reverse the pathophysiological phenotypes observed in INCL, it is unlikely that NHH carbamates, being irreversible inhibitors, would offer a potential treatment strategy for this disease. We believe, however, that ABC44 (17) and related NHH carbamates should serve as useful probes to characterize the function of PPT1 in cell and animal models. These pharmacological studies, which could also employ control probes that inhibit some of the off-targets of ABC44, but spare PPT1 activity (e.g., ABHD6-selective inhibitors, such as MJN193 described herein and others (Hsu et al., 2013; Janssen et al., 2014; Li et al., 2007; Marrs et al., 2011), may illuminate the endogenous substrate, products, and pathways affected by PPT1 inhibition and, through doing so, provide mechanistic insights into the biochemical basis for INCL.
Finally, the NHH carbamates explored herein furnished lead inhibitors for several additional SHs for which selective chemical probes are currently lacking, including ABHD12, PLA2G15, and CPVL (as well as the aforementioned ABHD3 and ABHD4). As we have shown for ABHD3, ABHD4, and PPT1, each SH appears to display a distinct structure-activity relationship (SAR) with the limited subset of NHH carbamates evaluated so far, suggesting that continued medicinal chemistry exploration of this chemotype has the potential to yield selective inhibitors for other SHs. We therefore conclude that NHH carbamates represent a versatile scaffold for the discovery and optimization of inhibitors for the pharmacological characterization of SHs in living systems.
SIGNIFICANCE
This study explores N-hydroxyhydantoin (NHH) carbamates as inhibitors of the serine hydrolase (SH) enzyme class using activity-based protein profiling (ABPP) methods. We show that NHH carbamates offer advantages over previously described classes of carbamate inhibitors of SHs, in particular by permitting the facile derivatization of both staying and leaving group components of the carbamate scaffold to improve inhibitor potency and selectivity. We discover selective and in vivo-active NHH carbamate inhibitors for SHs that heretofore have lacked chemical probes, including palmitoyl-protein thioesterase-1 (PPT1), mutations of which cause the neurological disease infantile neuronal ceroid lipofuscinosis (INCL). We further show that these inhibitors can be converted into tailored probes for profiling the activity and inhibition of PPT1 and others SHs in native biological systems. The selective inhibitors and activity probes described herein should facilitate the pharmacological analysis of PPT1 and other SHs in cell and animal models and, through doing so, illuminate the functions that these enzymes play in mammalian biology and disease.
EXPERIMENTAL PROCEDURES
Gel-Based ABPP Analysis
Gel-based ABPP analysis was performed as described previously (Chang et al., 2013). Briefly, tissue and cell proteomes (1 mg/mL, 50 µL) were treated with FP-rhodamine (1 µM, 30 min) or ABC45 (1 µM, 60 min) and chased with a rhodamine-N3 tag under CuAAC conditions (Chang et al., 2013). Samples were then quenched by adding 4× SDS-PAGE loading buffer (17 µL), resolved by SDS-PAGE, and visualized using an FMBio II Multiview flatbed fluorescence scanner (Hitachi) or a ChemiDoc MP (Bio-Rad). Signal intensity was then quantified with ImageJ 1.45s and IC50 values were determined with GraphPad Prism 5.01.
In Situ Inhibitor Treatment of Human Cell Lines
In situ inhibitor treatments of human cell lines were performed as described previously (Hsu et al., 2012). Briefly, PC3 or transfected HEK293T cells were grown to confluency in media (RPMI or DMEM, respectively) supplemented with serum. The culture media was then removed and replaced with fresh media containing DMSO or inhibitors at the desired concentration and incubated for 4 h in a tissue culture incubator (37 °C, 5% CO2). Cells were then washed with PBS, harvested, washed with PBS once more, and lysed with a probe sonicator in PBS.
In Vivo Administration of Inhibitors
Mice were injected with ABC44, ABC51, JJH254, or vehicle, as described previously (Chang et al., 2013; Hsu et al., 2012). Briefly, C57Bl/6 mice were injected intraperitoneally with 18:1:1 (v/v/v) solution of saline/ethanol/emulphor containing varying doses of compounds. After 4 h mice were anesthetized with isoflurane, euthanized by cervical dislocation, and harvested tissues were prepared for Gel-Based ABPP analysis. The studies were performed with the approval of the Institutional Animal Care and Use Committee at The Scripps Research Institute in accordance with the Guide for the Care and Use of Laboratory Animals.
ABPP-SILAC Sample Preparation and Data Analysis
ABPP-SILAC samples were prepared and analyzed as described previously (Hsu et al., 2012). In brief, isotopically heavy and light PC3 cells (or cell lysates) were treated with inhibitor or DMSO, respectively, and cell proteomes (500 µL, 2 mg/mL) were incubated with FP-biotin (2.5 µM, 1 h) or ABC45 (5 µM, 1 h) followed by conjugation to biotin-azide by CuAAC (Speers and Cravatt, 2004). Light and heavy cell lysates were combined, additional PBS was added (1 mL), and the resulting mixture was precipitated with 4:1 MeOH/CHCl3 (2.5 mL). Pellets were then washed with 1:1 MeOH/CHCl3 (1 mL, 2× vol), resuspended in 0.5 mL of 6 M urea in PBS, reduced using tris(2-carboxyethyl)phosphine (TCEP, 10 mM) for 30 min at 37 °C and then alkylated using iodoacetamide (40 mM) for 30 min at 25 °C in the dark. Biotinylated proteins were enriched with PBS-washed streptavidin beads (100 µL, Sigma-Aldrich) by shaking at 25 °C for 1.5 h in PBS (5.5 mL) with 2% SDS. The beads were washed sequentially with 10 ml PBS with 0.2% SDS (3×), 10 ml PBS (3×) and 10 ml DI H2O (3×). On-bead digestion was performed using sequence-grade trypsin (2 µg; Promega) in 2 M urea in PBS with 2 mM CaCl2 for 12–14 h at 37 °C (200 µL). Peptides obtained from this procedure were acidified using formic acid (5% final) and stored at −20 °C before analysis. Digested peptides were loaded on to a biphasic (strong cation exchange/reverse phase) capillary column and analyzed by 2D liquid chromatography in combination with tandem mass spectrometry on an LTQ-Orbitrap (Thermo Scientific). Samples were then searched using the ProLuCID algorithm against a human reverse-concatenated nonredundant (gene-centric) FASTA database that was assembled from the Uniprot database. SILAC ratios were quantified using in house CIMAGE software (Weerapana et al., 2010).
Additional Experimental Procedures can be found in Supplemental Information.
Supplementary Material
HIGHLIGHTS.
N-hydroxyhydantoin (NHH) carbamates inhibit diverse serine hydrolases
Inhibitors were discovered for palmitoyl-protein thioesterase-1 (PPT1)
PPT1 inhibitors can be converted into tailored activity-based probes
PPT1 inhibitors are in vivo-active
ACKNOWLEDGMENTS
This research was supported by the NIH (DA033760) and the Skaggs Institute for Chemical Biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
A.B.C., J.J.H., and B.F.C. conceived of research and wrote the paper. A.B.C., J.J.H., and M.J.N. synthesized compounds. A.B.C., J.J.H., and H.-C.L. designed experiments. A.B.C., J.J.H. H.-C.L., and M.L.M. performed experiments.
REFERENCES
- Adibekian A, Martin BR, Chang JW, Hsu KL, Tsuboi K, Bachovchin DA, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, et al. Confirming target engagement for reversible inhibitors in vivo by kinetically tuned activity-based probes. J Am Chem Soc. 2012;134:10345–10348. doi: 10.1021/ja303400u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adibekian A, Martin BR, Wang C, Hsu KL, Bachovchin DA, Niessen S, Hoover H, Cravatt BF. Click-generated triazole ureas as ultrapotent in vivo-active serine hydrolase inhibitors. Nat Chem Biol. 2011;7:469–478. doi: 10.1038/nchembio.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachovchin DA, Cravatt BF. The pharmacological landscape and therapeutic potential of serine hydrolases. Nat Rev Drug Discov. 2012;11:52–68. doi: 10.1038/nrd3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellizzi JJ, 3rd, Widom J, Kemp C, Lu JY, Das AK, Hofmann SL, Clardy J. The crystal structure of palmitoyl protein thioesterase 1 and the molecular basis of infantile neuronal ceroid lipofuscinosis. Proc Natl Acad Sci U S A. 2000;97:4573–4578. doi: 10.1073/pnas.080508097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biel M, Deck P, Giannis A, Waldmann H. Synthesis and evaluation of acyl protein thioesterase 1 (APT1) inhibitors. Chemistry. 2006;12:4121–4143. doi: 10.1002/chem.200501128. [DOI] [PubMed] [Google Scholar]
- Blankman JL, Simon GS, Cravatt BF. A Comprehensive Profile of Brain Enzymes that Hydrolyze the Endocannabinoid 2-Arachidonoylglycerol. Chem Biol. 2007;14:1347–1356. doi: 10.1016/j.chembiol.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp LA, Verkruyse LA, Afendis SJ, Slaughter CA, Hofmann SL. Molecular cloning and expression of palmitoyl-protein thioesterase. J Biol Chem. 1994;269:23212–23219. [PubMed] [Google Scholar]
- Chang JW, Cognetta AB, 3rd, Niphakis MJ, Cravatt BF. Proteome-Wide Reactivity Profiling Identifies Diverse Carbamate Chemotypes Tuned for Serine Hydrolase Inhibition. ACS chemical biology. 2013;8:1590–1599. doi: 10.1021/cb400261h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JW, Niphakis MJ, Lum KM, Cognetta AB, 3rd, Wang C, Matthews ML, Niessen S, Buczynski MW, Parsons LH, Cravatt BF. Highly selective inhibitors of monoacylglycerol lipase bearing a reactive group that is bioisosteric with endocannabinoid substrates. Chem Biol. 2012;19:579–588. doi: 10.1016/j.chembiol.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JW, Nomura DK, Cravatt BF. A potent and selective inhibitor of KIAA1363/AADACL1 that impairs prostate cancer pathogenesis. Chem Biol. 2011;18:476–484. doi: 10.1016/j.chembiol.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson G, Schroeder C, Dawson PE. Palmitoyl:protein thioesterase (PPT1) inhibitors can act as pharmacological chaperones in infantile Batten disease. Biochemical and biophysical research communications. 2010;395:66–69. doi: 10.1016/j.bbrc.2010.03.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker FJ, Rocks O, Vartak N, Menninger S, Hedberg C, Balamurugan R, Wetzel S, Renner S, Gerauer M, Scholermann B, et al. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat Chem Biol. 2010;6:449–456. doi: 10.1038/nchembio.362. [DOI] [PubMed] [Google Scholar]
- Goldman SD, Funk RS, Rajewski RA, Krise JP. Mechanisms of amine accumulation in, and egress from, lysosomes. Bioanalysis. 2009;1:1445–1459. doi: 10.4155/bio.09.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross RW. Purification of rabbit myocardial cytosolic acyl-CoA hydrolase, identity with lysophospholipase, and modulation of enzymic activity by endogenous cardiac amphiphiles. Biochemistry. 1983;22:5641–5646. doi: 10.1021/bi00293a028. [DOI] [PubMed] [Google Scholar]
- Hoover HS, Blankman JL, Niessen S, Cravatt BF. Selectivity of inhibitors of endocannabinoid biosynthesis evaluated by activity-based protein profiling. Bioorganic & medicinal chemistry letters. 2008;18:5838–5841. doi: 10.1016/j.bmcl.2008.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu KL, Tsuboi K, Adibekian A, Pugh H, Masuda K, Cravatt BF. DAGLbeta inhibition perturbs a lipid network involved in macrophage inflammatory responses. Nat Chem Biol. 2012;8:999–1007. doi: 10.1038/nchembio.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu KL, Tsuboi K, Chang JW, Whitby LR, Speers AE, Pugh H, Cravatt BF. Discovery and optimization of piperidyl-1,2,3-triazole ureas as potent, selective, and in vivo-active inhibitors of alpha/beta-hydrolase domain containing 6 (ABHD6) J Med Chem. 2013;56:8270–8279. doi: 10.1021/jm400899c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulce JJ, Joslyn C, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, Ferguson J, Cravatt BF, Hodder P, Rosen H. Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD): 2014. An in Vivo Active Carbamate-based Dual Inhibitor of Lysophospholipase 1 (LYPLA1) and Lysophospholipase 2 (LYPLA2) [PubMed] [Google Scholar]
- Janssen FJ, Deng H, Baggelaar MP, Allara M, van der Wel T, den Dulk H, Ligresti A, van Esbroeck AC, McGuire R, Di Marzo V, et al. Discovery of glycine sulfonamides as dual inhibitors of sn-1-diacylglycerol lipase alpha and alpha/beta-hydrolase domain 6. J Med Chem. 2014;57:6610–6622. doi: 10.1021/jm500681z. [DOI] [PubMed] [Google Scholar]
- Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, Gross RW. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem. 2004;279:48968–48975. doi: 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- Kyttala A, Lahtinen U, Braulke T, Hofmann SL. Functional biology of the neuronal ceroid lipofuscinoses (NCL) proteins. Biochimica et biophysica acta. 2006;1762:920–933. doi: 10.1016/j.bbadis.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Lee HC, Simon GM, Cravatt BF. ABHD4 regulates multiple classes of N-acyl phospholipids in the mammalian nervous system. Biochemistry. 2015 doi: 10.1021/acs.biochem.5b00207. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Blankman JL, Cravatt BF. A functional proteomic strategy to discover inhibitors for uncharacterized hydrolases. J Am Chem Soc. 2007;129:9594–9595. doi: 10.1021/ja073650c. [DOI] [PubMed] [Google Scholar]
- Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci USA. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JZ, Cisar JS, Milliken D, Niessen S, Wang C, Trauger SA, Siuzdak G, Cravatt BF. Metabolomics annotates ABHD3 as a physiologic regulator of medium-chain phospholipids. Nat Chem Biol. 2011;7:763–765. doi: 10.1038/nchembio.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JZ, Cravatt BF. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chemical reviews. 2011;111:6022–6063. doi: 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JZ, Li W, Booker L, Burston JJ, Kinsey SG, Schlosburg JE, Pavon FJ, Serrano AM, Selley DE, Parsons LH, et al. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat Chem Biol. 2009;5:37–44. doi: 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyly A, von Schantz C, Salonen T, Kopra O, Saarela J, Jauhiainen M, Kyttala A, Jalanko A. Glycosylation, transport, and complex formation of palmitoyl protein thioesterase 1 (PPT1)--distinct characteristics in neurons. BMC cell biology. 2007;8:22. doi: 10.1186/1471-2121-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- Marks KM, Jacobson IM. The first wave: HCV NS3 protease inhibitors telaprevir and boceprevir. Antiviral therapy. 2012;17:1119–1131. doi: 10.3851/IMP2424. [DOI] [PubMed] [Google Scholar]
- Marrs WR, Horne EA, Ortega-Gutierrez S, Cisneros JA, Xu C, Lin YH, Muccioli GG, Lopez-Rodriguez ML, Stella N. Dual inhibition of alpha/beta-hydrolase domain 6 and fatty acid amide hydrolase increases endocannabinoid levels in neurons. J Biol Chem. 2011;286:28723–28728. doi: 10.1074/jbc.M110.202853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Global profiling of dynamic protein palmitoylation. Nat Methods. 2012;9:84–89. doi: 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Sin N, Crews CM. The antiproliferative agent didemnin B uncompetitively inhibits palmitoyl protein thioesterase. Biochemistry. 1998;37:10488–10492. doi: 10.1021/bi9804479. [DOI] [PubMed] [Google Scholar]
- Navia-Paldanius D, Savinainen JR, Laitinen JT. Biochemical and pharmacological characterization of human alpha/beta-hydrolase domain containing 6 (ABHD6) and 12 (ABHD12) J Lipid Res. 2012;53:2413–2424. doi: 10.1194/jlr.M030411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RH, Miles JM. The use of orlistat in the treatment of obesity, dyslipidaemia and Type 2 diabetes. Expert Opin Pharmacother. 2005;6:2483–2491. doi: 10.1517/14656566.6.14.2483. [DOI] [PubMed] [Google Scholar]
- Niphakis MJ, Cognetta AB, 3rd, Chang JW, Buczynski MW, Parsons LH, Byrne F, Burston JJ, Chapman V, Cravatt BF. Evaluation of NHS carbamates as a potent and selective class of endocannabinoid hydrolase inhibitors. ACS chemical neuroscience. 2013;4:1322–1332. doi: 10.1021/cn400116z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niphakis MJ, Cravatt BF. Enzyme inhibitor discovery by activity-based protein profiling. Annu Rev Biochem. 2014;83:341–377. doi: 10.1146/annurev-biochem-060713-035708. [DOI] [PubMed] [Google Scholar]
- Patricelli MP, Giang DK, Stamp LM, Burbaum JJ. Direct visualization of serine hydrolase activities in complex proteome using fluorescent active site-directed probes. Proteomics. 2001;1:1067–1071. doi: 10.1002/1615-9861(200109)1:9<1067::AID-PROT1067>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Racchi M, Mazzucchelli M, Porrello E, Lanni C, Govoni S. Acetylcholinesterase inhibitors: novel activities of old molecules. Pharmacological Research. 2004;50:441–451. doi: 10.1016/j.phrs.2003.12.027. [DOI] [PubMed] [Google Scholar]
- Rostovtsev VV, Green JG, Fokin VV, Sharpless KB. A stepwise Huisgen cycloaddition process: copper(I)-catalyzed regioselective "ligation" of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry. 2004;43:14332–14339. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- Simon GM, Cravatt BF. Endocannabinoid biosynthesis proceeding through glycerophospho-N-acyl ethanolamine and a role for alpha/beta hydrolase 4 in this pathway. J Biol Chem. 2006;281:26465–26472. doi: 10.1074/jbc.M604660200. [DOI] [PubMed] [Google Scholar]
- Simon GM, Niphakis MJ, Cravatt BF. Determining target engagement in living systems. Nat Chem Biol. 2013;9:200–205. doi: 10.1038/nchembio.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Staub I, Sieber SA. Beta-lactams as selective chemical probes for the in vivo labeling of bacterial enzymes involved in cell wall biosynthesis, antibiotic resistance, and virulence. J Am Chem Soc. 2008;130:13400–13409. doi: 10.1021/ja803349j. [DOI] [PubMed] [Google Scholar]
- Thornberry NA, Weber AE. Discovery of JANUVIA (Sitagliptin), a selective dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes. Curr Top Med Chem. 2007;7:557–568. doi: 10.2174/156802607780091028. [DOI] [PubMed] [Google Scholar]
- van Diggelen OP, Keulemans JL, Winchester B, Hofman IL, Vanhanen SL, Santavuori P, Voznyi YV. A rapid fluorogenic palmitoyl-protein thioesterase assay: pre- and postnatal diagnosis of INCL. Molecular genetics and metabolism. 1999;66:240–244. doi: 10.1006/mgme.1999.2809. [DOI] [PubMed] [Google Scholar]
- Wang R, Borazjani A, Matthews AT, Mangum LC, Edelmann MJ, Ross MK. Identification of palmitoyl protein thioesterase 1 in human THP1 monocytes and macrophages and characterization of unique biochemical activities for this enzyme. Biochemistry. 2013;52:7559–7574. doi: 10.1021/bi401138s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.