Abstract
Background
Maraviroc activity against HIV-2, a virus naturally resistant to different HIV-1 antiretroviral drugs, has been recently demonstrated. The aim of this study was to assess HIV-2 susceptibility to cenicriviroc, a novel, once-daily, dual CCR5 and CCR2 antagonist that has completed Phase 2b development in HIV-1 infection.
Methods
Cenicriviroc phenotypic activity has been tested using a PBMC phenotypic susceptibility assay against four R5-, one X4- and one dual-tropic HIV-2 clinical primary isolates. All isolates were obtained by co-cultivation of PHA-activated PBMC from distinct HIV-2-infected CCR5-antagonist-naïve patients included in the French HIV-2 cohort and were previously tested for maraviroc susceptibility using the same protocol. HIV-2 tropism was determined by phenotypic assay using Ghost(3) cell lines.
Results
Regarding the 4 R5 HIV-2 clinical isolates tested, effective concentration 50% EC50 for cenicriviroc were 0.03, 0.33, 0.45 and 0.98 nM, similar to those observed with maraviroc: 1.13, 0.58, 0.48 and 0.68 nM, respectively. Maximum percentages of inhibition (MPI) of cenicriviroc were 94, 94, 93 and 98%, similar to those observed with maraviroc (93, 90, 82, 100%, respectively). The dual- and X4-tropic HIV-2 strains were resistant to cenicriviroc with EC50 >1000 nM and MPI at 33% and 4%, respectively.
Conclusions
In this first study assessing HIV-2 susceptibility to cenicriviroc, we observed an in vitro activity against HIV-2 R5-tropic strains similar to that observed with maraviroc. Thus, cenicriviroc may offer a once-daily treatment opportunity in the limited therapeutic arsenal for HIV-2. Clinical studies are warranted.
Introduction
Human Immunodeficiency Virus type 2 (HIV-2) was discovered few years after HIV type 1 (HIV-1). Currently, between one and two millions of people are estimated to be infected with HIV-2. HIV-2 is mainly present in West Africa including Guinea-Bissau, Gambia, Senegal, and Guinea. HIV-2 is also prevalent in some European countries, mainly in Portugal and France. HIV-2 is naturally resistant to non nucleoside reverse transcriptase inhibitors, the fusion inhibitor enfuvirtide, and has a decreased susceptibility to some of protease inhibitors including the once daily dosing atazanavir [1–3]. A limitation of CCR5 inhibitors use in HIV-2 may be the in vitro use of a broad range of coreceptors for this virus, including CCR5 and CXCR4 as well as other alternative coreceptors [4,5]. But some others studies demonstrated that these alternative coreceptors don’t seem to have a major role in viral entry [6–9]. Maraviroc, the only licensed CCR5 antagonist, has been recently shown to be active in vitro against HIV-2 R5-tropic viruses [10–12] and a limited number of case reports also showed an early in vivo virological response to maraviroc-based treatment [13–15]. Thus, CCR5 inhibitors may represent a new antiretroviral drug class in the HIV-2 limited arsenal as demonstrated in vitro with maraviroc.
Cenicriviroc is a new dual CCR5 and CCR2 antagonist. This new compound has completed Phase 2b development for treatment of HIV-1 infection (NCT01338883) [16–18]. Cenicriviroc is administered once daily and is suitable for single-tablet regimen combination products [19]. This point is of a particular interest, because of the complex and twice-daily dosing of maraviroc, and also because of the lack of once-daily dosing therapeutic regimen in HIV-2 due to the natural resistance of HIV-2 to non-nucleoside reverse transcriptase inhibitors and the decrease susceptibility to some protease inhibitors.
The aim of this study was to assess the in vitro HIV-2 phenotypic susceptibility to cenicriviroc.
Patients and Methods
Study Patients and Virus Stocks
Four R5-tropic, one dual-tropic and one X4-tropic HIV-2 strains, as well as one R5-tropic HIV-1 strain, have been assessed in this study. All these strains were primary clinical isolates issued from distinct HIV-2-infected maraviroc-naïve patients included in the French Agence Nationale de Recherches sur le SIDA et les hépatites virales (ANRS) HIV-2 cohort (CO5) and were isolated by co-cultivation of blood donor, phytohemagglutinin (PHA)-activated peripheral-blood mononuclear cells (PBMC) pool of healthy blood donors as previously described [20]. Written informed consent was provided by all patients at the time of inclusion in the French ANRS CO5 HIV-2 cohort. The ANRS CO5 cohort and substudies has been approved by the French institutional review board “Comité de Protection des Personnes (CPP)—Ile-de-France XI”.
Tropism Determination
Tropism was assessed for all strains by a tropism phenotypic assay (TPA) recently developed [6,21]. Briefly, this test use Ghost(3) cell lines, expressing CD4+CCR5 or CD4+CXCR4 coreceptors with HIV-2 Long Terminal Repeat-driven Green Fluorescent Protein (GFP) gene activated upon infection. Infection efficiency was quantified by flow cytometric analysis for GFP detection.
Phenotypic Susceptibility Determination
Phenotypic susceptibility of HIV-2 clinical isolates to cenicriviroc was determined using a slightly adapted ANRS PBMC method [22]. All of the strains have been previously tested for HIV-2 maraviroc susceptibility using the same protocol [4]. All susceptibility assays were performed in RPMI 1640 medium containing 10% (vol/vol) heat-inactivated fetal bovine serum (FBS), 2 mM L-glutamine, and 1000 U/mL penicillin/streptomycin. All PBMC were pooled from three healthy donors and incubated for 3 days in culture medium containing PHA. PHA-activated PBMC were washed and resuspended in medium containing human recombinant interleukin-2 immediately prior to use in an antiviral assay. Drug susceptibility assays were performed in 96-well tissue culture plates. Six-point dilution series of cenicriviroc (final concentrations at 0, 0.05, 0.5, 5, 50 and 500 nM) were prepared in culture medium. PHA-stimulated PBMC were pre-incubated with cenicriviroc at appropriate concentrations during 1 hour before viral infection. Cells were washed and infected with cell-free virus supernatant at 100 median tissue culture infective doses (TCID50) for 1 h at 37°C. Cells were subsequently washed once and 2.106 infected PBMC were added to each well of assay plates containing diluted compound. Plates were incubated for 4 days at 37°C in a humidified 5% CO2 (vol/vol) atmosphere. Each dilution was tested in quadruplicate. At day 4, viral supernatants were collected and virus replication was quantified by HIV-2 specific viral load measurement using the Generic HIV-2 RNA assay (Biocentric, Bandol, France). All viral supernatants were tested at 100 TCID50. The percentage of viral replication inhibition for each concentration of maraviroc was calculated to determine effective concentration 50% (EC50) and maximum percentage of inhibition (MPI).
V3 loop direct sequencing was performed on viral supernatants at time of phenotypic assay. No changes, between day 0 and day 4 of the adapted ANRS PBMC phenotypic susceptibility assay, were observed in gp105 V3 loop sequences in all tested HIV-2 clinical isolates.
Results
Dose response curves of HIV-2 strains with cenicriviroc are depicted in Fig 1.
Fig 1. Dose response curves for cenicriviroc-dependent inhibition (A) and maraviroc-dependent inhibition (B).
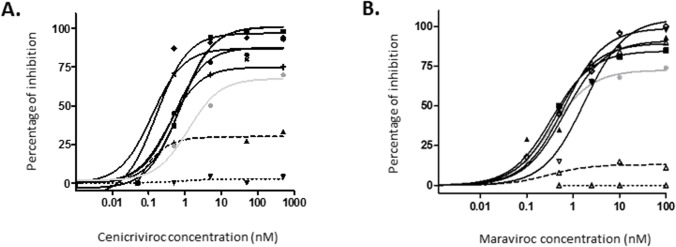
R5, dual and X4-tropic HIV-2 strains are depicted in continuous, dashed and dotted black lines, respectively. HIV-1 R5-tropic strain is depicted using the continuous gray line.
Phenotypic susceptibility testing showed, for the four R5-tropic HIV-2 isolates, a median EC50 for cenicriviroc of 0.39 nM (0.03, 0.33, 0.45 and 0.98 nM). These results were comparable to those observed in the same conditions with maraviroc with a median EC50 of 0.63 nM (respectively 1.13, 0.58, 0.48, 0.68 nM). Concerning MPI, R5-tropic HIV-2 strains presented a median MPI at 94% (94, 94, 93 and 98%), similar to those previously observed for maraviroc using the same protocol with a median MPI at 93% (respectively 93, 90, 82, 100%). The HIV-1 R5-tropic strain presented an EC50 at 5.06 nM with a MPI at 70%, similar to those observed for maraviroc at 3.69 nm and 74%.
The dual-tropic and the X4-tropic HIV-2 strains were resistant to cenicriviroc with EC50 at >1000 nM, and MPI at 33% and 4%, respectively. Similar results were observed for maraviroc with EC50 and MPI above 1000 nM, 12% and above 1000 nM, 0% for the dual-tropic and the X4-tropic, respectively.
Discussion
In the present study, we demonstrated, for the first time using PBMC assay, that cenicriviroc is active in vitro against HIV-2 R5-tropic strains with similar EC50 and MPI values to those obtained with maraviroc. HIV-2 EC50 for cenicriviroc were also similar to those previously described with HIV-1 (0.29 nM) [16].
Our study presents some limitations. The use of a PBMC phenotypic assay, with detection of viral replication at day 4, has already been depicted as unable to provide MPI at 100% (ref?). The lower MPI observed with this assay can be explained by the competitive fixation of cenicriviroc that allows a low amount of activated PBMC to be infected during the infection step. Thus, the low amount of infected cells is able to produce a non-negligible amount of viral particles at day 4 as observed with HIV-1 and HIV-2 in our previous study concerning maraviroc [10]. PBMC assays may have also intrinsic variability due to the PBMC healthy donors’ diversity. This point was taken into account in our assay by the use of a PBMC pool of three healthy donors at each experiment. Despite these limitations, PBMC assay was chosen for evaluation of HIV-2 susceptibility to entry inhibitors because of the possible use of a broad range of accessories coreceptors that is still debated in HIV-2 infection [4–9]. Thus, in our assay, the possible use of these accessories coreceptors doesn’t seem to compromise the cenicriviroc in vitro efficiency as results were similar to those obtained with HIV-1 for both maraviroc and cenicriviroc.
Cenicriviroc is a novel CCR5 inhibitor in development and clinical studies on cenicriviroc efficacy, tolerance and resistance profile in HIV-2 infection are warranted. However cenicriviroc has demonstrated in this study a similar efficiency in vitro against both HIV-2 and HIV-1. This new compound also presents a long half-life supporting once-daily dosing [23]. This point is of particular interest because of the complex dose management and the twice daily dosing of maraviroc, the only CCR5 inhibitor licensed to date. Cenicriviroc may offer new opportunities in the limited HIV-2 therapeutic arsenal.
Acknowledgments
We wish to thank all the investigators of the HIV-2 Cohort Study (ANRS CO5 VIH-2) listed below:
Investigator coordinator and lead coordinator: S. Matheron (sophie.matheron@aphp.fr, Hôpital Bichat-Claude Bernard, Paris)
Virological coordination: F. Brun-Vezinet, F. Damond, D. Descamps (Hôpital Bichat-Claude Bernard, Paris)
Methodological coordination: G. Chêne, C. Fagard (INSERM U897, Bordeaux)
Immunological coordination: B. Autran (Hôpital Pitié-Salpétrière, Paris)
Clinical investigators:
Marie Caroline Meyohas (Hôpital St Antoine—Paris); Salmon Dominique (Hôpital Cochin—Paris); Agathe Rami (Hôpital Lariboisière—Paris); Gilles Pialoux (Hôpital Tenon—Paris); Sophie Matheron (Hôpital Bichat Claude Bernard—Paris); Roland Tubiana (Hôpital Pitié Salpétrière—Paris); Pierre De Truchis (Hôpital Raymond Poincarré—Garches); Marie-Aude Khuong-Josses (Hôpital Delafontaine—Saint Denis); Ouda Derradji (Hôpital Bicêtre—Le Kremlin Bicêtre); Didier Troisvallet (CH de Gonesse—Gonesse); Françoise Timsit (Hôpital Saint Louis—Paris); Vincent Jeantils (Hôpital Jean Verdier—Bondy); Olivier Patey (CH de Villeneuve Saint Georges—Villeneuve Saint Georges); Cécile Goujard (Hôpital de Bicêtre—Le Kremlin Bicêtre); Laurence Gérard (Hôpital Saint Louis—Paris); Emmanuel Mortier (Hôpital Louis Mourier—Colombes); Eric Froguel (Hôpital de Lagny—Marne la Vallée—Jossigny); Yves Welker (Hôpital de Saint Germain en Laye—St Germain en Laye); Vincent Daneluzzi (Hôpital Max Fourestier—Nanterrre); Philippe Genet (CH Victor Dupouy—Argenteuil); Annie Leprêtre (CH d'Eaubonne-Montmorency—Eaubonne); David Zucman (Hôpital Foch—Suresnes); Gilles Force (Hôpital du Notre Dame du Perpétuel Secours—Levallois Perret); Anne Coutellier-Simon (Hôpital Pitié Salpétrière—Paris); Jean Deleuze (Hôpital Cochin—Paris); Laurence Weiss (Hôpital Européen Georges Pompidou—Paris Cédex 15); Valérie Garrait (CHIC de Créteil—Créteil); Hélène Masson Colin (Hôpital de Poissy—Poissy); Laurent Blum (CH René Dubos—Pontoise); Jean Daniel Lelièvre (Hôpital Henri Mondor—Créteil); Anne Dulioust (Centre Médico—Chirurgical de Bligny—Briis sous Forge); Véronique Perrone (Hôpital François Quesnay—Mantes la jolie); Cécile Winter (CHIC André Grégoire—Montreuil); Iram La Torre (CH de l'agglomération Montargeoise—Amilly—Montargis); Jean-Marc Zini (Hôpital Lariboisière—Paris); Jean Michel Molina (Hôpital Saint Louis—Paris); Patrick Brunet (Etablissement public de santé—Arpajon); Alain Devidas (CH Sud Francilien—Corbeil Essonnes); Elisabeth Rouveix (Hôpital Ambroise Paré—Boulogne Billancourt); Serge Kernbaum (Hôpital Américain de Paris—Neuilly sur Seine); Claudine Duvivier (Institut Pasteur—Paris); Jean-Luc Delassus (Hôpital Roger Balanger—Aulnay sous Bois); Loïc Bodard (Hôpital Mutualiste Montsouris—Paris); Patrick Imbert (Hôpital Bégin—Saint Mandé); François Boué (Hôpital Antoine Béclère—Clamart); Agnès Uludag (Hôpital Beaujon—Clichy); Fabrice Chaix (Hôpital de Longjumeau—Longjumeau); Olivier Bouchaud (Hôpital Avicenne—Avicenne); Jean Paul Viard (Hôpital Hôtel Dieu—Paris); Xavier Duval (Hôpital Bichat Claude Bernard—Paris); Alix Greder Belan (Hôpital André Mignot—Versailles); Marc Gatfossé (Hôpital René Arbeltier—Coulommiers); Geneviève Beck-Wirth (CH de Mulhouse—Mulhouse); François Raffi (Hôpital de l'Hôtel-Dieu—Nantes); Christine Drobacheff (Hôpital Minjoz—Besançon); Thierry Allègre (CH du Pays d'Aix—Aix en Provence); Renaud Verdon (Hôpital de la Côte de Nacre—Caen); Isabelle Poizot-Martin (Centre Hospitalier Régional—Marseille); Corinne Daniel (CHU de Saint Brieuc—Saint Brieuc); Dominique Merrien (CH de Compiègne—Compiègne); Bruno Marchou (CHU Toulouse—hôpital Purpan—Toulouse); Louis Bernard (Hôpital Bretonneau—Tours); Yasmine Debab (Hôpital Charles Nicolle—Rouen); Pascale Leclercq (Hôpital Michallon—Grenoble); Joelle Julien (Hôpital Jacques Cœur—Bourges); Jacques Moreau (Hôpital Nord—Marseille); Cédric Arvieux (Hôpital Pontchaillou—Rennes); Bertrand Héry (CH de Saint Nazaire—Saint Nazaire); Djamila Makhloufi (Hôpital Edouard Herriot—Lyon); Gwenael Lemoal (Hôpital Jean Bernard—Poitiers); Philippe Arsac (CHR d'Orléans—Orléans); Alain Lafeuillade (Hôpital Sainte Musse—Toulon); Didier Neau (Hôpital Pellegrin—Tripode—Bordeaux); Benoit Martha (CH William Morey—Chalon-sur-Saône); Philippe Perré (CHD de La Roche sur Yon—La Roche sur Yon); Faïza Ajana (Hôpital Gustave Dron—Tourcoing); Philippe Morlat (Hôpital Saint—André—Bordeaux); Elisabeth Brottier—Mansini (Hôpital de la Rochelle—La Rochelle); Michel Dupon (Hôpital Pellegrin—Bordeaux); Jean Luc Pellegrin (Hôpital du Haut—Lévêque—Bordeaux Pessac); Nathalie Montagne (Hôpital de Cannes—Cannes); Thierry May (CHU de Nancy Brabois—Vandoeuvre les Nancy); Wille Heidi (CH de la Côte basque—Bayonne); Douadi Youceff (CH de Saint Quentin—St Quentin); Jacques Reynes (CHU Montpellier—Montpellier); Jean Luc Schmit (CHU d'Amiens—Hôpital Nord—Amiens); Christine Rouger (Hôpital Robert Debré—Reims); Yves Poinsignon (CH Prosper Chubert—Vannes); Christine Jacomet (Hôpital de l'Hôtel Dieu—Clermont Ferrand); Lionel Piroth (Hôpital du Bocage—Dijon); Eric Rosenthal (Hôpital de l'Archet CHU de Nice—Nice); Daniel Garipuy (Hôpital Joseph Ducuing—Toulouse); Agnès Riché (CH d'Angoulême—Angoulême); François Prevoteau du Clary (Hôpital la Grave—Toulouse); Alissa Naqvi (Hôpital de l'Archet—Nice); Jacques Gaillat (CH Annecy-Genévois—Annecy); David Rey (Hôpital de jour—Strasbourg); Jean- François. Abino (Hôpital Eugénie—Ajaccio); Patrick Mercié (Hôpital St André—Bordeaux); Pierre Marie Roger (CH de Draguignan—Draguignan); Aïssi Emmanuelle (CH Dr Schaffner—Lens); Jean-Marie Chennebault (CHU d’Angers—Angers); Dominique Liné (CH de Soissons—Soissons); Narimamy Randrianasolo (Hôpital de Chaumont—Chaumont); Catherine Merle (Hôpital de Vesoul—Vesoul); Patricia Granet (CH de Dignes Les Bains—Dignes Les Bains); Laurence Caunègre (CH de Dax—Dax); Laurent Cotte (Hôpital de la Croix Rousse—Lyon); Julien Boileau (CH du Pays de Molaix—Morlaix); Pascale Perfezou (Hôpital Laennec—Quimper); Nicolas Lefebvre (Nouvel Hôpital Civil—CHU Strasbourg—Strasbourg); Jean-Philippe Talarmin (Hôpital Laennec—Quimper); Haffner-Mauvais (CHU Besançon-Hôpital Minjoz—Besançon); Eric Libbrecht (CH de Troyes—Service Infectiologie—Troyes); Ponceau Bénédicte (CH de Valence-—Valence)
Biologist and Virological investigators: F. Brun-Vézinet, F. Damond, D. Descamps (Bichat, Paris); F. Simon (St-Louis, Paris); L. Morand-Joubert (St-Antoine, Paris); D. Cottalorda (F. de médecine, Nice); JM Delarbre (Mulhouse);V. Ferre (Institut de biologie, Nantes); D Bettinger (Besançon); P. Barin (Bretonneau, Tours); TT Le THI (Université, Lyon); E Lagier (Aix-en-provence); A Vabret (Caen); C. Tamalet (Sainte Marguerite, Marseille); C Espanel (Compiegne); P Guinot (Bourg-en-Bresse); J Izopet, F Nicot (Purpan, Toulouse); Dr Barin (Tours); JC Plantier (C. Nicolle, Rouen); A Signoris-Schmuck (Grenoble); N Tivoli (La Timone, Marseille); A Maillard (Pontchaillou, Rennes); S Sachot-Ollivier (Saint Nazaire); P. Agius, G Giraudeau (Poitiers); C Poggi, M Chouraqui (Centre intercommunal, Toulon); P. Fleury (Pellegrin-Tripode, Bordeaux); B Martha, A Goux (Chalon-sur-soane); AS Poirier (LaRoche-Sur-Yon); M Marty (La Rochelle); V Vénard (Vandoeuvre-les-Nancy); MC Gemain (Bayonne); MT Albertini (Saint Quentin); B. Montes (St-Eloi, Montpellier); C Segard (Amiens); V Brodart (Reims); P Pouedras (Vannes); C Regagnon (Clermond-Ferrand); JB Bourg (Dijon); L Bocket (Lille); B Chanzy (Pringy); E Schvoerer (Nancy); S Fafi-Kremer (Strasbourg); P Valayer (Ajaccio); C Bouquigny (Soissons); MJ Carles (Nimes); C Alba-Sauviat (Chaumont); N Azencott (Dignes-les-bains).
Data management and statistical analysis: A Besseghir, N Chaghil-Boissière, M Naudin, L Pinoges, D Touchard (INSERM U897, Bordeaux).
Data Availability
All relevant data are within the paper.
Funding Statement
Tobira Pharmaceutics graciously provided cenicriviroc. All the other experimental costs were supported by the French National Agency for Research on AIDS and Viral Hepatitis (ANRS). Tobira Pharmaceutics provided support in the form of salaries for author E.L., but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific role of this author is articulated in the ‘author contributions’ section.
References
- 1. Poveda E, Briz V, Soriano V. Enfuvirtide, the first fusion inhibitor to treat HIV infection. AIDS Rev. 2005;7: 139–147. [PubMed] [Google Scholar]
- 2. Ren J, Bird LE, Chamberlain PP, Stewart-Jones GB, Stuart DI, Stammers DK. Structure of HIV-2 reverse transcriptase at 2.35-A resolution and the mechanism of resistance to non-nucleoside inhibitors. Proc Natl Acad Sci USA. 2002;99: 14410–14415. 10.1073/pnas.222366699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Desbois D, Roquebert B, Peytavin G, Damond F, Collin G, Bénard A, et al. In vitro phenotypic susceptibility of human immunodeficiency virus type 2 clinical isolates to protease inhibitors. Antimicrob Agents Chemother. 2008;52: 1545–1548. 10.1128/AAC.01284-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Visseaux B, Charpentier C, Hurtado-Nedelec M, Storto A, Antoine R, Peytavin G, et al. In vitro phenotypic susceptibility of HIV-2 clinical isolates to CCR5 inhibitors. Antimicrob Agents Chemother. 2012;56: 137–139. 10.1128/AAC.05313-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Espírito-Santo M, Santos-Costa Q, Calado M, Dorr P, Azevedo Pereira JM. Susceptibility of HIV-2 primary isolates to CCR5 and CXCR4 monoclonal antibodies, ligands and small molecule inhibitors. AIDS Research and Human Retroviruses. 2011; 10.1089/AID.2011.0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Borrego P, Calado R, Marcelino JM, Bártolo I, Rocha C, Cavaco-Silva P, et al. Baseline susceptibility of primary HIV-2 to entry inhibitors. Antivir Ther (Lond). 2012;17: 565–570. 10.3851/IMP1996 [DOI] [PubMed] [Google Scholar]
- 7. Armstrong-James D, Stebbing J, Scourfield A, Smit E, Ferns B, Pillay D, et al. Clinical outcome in resistant HIV-2 infection treated with raltegravir and maraviroc. Antiviral Res. 2010;86: 224–226. 10.1016/j.antiviral.2010.02.324 [DOI] [PubMed] [Google Scholar]
- 8. Stegmann S, Manea ME, Charpentier C, Damond F, Karmochkine M, Laureillard D, et al. Foscarnet as salvage therapy in HIV-2-infected patient with antiretroviral treatment failure. J Clin Virol. 2010;47: 79–81. 10.1016/j.jcv.2009.11.008 [DOI] [PubMed] [Google Scholar]
- 9. Caixas U, Ferreira J, Marinho AT, Faustino I, Grilo NM, Lampreia F, et al. Long-term maraviroc use as salvage therapy in HIV-2 infection. J Antimicrob Chemother. 2012;67: 2538–2539. 10.1093/jac/dks240 [DOI] [PubMed] [Google Scholar]
- 10. Calado M, Matoso P, Santos-Costa Q, Espirito-Santo M, Machado J, Rosado L, et al. Coreceptor usage by HIV-1 and HIV-2 primary isolates: the relevance of CCR8 chemokine receptor as an alternative coreceptor. Virology. 2010;408: 174–182. 10.1016/j.virol.2010.09.020 [DOI] [PubMed] [Google Scholar]
- 11. Mörner A, Björndal A, Albert J, Kewalramani VN, Littman DR, Inoue R, et al. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J Virol. 1999;73: 2343–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blaak H, Boers PHM, Gruters RA, Schuitemaker H, van der Ende ME, Osterhaus ADME. CCR5, GPR15, and CXCR6 are major coreceptors of human immunodeficiency virus type 2 variants isolated from individuals with and without plasma viremia. J Virol. 2005;79: 1686–1700. 10.1128/JVI.79.3.1686-1700.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McKnight A, Dittmar MT, Moniz-Periera J, Ariyoshi K, Reeves JD, Hibbitts S, et al. A broad range of chemokine receptors are used by primary isolates of human immunodeficiency virus type 2 as coreceptors with CD4. J Virol. 1998;72: 4065–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mörner A, Björndal A, Leandersson A- C, Albert J, Björling E, Jansson M. CCR5 or CXCR4 is required for efficient infection of peripheral blood mononuclear cells by promiscuous human immunodeficiency virus type 2 primary isolates. AIDS Res Hum Retroviruses. 2002;18: 193–200. 10.1089/08892220252781248 [DOI] [PubMed] [Google Scholar]
- 15. Zhang Y, Lou B, Lal RB, Gettie A, Marx PA, Moore JP. Use of inhibitors to evaluate coreceptor usage by simian and simian/human immunodeficiency viruses and human immunodeficiency virus type 2 in primary cells. J Virol. 2000;74: 6893–6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lalezari J, Gathe J, Brinson C, Thompson M, Cohen C, Dejesus E, et al. Safety, efficacy, and pharmacokinetics of TBR-652, a CCR5/CCR2 antagonist, in HIV-1-infected, treatment-experienced, CCR5 antagonist-naive subjects. J Acquir Immune Defic Syndr. 2011;57: 118–125. 10.1097/QAI.0b013e318213c2c0 [DOI] [PubMed] [Google Scholar]
- 17.Gathe J, Cade J, DeJesus E, Feinberg J, Lalezari J, Morales-Ramirez J, et al. Week 24 Primary Analysis of Cenicriviroc vs Efavirenz, in Combination with FTC/TDF, in Treatment-naïve HIV-1 Infected Adults with CCR5-tropic Virus. 20th Conference on Retroviruses and Opportunistic Infections (CROI), March 2013, Atlanta, GA, USA. Abstract 106LB. Available: http://www.retroconference.org/2013b/abstracts/48023.htm.
- 18.Feinberg J, Thompson M, Cade J, DeJesus E, Gathe J, Lalezari J, et al. Final Week 48 Analysis of Cenicriviroc (CVC) Compared to Efavirenz (EFV), in Combination with Emtricitabine/Tenofovir (FTC/TDF), in Treatment-Naïve HIV-1-Infected Adults with CCR5-Tropic Virus. 14th European AIDS Conference (EACS), October 2013, Brussels, Belgium. Abstract 202.
- 19. Menning MM, Dalziel SM. Fumaric acid microenvironment tablet formulation and process development for crystalline cenicriviroc mesylate, a BCS IV compound. Mol Pharm. 2013;10: 4005–4015. 10.1021/mp400286s [DOI] [PubMed] [Google Scholar]
- 20. Roquebert B, Damond F, Collin G, Matheron S, Peytavin G, Bénard A, et al. HIV-2 integrase gene polymorphism and phenotypic susceptibility of HIV-2 clinical isolates to the integrase inhibitors raltegravir and elvitegravir in vitro. J Antimicrob Chemother. 2008;62: 914–920. 10.1093/jac/dkn335 [DOI] [PubMed] [Google Scholar]
- 21. Visseaux B, Hurtado-Nedelec M, Charpentier C, Collin G, Storto A, Matheron S, et al. Molecular determinants of HIV-2 R5–X4 tropism in the V3 loop: development of a new genotypic tool. Journal of infectious diseases. 2012. [DOI] [PubMed] [Google Scholar]
- 22. Brun-Vézinet F, Ingrand D, Deforges L, Gochi K, Ferchal F, Schmitt MP, et al. HIV-1 sensitivity to zidovudine: a consensus culture technique validated by genotypic analysis of the reverse transcriptase. J Virol Methods. 1992;37: 177–188. [DOI] [PubMed] [Google Scholar]
- 23. Marier J-F, Trinh M, Pheng LH, Palleja SM, Martin DE. Pharmacokinetics and pharmacodynamics of TBR-652, a novel CCR5 antagonist, in HIV-1-infected, antiretroviral treatment-experienced, CCR5 antagonist-naïve patients. Antimicrob Agents Chemother. 2011;55: 2768–2774. 10.1128/AAC.00713-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.


