Abstract
Aim:
Determine the effect of the genetic variants beyond CYP3A5*3 on tacrolimus disposition.
Patients & methods:
We studied genetic correlates of tacrolimus trough concentrations with POR*28, CYP3A4*22 and ABCC2 haplotypes in a large, ethnically diverse kidney transplant cohort (n = 2008).
Results:
Subjects carrying one or more CYP3A5*1 alleles had lower tacrolimus trough concentrations (p = 9.2 × 10-75). The presence of one or two POR*28 alleles was associated with a 4.63% reduction in tacrolimus trough concentrations after adjusting for CYP3A5*1 and clinical factors (p = 0.037). In subset analyses, POR*28 was significant only in CYP3A5*3/*3 carriers (p = 0.03). The CYP3A4*22 variant and the ABBC2 haplotypes were not associated.
Conclusion:
This study confirmed that CYP3A5*1 was associated with lower tacrolimus trough concentrations. POR*28 was associated with decreased tacrolimus trough concentrations although the effect was small possibly through enhanced CYP3A4 enzyme activity. CYP3A4*22 and ABCC2 haplotypes did not influence tacrolimus trough concentrations.
Keywords: : ABCC2, calcineurin inhibitor, cytochrome P450 3A4 and 3A5, kidney transplant, pharmacogenomics, pharmacokinetics, POR, tacrolimus
Tacrolimus is the most commonly used calcineurin inhibitor. More than 90% of kidney transplants are performed with tacrolimus as the cornerstone immunosuppressive agent. Tacrolimus has a narrow therapeutic range where transplant recipients with tacrolimus concentrations above the therapeutic range are at greater risk for toxicity and those below the range at greater risk of acute rejection (AR), which is a major risk factor for graft loss. Tacrolimus displays wide interpatient pharmacokinetic variability necessitating therapeutic drug monitoring with dose adjustments to achieve the therapeutic trough range (typically 6–12 ng/ml although this varies with time post-transplant and center). Although therapeutic monitoring of blood concentrations ultimately achieves desired concentrations, many patients spend time out of range in the critical early transplant period increasing their risk for AR. One of the drawbacks to using therapeutic drug monitoring as a method to determine dose is that it cannot be used to select the initial dose. Therefore all patients are started on a dose optimal for the average individual and then modified when drug concentration data are available. However substantial number of patients fall outside the average dose requirements. Defining variables to personalize the first dose of tacrolimus may reduce the number of days spent out of range and reduce the amount of needed therapeutic drug monitoring.
There are many clinical variables that affect tacrolimus pharmacokinetics and blood concentrations. Tacrolimus is metabolized by CYP3A4 and CYP3A5 enzymes to active and inactive metabolites [1]. However, CYP3A5 has twice the intrinsic clearance for tacrolimus 13-demethylation and 12-hydroxylation than CYP3A4 [2]. Therefore, in carriers of the CYP3A5*1 allele (3A5 expressors), 60% of the estimated hepatic oxidative metabolism of tacrolimus is through CYP3A5 [2]. The CYP3A5*3 variant is therefore an important determinant of tacrolimus pharmacokinetic variability. The formation rates of the primary tacrolimus metabolites are significantly higher in human liver microsomes from individuals with the CYP3A5*1/*3 or *1/*1 genotypes [2].
Genotype guided dosing of tacrolimus has been studied in a randomized controlled trial of kidney transplant recipients [3]. CYP3A5 genotype directed dosing was compared with a control group where the dose was based on body weight alone. A greater proportion of patients in the genotype dosed arm achieved blood concentrations in the therapeutic range (10–15 ng/ml after 3 days) compared with the control arm (43.2 vs 29.1%; p = 0.03). The therapeutic target was achieved by 75% of subjects in the genotype dosed arm by day 8 and by 25% in the control group. The genotype guided subjects had fewer dose changes than control group (281 vs 420; p = 0.004). Genotype guided dosing improved care in 43.2% of individuals but had no positive or negative effects in the remaining. Genotype guided dosing may have been improved in this study with the inclusion of additional influential genotypes into dosing algorithm. We previously developed (n = 681) and validated (n = 795) a tacrolimus dosing algorithm in a multicenter kidney transplant consortium study that incorporated clinical factors and CYP3A5*3 genotype [4,5]. The dosing algorithm represented an improvement over standard weight-based dosing protocols, although it did not fully explain all the variability in tacrolimus pharmacokinetics. The dosing algorithm was tested retrospectively in 255 kidney transplant recipients. The algorithm predicted a higher tacrolimus clearance at day 7 post-transplant than the observed clearance [6]. This may be due to differences in calcium channel blocker or steroid use, or the presence of important clinical factors and/or additional genetic variants present in their cohort and not accounted for in our algorithm. Our algorithm was also retrospectively studied in 185 subjects enrolled on the mycophenolate fixed dose versus concentration controlled trial [7]. The algorithm was predictive with a slight, but not significant, overestimation of the trough concentrations. The authors suggested that the algorithm may have overestimated the concentrations because the low activity CYP3A4*22 variant was not accounted for in our algorithm. We hypothesized that missing pharmacokinetic variability may be explained by additional genetic variants. Therefore our objective was to test additional variants toward tacrolimus trough concentrations in our large multicenter population. Variants with a significant association could in the future be incorporated into a refined tacrolimus dosing algorithm.
Patients & methods
Study design & population
Data for this analysis were obtained from subjects enrolled in the Deterioration of Kidney Allograft Function (DeKAF) Genomics study. This is a seven-center prospective, observational study of 2008 recipients undergoing kidney or simultaneous kidney-pancreas transplantation. Subjects were selected for this analysis if they were 18 years and older, received tacrolimus and had tacrolimus trough concentrations available in the first 6 months post-transplant. This study is registered [8]. Subjects were enrolled at time of transplant. Signed informed consent was obtained from each subject. The study protocol and consent form was approved by the Institutional Review Boards each of the enrolling centers.
Participants received tacrolimus and mycophenolate maintenance with standard dose prednisone or a steroid sparing course. Induction therapy was per transplant center preference. Donor and recipient characteristics, race, serum creatinine (SCr) and estimated creatinine clearance (CrCl), concomitant medications at time of each trough measurement were obtained from the medical record. Tacrolimus trough concentrations (n = 35,043) were measured from whole blood and were obtained as part of clinical care. Two measurements, if available, were obtained in each of weeks 1–8 and in each of months 3, 4, 5 and 6 post-transplant, for a maximum of 24 measurements per patient. Tacrolimus doses were adjusted based on trough concentrations to reach institution specific trough goals based on time post-transplant (generally 8–12 ng/ml in months 0 to 3 and 6–10 ng/ml in months 4 to 6). Trough values were normalized for dose (ng/ml per total daily dose in mg) prior to statistical analysis. Whole blood tacrolimus concentrations were measured by each institutions preferred analytical technique. Liquid chromatography-mass spectrometry was used to measure 32,402 (92.5%) of the 35,043 concentrations.
Genotyping
Pretransplant recipient DNA was isolated at time of transplant from peripheral blood lymphocytes. Lymphocytes were isolated by centrifugation after red blood cell lysis and the DNA isolated. DNA was quantified by measuring the absorbance at 260 nm. DNA was genotyped for POR*28 (rs1057868), CYP3A4*22 (rs35599367), CYP3A5*3 (rs776746) and three ABCC2 (rs717620, rs2273697 and rs3740066) variants. The POR*28 and CYP3A4*22 genotypes were determined using Taqman methods using a Prism 7500 (Life Technologies, NY, USA). Data quality was assessed by negative controls and duplicate samples. For POR*28, 19 of the 1458 samples did not pass quality control and were excluded from analysis for a 98.49% success rate. For CYP3A4*22, 42 of 1458 samples did not pass quality control and were excluded from analysis for a 96.91% success rate. Genotyping for CYP3A5*3 and the ABCC2 variants was previously performed on these subjects on an Affymetrix Gene Chip (CA, USA) and the Illumina VeraCode (CA, USA) platforms and was previously described [9,10]. None of the genotypes showed strong evidence of being out of Hardy-Weinberg equilibrium. Allele frequencies in all subjects and by race are shown in Table 1.
Table 1. . Minor allele frequencies in African–Americans and non-African–American subjects.
| Variant | All subjects | Non-AA | AA |
|---|---|---|---|
| POR*28, rs1057868, 1508C > T |
0.262 |
0.265 |
0.254 |
| CYP3A4*22, rs35599367, c522-191 C > T |
0.039 |
0.040 |
0.036 |
| CYP3A5*1, rs776746, 6986 G > A, *1 = A, *3 = G |
0.20 |
0.08 |
0.70 |
| ABCC2, rs717620, -24C > T |
0.16 |
0.19 |
0.06 |
| ABCC2, rs2273697, 1249G > A |
0.20 |
0.21 |
0.15 |
| ABCC2, rs3740066, 3972C > T | 0.34 | 0.35 | 0.26 |
AA: African–American; non-AA: Non-African–American.
Statistical analysis
ABCC2 haplotype formation
The ABCC2 haplotypes were determined based on three variants within the ABCC2 gene locus. Haplotype inference was carried out with the PHASE program for non-African–Americans and African–Americans separately [11]. Haplotypes were then categorized as high, wild-type/average, low or unknown ABCC2 expression groups (Table 2) [12,13].
Table 2. . Predicted haplotypes and diplotypes of ABCC2 variants.
| Haplotype | rs717620, -24C > T | rs2273697, 1249G > A | rs3740066, 3972C > T | No. of haplotypes | Diplotype | No. with diplotype |
|---|---|---|---|---|---|---|
| H1 (wild-type, average expression, reference) |
C |
G |
C |
1876 |
H1/H1 (wild-type, average expression, reference) |
460 |
| H2 (high expression) |
C |
A |
C |
779 |
H1/H2 (high expression) |
348 |
| H9 (low expression) |
C |
G |
T |
685 |
H1/H9 (low expression) |
87 |
| H10 (low expression) |
T |
G |
C |
11 |
H1/H10 (low expression) |
6 |
| H12 (low expression) |
T |
G |
T |
639 |
H1/H12 (low expression) |
293 |
| HX (unknown) |
T |
A |
C |
2 |
H2/H2 (high expression) |
71 |
| HY (unknown) | C | A | T | 24 | All others (unknown expression) | 743 |
Association testing of variants toward trough concentrations
Simple time-trend and multivariable linear mixed effects regression models were used to test for associations between natural log (ln) transformed dose normalized tacrolimus trough concentrations and genotypes. We used the multivariable model that was developed by Jacobson et al. [9] which adjusted for CYP3A5*3 genotype, recipient race (African–American vs non-African–American) and weight, enrolling center, recipient and donor age, gender, donor type (living or deceased), diabetes at transplant, antibody induction and concomitant medications (antiviral, calcium channel blocker and steroid use at time of measurement) as time varying covariates. The correlation structure consisted of random slopes and intercept per individual and a model correlation between trough concentrations within each individual. Visual inspection showed that dose normalized trough concentrations initially started low, rose and then plateaued at day 9 post-transplant. Therefore, a simple spline method was used to model the effect of time on trough concentrations, with the change in slope occurring at day 9. POR*28 and CYP3A4*22 genotypes and the ABBC2 diplotypes were tested separately for association. The association analyses for ABCC2, POR*28 and CYP3A4*22 genotypes were conducted in 2008, 1429 and 1407 subjects, respectively, since not all genotypes and phenotypes were available for each subject. SNPs were modeled with an additive genetic model except for POR*28 which was modeled by a dominant genetic model after visual inspection of the tacrolimus trough concentration versus days post-transplant plots by POR*28 genotypes. For the ABCC2 analysis, high expression diplotypes (H2/H2 and H1/H2) were tested versus all other diplotypes as described by Ogasawara et al. [12] Finally, subset analyses were conducted in CYP3A5*3 genotype groups and by race groups. Analyses were conducted with SAS version 9.2 software (SAS Institute, NC, USA).
Results
Population characteristics
A total of 2008 adult recipients of living or deceased donor kidneys were studied. Demographic and clinical characteristics of the patient population are shown in Table 3. Tacrolimus doses, trough concentrations and concomitant medications at time of trough are shown in Table 4.
Table 3. . Patient demographics (n = 2008).
| Number with characteristic (%) | |
|---|---|
| Recipient age (years): | |
| – 18–34 | 276 (13.8) |
| – 35–64 | 1447 (72.1) |
| – 65–84 |
285 (14.2) |
| Recipient weight (kg) at transplant |
81.8 (69.0–95.5) |
| Male sex |
1261 (62.8) |
| Race (self-reported): | |
| – White/Caucasian | 1533 (76.3) |
| – African–American | 373 (18.6) |
| – Asian | 55 (2.8) |
| – Native American/Aleutian Islander | 31 (1.5) |
| – Multiracial | 10 (0.5) |
| – Hawaiian/Pacific Islander | 4 (0.2) |
| – Not specified |
2 (0.1) |
| Cause of kidney disease: | |
| – Diabetes | 606 (30.2) |
| – Glomerular disease | 441 (22.0) |
| – Other | 359 (17.9) |
| – Hypertension | 275 (13.7) |
| – Polycystic kidney disease | 262 (13.1) |
| – Unknown |
65 (3.2) |
| Diabetes at time of transplant |
780 (38.8) |
| Prior kidney transplant |
295 (14.7) |
| Simultaneous kidney/pancreas |
157 (7.8) |
| Donor age (years): | |
| – 0–34 | 696 (34.7) |
| – 35–64 | 1254 (62.5) |
| – 65–84 |
58 (2.9) |
| Deceased donor |
838 (41.7) |
| Cytomegalovirus antibody status pretransplant: | |
| – Negative recipient/negative donor | 415 (21.3) |
| – Positive recipient/negative or positive donor | 1217 (62.6) |
| – Negative recipient/positive donor |
313 (16.1) |
| Induction immunosuppression: | |
| – Combination | 54 (2.7) |
| – IL-2 receptor antagonists (basiliximab, daclizumab) | 410 (20.4) |
| – Monoclonal antibodies (OKT3, alemtuzumab, rituximab) | 402 (20.0) |
| – None | 72 (3.6) |
| – Polyclonal antibody (antithymocyte globulin) |
1070 (53.3) |
| Dialysis after transplant | 179 (8.9) |
Table 4. . Tacrolimus doses, trough concentrations and concomitant medication use.
| Median (IQR) or number with (%) | |
|---|---|
| Number of trough concentrations |
35,043 |
| Total daily dose (mg) |
6.0 (4–8) |
| Trough concentration (ng/ml) |
8.1 (6.1–10.1) |
| Trough dose-normalized (ng/ml per mg/day) |
1.38 (0.87–2.18) |
| Dosing interval: | |
| – Twice daily | 34,553 (98.6%) |
| – Once daily | 414 (1.2%) |
| – Three-times a day |
75 (0.2%) |
| Trough concentrations with ACE inhibitor |
4592 (13.1%) |
| Trough concentrations with calcium channel blocker |
14,002 (40.0%) |
| Trough concentrations with corticosteroids |
21,379 (61.0%) |
| Trough concentrations with antiviral drug | 19,658 (56.1%) |
IQR: Interquartile range.
Association between variants & tacrolimus trough concentrations
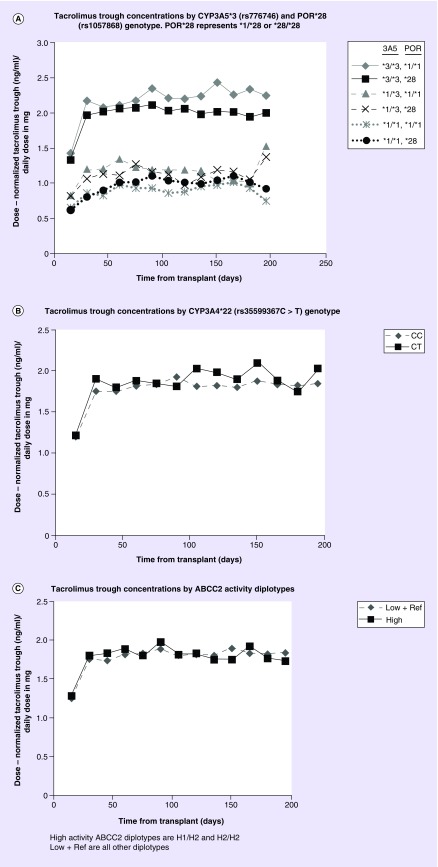
There was no association between POR*28 and ln transformed dose normalized trough concentrations in simple time-trend analysis adjusting only for CYP3A5*1 status (p = 0.0502, data not shown). However, in the multivariable model adjusting for CYP3A5*1 status and clinical factors, one or two POR*28 alleles were associated with a 4.63% (p = 0.037) reduction in trough concentrations (Table 5 & Figure 1A). The CYP3A5*1 genotype had a large and highly significant effect on ln transformed dose normalized trough concentrations (one *1 allele reduced trough concentrations by 34.8% and two *1 alleles were associated with 57.5% reduction p = 9.2 × 10-75). On average, trough concentrations increased in the first 9 days post-transplant and then became mostly unchanged after day 9. Younger recipient age and increasing weight were also associated with lower trough concentrations, whereas diabetes at time of transplant, calcium channel blocker use and antiviral drug use were associated with higher trough concentrations. The median (IQR) tacrolimus trough concentrations over the first 6 months in recipients carrying zero, one or two POR*28 alleles was 8.0 (6.1–10.2), 8.2 (6.2–10.3) and 8.1 (6.0–10.2) ng/ml, respectively. A plot of mean dose normalized trough concentrations over time by POR*28 and CYP3A5*1 genotypes is shown in Figure 1A. In a subset of CYP3A5 nonexpressors (*3/*3; n = 997 subjects) with one or two POR*28 alleles, dose normalized tacrolimus trough concentrations were reduced by 5.6% after adjustment for clinical factors (p = 0.03). In the subset of CYP3A5 expressors (*1/*3 or *1/*1; n = 432), with adjustment for clinical factors the POR*28 alleles were not associated with trough concentrations (p = 0.68). The minor allele frequency of POR*28 was 26.2% in all subjects and was similar between African–American and non-African–Americans (Table 1).
Table 5. . Multivariable models for the association of POR*28, CYP3A4*22 and ABCC2 diplotypes with ln transformed dose-normalized tacrolimus trough concentrations.
| Variables | Effect on tacrolimus trough concentrations (95% CI) | p-value |
|---|---|---|
|
POR*28 (rs1057868) model (n = 1429 subjects) | ||
| POR*28† |
-0.0463 (-0.090– -0.0027) |
0.037 |
| For each day post-transplant‡ |
0.071 (0.064–0.078) |
1.9 × 10-86 |
| Additional effect for each day after day 9 post-transplant‡ |
-0.071 (-0.077– -0.064) |
1.9 × 10-83 |
| CYP3A5*1 |
-0.428 (-0.474– -0.380) |
9.2 × 10-75 |
| Age, recipient (years): | ||
| – 18–34 vs 65–84 | -0.295 (-0.385– -0.206) | 6.6 × 10-10 |
| – 35–64 vs 65–84 |
-0.139 (-0.206– -0.071) |
|
| Age, donor (years): | ||
| – 0–34 vs 65–84 | 0.111 (-0.025–0.247) | 0.25 |
| – 35–64 vs 65–84 |
0.112 (-0.021–0.245) |
|
| African–American |
0.078 (-0.007–0.164) |
0.073 |
| Male recipient |
-0.011 (-0.059–0.037) |
0.66 |
| Diabetes at transplant |
0.097 (0.051–0.144) |
4.0 × 10-5 |
| Deceased donor vs living donor |
0.034 (-0.016–0.085) |
0.19 |
| Steroid use§ |
-0.020 (-0.052–0.011) |
0.21 |
| Calcium channel blocker use§ |
0.050 (0.033–0.066) |
4.1 × 10-9 |
| Antiviral use§ |
0.055 (0.043–0.066) |
1.8 × 10-21 |
| Induction immunosuppression: | ||
| – Combination vs polyclonal | -0.162 (-0.294– -0.030) | 1.8 × 10-5 |
| – Monoclonal vs polyclonal | 0.049 (-0.006–0.105) | |
| – None vs polyclonal |
0.279 (0.148–0.410) |
|
| Recipient weight (kg) |
-0.0023 (-0.003– -0.001) |
1.6 × 10-5 |
|
CYP3A4*22 (rs35599367) model (n = 1407) | ||
| CYP3A4*22 |
0.044 (-0.038–0.125) |
0.29 |
| For each day post-transplant‡ |
0.070 (0.063–0.076) |
8.8 × 10-84 |
| Additional effect for each day after day 9 post-transplant‡ |
-0.069 (-0.076– -0.063) |
7.6 × 10-81 |
| CYP3A5*1 |
-0.43 (-0.480– -0.388) |
7.1 × 10-75 |
| Age, recipient (years): | ||
| – 18–34 vs 65–84 | -0.296 (-0.386– -0.207) | 6.7 × 10-10 |
| – 35–64 vs 65–84 |
-0.142 (-0.210– -0.074) |
|
| Age, donor (years): | ||
| – 0–34 vs 65–84 | 0.114 (-0.022–0.250) | 0.24 |
| – 35–64 vs 65–84 |
0.114 (-0.019–0.247) |
|
| African–American |
0.098 (0.011–0.185) |
0.028 |
| Male recipient |
-0.015 (-0.064–0.034) |
0.55 |
| Diabetes at transplant |
0.096 (0.050–0.143) |
5.5 × 10-5 |
| Deceased donor vs living donor |
0.034 (-0.017–0.085) |
0.2 |
| Steroid use§ |
-0.018 (-0.050–0.013) |
0.26 |
| Calcium channel blocker use§ |
0.049 (0.032–0.066) |
8.0 × 10-9 |
| Antiviral use§ |
0.053 (0.042–0.064) |
5.5 × 10-20 |
| Induction immunosuppression: | ||
| – Combination vs polyclonal | -0.17 (-0.302– -0.038) | 1.6 × 10-5 |
| – Monoclonal vs polyclonal | 0.040 (-0.016–0.096) | |
| – None vs polyclonal |
0.279 (0.148–0.409) |
|
| Recipient weight (kg) |
-0.002 (-0.003– -0.001) |
1.4 × 10-5 |
|
ABCC2 diplotype model (n = 2008) | ||
| High ABCC2 (H2/H2 or H1/H2 vs other) |
0.029 (-0.016–0.074) |
0.20 |
| For each day post-transplant‡ |
0.063 (0.057–0.068) |
6.3 × 10-95 |
| Additional effect for each day after day 9 post-transplant‡ |
-0.062 (-0.068– -0.057) |
6.6 × 10-92 |
| CYP3A5*1 |
-0.45 (-0.490– -0.407) |
1.3 × 10-106 |
| Age, recipient (years): | ||
| – 18–34 vs 65–84 | -0.289 (-0.362– -0.216) | 9.1 × 10-14 |
| – 35–64 vs 65–84 |
-0.129 (-0.184– -0.073) |
|
| Age, donor (years): | ||
| – 0–34 vs 65–84 | 0.146 (0.032–0.260) | 0.039 |
| – 35–64 vs 65–84 |
0.125 (0.014–0.236) |
|
| African–American |
0.0738 (-0.0003–0.148) |
0.051 |
| Male recipient |
-0.009 (-0.049–0.031) |
0.66 |
| Diabetes at transplant |
0.089 (0.050–0.128) |
7.2 × 10-6 |
| Deceased donor vs living donor |
0.035 (-0.007–0.078) |
0.10 |
| Steroid use§ |
-0.026 (-0.053–0.002) |
0.067 |
| Calcium channel blocker use§ |
0.047 (0.033–0.062) |
1.4 × 10-10 |
| Antiviral use§ |
0.050 (0.041–0.060) |
6.0 × 10-25 |
| Induction immunosuppression: | ||
| – Combination vs polyclonal | -0.153 (-0.270– -0.036) | 2.3 × 10-6 |
| – Monoclonal vs polyclonal | 0.028 (-0.019–0.074) | |
| – None vs polyclonal |
0.270 (0.155–0.385) |
|
| Recipient weight (kg) | -0.002 (-0.003– -0.002) | 3.3 × 10-8 |
Data are adjusted for enrolling center.
†Carrying one or two POR*28 alleles was associated with a 4.63% reduction in ln dose normalized tacrolimus trough concentrations.
‡For each day post-transplant (day 1 to 180) there is a daily increase in ln transformed dose-normalized tacrolimus troughs. There is an additional effect for each day after day 9 (day 10–180) where troughs are reduced.
§Concomitant drug use at the time trough was measured.
Figure 1. . Mean tacrolimus trough concentrations by genotypes in 15 day intervals.
(A) Tacrolimus trough concentrations by CYP3A5*3 (rs776746) and POR*28 (rs1057868) genotype. POR*28 represents *1/*28 or *28/*28. (B) Tacrolimus trough concentrations by CYP3A4*22 (rs35599367 C > T genotype). (C) Tacrolimus trough concentrations by ABCC2 activity diplotypes. High activity ABCC2 diplotypes are H1/H2 and H2/H2. Low + ref are all other diplotypes.
No associations were observed between CYP3A4*22 genotype or ABCC2 diplotypes and tacrolimus trough concentrations in simple time-trend analyses after adjustment for CYP3A5*1 status or after multivariable analysis adjusting for CYP3A5*1 status and clinical factors (Table 5 & Figure 1B & C). The CYP3A4*22 variant was infrequent with a minor allele frequency of 3.9% and was similar between African–American and non-African–Americans (Table 1). ABCC2 haplotype and diplotype frequencies results are shown in Table 1. The estimated haplotype frequencies in our study population are similar to those described by Ogasawara et al. [12]
Discussion
This is the largest pharmacogenomic study of tacrolimus pharmacokinetics in kidney transplantation published and includes 2008 patients from seven centers with 35,043 tacrolimus trough concentrations. Because of the large sample size the effect of genetic variants which are infrequent or have small effect sizes can be characterized with greater certainty. We previously described a tacrolimus dosing algorithm incorporating clinical factors and CYP3A5*3 genotype status which showed an improvement in predicting trough concentrations over weight based dosing [4,5]. However, there remained individuals for which the algorithm was of modest benefit. We hypothesized that there are individuals with genetic variation in transporters or drug metabolizing enzymes which contribute additive or opposing effects of the CYP3A5*3 variant. Other candidate variants have been tested; however, there is conflicting data as to their association with tacrolimus. Therefore we tested these variants in our large multicenter cohort. We found that POR*28 reduced tacrolimus trough concentrations, although the effect was small, accounting for a 4.63% reduction in trough values. We did not find a significant associations between the CYP3A4*22 variant or the ABCC2 diplotypes with tacrolimus trough concentrations.
P450 oxidoreductase (POR) is a membrane-bound co-enzyme that is essential to the oxidative activation of cytochrome P450 enzymes. POR supplies microsomal P450 enzymes with electrons from reduced nicotinamide adenine dinucleotide phosphate (NADPH) for catalytic functions critical to the oxidative metabolism of drugs, and biosynthesis of steroids, fatty acids and bile salts and variants result in complex human disorders [14–17]. There are significant associations between several nonsynonymous coding region mutations in the POR gene and altered cytochrome P450 activity particularly for the CYP3A4/5, 2E1, 2C9 and 2C8 enzymes [18]. Although POR works primarily through activation of cytochrome P450, it may directly induce transformation of some anticancer substrates [19]. The POR gene is highly polymorphic with over 45 variants and may increase or decrease the metabolism of drugs [20]. In vitro, catalytic activities of different P450 enzymes with POR genetic variants appear to be enzyme and substrate specific [21]. The POR*28 is a common variant on chromosome 7 (rs1057868, c.1508 C > T, p.A503V) which when reconstituted in vitro with cytochrome P450 enzymes along with phospholipids results in either increased or decreased substrate oxidation activity [22–25]. The POR*28 variant was associated with an increased risk of new onset diabetes after transplantation possibly due to the effects of altered cytochrome P450 activity on glucocorticoid and/or steroids [26]. Homozygous carriers of POR*28 display a 1.6-fold increase in midazolam metabolic rate – a marker for CYP3A4/5 activity [27]. The initial analysis of POR*28 in kidney transplant recipients reported that it was associated with lower tacrolimus dose-normalized trough concentrations in patients who expressed CYP3A5*1 [28]. The effect was unstable and was observed only on days 1, 2 and 3 but at no other times out to one year post-transplant. The authors hypothesized that POR*28 affected tacrolimus metabolism primarily through an increase in CYP3A5 enzyme activity which would explain the lack of effect in the CYP3A5 nonexpressors. This hypothesis was supported by a subsequent association study in healthy Chinese volunteers [29]. However, both of these studies had limited numbers of CYP3A5 expressors. Other groups also reported that POR*28 lowers dose normalized tacrolimus trough concentrations, though subset analysis by CYP3A5 status was either not performed or not significant [30–32]. Recently POR*28 was associated with reduced tacrolimus trough concentrations but only in CYP3A5 nonexpressors which is consistent with our findings [33]. In our population, we found POR*28 to be significant but only after adjusting for CYP3A5 status and clinical factors. In subset analyses, the effect was present only in CYP3A5 nonexpressors who carried one or two POR*28 alleles (Figure 1A). Therefore, we speculate that the POR*28 variant may effect tacrolimus metabolism possibly through enhanced CYP3A4 enzyme activity.
CYP3A4*22 (rs35599367, c522-191 C > T) is an infrequent single nucleotide polymorphism located in intron 6 of the CYP3A4 gene on chromosome 7. Carriers of the T allele have decreased CYP3A4 mRNA hepatic expression and reduced CYP3A4 enzymatic activity, and require lower statin doses for optimal lipid control [34]. In a small bank of Caucasians liver microsomes, microsomal samples that were CYP3A4*1/*22, CYP3A5*3/*3 (n = 4) showed significantly lower midazolam 1’-hydroxylation and testosterone 6-beta-hydroxylation activity and lower CYP3A4 protein content [35]. Presence of one CYP3A4*22 allele was found to be a risk factor for delayed graft function and lower creatinine clearance in cyclosporine treated patients after kidney transplant [36]. This polymorphism has been most extensively evaluated toward tacrolimus pharmacokinetics in adults by the Rotterdam group in The Netherlands where they have reported that CYP3A4*22 is associated with higher tacrolimus concentrations [7,37–40]. In 60 pediatric heart recipients, CYP3A4*22 was also associated with reduced tacrolimus dose requirements compared with noncarriers although the effect was only found at day 3 post-transplant [41]. A subsequent report in a Brazilian cohort did not demonstrate an effect of CYP3A4*22 on tacrolimus metabolism [42]. This difference may be due to genetic differences between the primarily Caucasian cohorts from The Netherlands compared with the Brazilian cohort which has strong African ancestry. The Brazilian cohort likely has other important low activity or nonfunctional CYP3A5 variants not present in Caucasians which may confound the analysis [43]. Recently, another group in The Netherlands reported CYP3A4*22 in 101 kidney transplant recipients receiving tacrolimus and found a trend toward reduced tacrolimus clearance but considered the effect too small to be clinically important [44]. However, they found a significant association with cyclosporine clearance. We did not identify an association between CYP3A4*22 and tacrolimus trough concentrations. Our study includes about 19% African–American and 5% non-European non-African–American subjects which may result in population stratification differences relative to other studies. Additionally, differences in standard post-transplant drug protocols such as calcium channel blockers, steroids and anti-infectives resulting in drug–drug interactions that may obscure the genetic effect.
Tacrolimus is a substrate for P-glycoprotein transporter which is encoded by the ABCB1 gene. P-glycoprotein has been studied extensively for its association with tacrolimus pharmacokinetics since it affects absorption from the gut, distribution in the body compartments and excretion. The P-glycoprotein associations are controversial and data are conflicting [45,46]. Data suggest that other transporters may be important to the disposition of tacrolimus [47,48]. Therefore, the multidrug resistance-associated protein 2 (MRP2) encoded by the ABCC2 gene and its variants have also been evaluated for their association with calcineurin inhibitor and mycophenolic acid pharmacokinetics [12,13,49]. Previous data showed a haplotype-dependent influence on protein expression and transport capacity of ABCC2 variants. Three well-studied variants of ABCC2 (rs717620, -24 C > T; rs2273697, 1249 G > A and rs3740066, 3972 C > T) create haplotypes conveying high, low and reference expression and transport activity. Four ABCC2 haplotypes were studied in 102 kidney transplant recipients [12]. Individuals with at least one high activity haplotype (H2/H2 or H1/H2) who were also CYP3A5 expressors had significantly lower tacrolimus trough concentrations. We were unable to confirm this finding in our population. This may be due to differences in the size of the populations (we had 2008 subjects compared with their 102) and the number who were CYP3A5 expressors. Because of our larger population we identified seven ABCC2 haplotypes relative to their four and we controlled for a large number of clinical factors all of which may contribute to the differences in findings. Interestingly, cyclosporine is an inhibitor of MRP2 which may obscure or reduce the activity of the high expression/activity ABCC2 haplotypes [50]. Therefore, observations between cyclosporine trough concentrations and ABCC2 variants may be different than what we have observed here with tacrolimus.
Conclusion
We could not identify the previously observed associations between CYP3A4*22 and ABCC2 haplotypes but did confirm that POR*28 is associated with lower tacrolimus concentrations possibly through enhanced cytochrome CYP3A4 activity. The large size of our cohort allows us to evaluate genetic associations for the infrequent variants such as CYP3A4*22 with greater confidence. Although we cannot rule out that differences in post-transplant drug protocols and/or population specific variants not present in our population may account for positive association in other studies [51]. These data demonstrate that tacrolimus disposition is influenced by variation beyond CYP3A5*3 namely POR*28; however, the effect is small and alone this effect does not justify genotyping but may in combined panels. In the future developing models that incorporate multiple variants including those with small effects along with clinical factors may accurately explain tacrolimus trough concentrations. Large samples sizes though are needed to accurately identify these small effects. Future efforts should be placed on conducting large, multicenter studies such that variants with small effects or infrequent variants can be identified. There are also other clinical factors that are likely important including hematocrit and antifungal therapies that we were unable to evaluate and future trials must include these effects too. Identifying all factors even those with small effect sizes and infrequent are critical if we are to develop robust pharmacogenomic tools for individualizing therapy.
Future perspective
Clinical application of pharmacogenomic information has the potential to enhance patient outcomes by improving efficacy, reducing toxicity or both. Specifically the application of genotype information in transplantation may reduce the risk of donor allograft rejection and/or decrease the frequency of the many adverse effects associated with immune suppressant drugs. Initial immunosuppressant doses are given as a one size fits all however knowing an individual's capacity for metabolism through pretransplant genotyping could lead to personalized dosing. This may reduce the amount of time an individual is out of the therapeutic range, reduce the number of dose changes and reduce the frequency of therapeutic drug monitoring.
Executive summary.
CYP3A5*3 genotype is associated with lower tacrolimus clearance.
Individuals may have other variants that influence tacrolimus metabolism.
Accounting for these additional variants may improve the precision of genotype guided dosing.
We evaluated the effect of CYP3A4*22, POR*28 and ABCC2 haplotypes on tacrolimus trough concentrations while controlling for CYP3A5*3 in a large ethnically diverse cohort of kidney transplant recipients.
POR*28 decreased tacrolimus trough concentrations by approximately 5% but only in CYP3A5 nonexpressors (CYP3A5*3/*3). POR*28 is common with a minor allele frequency of 26.3%.
CYP3A4*22 and ABCC2 haplotype did not influence trough concentrations.
Acknowledgements
The authors thank the dedication and hard work of their coordinators: University of Alberta: N Bobocea, T Wong, A Geambasu and A Sader; University of Manitoba: M Ross and K Peters; University of Minnesota: M DeGrote and R Brown; Hennepin County Medical Center: L Berndt; Mayo Clinic: T DeLeeuw; University of Iowa: W Wallace and T Lowe; University of Alabama: T Hailey. The authors also thank the research scientists for dedicated work: M Brott and A Muthusamy.
Footnotes
DeKAF investigators
Arthur Matas, MD, Department of Surgery, University of Minnesota, Minneapolis, MN, USA, E-mail: matas001@umn.edu; J Michael Cecka, MD, UCLA Immunogenetics Center, Los Angeles, CA, USA, E-mail: mcecka@ucla.edu; John Connett, PhD, Division of Biostatistics, University of Minnesota, Minneapolis, MN, USA, E-mail: john-c@biostat.umn.edu; Fernando G Cosio, MD, Division of Nephrology, Mayo Clinic, Rochester, MN, USA, E-mail: Cosio.Fernando@mayo.edu; Robert Gaston, MD, Division of Nephrology, University of Alabama, Division of Nephrology, Birmingham, AL, USA, E-mail: rgaston@uab.edu; Rosalyn Mannon, MD, Division of Nephrology, University of Alabama, Division of Nephrology, Birmingham, AL, USA, E-mail: rmannon@uab.edu; Sita Gourishankar, MD, Division of Nephrology and Immunology, University of Alberta, Edmonton, Alberta, Canada, E-mail: sitag@ualberta.ca; Joseph P Grande, MD, PhD, Mayo Clinic College of Medicine, Rochester, MN, USA, E-mail: Grande.Joseph@mayo.edu; Lawrence Hunsicker, MD, Nephrology Division, Iowa City, IA, USA, E-mail: lawrencehunsicker@uiowa.edu; Bertram Kasiske, MD, Division of Nephrology, Hennepin County Medical Center, Minneapolis, MN, USA, E-mail: kasis001@umn.edu; and David Rush, MD, Health Sciences Center, Winnipeg MB, Canada, E-mail: drush@exchange.hsc.mb.ca.
Participating centers
Participating transplant centers were University of Alberta, Edmonton, Canada; University of Manitoba, Winnipeg, Canada; University of Minnesota, Minneapolis, MN, USA; Hennepin County Medical Center, Minneapolis, MN, USA; Mayo Clinic, Rochester, MN, USA; University of Iowa, Iowa City, IA, USA and University of Alabama, Birmingham, AL, USA.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Disease or the NIH.
Financial & competing interests disclosure
This project was supported by grant number (5U19-AI070119 and 5U01-AI058013) from the National Institute of Allergy and Infectious Disease. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
- 1.Iwasaki K. Metabolism of tacrolimus (FK506) and recent topics in clinical pharmacokinetics. Drug Metab. Pharmacokinet. 2007;22(5):328–335. doi: 10.2133/dmpk.22.328. [DOI] [PubMed] [Google Scholar]
- 2.Dai Y, Hebert MF, Isoherranen N, et al. Effect of CYP3A5 polymorphism on tacrolimus metabolic clearance in vitro . Drug Metab. Dispos. 2006;34(5):836–847. doi: 10.1124/dmd.105.008680. [DOI] [PubMed] [Google Scholar]
- 3.Thervet E, Loriot MA, Barbier S, et al. Optimization of initial tacrolimus dose using pharmacogenetic testing. Clin. Pharmacol. Ther. 2010;87(6):721–726. doi: 10.1038/clpt.2010.17. [DOI] [PubMed] [Google Scholar]
- 4.Passey C, Birnbaum AK, Brundage RC, Oetting WS, Israni AK, Jacobson PA. Dosing equation for tacrolimus using genetic variants and clinical factors. Br. J. Clin. Pharmacol. 2011;72(6):948–957. doi: 10.1111/j.1365-2125.2011.04039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Passey C, Birnbaum AK, Brundage RC, et al. Validation of tacrolimus equation to predict troughs using genetic and clinical factors. Pharmacogenomics. 2012;13(10):1141–1147. doi: 10.2217/pgs.12.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boughton O, Borgulya G, Cecconi M, Fredericks S, Moreton-Clack M, MacPhee IA. A published pharmacogenetic algorithm was poorly predictive of tacrolimus clearance in an independent cohort of renal transplant recipients. Br. J. Clin. Pharmacol. 2013;76(3):425–431. doi: 10.1111/bcp.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elens L, Hesselink DA, van Schaik RH, van Gelder T. The CYP3A4*22 allele affects the predictive value of a pharmacogenetic algorithm predicting tacrolimus predose concentrations. Br. J. Clin. Pharmacol. 2013;75(6):1545–1547. doi: 10.1111/bcp.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.www.clinicaltrials.gov Clinicaltrials.gov: NCT01714440 (2015)
- 9.Jacobson PA, Oetting WS, Brearley AM, et al. Novel polymorphisms associated with tacrolimus trough concentrations: results from a multicenter kidney transplant consortium. Transplantation. 2011;91(3):300–308. doi: 10.1097/TP.0b013e318200e991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobson PA, Schladt D, Oetting WS, et al. Lower calcineurin inhibitor doses in older compared with younger kidney transplant recipients yield similar troughs. Am. J. Transplant. 2012;12(12):3326–3336. doi: 10.1111/j.1600-6143.2012.04232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68(4):978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogasawara K, Chitnis SD, Gohh RY, Christians U, Akhlaghi F. Multidrug resistance-associated protein 2 (MRP2/ABCC2) haplotypes significantly affect the pharmacokinetics of tacrolimus in kidney transplant recipients. Clin. Pharmacokinet. 2013;52(9):751–762. doi: 10.1007/s40262-013-0069-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laechelt S, Turrini E, Ruehmkorf A, Siegmund W, Cascorbi I, Haenisch S. Impact of ABCC2 haplotypes on transcriptional and posttranscriptional gene regulation and function. Pharmacogenomics J. 2011;11(1):25–34. doi: 10.1038/tpj.2010.20. [DOI] [PubMed] [Google Scholar]
- 14.Shen AL, O'Leary KA, Kasper CB. Association of multiple developmental defects and embryonic lethality with loss of microsomal NADPH-cytochrome P450 oxidoreductase. J. Biol. Chem. 2002;277(8):6536–6541. doi: 10.1074/jbc.M111408200. [DOI] [PubMed] [Google Scholar]
- 15.Gu J, Weng Y, Zhang QY, et al. Liver-specific deletion of the NADPH-cytochrome P450 reductase gene: impact on plasma cholesterol homeostasis and the function and regulation of microsomal cytochrome P450 and heme oxygenase. J. Biol. Chem. 2003;278(28):25895–25901. doi: 10.1074/jbc.M303125200. [DOI] [PubMed] [Google Scholar]
- 16.Henderson CJ, Otto DM, Carrie D, et al. Inactivation of the hepatic cytochrome P450 system by conditional deletion of hepatic cytochrome P450 reductase. J. Biol. Chem. 2003;278(15):13480–13486. doi: 10.1074/jbc.M212087200. [DOI] [PubMed] [Google Scholar]
- 17.Scott RR, Miller WL. Genetic and clinical features of p450 oxidoreductase deficiency. Horm. Res. 2008;69(5):266–275. doi: 10.1159/000114857. [DOI] [PubMed] [Google Scholar]
- 18.Hart SN, Wang S, Nakamoto K, Wesselman C, Li Y, Zhong XB. Genetic polymorphisms in cytochrome P450 oxidoreductase influence microsomal P450-catalyzed drug metabolism. Pharmacogenet. Genomics. 2008;18(1):11–24. doi: 10.1097/FPC.0b013e3282f2f121. [DOI] [PubMed] [Google Scholar]
- 19.Hu L, Zhuo W, He YJ, Zhou HH, Fan L. Pharmacogenetics of P450 oxidoreductase: implications in drug metabolism and therapy. Pharmacogenet. Genomics. 2012;22(11):812–819. doi: 10.1097/FPC.0b013e328358d92b. [DOI] [PubMed] [Google Scholar]
- 20.www.cypalleles.ki.se/por.htm P450 oxidoreductase (POR) allele nomenclature (2015)
- 21.Huang N, Agrawal V, Giacomini KM, Miller WL. Genetics of P450 oxidoreductase: sequence variation in 842 individuals of four ethnicities and activities of 15 missense mutations. Proc. Natl Acad. Sci. USA. 2008;105(5):1733–1738. doi: 10.1073/pnas.0711621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandee D, Morrissey K, Agrawal V, et al. Effects of genetic variants of human P450 oxidoreductase on catalysis by CYP2D6 in vitro . Pharmacogenet. Genomics. 2010;20(11):677–686. doi: 10.1097/FPC.0b013e32833f4f9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian M, Agrawal V, Sandee D, Tam HK, Miller WL, Tracy TS. Effect of P450 oxidoreductase variants on the metabolism of model substrates mediated by CYP2C9.1, CYP2C9.2, and CYP2C9.3. Pharmacogenet. Genomics. 2012;22(8):590–597. doi: 10.1097/FPC.0b013e3283544062. [DOI] [PubMed] [Google Scholar]
- 24.Agrawal V, Choi JH, Giacomini KM, Miller WL. Substrate-specific modulation of CYP3A4 activity by genetic variants of cytochrome P450 oxidoreductase. Pharmacogenet. Genomics. 2010;20(10):611–618. doi: 10.1097/FPC.0b013e32833e0cb5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosal A, Hapangama N, Yuan Y, et al. Rapid determination of enzyme activities of recombinant human cytochromes P450, human liver microsomes and hepatocytes. Biopharm. Drug Dispos. 2003;24(9):375–384. doi: 10.1002/bdd.374. [DOI] [PubMed] [Google Scholar]
- 26.Elens L, Sombogaard F, Hesselink DA, van Schaik RH, van Gelder T. Single-nucleotide polymorphisms in P450 oxidoreductase and peroxisome proliferator-activated receptor-alpha are associated with the development of new-onset diabetes after transplantation in kidney transplant recipients treated with tacrolimus. Pharmacogenet. Genomics. 2013;23(12):649–657. doi: 10.1097/FPC.0000000000000001. [DOI] [PubMed] [Google Scholar]
- 27.Oneda B, Crettol S, Jaquenoud Sirot E, Bochud M, Ansermot N, Eap CB. The P450 oxidoreductase genotype is associated with CYP3A activity in vivo as measured by the midazolam phenotyping test. Pharmacogenet. Genomics. 2009;19(11):877–883. doi: 10.1097/FPC.0b013e32833225e7. [DOI] [PubMed] [Google Scholar]
- 28.de Jonge H, Metalidis C, Naesens M, Lambrechts D, Kuypers DR. The P450 oxidoreductase *28 SNP is associated with low initial tacrolimus exposure and increased dose requirements in CYP3A5-expressing renal recipients. Pharmacogenomics. 2011;12(9):1281–1291. doi: 10.2217/pgs.11.77. [DOI] [PubMed] [Google Scholar]
- 29.Zhang JJ, Zhang H, Ding XL, Ma S, Miao LY. Effect of the P450 oxidoreductase 28 polymorphism on the pharmacokinetics of tacrolimus in Chinese healthy male volunteers. Eur. J. Clin. Pharmacol. 2013;69(4):807–812. doi: 10.1007/s00228-012-1432-1. [DOI] [PubMed] [Google Scholar]
- 30.Elens L, Hesselink DA, Bouamar R, et al. Impact of POR*28 on the pharmacokinetics of tacrolimus and cyclosporine A in renal transplant patients. Ther. Drug Monit. 2014;36(1):71–79. doi: 10.1097/FTD.0b013e31829da6dd. [DOI] [PubMed] [Google Scholar]
- 31.Lesche D, Sigurdardottir V, Setoud R, et al. CYP3A5*3 and POR*28 genetic variants influence the required dose of tacrolimus in heart transplant recipients. Ther. Drug Monit. 2014;36(6):710–715. doi: 10.1097/FTD.0000000000000080. [DOI] [PubMed] [Google Scholar]
- 32.Kuypers DR, de Loor H, Naesens M, Coopmans T, de Jonge H. Combined effects of CYP3A5*1, POR*28, and CYP3A4*22 single nucleotide polymorphisms on early concentration-controlled tacrolimus exposure in de-novo renal recipients. Pharmacogenet. Genomics. 2014;24(12):597–606. doi: 10.1097/FPC.0000000000000095. [DOI] [PubMed] [Google Scholar]
- 33.Lunde I, Bremer S, Midtvedt K, et al. The influence of CYP3A, PPARA, and POR genetic variants on the pharmacokinetics of tacrolimus and cyclosporine in renal transplant recipients. Eur. J. Clin. Pharmacol. 2014;70(6):685–693. doi: 10.1007/s00228-014-1656-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J. 2011;11(4):274–286. doi: 10.1038/tpj.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okubo M, Murayama N, Shimizu M, Shimada T, Guengerich FP, Yamazaki H. CYP3A4 intron 6 C>T polymorphism (CYP3A4*22) is associated with reduced CYP3A4 protein level and function in human liver microsomes. J. Toxicol. Sci. 2013;38(3):349–354. doi: 10.2131/jts.38.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elens L, Bouamar R, Hesselink DA, Haufroid V, van Gelder T, van Schaik RH. The new CYP3A4 intron 6 C>T polymorphism (CYP3A4*22) is associated with an increased risk of delayed graft function and worse renal function in cyclosporine-treated kidney transplant patients. Pharmacogenet. Genomics. 2012;22(5):373–380. doi: 10.1097/FPC.0b013e328351f3c1. [DOI] [PubMed] [Google Scholar]
- 37.Elens L, Bouamar R, Hesselink DA, et al. A new functional CYP3A4 intron 6 polymorphism significantly affects tacrolimus pharmacokinetics in kidney transplant recipients. Clin. Chem. 2011;57(11):1574–1583. doi: 10.1373/clinchem.2011.165613. [DOI] [PubMed] [Google Scholar]
- 38.Elens L, van Schaik RH, Panin N, et al. Effect of a new functional CYP3A4 polymorphism on calcineurin inhibitors’ dose requirements and trough blood levels in stable renal transplant patients. Pharmacogenomics. 2011;12(10):1383–1396. doi: 10.2217/pgs.11.90. [DOI] [PubMed] [Google Scholar]
- 39.Elens L, van Gelder T, Hesselink DA, Haufroid V, van Schaik RH. CYP3A4*22: promising newly identified CYP3A4 variant allele for personalizing pharmacotherapy. Pharmacogenomics. 2013;14(1):47–62. doi: 10.2217/pgs.12.187. [DOI] [PubMed] [Google Scholar]
- 40.Elens L, Capron A, van Schaik RH, et al. Impact of CYP3A4*22 allele on tacrolimus pharmacokinetics in early period after renal transplantation: toward updated genotype-based dosage guidelines. Ther. Drug Monit. 2013;35(5):608–616. doi: 10.1097/FTD.0b013e318296045b. [DOI] [PubMed] [Google Scholar]
- 41.Gijsen VM, van Schaik RH, Elens L, et al. CYP3A4*22 and CYP3A combined genotypes both correlate with tacrolimus disposition in pediatric heart transplant recipients. Pharmacogenomics. 2013;14(9):1027–1036. doi: 10.2217/pgs.13.80. [DOI] [PubMed] [Google Scholar]
- 42.Santoro AB, Struchiner CJ, Felipe CR, Tedesco-Silva H, Medina-Pestana JO, Suarez-Kurtz G. CYP3A5 genotype, but not CYP3A4*1b, CYP3A4*22, or hematocrit, predicts tacrolimus dose requirements in Brazilian renal transplant patients. Clin. Pharmacol. Ther. 2013;94(2):201–202. doi: 10.1038/clpt.2013.68. [DOI] [PubMed] [Google Scholar]
- 43.Suarez-Kurtz G, Vargens DD, Santoro AB, et al. Global pharmacogenomics: distribution of CYP3A5 polymorphisms and phenotypes in the Brazilian population. PLoS ONE. 2014;9(1):e83472. doi: 10.1371/journal.pone.0083472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moes DJ, Swen JJ, den Hartigh J, et al. Effect of CYP3A4*22, CYP3A5*3, and CYP3A Combined Genotypes on Cyclosporine, Everolimus, and Tacrolimus Pharmacokinetics in Renal Transplantation. CPT Pharmacometrics Syst. Pharmacol. 2014;3:e100. doi: 10.1038/psp.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Staatz CE, Goodman LK, Tett SE. Effect of CYP3A and ABCB1 single nucleotide polymorphisms on the pharmacokinetics and pharmacodynamics of calcineurin inhibitors: part I. Clin. Pharmacokinet. 2010;49(3):141–175. doi: 10.2165/11317350-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 46.Staatz CE, Goodman LK, Tett SE. Effect of CYP3A and ABCB1 single nucleotide polymorphisms on the pharmacokinetics and pharmacodynamics of calcineurin inhibitors: part II. Clin. Pharmacokinet. 2010;49(4):207–221. doi: 10.2165/11317550-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 47.Takano M, Yumoto R, Murakami T. Expression and function of efflux drug transporters in the intestine. Pharmacol. Ther. 2006;109(1–2):137–161. doi: 10.1016/j.pharmthera.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 48.Lampen A, Christians U, Gonschior AK, et al. Metabolism of the macrolide immunosuppressant, tacrolimus, by the pig gut mucosa in the Ussing chamber. Br. J. Pharmacol. 1996;117(8):1730–1734. doi: 10.1111/j.1476-5381.1996.tb15346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naesens M, Kuypers DR, Verbeke K, Vanrenterghem Y. Multidrug resistance protein 2 genetic polymorphisms influence mycophenolic acid exposure in renal allograft recipients. Transplantation. 2006;82(8):1074–1084. doi: 10.1097/01.tp.0000235533.29300.e7. [DOI] [PubMed] [Google Scholar]
- 50.Hesselink DA, van Hest RM, Mathot RA, et al. Cyclosporine interacts with mycophenolic acid by inhibiting the multidrug resistance-associated protein 2. Am. J. Transplant. 2005;5(5):987–994. doi: 10.1046/j.1600-6143.2005.00779.x. [DOI] [PubMed] [Google Scholar]
- 51.Gomes AM, Winter S, Klein K, et al. Pharmacogenomics of human liver cytochrome P450 oxidoreductase: multifactorial analysis and impact on microsomal drug oxidation. Pharmacogenomics. 2009;10(4):579–599. doi: 10.2217/pgs.09.7. [DOI] [PubMed] [Google Scholar]