Abstract
Studying of the effects of low doses of γ-irradiation is a crucial issue in different areas of interest, from environmental safety and industrial monitoring to aerospace and medicine. The goal of this work is to identify changes of lifespan and expression stress-sensitive genes in Drosophila melanogaster, exposed to low doses of γ-irradiation (5 – 40 cGy) on the imaginal stage of development. Although some changes in life extensity in males were identified (the effect of hormesis after the exposure to 5, 10 and 40 cGy) as well as in females (the effect of hormesis after the exposure to 5 and 40 cGy), they were not caused by the organism “physiological” changes. This means that the observed changes in life expectancy are not related to the changes of organism physiological functions after the exposure to low doses of ionizing radiation. The identified changes in gene expression are not dose-dependent, there is not any proportionality between dose and its impact on expression. These results reflect nonlinear effects of low dose radiation and sex-specific radio-resistance of the postmitotic cell state of Drosophila melanogaster imago.
Introduction
Throughout the history of living things, the natural background radiation of the Earth and cosmic rays have been one of the key environmental factors that have affected the rate of evolutionary processes [1, 2]. As a result of nuclear weapons testing, nuclear accidents and the activities of the nuclear fuel cycle, large areas were contaminated with artificial radionuclides [3–5]. Furthermore, additional sources of irradiation are present in medical procedures, air travel and certain manufacturing [6–9]. Thus, the problem of biological effects of low doses of ionizing radiation is becoming increasingly important.
Although there are many common mechanisms of response of organism and cell to irradiation and other stresses (thermal, oxidative etc.) [10], their principal difference is a significant role of DNA damage on the biological effects of ionizing radiation [11, 12]. However, these differences are attributed mostly to high dose rates. In the case of low dose radiation, direct effects of irradiation such as clustered DNA damage and DNA double strand breaks are minimal, whereas indirect DNA damages caused by the induction of reactive oxygen species become the primary result [11, 13]. In high doses, adverse effects accumulate in the tissues in a deterministic manner that depends linearly on the dose, but in low doses the effects are stochastic, non-linear on the dose, and depend mainly on the efficiency of the stress response’s protective mechanisms [14]. Therefore, low doses of radiation can be regarded as moderate stress, which is known to induce hormesis [15]. Indeed, in our previous work [14, 16], and in the work of other authors [17] it has been revealed, that relatively low dose exposure (20–75 cGy) of fruit flies on immature preimaginal stages in some cases has long-term effects that lead to an increased life span and resistance to other stresses, such as hyperthermia [18, 19]. It is known that preimaginal stages of Drosophila have comparable radiosensitivity to mammals [20]. At the same time, adult individuals, due to the postmitotic state of most tissues, are about 100 times more radioresistant [21]. In their recent work, Antosh et al. revealed that irradiation of Drosophila individuals in the imago stage in doses from 0.1 to 400 Gy causes a statistically significant effect on lifespan and gene expression only if the dose is higher than 100 Gy [22]. At the same time, in our recent work on comparing the effects of irradiation in the adult Drosophila male and female at the 20 cGy dose rate, we observed some differentially expressed genes [23].
Therefore, the goal of this work was to identify changes of lifespan and expression of several previously identified low dose radiation-induced genes in Drosophila melanogaster, exposed to low doses of γ-irradiation (5–40 cGy) at the imaginal stage of development.
Materials and Methods
Experimental design
In our experiments, we used laboratory wild-type (Canton-S) males and females. The line was obtained from the collection at the Bloomington Drosophila Stock Center at Indiana University (Bloomington, USA).
The control- and experimental flies were maintained at T 25±0.5°C and a 12 hour light regime on a sugar-yeast medium containing 7 g of agar, 30 g of sugar, 8 g of dry yeast, 30 g of semolina, 4 ml of propionic acid, and 1 liter of water. Males and females were kept separately at densities of 30 flies of the same sex and age per 120 mL vials.
For analyzes of the expression profiles, the flies in the imago stage of development were used for each control- and experimental variant. For each variant, 3 biological replicates were pooled. Experimental flies were exposed to gamma-irradiation from 226Ra source with the dose rate of 36 mGy/h. The source had metal casing (aluminum filter) impervious to alpha particles, so the spectrum of ionizing radiation had been exposed to gamma irradiation. The exposure time was 1 h 23 min, 2 h 47 min, 5 h 34 min and 11 h 8 min, and the absorbed dose was 5, 10, 20 and 40 cGy, respectively. The control flies were maintained in the same conditions excluding irradiation factor. The flies in the control- and experimental groups were fixed by liquid nitrogen after a specific time following irradiation: immediately after the radiation impact, after 6, 24, 48 and 72 hours and stored in a freezer at -86°C.
The lifespan replicates and the gene expression samples were in one pool, from which the gene expression samples were extracted at fixed time points (0, 6, 12, 48, 72 hours after the exposure).
Lifespan analysis
For the analysis of the lifespan alterations, 150–170 individuals (males and females were kept separately) were used. Flies were transferred to a fresh medium two times a week. Dead flies were counted daily. For each experimental variant 3 biological replicates were pooled. Two control groups (one–for 5 and 10 cGy, another–for 20 and 40 cGy) for males as well as for females were used, due to the large exposure time difference (1 h 23 min and 2 h 47 min–for 5 and 10 cGy; 5 h 34 min and 11 h 8 min–for 20 and 40 cGy respectively). These replicates were merged, since flies were kept in the same conditions and the similar effects in the same variants were observed.
Survival functions were estimated using the Kaplan–Meier procedure and plotted as survival curves [24]. Median lifespan and the age of 90% mortality were calculated. The statistical analysis of survival data was conducted using nonparametric methods. Comparison of survival functions was done using the modified Kolmogorov–Smirnov test [25]. The statistical significance of differences between the mean lifespans for the experimental and control variants was determined using the Gehan–Breslow–Wilcoxon test [26]. To test the statistical significance of differences in maximum lifespan (age of 90% mortality), the Wang–Allison test was used [27]. Results of the log rank test are presented in the S1 Table.
It is well known that the Gompertz function is applicable for describing Drosophila lifespan alterations [28], so we approximate all survival curves with Gompertz equation: µ(x) = exp(αx) R0 [29]. We calculated parameters α and of the Gompertz equation, coefficients of determination that characterize the quality of the Gompertz function approximation [30] and the mortality rate doubling time (MRDT) [30]. Maximum likelihood method was used to evaluate the significance of differences in the intensity of mortality [31]. It's well known that there is a Strehler-Mildvan correlation between α and R0 parameters of the Gompertz equation [32]: [32]: ln(R0) = γ-βα (α and R0 – parameters of Gompertz equation, γ and β –regression parameters).
The Kaplan-Meier curves were plotted using STATISTICA, version 6.1 (StatSoft Inc, USA). Calculation of lifespan parameters and their statistical analysis were performed in the R software environment for statistical computing and graphics (http://www.r-project.org/). WinModest Version 1.0.2. [31] was used to calculate the parameters of the intensity of mortality.
RNA isolation and cDNA synthesis
Total RNA was isolated from homogenized samples (five flies from every sample) by QIAzol Lysis Reagent (Qiagen, Netherlands) and further isopropanol precipitation. The RNA concentration was determined using a NanoDrop® ND-1000 spectrophotometer (NanoDrop Technologies Inc., USA). The A260/A280 ratio of the RNA samples was 1.8–2.0. The integrity of the isolated RNA (RNA integrity number, RIN) was determined using the Bioanalyzer Agilent 2100 (Agilent Technologies, USA). Only the samples with an RIN value not less than 8.0 were used. Single-strand cDNA was synthesized using 1 μg of total RNA pretreated with DNase I (Fermentas, Lithuania), hexanucleotide primers, and M-MuLV reverse transcriptase (Fermentas, Lithuania) by the following scheme: 10 min at 25°C, 60 min at 42°C, 10 min at 50°C, and 10 min at 70°C.
qPCR
Real-time PCR was carried out on the 7500 Real-Time PCR System (Applied Biosystems, USA) by using modified short 6-carboxyfluorescein (FAM)-labeled probes from the Universal Probe Library (UPL, Roche, Switzerland). Pairs of primers were selected for every gene with the estimation of probability of primer dimers and heterodimers using OligoAnalyzer (http://eu.idtdna.com/calc/analyzer). The primer sequences are listed in the S2 Table. Each reaction was run 3 times with 10 μL mix, containing PCR-buffer, dNTPs in concentration 250 nM, primers– 300 nM, UPL, ROX, DNA polymerase 1 unit and cDNA diluted 17.5 times. The threshold cycle Ct was determined (7500 Software v2.0.5, Applied Biosystems, USA). The amplification efficiency values were calculated as described earlier [33]. The primers and probes proved to be specific by electrophoresis using Bioanalyzer Agilent 2100 (Agilent Technologies, USA); the size of amplification products were as expected.
Statistical analysis of qPCR data
The first step of the analysis of qPCR data is the evaluation of the stability of reference genes by four methods ΔCT [34], BestKeeper [35], Normfinder [36], Genorm [37]. The stability of all genes was analyzed relative to each other so the average rating of all genes was obtained by using all four methods. This rating showed the stability of all genes relative to each other in the certain experimental conditions. Only genes with high stability ratings were used as reference genes for expression normalization. The expression of four reference genes Actin, RpL32, EF1alpha, betaTub [38] was analyzed. Analysis of expression stability revealed that genes Actin, betaTub are very variable in this experiment. So only genes RpL32, EF1alpha were used as reference for expression normalization.
Ct values obtained for each gene in each sample were normalized to the reference gene Ct values for the calculation of the relative gene expression according to the formula:
, where Rij – relative gene expression of i gene in j sample, Ei, Er1j, Ernj−efficiency of reaction for gene and reference gene respectively, Ctij, Ctr1j, Ctrnj−threshold cycle of gene and reference gene respectively. All efficiencies were more than 90%. The expression change compared with control was log2FC (Fold Change), where FC = Riexp/Ricontrol for each biological replicates, then mean log2FC was calculated for all biological replicates. All calculations were performed using statistical computing programming language R (version 2.15.1). At least 2-fold mRNA level changes were considered as significant because of reference genes mRNA level variability.
Results and Discussion
Lifespan alterations in Drosophila melanogaster wild-type Canton-S individuals after the exposure to low doses of γ-irradiation
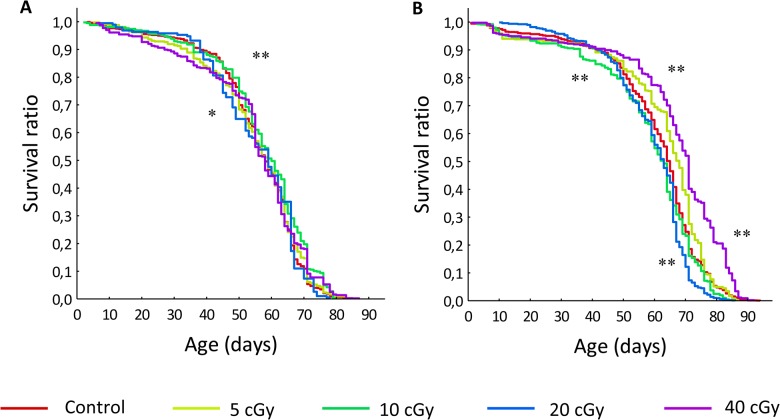
In Drosophila melanogaster wild-type Canton-S males, after exposure to low doses of ionizing radiation, we have observed the effect of hormesis: after the influence of γ-irradiation at a dose of 10 cGy, median lifespan increased by 3.4% (p<0.01, Gehan-Breslow-Wilcoxon test), the maximum lifespan increased by 4.2% (p<0.01, Wang-Allison test), exposure to γ-irradiation at doses of 5 and 40 cGy caused the extension of MRDT by 11.4 and 22.5% (p <0.01 maximum likelihood method), respectively (Table 1, Fig 1A).
Table 1. Alterations of the lifespan parameters in Drosophila melanogaster after exposure to low doses of ionizing radiation.
| Sex | Dose | М (day) | ΔM (%) | 90% (day) | Δ90% | MRDT (day) | Δ MRDT (%) | α (day−1) | R0 (day−1) | R2 | N |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ♂ | Control | 58 | - | 71 | - | 7.52 | - | 0.092 | 0.00031 | 0.805 | 1044 |
| 5 cGy | 59 | 1.7 | 71 | 0 | 8.38 | 11.4 (*) | 0.083 (*) | 0.0005 (*) | 0.718 | 423 | |
| 10 cGy | 60 | 3.4 (**) | 74 | 4.2 (**) | 7.88 | 4.8 | 0.088 | 0.00032 | 0.703 | 426 | |
| 20 cGy | 59 | 1.7 | 70 | -1.4 | 7.35 | -2.3 | 0.094 | 0.00029 | 0.743 | 391 | |
| 40 cGy | 58 | 0 | 71 | 0 | 9.21 | 22.5 (**) | 0.075 (**) | 0.00071 (**) | 0.563 | 438 | |
| ♀ | Control | 66 | - | 79 | - | 8.64 | - | 0.08 | 0.00032 | 0.77 | 1017 |
| 5 cGy | 69 | 4.5 (*) | 78 | -1.3 | 7.87 | -8.9 | 0.088 | 0.00019 | 0.57 | 381 | |
| 10 cGy | 63 | -4.5 (**) | 76 | -3.8 (**) | 9.06 | 4.9 | 0.076 | 0.00051 (*) | 0.63 | 318 | |
| 20 cGy | 63 | -4.5 (**) | 71 | -10.1 (**) | 7 | -19 (**) | 0.099 (**) | 0.00016 (**) | 0.82 | 457 | |
| 40 cGy | 71 | 7.6 (**) | 84 | 6.3 (**) | 8.04 | -2.8 | 0.082 | 0.00018 | 0.64 | 438 |
Table 1 legend: M–median lifespan, 90%–age of death of 90% of the sample (maximum lifespan), MRDT–mortality rate doubling time, ΔM, Δ90% and ΔMRDT–differences with the control for M, 90% and MRDT, α and R0 – parameters α and of Gompertz equation, R2 – determination coefficient of Gompertz approximation, N–number of individuals in the sample.
*—p<0.05
**—p<0.01, (Wang–Allison test for Δ90%; Gehan–Breslow–Wilcoxon test for ΔM; maximum likelihood method for α and ΔMRDT).
Fig 1. Influence of low doses of γ-irradiation on the lifespan of Drosophila melanogaster, wild-type line Canton-S.
A–males, B–females, *—p<0.05, **—p<0.01, (Kolmogorov-Smirnov test).
In Drosophila melanogaster wild-type Canton-S females, after exposure to γ-irradiation at doses of 5 and 40 cGy, an increase of median lifespan was observed (by 4.5 (p <0.05, Gehan-Breslow-Wilcoxon test) and 7.6% (p <0.01, Gehan-Breslow-Wilcoxon test) respectively). The impact of radiation at doses of 10 and 20 cGy leads to a decrease in this index by 4.5% (in both cases) (p <0.01, Gehan-Breslow-Wilcoxon test). The maximum lifespan increased by 6.3% after the influence of irradiation at a dose of 40 cGy and decreased after the impact at doses of 10 and 20 cGy by 3.8 and 10.1% (p <0.01, Wang-Allison test). The impact of irradiation at a dose of 20 cGy has revealed itself in decreased MRDT by 19% (p <0.01, maximum likelihood method). According to the above results, we can conclude that hormesis appears in Drosophila melanogaster females during the exposition doses of 5 and 40 cGy, and the opposite effect of hyperradiosensivity is demonstrated after irradiation treatment at doses of 10 cGy and 20 cGy (Table 1, Fig 1B).
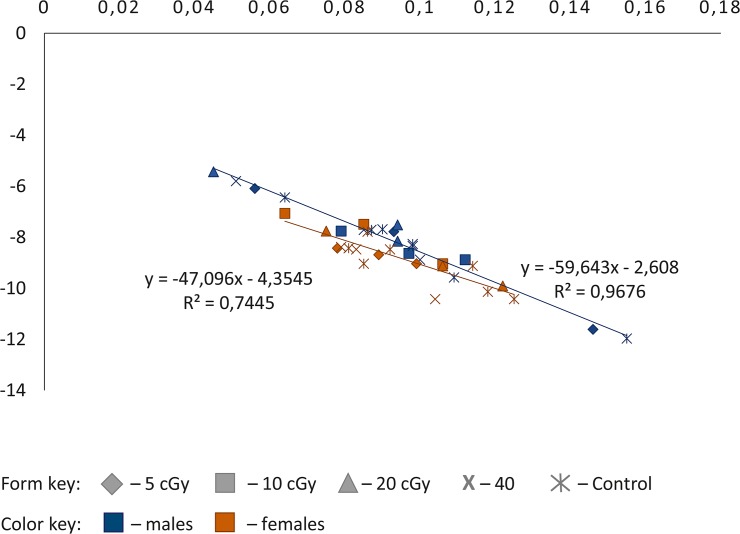
Fig 2 demonstrates the presence of the Strehler-Mildvan correlation between the parameters α and R0 of the Gompertz equation in Drosophila melanogaster wild-type line Canton-S males and females after the studied exposure doses. Each point on this parametric plane corresponds to the specific survival curve (three replicates per each exposure dose for male as well as for female). Correlation coefficients are equal to—0.98 (р < 0.0001) and—0.93 (р < 0.0001) in males and females respectively. It is known that the link between the parameters of the Gompertz function is equivalent to the presence of the intersection point of the survival curves. Moreover, the abscissa of this point is equal to the regression parameter β of the Strehler-Mildvan correlation equation, that is, the meaning of "typical life expectancy of the population" can be attributed to the value of this parameter [39]. In Fig 2, it is well shown that parameters of the Gompertz equation are approximated by the regression line, which is usual for “normal” physiological conditions [40]. In addition, the α and R0 of the Gompertz equation for all groups in males as well as in females do not significantly diverge from the regression line, thus, we can conclude that there are no differences in the "typical life expectancy of the population" between treated and control flies.
Fig 2. Strehler-Mildvan correlation between the parameters of the Gompertz function in Drosophila melanogaster wild-type Canton-S individuals exposed to low doses of ionizing radiation.
We have thus demonstrated the presence of hormesis in Drosophila melanogaster wild-type strain Canton-S male and female animals after exposure to γ-irradiation at doses of 5 and 40 cGy (according to various criteria). Females have also revealed the effect of hyperradiosensivity after irradiation doses of 10 and 20 cGy. However, it should be noted that because of calculation of the Strehler-Mildvan correlation, it was demonstrated that there are not deviations from the normal organism's physiological functions in treated male and female Drosophila melanogaster relative to the control.
Any change in lifespan relates to complex interactions of genetic and physiological factors [41, 42]. It is known that the effect of ionizing radiation in low doses can deviate in the direction of increasing negative consequences (hyperradiosensivity) [43] as well as in the direction of reducing negative consequences (radiation hormesis) [44]. Speaking about the possible mechanisms of radiation-induced changes, we should note that the effects of low doses of ionizing radiation affect the development of the organism, the immune response, lead to a change in the metabolism of proteins, amino acids, lipids, fatty acids, and hormones, alter energy metabolism, lead to tumor necrosis factors induction, cause changes in the cell cycle, in the processes of cell proliferation and differentiation, cause DNA damage, apoptosis, proteolytic degradation, autophagy and oxidative stress [17, 45–48].
For this reason, we investigated the time- and dose-response dependence of the alterations in differential expression of 29 genes involved in the cell stress response, DNA repair, apoptosis, antioxidant protection, and detoxification of xenobiotics using qPCR method.
Gene expression analysis after low dose radiation exposure
In this work, the dynamics of changes in the expression of stress sensitive genes (Table 2) in response to irradiation by low doses of 5–40 cGy in the Drosophila melanogaster wild-type strain Canton-S were analyzed.
Table 2. The genes selected for expression analysis in the samples of Drosophila melanogaster wild-type strain Canton-S 72 hours after radiation exposure in doses from 5 cGy to 40 cGy.
| Gene | Function | Reference |
|---|---|---|
| Hus1-like | DNA-damage-induced checkpoint response, activation of an S-phase checkpoint, oocyte DNA organization | [49] |
| foxo | Insulin signaling, resistance against oxidativestress | [50] |
| spn-B | RAD52 DNA repair pathway, double-strand DNA break (DSB) repair, meiotic checkpoint activation | [51, 52] |
| p53 | G1 growth arrest, induction of apoptosis, radiation-induced apoptosis | [53, 54] |
| mei-41 | Cell-cycle control, post-replication repair | [55, 56] |
| DJNK | Immune response activated by bacterial infection, wound healing, morphogenetic movement during embryogenesis | [57–59] |
| tefu | Spontaneous apoptosis suppression, female fertility, protection from telomere fusion, activation of checkpoint signaling in response to DNA double-stranded breaks induced by low-dose ionizing radiation | [60–62] |
| Clk | Master transcriptional regulator of the circadian clock | [63] |
| PCNA | Control of eukaryotic DNA replication by increasing the polymerase's processability | [64, 65] |
| hpo | Hippo/SWH (Sav/Wts/Hpo) signaling pathway, organ size control, tumor suppression, inhibition of transcriptional complex activity, regulation of Th/DIAP1 apoptosis inhibitor | [66, 67] |
| Sod | Radical detoxification | [68] |
| Brca2 | Double-strand break repair by meiotic and mitotic homologous recombination | [69] |
| mei-9 | Meiosis recombination events, Holliday junctions within recombination intermediates, repair of mismatches within meiotic heteroduplex DNA, nucleotide excision repair | [70, 71] |
| RAD54 | Mitotic DNA repair, meiotic recombination, recombinational DNA repair pathway | [72] |
| mus309 | DNA replication, DNA repair, exhibition of a magnesium-dependent ATP-dependent DNA-helicase activity | [73, 74] |
| wrinkled | Apoptosis activation | [75] |
| Cyp6a20 | Monooxygenase, oxidoreductase, electron carrier activity, heme binding, iron ion binding, takes part in aggressive behavior and defense response to Gram-negative bacterium | [76–78] |
| CG13323 | Unknown function | http://www.uniprot.org |
| GstE3 | Glutathione transferase activity, response to oxidative stress, resistance to insecticides | [79–81] |
| CG18180 | Serine-type endopeptidase activity, proteolysis with a possible role in immune function | [82–85] |
| Keap1 | Actin binding, defends organisms against the detrimental effects of oxidative stress | [86, 87] |
| CG42751 | Unknown function | http://www.uniprot.org |
| CG6295 | Hydrolase, lipid metabolic process | [88, 89] |
| CG6675 | Hydrolase, lipid metabolic process | http://www.uniprot.org |
| Fer3 | Transcription factor that binds to the E-box and functions as inhibitor of transcription. DNA binding requires dimerization with an E protein. Inhibits transcription activation by ASCL1/MASH1 by sequestering E proteins | [90, 91] |
| CG9360 | Oxidoreductase activity | http://www.uniprot.org |
| Cyp4e2 | Metabolism of insect hormones | [92] |
| Hsp70Aa | Recognition of sequences of hydrophobic amino acid residues, transmembrane transport of proteins, cell protection from thermal or oxidative stress, disposal of damaged or defective proteins, apoptosis inhibition | [93–97] |
| per | Period length of circadian and ultradian rhythms, eclosion behavior, male courtship song, circadian transcriptional loop | [98] |
The genes CG13323, GstE3, CG18180, Keap1, CG42751, CG6295, CG6675, Fer3, CG9360, Cyp4e2, Hsp70Aa, Cyp6a20, per were included in this analysis because previously in our laboratory differential expression of these genes in response to different stress factors including radiation was identified [23]. Other genes, including Hus1-like, foxo, spn-B, p53, mei-41, tefu, PCNA, hpo, DJNK, Sod, Brca2, mei-9, RAD54, mus309, whose expression were analyzed, are very important in response to stress impact. The regulation of circadian rhythm [99] and apoptosis [100] are also known to be changed by genotoxic stress, and, therefore, the genes Clk and wrinkled were included in this analysis.
Data about up- and down expression and p-value obtained for each gene in each sample are shown in Table 3 and raw qPCR data is presented in S1 File. The values were considered as statistically significant if appropriate p-value was less than 0.05. Ct values from qPCR for three biological replicates after the radiation exposure are performed in the S3 Table. The data of the relative expression, log2FC, the mean and the standard deviation are performed on the graphics in S1 File. These graphics show that for some genes under a certain radiation dose and at a certain time after the impact, the standard deviation is very high (more than 5% of mean value) or log2FC is less than 1 (FC is less than 2-fold change in this case). Such genes were identified as non-differentially expressed at this experimental point. In this way, the set of differentially expressed genes for every irradiation dose was obtained (Fig 3). The highest FC effects were performed 48 hours after the impact, but there are very few statistically significant values at this point. Under the radiation impact of a 40 cGy dose, the data are comparable with the biological variability in most cases.
Table 3. Analysis of the gene expression by the qPCR in the samples of Drosophila melanogaster wild-type strain Canton-S 72 hours after radiation exposure in doses from 5 cGy to 40 cGy (Female/male).
| Irradiation dose, cGy | 5 | 10 | ||||||||
| Analysis time, hours after exposure | 0 | 6 | 24 | 48 | 72 | 0 | 6 | 24 | 48 | 72 |
| CG6295 | n/n | n/n | -/- | -/n | n/n | n/n | + */n | n/n | n/n* | +/n |
| CG18180 | n/+ * | n/+ | + */n | n/n | n/+ | n/+ * | + */+ | + */n* | n/n | n/- |
| CG42751 | n*/n | n*/+ | n/n* | n/- * | n*/n* | n/n | n/+ | n/n | n/- * | n/n |
| Clk | n*/n | n*/n | n/n | n/n | n/n | n*/n | n/n | n*/n* | n/n | n/n |
| Cyp4e2 | n/n* | n*/n* | n/n* | n*/n* | n*/+ * | n*/n* | n/n* | n*/n | n*/n* | n*/+ * |
| Cyp6a20 | n/+ * | n/n* | n/n* | n*/n* | n/n | n/- * | n/n* | n/n | n/n* | n/+ |
| Fer3 | -/- | +/- | n/n | n/n | +/+ | n/- | n*/- | n/n | n/n | +/+ |
| foxo | -/n | n/n | n/n | n/n | n/n | n/n | n/n | n*/n | n*/n | n/n* |
| GstE3 | n/n | n/n | +/n | n*/n | n/n | n*/n | n/n | +/n | n*/n | n/n |
| hpo | n/n | n*/n | n/n | n/n | n/n | n/n | n/n | n*/n* | n*/n | n/n |
| Hsp70Aa | n*/n | n/+ | -/n | n/- | n/n | n/+ | - */- | n/n | -/- | n/n |
| Hus1-like | n/n* | n/n | n/n | n/n | n/n | n/n | n/n | n*/n* | n*/n | n/n* |
| DJNK | n/n | n/n | n/n | n/n | n/n* | n/n | n*/n | n/n | n*/n | n/+ |
| Keap1 | n/n | n/n | -/n | +/n | n/n | n/n | -/n | n/n | +/n | +/n |
| mei-9 | n/n | n/n | n/n | n/n | n/n | n/n | n/n | n/n* | n*/n | n/n |
| mei-41 | n/n | n/n | -/n | n/n | n/+ | n/n | -/+ | n/n* | n/n | n/+ * |
| PCNA | n/n* | n/n | n/n | n/+ | n/n | n/n | n/n | n/n* | +/+ | n/n |
| mus 309 | + */n | n/n | n/n | n/n | n/n | n/n | n/n | n/- | n/n | n/n |
| p53 | n/n | n/n | -/- | -/- | n/n | n/n | n/n | -/- | -/n | n/n |
| per | n/n | n/n* | n/n | +/n | n*/n* | n/n | -/n* | n/n* | n/n* | n/n |
| RAD54 | n/n | n/n | n*/n | n/n | n/n | n*/n | n*/n | n/- | n*/n | n/n |
| Sod | n/n | n/n* | n/n | n/n | n/n | n/n | +/n | n/n* | n/n | n/n |
| spn-B | n/n | n/+ | n/n | +/- * | n/+ | n/n* | n/+ | n/n | +/- * | n/+ |
| tefu | n/n | n/n | -/n | n/n | n/n | n/n | n*/n | -/n* | n/n | n/n |
| wrinkled | n/n | n*/n* | n/+ | n/+ | n/+ | n*/- | n/- | n/+ | n/+ | n/n |
| CG13323 | n*/n | n/n | n*/n | n*/n | +/n | n/n | + */n | n/n | n/n | n*/n |
| Brca2 | n/n | n*/n | n/n | n*/n | n/+ | n/n | n*/n | n*/n | n/n | n*/+ |
| CG6675 | n/n | n/n | n/n | n/n | n/n | n*/n | n*/n | n/n | n/n | n/n |
| CG9360 | n/n | n/n | n/n | n/n | n/n | n/n | n/n | n/n | n/n | n/n |
| Irradiation dose, cGy | 20 | 40 | ||||||||
| Analysis time, hours after exposure | 0 | 6 | 24 | 48 | 72 | 0 | 6 | 24 | 48 | 72 |
| CG6295 | n*/n | n*/n | -/- | n/n* | n/n | +/n | +/- | n/- | +/- | n/+ |
| CG18180 | n/n | n/n* | n/n | n/n | n/- | n/n | n/+ | +/n | n/n | n/+ |
| CG42751 | n/n | n/+ | n/n | n/- * | n/n | n*/n | n/+ | n/n | n*/- | n/+ |
| Clk | n/n | n/+ | n*/n | n/n | n/n | n*/- | n*/n* | +/n | n/n | n/n |
| Cyp4e2 | n*/n* | n*/n* | n*/n* | n*/n* | n*/+ * | n*/n* | n*/n | n*/n* | n*/n* | n*/+ * |
| Cyp6a20 | n/+ * | n/n* | n/n* | n/n* | n/n* | n/- * | +/n | n*/n* | -/n* | n/n* |
| Fer3 | n*/- | n/n | +/n | n/+ | n*/+ | +/n | n*/- | n/n | n/n | n*/n |
| foxo | n/n* | n/n | n*/n | n*/n | n*/n | n/n | n/+ | n*/n* | n*/n | n/+ |
| GstE3 | n/n | n/n | n*/n | n/n | n/n | n*/n | n/+ | + */n | n*/n | n/+ |
| hpo | n/n | n/n | n*/n | n*/n | n/n | n/n | n/n* | n/n | n*/n | n/n |
| Hsp70Aa | n/+ * | n/n | -/+ * | n*/- | +/+ * | +/+ | +/- | n*/- | + */- | +/n |
| Hus1-like | n*/n | n/n* | n/n | n*/n | n/n | n/n | n/+ | n/n | n*/n | n/n |
| DJNK | n/n* | n/n | n*/n | n*/n | n/n* | n/n | n/+ | n*/n | n*/n | n/n* |
| Keap1 | n/n* | n/n* | n/n | n*/n | +/n | n/n | n*/n | n/n | n*/n | n/n |
| mei-9 | n/n | n/+ * | n/n | n*/n | n/+ * | n/n | n/n* | n/n | n*/n | n/+ |
| mei-41 | n/n | n/n | n/n | n/n | n/n | n/n | n/n | -/n | n/n | n/+ |
| PCNA | n/n | n/n | n/+ * | n*/+ * | n/n* | n/n | n/n | n/+ | n*/n* | n/n* |
| mus 309 | n/n | n/+ | n/n | n/n | n/+ * | n/n | n/n | n/n | n/n | n/+ |
| p53 | n/- | n/n | -/- | n*/- | n/n | n/n | n/n | n*/n | n/- | -/+ |
| per | n/n | n/n | n/n | n/n* | n/n | n/n | +/n* | n/n | n/n* | -/+ |
| RAD54 | n*/- | n*/n | n*/n | n*/n | n*/n | n/n | +/n | n/- | n*/- | n/n |
| Sod | n/n | n/n | n/n | n/n* | n/n | n/n | -/n | n/n | -/n | n/+ |
| spn-B | n/- | n/+ | n/n* | n*/- * | n/n* | n/n | n/n | n/n | n*/- | n/n |
| tefu | n/n | n/n* | -/n | n/n | n/n | n/n | n/n | n/n | n/n | n/n |
| wrinkled | n*/n | n/n | n*/+ | n/+ | n/n | n*/- | +/- | n*/n | -/- | n/+ |
| CG13323 | n/n | n*/n | n/n | n*/n | n*/n | n/n | + */n | n/n | n/n | + */n |
| Brca2 | n/n | n/n | n*/n | -*/- | n/+ | n/n | n*/n* | n*/n | - */- | n/n |
| CG6675 | n*/n | n/n | n*/n | n/+ | n/n* | n/n | n/+ | n/n | n/n | n/+ |
| CG9360 | n/n | n/n | n/n | n/n | n/n | n/n | n/+ | n/n | n/n | n/n |
n–FC absolute value < 2; ǀLog2FCǀ<1
+–Log2FC > 1
-–Log2FC < -1
*—p-value < 0.05
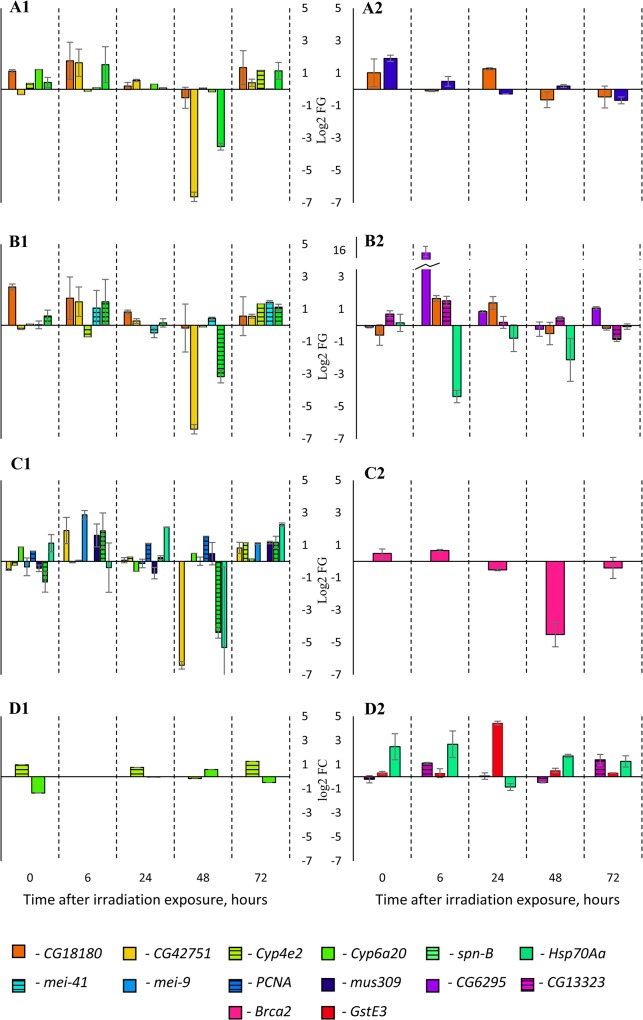
Fig 3. The differentially expressed genes in Drosophila melanogaster males and females after the radiation exposure.
A– 5 cGy, B– 10 cGy, C– 20 cGy, D– 40 cGy, 1 –males, 2 –females. Only gene changes with Log2FC > 1 and p-value < 0.05 during at least one time range are presented.
Analysis of differential expression in male samples
The analysis of differentially expressed genes of males revealed further changes (Fig 3A1, 3B1, 3C1 and 3D1). The genes CG42751 (more than 84 times down), spn-B (more than 8.6 times down) and the genes mei-9 (2 times up), mei-41 (2.6 times up), mus309 (2 times up), Cyp4e2 (more than 2.2 up) are differentially expressed 48 and 72 hours after the exposure respectively. This effect was observed only after 5 cGy, 10 cGy and 20 cGy dose irradiation. Such extended expression changes may reflect the fact that these genes are genes of late response to stress. For example, the expression of the gene mei-9 encoding the protein of nucleotide excision repair and DNA mismatch repair is shown to be activated in response to UV radiation 12 hours after impact and later [101]. Overexpression of gene Cyp4e2 and down-regulation of gene CG42751 revealed in this study are matched with results of analysis of response to different stressors by Drosophila melanogaster transcriptome sequencing [23]. Although the function of the gene CG42751 is still unknown, its expression changes were identified in response to oxidative stress [102], and it is known that indirect effects of the ionizing radiation are mediated by the induction of free radicals [103]. Gene Cyp4e2 of the cytochrome P450 gene family plays a role in the regulation of circadian rhythms [104] and in response to different stresses, mostly chemical stressors. For example, overexpression of this gene is identified in different stress-resistant Drosophila melanogaster strains [105]. The genes mei-9 and mei-41 regulate DNA repair in somatic cells [106, 107], moreover, gene mei-41 is required for hormesis, since lack of the hormetic effect was shown in mutants with inactive mei-41 [14]. In our experiments, this gene is overexpressed (2.6 times) in response to 10 cGy dose radiation. The overexpression of the gene spn-B participating in the double-strand break DNA repair is not necessary for increase of the lifespan in response to low dose irradiation (30 cGy) [108], and it is downregulated 8.6–39 times after 48 hours in response to 5 cGy, 10 cGy and 20 cGy irradiation.
Gene Cyp6a20, encoding protein cytochrome P450 6a20, which plays a role in immune response and regulating fly behavior [78], is overexpressed immediately after impact of 5 cGy irradiation (2.3 times) and down-regulated in response to 40 cGy irradiation (2.5 times). Probably, effects of genes of rapid reaction to radiation differ among samples exposed to different doses of radiation because at the moment of measurement of higher cumulative radiation doses these gene expressions are already inversely compared with lower cumulative radiation doses, and consequently, shorter exposure time.
Similar regularity of gene CG18180 is observed in male samples. This gene is overexpressed immediately after 5 and 10 cGy exposure, but there are no expression changes in response to 20, 40 cGy irradiation. The difference between the time of the start of exposure and the measurement may also explain the mismatch between these results and the results of gene expression analysis by RNA-Seq, which identified down regulation of Cyp6a20 and CG18180 genes [23]. The gene PCNA, participating in DNA repair (nucleotide-excision repair, mismatch repair) [109] and DNA replication [65], is overexpressed in response to 20 cGy irradiation 24 hours (2.1 times) and 48 hours (2.8 times) after the impact. This result may characterize this gene as a gene of long-term radiation stress response. The expression of gene Hsp70Aa is upregulated immediately after 20 cGy radiation exposure (2 times) and then 72 hours (4.8 times) after the impact.
Analysis of differential expression in female samples
The analysis of differentially expressed genes in female samples in response to low dose radiation exposure did not reveal any clear effects (S1 File). Most of them have high standard deviation and are very low, although they are higher than the biological variability.
The gene mus309 responsible for DNA damage signaling and DNA repair [74] is overexpressed (3.7 times) immediately after the 5 cGy dose irradiation. The gene CG13323 with unknown function is overexpressed in response to radiation exposure at dose 10 cGy (2.8 times) and 40 cGy (2.2 times) 6 hours after impact. This fact may reflect the participation of the CG13323 gene in radiation response. The gene CG6295, which plays a role in lipid metabolism, [89] is highly overexpressed (more than 15000 times) in response to 10 cGy irradiation 6 hours after exposure. The radiation induced production of ROS and RNS is known to lead to lipid metabolism disturbance [110]. But the expression of the CG6295 gene was down regulated in response to 20 cGy irradiation in other research [23]. Such a mismatch may be explained by the difference in time of exposure and analysis. The low expression level (22.6 times down) of the gene Brca2 participating in DNA repair [68] is observed after the 20 cGy exposure after 48 hours, but standard deviation is high. The gene Hsp70Aa is down-regulated (21 times) in response to 10 cGy irradiation and overexpressed (3.2 times up) in response to 40 cGy irradiation after 6 hours and 48 hours respectively. The expression changes of this gene involved in heat shock response [97] in both males and females after radiation exposure may confirm the existence of a non-specific stress response mechanism. It is interesting to mention that gene CG18180 is overexpressed in response to 5 cGy irradiation after 24 hours (2.5 times) and to 10 cGy irradiation 6 and 24 (more than 2.6 times up) hours after exposure respectively in females, whereas it is overexpressed just after exposure in the male samples. Although the function of gene CG18180 is still unknown, there is an assumption that this gene participates in immune response [83] and in response to different types of stresses [84, 85]. Perhaps the differences in the dynamics of CG18180 gene expression are responsible for sex-specific changes of lifespan of wild-type Drosophila melanogaster individuals after low dose radiation exposure. Gene GstE3, the glutathione S‐transferase playing a role in detoxification phase II [81], is overexpressed more than 20 times in response to 40 cGy irradiation after 24 hours, but in other research [23], the downregulation of this gene expression after 20 cGy radiation impact in males and females was observed, which may be explained by the difference in analysis time after exposure.
Sex-specific responses to different stimulus have been confirmed by many experiments. Also, hormesis effects of the same stresses depend on sex [111]. This difference may be explained by the fact that the same genes in individuals of different sexes have to act in various environments, although their functions are identical. For example, increased sexual activity reduces immunity in males [112]. Such a specific immune response regulation may be revealed also under other conditions, and sex-specific expression changes of CG18180 may be the consequence. Also, expression of this gene was shown to change in response to starvation and cold impact [85].
To sum up, we revealed that expression profiles of the 29 genes under research 72 hours after low dose irradiation from 5 cGy to 40 cGy are different in males and females of Drosophila melanogaster wild-type strain Canton-S. The gene Clk, responsible for circadian rhythm regulation, is not differentially expressed under experiment conditions, although previously, expression changes of the gene of this pathway in response to 20 cGy dose irradiation by enrichment analysis in 5-day-old flies were shown [23]. The genes spn-B, mei-9, mei-41, mus309 participating in the DNA repair and the response to different stresses are overexpressed in males 48 and 78 hours after radiation exposure, which may confirm their late transcriptional activation in response to radiation stress, and probably plays key role in extension of lifespan after the exposure to low doses of γ-irradiation. The expression of the gene mus309 is changed in both males and females, but the expression profiles are different: this gene, after 72 hours, is overexpressed in males more than twofold in response to 20 cGy irradiation and in females fourfold immediately after exposure of 5 cGy radiation impact. Reduced expression of the gene CG42751 with unknown function may be evidence of its role in changed lifespan and in the stress-response reaction to radiation. The expression changes of the gene Hsp70Aa (overexpressed more than threefold in response to 40 and 20 cGy after 6, 48 hours in females, and 24, 72 hours in males respectively, and down-regulated six hours after 10 cGy irradiation in females by 20 times) involved in heat shock response [97] in both males and females after radiation exposure may confirm the existence of a non-specific stress response mechanism. The dynamics of the expression change of gene CG18180, playing a role in immune response, differs in males and females (overexpressed immediately in males and after 6–24 hours in females after 5 and 10 cGy dose irradiation), which may play a role in reducing median and maximal lifespan of females after this impact. The differences in gene expression profile reflect a sex-specific stress response and lifespan features in Drosophila melanogaster wild strain Canton-S.
Conclusions
Although there were changes in various indicators of life expectancy after exposure to 5, 10, 20 and 40 cGy, according to our analyses, they were not caused by the changes of organism physiological functions in the Drosophila melanogaster individuals after treatment, and furthermore there were not dose-dependent changes in the expression profile of stress-response genes chosen for the present study. It also should be noted that the cases of low dose irradiation expression changes are characterized by high biological variability, displaying a stochastic nature of low dose radiation effects. These results demonstrate the nonlinear character of low dose radiation effects on the Drosophila melanogaster imago and reveal a possible role of the gene CG18180 in sex-specific stress response and lifespan features.
Supporting Information
Results for 5 cGy irradiation of males (Figure A1). Results for 10 cGy irradiation of males (Figure B1). Results for 20 cGy irradiation of males (Figure C1). Results for 40 cGy irradiation of males (Figure D1). Results for 5 cGy irradiation of females (Figure A2). Results for 10 cGy irradiation of females (Figure B2). Results for 20 cGy irradiation of females (Figure C2). Results for 40 cGy irradiation of females (Figure D2).
(DOC)
(DOCX)
(XLS)
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by RFBR grant N 14-04-01596 (http://www.rfbr.ru/rffi/eng) and the President Grant for Government Support of Young Russian Scientists MD-1090.2014.4. (http://grants.extech.ru). Part of this work (qPCR analysis) was performed using the equipment of EIMB RAS "Genome" center (http://www.eimb.ru/RUSSIAN_NEW/INSTITUTE/ccu_genome_c.php) under the financial support by the Ministry of Education and Science of the Russian Federation (Contract 14.621.21.0001, project's unique identifier RFMEFI62114X0001, http://www.eimb.ru/RUSSIAN_NEW/INSTITUTE/ccu_genome_c.php).
References
- 1. Moller AP, Mousseau TA. The effects of natural variation in background radioactivity on humans, animals and other organisms. Biol Rev Camb Philos Soc. 2013. February; 88(1): p. 226–54. 10.1111/j.1469-185X.2012.00249.x [DOI] [PubMed] [Google Scholar]
- 2. Shahbazi-Gahrouei D, Gholami M, Setayandeh S. A review on natural background radiation. Adv Biomed Res. 2013. 2: p. 65 10.4103/2277-9175.115821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nies H, Harms IH, Karcher MJ, Dethleff D, Bahe C. Anthropogenic radioactivity in the Arctic Ocean—review of the results from the joint German project. Sci Total Environ. 1999. September 30; 237–238: p. 181–91. [DOI] [PubMed] [Google Scholar]
- 4. Lou Y, Wan L, Ma Y, Li H, Meng Q, Kong Y, et al. Survey on radioactive contamination in Beijing following the Japanese Fukushima nuclear accident. J Radiol Prot. 2013. September; 33(3): p. 661–8. 10.1088/0952-4746/33/3/661 [DOI] [PubMed] [Google Scholar]
- 5. Kozhakhanov TE, Lukashenko SN, Larionova NV. Accumulation of artificial radionuclides in agricultural plants in the area used for surface nuclear tests. J Environ Radioact. 2014. November; 137: p. 217–26. 10.1016/j.jenvrad.2014.06.026 [DOI] [PubMed] [Google Scholar]
- 6. McGeoghegan D, Binks K. The mortality and cancer morbidity experience of workers at the Springfields uranium production facility, 1946–95. J Radiol Prot. 2000. June; 20(2): p. 111–37. [DOI] [PubMed] [Google Scholar]
- 7. Leszczynski D, Nylund R, Joenvaara S, Reivinen J. Applicability of discovery science approach to determine biological effects of mobile phone radiation. Proteomics. 2004. February; 4(2): p. 426–31. [DOI] [PubMed] [Google Scholar]
- 8. Amis ES Jr., Butler PF, Applegate KE, Birnbaum SB, Brateman LF, Hevezi JM, et al. American College of Radiology white paper on radiation dose in medicine. J Am Coll Radiol. 2007. May; 4(5): p. 272–84. [DOI] [PubMed] [Google Scholar]
- 9. Smith-Bindman R, Lipson J, Marcus R, Kim KP, Mahesh M, Gould R, et al. Radiation dose associated with common computed tomography examinations and the associated lifetime attributable risk of cancer. Arch Intern Med. 2009. December 14; 169(22): p. 2078–86. 10.1001/archinternmed.2009.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dent P, Yacoub A, Contessa J, Caron R, Amorino G, Valerie K, et al. Stress and radiation-induced activation of multiple intracellular signaling pathways. Radiat Res. 2003. March; 159(3): p. 283–300. [DOI] [PubMed] [Google Scholar]
- 11. Ravanat JL, Douki T, Cadet J. Direct and indirect effects of UV radiation on DNA and its components. J Photochem Photobiol B. 2001. October; 63(1–3): p. 88–102. [DOI] [PubMed] [Google Scholar]
- 12. Lavelle C, Foray N. Chromatin structure and radiation-induced DNA damage: from structural biology to radiobiology. Int J Biochem Cell Biol. 2014. April; 49: p. 84–97. 10.1016/j.biocel.2014.01.012 [DOI] [PubMed] [Google Scholar]
- 13. Feinendegen LE. Evidence for beneficial low level radiation effects and radiation hormesis. Br J Radiol. 2005. January; 78(925): p. 3–7. [DOI] [PubMed] [Google Scholar]
- 14. Moskalev AA, Plyusnina EN, Shaposhnikov MV. Radiation hormesis and radioadaptive response in Drosophila melanogaster flies with different genetic backgrounds: the role of cellular stress-resistance mechanisms. Biogerontology. 2011. June; 12(3): p. 253–63. 10.1007/s10522-011-9320-0 [DOI] [PubMed] [Google Scholar]
- 15. Calabrese EJ, Baldwin LA. Defining hormesis. Hum Exp Toxicol. 2002. February; 21(2): p. 91–7. [DOI] [PubMed] [Google Scholar]
- 16. Moskalev A. Radiation-induced life span alteration of Drosophila lines with genotype differences. Biogerontology. 2007. October; 8(5): p. 499–504. [DOI] [PubMed] [Google Scholar]
- 17. Seong KM, Kim CS, Seo SW, Jeon HY, Lee BS, Nam SY, et al. Genome-wide analysis of low-dose irradiated male Drosophila melanogaster with extended longevity. Biogerontology. 2011. April; 12(2): p. 93–107. 10.1007/s10522-010-9295-2 [DOI] [PubMed] [Google Scholar]
- 18. Moskalev A, Shaposhnikov M, Turysheva E. Life span alteration after irradiation in Drosophila melanogaster strains with mutations of Hsf and Hsps. Biogerontology. 2009. February; 10(1): p. 3–11. 10.1007/s10522-008-9147-5 [DOI] [PubMed] [Google Scholar]
- 19. Vaiserman AM, Koshel NM, Litoshenko AY, Mozzhukhina TG, Voitenko VP. Effects of X-irradiation in early ontogenesis on the longevity and amount of the S1 nuclease-sensitive DNA sites in adult Drosophila melanogaster. Biogerontology. 2003. 4(1): p. 9–14. [DOI] [PubMed] [Google Scholar]
- 20. Nakamura N, Suyama A, Noda A, Kodama Y. Radiation effects on human heredity. Annu Rev Genet. 2013. 47: p. 33–50. 10.1146/annurev-genet-111212-133501 [DOI] [PubMed] [Google Scholar]
- 21. Ogaki M, Nakashima-Tanaka E. Inheritance of radioresistance in Drosophila. I. Mutat Res. 1966. October; 3(5): p. 438–43. [DOI] [PubMed] [Google Scholar]
- 22. Antosh M, Fox D, Hasselbacher T, Lanou R, Neretti N, Cooper LN. Drosophila melanogaster show a threshold effect in response to radiation. Dose Response. 2014. December; 12(4): p. 551–81. 10.2203/dose-response.13-047.Antosh [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moskalev A, Shaposhnikov M, Snezhkina A, Kogan V, Plyusnina E, Peregudova D, et al. Mining gene expression data for pollutants (dioxin, toluene, formaldehyde) and low dose of gamma-irradiation. PLoS One. 2014. 9(1): p. e86051 10.1371/journal.pone.0086051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaplan EL, Meier P. Nonparametric Estimation from Incomplete Observations. Journal of the American Statistical Association. 1958. 53: p. 457–481. [Google Scholar]
- 25. Fleming TR, O’Fallon JR, O’Brien PC, Harrington DP. Modifid Kolmogorov-Smirnov test procedures with application to arbitrarily right-censored data. Biometrics. 1980. 36(4): p. 607–625. [Google Scholar]
- 26. Breslow N. A generalized Kruskal-Wallis test for comparing K samples subject to unequal patterns of censorship. Biometrika. 1970. 57(3): p. 579–594. [Google Scholar]
- 27. Wang C, Li Q, Redden DT, Weindruch R, Allison DB. Statistical methods for testing effects on "maximum lifespan". Mech Ageing Dev. 2004. September; 125(9): p. 629–32. [DOI] [PubMed] [Google Scholar]
- 28. Gavrilov LA, Gavrilova NS, The biology of life span: a quantitative approach. Rev. and updated English ed. 1991, Chur—New York: Harwood Academic Publishers. 385. [Google Scholar]
- 29. Gompertz B. On the nature of the function expressive of the law of human mortality and on a new mode of determining life contingencies Philos. Trans. Roy. Soc. London. A. 1825. 155: p. 513–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tollefsbol TO, Biological aging: methods and protocols. Second edition. ed. Methods in molecular biology,. 2013. 354.
- 31. Pletcher SD. Model fitting and hypothesis testing for age-specific mortality data. J. Evol. Biol. 1999. 12(3): p. 430–439. [Google Scholar]
- 32. Strehler BL, Mildvan AS. General theory of mortality and aging. Science. 1960. July 1; 132(3418): p. 14–21. [DOI] [PubMed] [Google Scholar]
- 33. Anedchenko EA, Kiseleva NP, Dmitriev AA, Kiselev FL, Zabarovskii ER, Senchenko VN. [Tumor suppressor gene RBSP3 in cervical carcinoma: copy number and transcriptional level]. Mol Biol (Mosk). 2007. Jan-Feb; 41(1): p. 86–95. [PubMed] [Google Scholar]
- 34. Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006. 7: p. 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004. March; 26(6): p. 509–15. [DOI] [PubMed] [Google Scholar]
- 36. Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004. August 1; 64(15): p. 5245–50. [DOI] [PubMed] [Google Scholar]
- 37. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002. June 18; 3(7): p. Research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ponton F, Chapuis MP, Pernice M, Sword GA, Simpson SJ. Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J Insect Physiol. 2011. 57(6): p. 840–850. 10.1016/j.jinsphys.2011.03.014 [DOI] [PubMed] [Google Scholar]
- 39. Hirsch HR. Do intersections of mortality-rate and survival functions have significance? Exp Gerontol. 1995. Mar-Apr; 30(2): p. 147–67. [DOI] [PubMed] [Google Scholar]
- 40. Strehler BL. Origin and comparison of the effects of time and high energy radiations on living systems. Quart. Rev. Biol. 1959. 34. [DOI] [PubMed] [Google Scholar]
- 41. Garinis GA, van der Horst GT, Vijg J, Hoeijmakers JH. DNA damage and ageing: new-age ideas for an age-old problem. Nat Cell Biol. 2008. November; 10(11): p. 1241–7. 10.1038/ncb1108-1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kirkwood TB. Understanding the odd science of aging. Cell. 2005. February 25; 120(4): p. 437–47. [DOI] [PubMed] [Google Scholar]
- 43. Enns L, Bogen KT, Wizniak J, Murtha AD, Weinfeld M. Low-dose radiation hypersensitivity is associated with p53-dependent apoptosis. Mol Cancer Res. 2004. October; 2(10): p. 557–66. [PubMed] [Google Scholar]
- 44. Moskalev AA, Pliusnina EN, Zainullin VG. [The influence of low doze gamma-irradiation on life span of Drosophila mutants with defects of DNA damage sensation and repair]. Radiats Biol Radioecol. 2007. Sep-Oct; 47(5): p. 571–3. [PubMed] [Google Scholar]
- 45. Fachin AL, Mello SS, Sandrin-Garcia P, Junta CM, Donadi EA, Passos GA, et al. Gene expression profiles in human lymphocytes irradiated in vitro with low doses of gamma rays. Radiat Res. 2007. December; 168(6): p. 650–65. [DOI] [PubMed] [Google Scholar]
- 46. Rudqvist N, Parris TZ, Schuler E, Helou K, Forssell-Aronsson E. Transcriptional response of BALB/c mouse thyroids following in vivo astatine-211 exposure reveals distinct gene expression profiles. EJNMMI Res. 2012. 2(1): p. 32 10.1186/2191-219X-2-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Saini D, Shelke S, Mani Vannan A, Toprani S, Jain V, Das B, et al. Transcription profile of DNA damage response genes at G(0) lymphocytes exposed to gamma radiation. Mol Cell Biochem. 2012. May; 364(1–2): p. 271–81. 10.1007/s11010-012-1227-9 [DOI] [PubMed] [Google Scholar]
- 48. Wyrobek AJ, Manohar CF, Krishnan VV, Nelson DO, Furtado MR, Bhattacharya MS, et al. Low dose radiation response curves, networks and pathways in human lymphoblastoid cells exposed from 1 to 10cGy of acute gamma radiation. Mutat Res. 2011. June 17; 722(2): p. 119–30. 10.1016/j.mrgentox.2011.03.002 [DOI] [PubMed] [Google Scholar]
- 49. Song YH. Drosophila melanogaster: a model for the study of DNA damage checkpoint response. Mol Cells. 2005. April 30; 19(2): p. 167–79. [PubMed] [Google Scholar]
- 50. Junger MA, Rintelen F, Stocker H, Wasserman JD, Vegh M, Radimerski T, et al. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J Biol. 2003. 2(3): p. 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ghabrial A, Ray RP, Schupbach T. okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes Dev. 1998. September 1; 12(17): p. 2711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Abdu U, Gonzalez-Reyes A, Ghabrial A, Schupbach T. The Drosophila spn-D gene encodes a RAD51C-like protein that is required exclusively during meiosis. Genetics. 2003. September; 165(1): p. 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jin S, Martinek S, Joo WS, Wortman JR, Mirkovic N, Sali A, et al. Identification and characterization of a p53 homologue in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2000. June 20; 97(13): p. 7301–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000. March 31; 101(1): p. 103–13. [DOI] [PubMed] [Google Scholar]
- 55. Brodsky MH, Sekelsky JJ, Tsang G, Hawley RS, Rubin GM. mus304 encodes a novel DNA damage checkpoint protein required during Drosophila development. Genes Dev. 2000. March 15; 14(6): p. 666–78. [PMC free article] [PubMed] [Google Scholar]
- 56. LaRocque JR, Jaklevic B, Su TT, Sekelsky J. Drosophila ATR in double-strand break repair. Genetics. 2007. March; 175(3): p. 1023–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ramet M, Lanot R, Zachary D, Manfruelli P. JNK signaling pathway is required for efficient wound healing in Drosophila. Dev Biol. 2002. January 1; 241(1): p. 145–56. [DOI] [PubMed] [Google Scholar]
- 58. Sluss HK, Han Z, Barrett T, Goberdhan DC, Wilson C, Davis RJ, et al. A JNK signal transduction pathway that mediates morphogenesis and an immune response in Drosophila. Genes Dev. 1996. November 1; 10(21): p. 2745–58. [DOI] [PubMed] [Google Scholar]
- 59. Zeitlinger J, Bohmann D. Thorax closure in Drosophila: involvement of Fos and the JNK pathway. Development. 1999. September; 126(17): p. 3947–56. [DOI] [PubMed] [Google Scholar]
- 60. Bi X, Gong M, Srikanta D, Rong YS. Drosophila ATM and Mre11 are essential for the G2/M checkpoint induced by low-dose irradiation. Genetics. 2005. October; 171(2): p. 845–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bi X, Wei SC, Rong YS. Telomere protection without a telomerase; the role of ATM and Mre11 in Drosophila telomere maintenance. Curr Biol. 2004. August 10; 14(15): p. 1348–53. [DOI] [PubMed] [Google Scholar]
- 62. Song YH, Mirey G, Betson M, Haber DA, Settleman J. The Drosophila ATM ortholog, dATM, mediates the response to ionizing radiation and to spontaneous DNA damage during development. Curr Biol. 2004. August 10; 14(15): p. 1354–9. [DOI] [PubMed] [Google Scholar]
- 63. Abruzzi KC, Rodriguez J, Menet JS, Desrochers J, Zadina A, Luo W, et al. Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev. 2011. November 15; 25(22): p. 2374–86. 10.1101/gad.174110.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ng L, Prelich G, Anderson CW, Stillman B, Fisher PA. Drosophila proliferating cell nuclear antigen. Structural and functional homology with its mammalian counterpart. J Biol Chem. 1990. July 15; 265(20): p. 11948–54. [PubMed] [Google Scholar]
- 65. Strzalka W, Ziemienowicz A. Proliferating cell nuclear antigen (PCNA): a key factor in DNA replication and cell cycle regulation. Ann Bot. 2011. May; 107(7): p. 1127–40. 10.1093/aob/mcq243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003. October; 5(10): p. 914–20. [DOI] [PubMed] [Google Scholar]
- 67. Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003. August 22; 114(4): p. 457–67. [DOI] [PubMed] [Google Scholar]
- 68. Seto NO, Hayashi S, Tener GM. The sequence of the Cu-Zn superoxide dismutase gene of Drosophila. Nucleic Acids Res. 1987. December 23; 15(24): p. 10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Brough R, Wei D, Leulier S, Lord CJ, Rong YS, Ashworth A. Functional analysis of Drosophila melanogaster BRCA2 in DNA repair. DNA Repair (Amst). 2008. January 1; 7(1): p. 10–9. [DOI] [PubMed] [Google Scholar]
- 70. Sekelsky JJ, McKim KS, Chin GM, Hawley RS. The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics. 1995. October; 141(2): p. 619–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Joyce EF, Tanneti SN, McKim KS. Drosophila hold'em is required for a subset of meiotic crossovers and interacts with the dna repair endonuclease complex subunits MEI-9 and ERCC1. Genetics. 2009. January; 181(1): p. 335–40. 10.1534/genetics.108.093104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kooistra R, Vreeken K, Zonneveld JB, de Jong A, Eeken JC, Osgood CJ, et al. The Drosophila melanogaster RAD54 homolog, DmRAD54, is involved in the repair of radiation damage and recombination. Mol Cell Biol. 1997. October; 17(10): p. 6097–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kusano K, Berres ME, Engels WR. Evolution of the RECQ family of helicases: A drosophila homolog, Dmblm, is similar to the human bloom syndrome gene. Genetics. 1999. March; 151(3): p. 1027–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Adams MD, McVey M, Sekelsky JJ. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science. 2003. January 10; 299(5604): p. 265–7. [DOI] [PubMed] [Google Scholar]
- 75. Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995. July 15; 9(14): p. 1694–708. [DOI] [PubMed] [Google Scholar]
- 76. Chung H, Sztal T, Pasricha S, Sridhar M, Batterham P, Daborn PJ. Characterization of Drosophila melanogaster cytochrome P450 genes. Proc Natl Acad Sci U S A. 2009. April 7; 106(14): p. 5731–6. 10.1073/pnas.0812141106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dahanukar A, Ray A. Courtship, aggression and avoidance: pheromones, receptors and neurons for social behaviors in Drosophila. Fly (Austin). 2011. Jan-Mar; 5(1): p. 58–63. [DOI] [PubMed] [Google Scholar]
- 78. Robin C, Daborn PJ, Hoffmann AA. Fighting fly genes. Trends Genet. 2007. February; 23(2): p. 51–4. [DOI] [PubMed] [Google Scholar]
- 79. Saisawang C, Wongsantichon J, Ketterman AJ. A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster. Biochem J. 2012. February 15; 442(1): p. 181–90. 10.1042/BJ20111747 [DOI] [PubMed] [Google Scholar]
- 80. Ortelli F, Rossiter LC, Vontas J, Ranson H, Hemingway J. Heterologous expression of four glutathione transferase genes genetically linked to a major insecticide-resistance locus from the malaria vector Anopheles gambiae. Biochem J. 2003. August 1; 373(Pt 3): p. 957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tu CP, Akgul B. Drosophila glutathione S-transferases. Methods Enzymol. 2005. 401: p. 204–26. [DOI] [PubMed] [Google Scholar]
- 82. Yang W, Hekimi S. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biol. 2010. 8(12): p. e1000556 10.1371/journal.pbio.1000556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci U S A. 2001. October 23; 98(22): p. 12590–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kalajdzic P, Oehler S, Reczko M, Pavlidi N, Vontas J, Hatzigeorgiou AG, et al. Use of mutagenesis, genetic mapping and next generation transcriptomics to investigate insecticide resistance mechanisms. PLoS One. 2012. 7(6): p. e40296 10.1371/journal.pone.0040296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Telonis-Scott M, Hallas R, McKechnie SW, Wee CW, Hoffmann AA. Selection for cold resistance alters gene transcript levels in Drosophila melanogaster. J Insect Physiol. 2009. June; 55(6): p. 549–55. 10.1016/j.jinsphys.2009.01.010 [DOI] [PubMed] [Google Scholar]
- 86. Sykiotis GP, Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell. 2008. January; 14(1): p. 76–85. 10.1016/j.devcel.2007.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Goldstein LS, Gunawardena S. Flying through the drosophila cytoskeletal genome. J Cell Biol. 2000. July 24; 150(2): p. F63–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Karpac J, Biteau B, Jasper H. Misregulation of an adaptive metabolic response contributes to the age-related disruption of lipid homeostasis in Drosophila. Cell Rep. 2013. September 26; 4(6): p. 1250–61. 10.1016/j.celrep.2013.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Horne I, Haritos VS, Oakeshott JG. Comparative and functional genomics of lipases in holometabolous insects. Insect Biochem Mol Biol. 2009. August; 39(8): p. 547–67. 10.1016/j.ibmb.2009.06.002 [DOI] [PubMed] [Google Scholar]
- 90. Segev E, Halachmi N, Salzberg A, Ben-Arie N. Nato3 is an evolutionarily conserved bHLH transcription factor expressed in the CNS of Drosophila and mouse. Mech Dev. 2001. August; 106(1–2): p. 197–202. [DOI] [PubMed] [Google Scholar]
- 91. Verzi MP, Anderson JP, Dodou E, Kelly KK, Greene SB, North BJ, et al. N-twist, an evolutionarily conserved bHLH protein expressed in the developing CNS, functions as a transcriptional inhibitor. Dev Biol. 2002. September 1; 249(1): p. 174–90. [DOI] [PubMed] [Google Scholar]
- 92. Pittendrigh BR, Mocelin G, Andreev O, ffrench-Constant RH. The sequence of a Drosophila Cyp4e2 cytochrome P450-encoding cDNA. Gene. 1996. November 14; 179(2): p. 295–6. [DOI] [PubMed] [Google Scholar]
- 93. Mukhopadhyay I, Siddique HR, Bajpai VK, Saxena DK, Chowdhuri DK. Synthetic pyrethroid cypermethrin induced cellular damage in reproductive tissues of Drosophila melanogaster: Hsp70 as a marker of cellular damage. Arch Environ Contam Toxicol. 2006. November; 51(4): p. 673–80. [DOI] [PubMed] [Google Scholar]
- 94. Gupta S, Deepti A, Deegan S, Lisbona F, Hetz C, Samali A. HSP72 protects cells from ER stress-induced apoptosis via enhancement of IRE1alpha-XBP1 signaling through a physical interaction. PLoS Biol. 2010. 8(7): p. e1000410 10.1371/journal.pbio.1000410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Bienz M, Pelham HR. Heat shock regulatory elements function as an inducible enhancer in the Xenopus hsp70 gene and when linked to a heterologous promoter. Cell. 1986. June 6; 45(5): p. 753–60. [DOI] [PubMed] [Google Scholar]
- 96. Kelty JD, Lee RE Jr. Rapid cold-hardening of Drosophila melanogaster (Diptera: Drosophiladae) during ecologically based thermoperiodic cycles. J Exp Biol. 2001. May; 204(Pt 9): p. 1659–66. [DOI] [PubMed] [Google Scholar]
- 97. Gong WJ, Golic KG. Loss of Hsp70 in Drosophila is pleiotropic, with effects on thermotolerance, recovery from heat shock and neurodegeneration. Genetics. 2006. January; 172(1): p. 275–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Bae K, Lee C, Sidote D, Chuang KY, Edery I. Circadian regulation of a Drosophila homolog of the mammalian Clock gene: PER and TIM function as positive regulators. Mol Cell Biol. 1998. October; 18(10): p. 6142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gotoh T, Vila-Caballer M, Liu J, Schiffhauer S, Finkielstein CV. Association of the circadian factor Period 2 to p53 influences p53's function in DNA-damage signaling. Mol Biol Cell. 2015. January 15; 26(2): p. 359–72. 10.1091/mbc.E14-05-0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Payne CM, Bjore CG Jr., Schultz DA. Change in the frequency of apoptosis after low- and high-dose X-irradiation of human lymphocytes. J Leukoc Biol. 1992. October; 52(4): p. 433–40. [DOI] [PubMed] [Google Scholar]
- 101. Karpac J, Younger A, Jasper H. Dynamic coordination of innate immune signaling and insulin signaling regulates systemic responses to localized DNA damage. Dev Cell. 2011. June 14; 20(6): p. 841–54. 10.1016/j.devcel.2011.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Weber AL, Khan GF, Magwire MM, Tabor CL, Mackay TF, Anholt RR . Genome-wide association analysis of oxidative stress resistance in Drosophila melanogaster. PLoS One. 2012. 7(4): p. e34745 10.1371/journal.pone.0034745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Riley PA. Free radicals in biology: oxidative stress and the effects of ionizing radiation. Int J Radiat Biol. 1994. January; 65(1): p. 27–33. [DOI] [PubMed] [Google Scholar]
- 104. Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000. March 10; 287(5459): p. 1834–7. [DOI] [PubMed] [Google Scholar]
- 105. Giraudo M, Unnithan GC, Le Goff G, Feyereisen R. Regulation of cytochrome P450 expression in Drosophila: Genomic insights. Pestic Biochem Physiol. 2010. June 1; 97(2): p. 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sarantseva SV, Khromykh Iu M. [Effect of gamma-irradiation on oogenesis of Drosophila mutants defective for reparation and meiotic recombination]. Genetika. 2001. June; 37(6): p. 770–8. [PubMed] [Google Scholar]
- 107. Sekelsky JJ, Brodsky MH, Burtis KC. DNA repair in Drosophila: insights from the Drosophila genome sequence. J Cell Biol. 2000. July 24; 150(2): p. F31–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Shilova LA, Pliusnina EN, Moskalev AA. Influence of conditionally ubiquitous overexpression of DNA repair genes on resistance of drosophila melanogaster individuals to different stress factors (oxidative stress, heat shock, starvation). Proc of the KSC Ural Branch RAS. 2014. 2(18): p. 41–45. [Google Scholar]
- 109. Nichols AF, Sancar A. Purification of PCNA as a nucleotide excision repair protein. Nucleic Acids Res. 1992. July 11; 20(13): p. 2441–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Schmidt-Ullrich RK, Dent P, Grant S, Mikkelsen RB, Valerie K. Signal transduction and cellular radiation responses. Radiat Res. 2000. March; 153(3): p. 245–57. [DOI] [PubMed] [Google Scholar]
- 111. Burger JM, Promislow DE. Sex-specific effects of interventions that extend fly life span. Sci Aging Knowledge Environ. 2004. July 14; 2004(28): p. pe30. [DOI] [PubMed] [Google Scholar]
- 112. McKean KA, Nunney L. Increased sexual activity reduces male immune function in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2001. July 3; 98(14): p. 7904–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Results for 5 cGy irradiation of males (Figure A1). Results for 10 cGy irradiation of males (Figure B1). Results for 20 cGy irradiation of males (Figure C1). Results for 40 cGy irradiation of males (Figure D1). Results for 5 cGy irradiation of females (Figure A2). Results for 10 cGy irradiation of females (Figure B2). Results for 20 cGy irradiation of females (Figure C2). Results for 40 cGy irradiation of females (Figure D2).
(DOC)
(DOCX)
(XLS)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.