Abstract
Interspecies transmission of prions is a well-established phenomenon, both experimentally and under field conditions. Upon passage through new hosts, prion strains have proven their capacity to change their properties and this is a source of strain diversity which needs to be considered when assessing the potential risks associated with consumption of prion contaminated protein sources. Rabbits were considered for decades to be a prion resistant species until proven otherwise recently. To determine the extent of rabbit susceptibility to prions and to assess the effects of passage of different prion strains through this species a transgenic mouse model overexpressing rabbit PrPC was developed (TgRab). Intracerebral challenges with prion strains originating from a variety of species including field isolates (ovine SSBP/1 scrapie, Nor98- scrapie; cattle BSE, BSE-L and cervid CWD), experimental murine strains (ME7 and RML) and experimentally obtained ruminant (sheepBSE) and rabbit (de novo NZW) strains were performed. On first passage TgRab were susceptible to the majority of prions (Cattle BSE, SheepBSE, BSE-L, de novo NZW, ME7 and RML) tested with the exception of SSBP/1 scrapie, CWD and Nor98 scrapie. Furthermore, TgRab were capable of propagating strain-specific features such as differences in incubation periods, histological brain lesions, abnormal prion (PrPd) deposition profiles and proteinase-K (PK) resistant western blotting band patterns. Our results confirm previous studies proving that rabbits are not resistant to prion infection and show for the first time that rabbits are susceptible to PrPd originating in a number of other species. This should be taken into account when choosing protein sources to feed rabbits.
Author Summary
Prions, the infectious agents responsible for causing mad cow disease, amongst other diseases, can transmit from one species to another. For example, Bovine Spongiform Encephalopathy can transmit to humans resulting in invariably fatal variant Creutzfeldt-Jakob Disease. We wanted to study the susceptibility of rabbits as, until recently, they were considered a prion resistant species. Once proven otherwise, we wanted to know which particular prions rabbits were susceptible to. With this aim, a transgenic mouse was designed expressing the rabbit prion protein gene instead of the corresponding mouse gene to model the transmission barrier between rabbits and other species. The resultant mice where challenged with several field prion isolates including classical and atypical strains of Bovine Spongiform Encephalopathy, sheep Scrapie and cervid Chronic Wasting disease. The transgenic mice were susceptible to classical and atypical Bovine Spongiform Encephalopathy prions and also to mouse-adapted Scrapie prions. This information must be taken into account when assessing the risk of using ruminant derived protein as a protein source to feed rabbits.
Introduction
Prions are protein based, genome devoid, infectious agents causing Transmissible Spongiform Encephalopathies (TSEs), a group of diseases classified as transmissible protein misfolding disorders [1,2]. Prions show a remarkable ability for interspecies transmission. Initially, a species barrier was defined, but extensive field and experimental evidence has been published proving that interspecies prion transmission is not an isolated phenomenon [3–6]. Interspecies transmission of prions has resulted in the generation of significant prion strain diversity and its incidence has been documented worldwide [3,4,7–10].
The existence of prion diseases has been documented for centuries with the earliest reports of scrapie cases dating back to 1732 [11]. In the last seven decades prions were also reported in other animal species, usually in the form of outbreaks, which somehow involved human intervention. Namely classical bovine spongiform encephalopathy (BSE-C) [12], feline spongiform encephalopathy (FSE) [13] and transmissible mink encephalopathy (TME) [14]. Humans can also be included in the list of TSE susceptible species due to the Fore tribe from Papua New Guinea suffering from Kuru [15] or the relatively newly created variant Creutzfeldt-Jakob disease (vCJD)[3]. Cervidae is another family of animals currently affected by a, yet uncontrolled, epizooty: chronic wasting disease (CWD) [16]. Although classical animal prion disease strains, as opposed to the so called atypical prion disease strains [17–20], have been documented for at least three centuries [11], sporadic spontaneous generation of atypical prions has probably existed for as long as susceptible species have been present in large enough numbers for the spontaneous event to occur. Currently there is no evidence to suggest that any mammalian species cannot undergo a spontaneous disease-linked prion protein misfolding event [21] as long as there are sufficient numbers of individuals with the necessary lifespan.
Although the mechanisms of interspecies prion transmission remain unknown, in vitro and in vivo studies have shown that species particularly susceptible to certain prion strains can actually be resistant to others which originated in the same or different species [9,21–28]. The ability of prions to adapt to new species and even generate new strains with pathobiological properties different from the original one is not an isolated phenomenon [9,27,29,30]. Therefore new prion strains may arise with the ability to infect new species previously considered resistant.
Normal cellular prion protein (PrPC) is a host encoded protein, particularly abundant in nerve cells, which when misfolded is believed to acquire pathological properties leading to TSE neurodegenerative disease [1]. Several studies argue that certain species specific amino acid sequences of PrPC may render some species less susceptible to TSE [31–33] due to them being less prone to misfolding. This, along with absence of experimental evidence or TSE field cases described, led to belief that dogs, horses and rabbits (leporids) were more resistant to prion infection than other mammalian species [34,35].
Leporids have been the most intensely studied, both in vivo and in vitro, of the presumed prion disease resistant species. This is probably because rabbits are consumed by humans and also due to their comparatively small size and long lifespan which facilitates their use as experimental animals. Our group has proved recently, in contrast with the last three decades of reports, that rabbits are susceptible to prion diseases. Using protein misfolding cyclic amplification (PMCA), inocula where generated in vitro which were infectious and transmissible in this species [23] and more recent studies have proven that rabbit PrPC has a misfolding ability comparable to other species as BSE prions have been shown to retain their in vivo strain properties after misfolding rabbit PrPC [36]. Houdebine’s group studied whether the genetic background of rabbits was responsible for their apparent prion resistance generated transgenic rabbits expressing ovine PrPC. Upon inoculation with scrapie, these rabbits succumbed to prion disease further proving that leporids are not resistant to prion disease [37].
In the present paper we report an extensive evaluation of the susceptibility of TgRab mice to a variety of prion strains by means of in vivo experiments. A transgenic murine model has been generated ad hoc for this purpose which overexpresses the leporid PRNP on a mouse Prnp-null background. This model, denoted TgRab, has already been shown to correlate well with the rabbit model [23]. Our results show the susceptibility of rabbits has been vastly underestimated previously and that they behave similarly to other species whose vulnerability and/or resistance to prion disease also varies depending upon the prion disease strain encountered.
Results
Generation of TgRab: an in vivo model to evaluate rabbit susceptibility to prion infection
Even though the actual rabbit model would be more suitable for this purpose there are several significant limitations (size, cage space in biocontainment conditions, lifespan, expression levels, and budget required) that are easily overcome by using a transgenic mouse model and such models have been of great use within the field of prion research. Based on our previous experience a new mouse line was generated by pronuclear injection of a construct consisting of the moPrP promoter and the rabbit PrP sequence. From a total of seven positive animals identified from the 83 pups obtained, five animal founders transmitted the transgene to their progeny. After backcrossing to a line that did not express endogenous PrP (STOCK-Prnptm2Edin), expression levels of the transgene were analyzed by western blot. Two out of five transgenic lines expressed PrP at higher levels than the endogenous gene. However, only hemizygous line 58 showed a consistent expression pattern of 5x-6x that of the endogenous rabbit prion protein level and 10x-12x that of the endogenous mouse prion protein level (S2 Fig). This line was selected for further studies. The low expressing lines were discarded since PrPC expression levels were lower than those found in WT rabbits and this would predictably diminish their susceptibility to prions.
Preliminary in vitro challenge to assess rabbit susceptibility
In previous experiments normal rabbit brain homogenate was seeded in vitro with different prion strains before applying serial automated PMCA (saPMCA) to determine the ability of rabbit PrPC to be converted by different PrPres conformations. The results of some of these experiments have been reported previously such as seeding with cattle BSE which generated BSE-RaPrPres [36]. Additional prion isolates were included in the present work, which successfully misfolded rabbit PrPC in vitro including SSBP/1 sheep scrapie, ME7 and RML murine adapted scrapie strains and CWD. The following rabbit adapted strains were generated respectively: SSBP/1-RaPrPres, ME7-RaPrPres, RML-RaPrPres and CWD-RaPrPres (S1 Fig) [23]. Spontaneously misfolded PrPres was also obtained from unseeded normal rabbit brain homogenates and named de novo RaPrPres. This spontaneous strain has been demonstrated to be infectious to rabbits [23]. Despite saPMCA not being a quantitative method, rabbit PrPC appeared to be quite susceptible to misfolding since all seeds tested were able to generate PK-resistant RaPrPres by or before round 7 and the unseeded homogenate produced RaPrPres by round 13 [23]. All in vitro-derived RaPrPres products were easily amplified further in vitro.
The western blotting migration pattern of the obtained RaPrPres, particularly the unglycosylated band, was similar to the strains of origin used in the bovine and ovine strains tested.
Accordingly, the following isolates were selected for in vivo challenge: BSE-C, SSBP/1, ME7, RML and CWD. Additionally, we included L-type atypical BSE (BSE-L) and Nor98 Atypical scrapie and the PMCA obtained de novo RaPrPres. The rationale for including the latter, in vitro generated, PrPres was that it was able to infect the natural host i.e. rabbits, our species of study [23], de novo RaPrPres was the only positive control available. Finally, de novo NZW prions (obtained from rabbits infected with de novo RaPrPres) were also inoculated into this transgenic mouse model and even though most of these results have already been published [23], some of them are discussed in the present paper.
A valid model to study prion susceptibility and transmission in rabbits
As reported for rabbits showing very long incubation times (766 dpi) and a 33% attack rate (1/3) [23], TgRab mice were also susceptible to de novo RaPrPres with a low attack rate (1/11) and a rather long incubation period of 604 dpi. However, upon inoculation with rabbit in vivo-adapted de novo RaPrPres (de novo NZW) the TgRab mice developed a 100% attack rate (8/8) with a shortened incubation period of 256 (±5) dpi (Table 1 and Fig 1). The same rate was obtained by Chianini et al. [23] in rabbits inoculated (in second passage) with this prion strain (Table 1). In vivo experimental challenges in rabbits and TgRab mice have shown a good correlation making the transgenic mouse model overexpressing rabbit PrPC a valid model to study rabbit prion susceptibility.
Table 1. Comparison of TgRab mice and actual rabbit bioassays.
| Animal model inoculated | PrP species of origin in inoculum | Strain origin | Attack rate | Survival time of positive animals (dpi) (±SEM) | |
|---|---|---|---|---|---|
| de novo RaPrPres | tgRab | Rabbit | PMCA | 1/11 | 604 |
| de novo—NZW | tgRab | Rabbit | Experimental | 8/8 | 265 (±5) |
| de novo RaPrPres | NZW Rabbit a | Rabbit | PMCA | 1/3 | 766 |
| de novo—NZW | NZW Rabbit | Rabbit | Experimental | 10/10 | 612 (±7) |
a Result published in Chianini et al. 2012. NZW: New Zealand White.
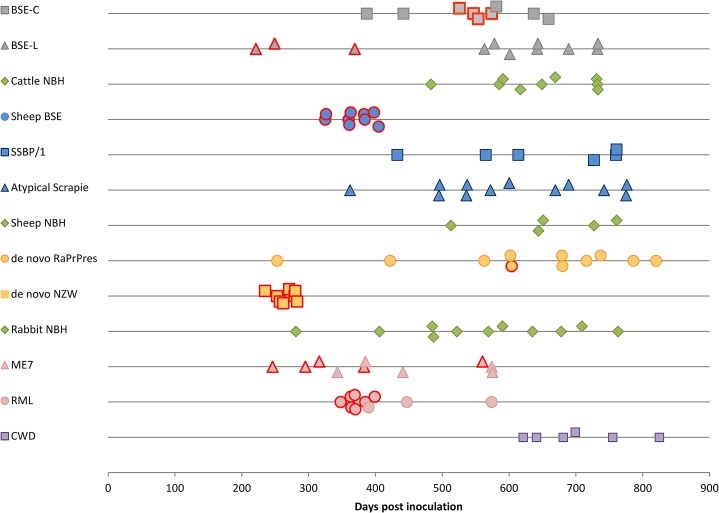
Fig 1. TgRab survival times after inoculation.
Each dot corresponds to a mouse. Dots with a red margin represent TSE positive animals. Green dots are negative controls; grey dots correspond to cattle strains; blue dots to sheep strains; yellow dots to rabbit originating strains; pink dots are murine strains and purple dots to CWD strain.
A model susceptible to bovine, ovine and murine prions
Even though rabbits had been considered resistant to prion infection until recently [23], TgRab mice could be infected with a number of the prions tested. Prions originating from BSE, i.e. cattle BSE-C and sheep BSE-C, were both infectious (Table 2). Cattle BSE-C showed an attack rate of 44.4% with an incubation period of 551(±10) dpi. Interestingly Sheep BSE-C showed a 100% attack rate and a significantly shortened incubation period of 368(±12) dpi (P = 0.0069, Mann-Whitney test) without previous adaptation to rabbit compared to cattle BSE-C (Fig 1). This supports, once again, the idea that after passage through sheep BSE-C shows enhanced virulence [29].
Table 2. Attack rates and survival times (±SEM) of the inoculated TgRab mice.
| PrP species of origin in inoculum | Strain origin | Attack rate a | Survival time of positive animals (dpi) (mean±SEM) | |
|---|---|---|---|---|
| BSE-C | Cattle | Field isolate | 4/9 (44%) | 551 (±10) |
| Sheep BSE | Sheep | Experimental | 9/9 (100%) | 368 (±10) |
| BSE-L | Cattle | Field isolate | 3/11(27%) | 280 (±26) |
| SSBP/1 | Sheep | Field isolate | 0/6 | 775 b |
| Atypical Scrapie | Sheep | Field isolate | 0 c /12 | 775 b |
| CWD | Cervid | Field isolate | 0/6 | 825 b |
| ME7 | Murine | Experimental | 5/10 (50%) | 360 (±41) |
| RML | Murine | Experimental | 7/10 (70%) | 371 (±6) |
| Cattle NBH | Cattle | Negative Control | 0/9 | 810 b |
| Rabbit NBH | Rabbit | Negative Control | 0/11 | 760 b |
| Sheep NBH | Sheep | Negative Control | 0/5 | 761 b |
aAnimals were considered TSE positive when spongiform lesions and/or PrPd was detected, either through IHC, WB or ELISA. Inconclusive results were confirmed by using more than one PrPd detection method. ND: Not determined, SEM: Standard Error of the Mean, NBH: Normal Brain Homogenate.
bAll animals TSE negative. Age at death of the oldest animal of the group (spontaneous deaths or humanely euthanized due to non-TSE causes). Individual survival times are shown in Fig 1.
cOne animal, euthanized at 742dpi, showed a positive result to PrPd ELISA but negative to the remaining tests (WB, IHC, HP) a second passage is ongoing.
The picture with scrapie-originating prion isolates was quite different. SSBP/1 prions were not able to infect TgRab (mice survived for longer than 750 dpi). Two other murine adapted classical scrapie prion sources were tested, ME7 and RML, and both strains readily infected TgRab mice with attack rates of 50% and 70% and incubation periods of 360(±41) and 371(±6) dpi, respectively (Fig 1). Therefore, prion strains originating from classical scrapie were transmissible to TgRab mice but only after being adapted previously to rodents. This situation is similar to that found with CWD which will infect hamsters readily after passage through ferrets [9].
Challenge with atypical ovine and bovine prions
The new TgRab model was further characterized by testing its susceptibility to atypical prion strains using the more frequent isolates for each species, BASE (BSE-L, cattle) and Nor89 (sheep). TgRab mice were resistant to infection on first passage with atypical scrapie prions (living up to 775 dpi) (Fig 1) with one exception: a single animal (euthanized at 742 dpi) showed a positive result for PrPd by ELISA but was negative when examined by western blotting and IHC and showed no TSE related spongiform change. A second passage is ongoing to determine if this animal was truly infected.
A 27% attack rate was present in the group inoculated with BSE-L with a mean incubation period of 280(±26) dpi, a similar rate to that of cattle BSE-C but with a much shorter incubation period (the number of positive animals per group was too low to assess statistical significance).
TgRab are not susceptible to CWD on first passage
As mentioned before, in vitro adaptation of CWD prion to rabbit PrPc was successful which indicated a potential susceptibility of rabbits in vivo. However, TgRab mice inoculated with CWD did not show any indication of a TSE on first passage, living up to 825 dpi (Fig 1 and Table 2). A second passage is ongoing to confirm these results.
TgRab mice: a model able to propagate distinct prion strains
Biochemical and neuropathological characterization of the brains of the inoculated mice strongly suggests that TgRab mice are not only susceptible to multiple prion strains but are also able to maintain their strain features.
Western blotting analysis of TgRab brain homogenates after protease K digestion revealed the characteristic three-band pattern with a predominant diglycosylated band and a 19-20kDa unglycosylated band in mice inoculated with BSE-derived strains (Fig 2). The brains of mice inoculated with RML showed a typical predominance of the monoglycosylated band and a 21kDa unglycosylated band. As shown in the 12B2 antibody developed membrane only ME7 and RML inoculated mice fully maintained the N-terminus specific epitope after PK digestion (Fig 2). The migration pattern of the bands from mice inoculated with de novo strains, both with the in vitro generated (de novo-RaPrPres) and the one obtained from NZW rabbits (de novo NZW), was constant and showed a similar pattern to BSE-C even though, as shown later, the immunohistochemical features differed completely. No bands were observed in western blots of brains of mice inoculated with SSBP/1, Atypical scrapie nor CWD or in any of the negative controls.
Fig 2. Biochemical analysis of brain homogenates from TgRab mice inoculated with different prion strains.
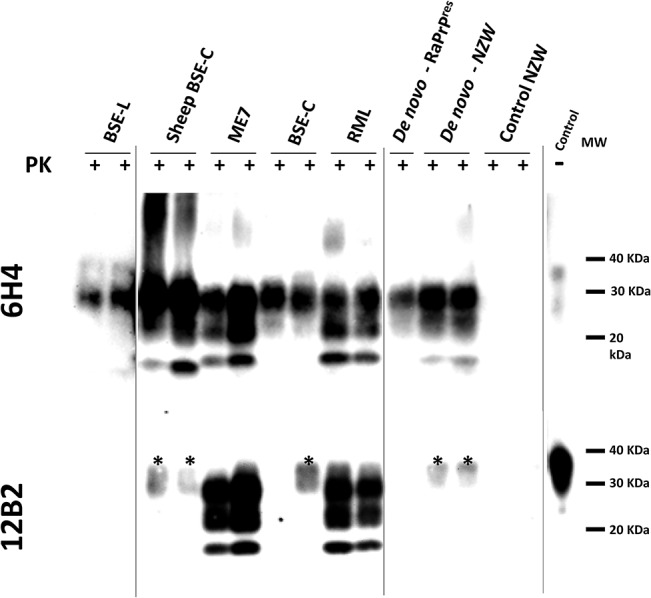
Two representative brain homogenates (per group) from TgRab mice inoculated with different prion strains [cattle: BSE-L and BSE-C; mouse: RML and ME7; sheep: SSPB/1, sheep BSE-C and atypical scrapie; deer: CWD; rabbit: de novo–RaPrPres (in vitro sample) and de novo–NZW (in vivo sample)] were digested with 100 μg/ml of proteinase K (PK) and analyzed by western blot using two different monoclonal antibodies (upper blot- 6H4 and lower blot– 12B2). Differential electrophoretic migrations and glycosylation patterns observed are consistent with the origin of the prion strains used for inoculation. Control NZW: Normal rabbit brain homogenate. MW: Molecular weight. Vertical lines separate blots with different exposition times.
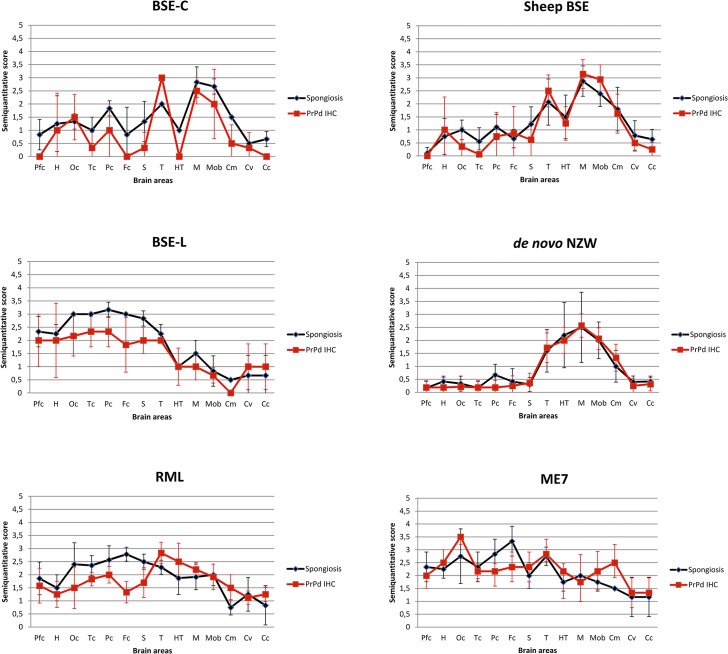
Spongiform change and PrPd distribution throughout the brain was semi-quantitatively assessed in histological sections of the inoculated brains of TgRab mice (Figs 3 and 4). Classical BSE-derived strains, namely BSE-C and sheep BSE, yielded very similarly shaped curves characterized by a strong involvement of the medulla oblongata, mesencephalon and thalamus but sparing of the hypothalamus. Involvement of the cortices and hippocampus was less intense but present, particularly at the deeper layers of the parietal cortex, involving the corpus callosum and sometimes extending to the oriens layer of the hippocampal formation. This pattern is equivalent to the one observed for BSE-C in the botg110 mouse model previously published by our group [36]. Mice inoculated with BSE-L, in contrast, showed a widespread involvement of the neocortex and less so in the diencephalon, mesencephalon and medulla oblongata in accordance with the brain PrPd distribution observed in natural and experimental cases of BSE-L in cattle [18,38].
Fig 3. Brain lesion and PrPd deposit distribution of the first passage of several prion strains in TgRab mice.
Brain lesion profiles and PrPd deposition profiles represent the mean semi-quantitative scoring (0–4, vertical axis) of the spongiform lesions (black) and the immunohistochemical labelling of PrPd deposits (red) against 14 brain regions (Pfc: piriform cortex, H: hippocampus, Oc: occipital cortex, Tc: temporal cortex, Pc: parietal cortex, Fc: frontal cortex, S: striatum, T: thalamus, HT: hypothalamus, M: mesencephalon, Mob: medulla oblongata, Cm: cerebellar nuclei, Cv: cerebellar vermis, Cc: cerebellar cortex). Note that classical BSE-originating strains (BSE-C and Sheep BSE) generate very similar shaped brain profiles with low scores in the hypothalamus and strong involvement of the brain stem, clearly distinguishable from BSE-L (which maintains its affinity towards cortices as seen in cattle field cases). Scrapie derived strain show a brain profile with cortical and hypothalamic tropisms. The de novo NZW strain shows a distinct tropism for the diencephalon (particularly hypothalamus) and brainstem while sparing the cortices.
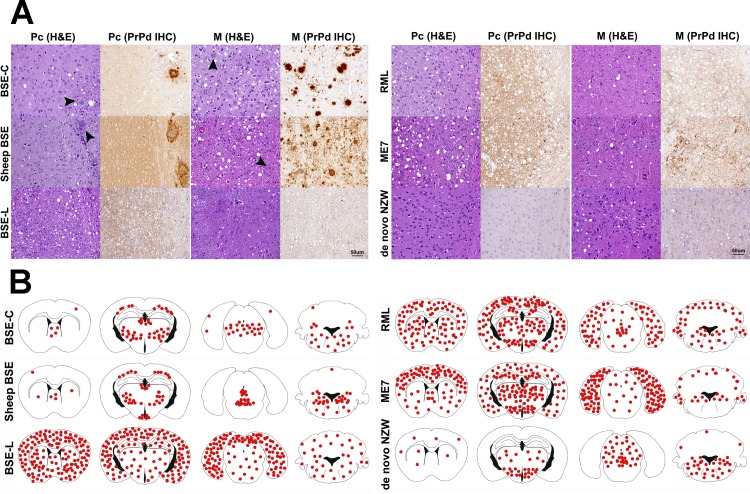
Fig 4. A: Histopathological characterization of several prion strains in TgRab mice.
Classical BSE-derived strains show a remarkably similar lesion and PrPd deposition patterns. PrPd plaques are readily conspicuous in haematoxylin and eosin (H&E) stained sections (arrowheads). Upon immunohistochemical (IHC) staining with 6H4 antibody against prion protein, the predominant staining pattern consists of intensely stained round-shaped plaques which can coalesce and form large aggregates. Scrapie derived strains, on the other hand, as well as BSE-L and de novo NZW show a rather discreet, plaqueless, punctiform immunolabelling pattern found in the neuropil within and around neuronal bodies. Linear and stellate shaped PrPd immunolabelling patterns are observed occasionally. In the case of RML, ME7 and BSE-L this pattern is also observed in the cortices while in de novo NZW the immunolabelling is restricted to the thalamus and brainstem. All images were taken at the same magnification. Bar 50 μm. B: Brain schematic mapping summary of the distribution of spongiform lesions and PrPd deposits in the brains of TgRab mice. The red dots depict the areas where spongiosis and/or PrPd deposits were mostly found in each group of infected mice. Note that the consistent picture yielded by BSE-C derived strains strongly involved the medulla oblongata, ventral mesencephalon, thalamus and deep parietal cortex while sparing the remaining cortices and the hypothalamus. This is clearly different from BSE-L which shows a remarkable tropism for the cerebral cortices and a lesser involvement of the brainstem structures. Scrapie derived strains, instead, show a clear tropism for the hypothalamus and a strong involvement of the cerebral cortices also. The spontaneous rabbit strain de novo NZW spares the cerebral cortices but shows a clear tropism for the hypothalamus.
The type of PrPd deposits seen by immunohistochemistry was also distinct in all mice inoculated with classical BSE-derived strains and consisted of amyloid-like rounded plaques, often confluent, which were readily visible on HE stained sections and positively stained in sections subjected to immunohistochemistry for PrPd (Fig 4A). BSE-L inoculated mice lacked plaque type deposits and showed a very different punctate immunolabelling pattern in the neuropil and perikarya (Fig 4A). This was consistent with the pattern obtained in tgBov mice when inoculated with BSE-L.
The scrapie-derived strains RML and ME7 showed PrPd deposits with a tropism for the diencephalon, including a consistent involvement of the hypothalamus (distinct from BSE strains), the mesencephalon and the medulla oblongata and also showed tropisms for the neocortex and cerebellar cortex. The PrPd type, on immunohistochemistry, was distinguishable from that of BSE infected mice, as it was comprised of a fine punctate pattern in the neuropil and perikarya (Fig 4).
The lesion and PrPd distribution of the rabbit-obtained de novo NZW strain showed a tropism confined to the diencephalon, including a consistent involvement of the hypothalamus, mesencephalon and medulla oblongata while sparing the cortices and hippocampus. The PrPd type, on immunohistochemistry, consisted of a fine punctate pattern in the neuropil and perikarya resembling that observed in ME7 and RML infected mice.
A model with spontaneous phenotype
The data presented validate the TgRab model to study rabbit susceptibility to prion strains. However, the TgRab line 058, chosen because it was the transgenic line showing the highest PrPC expression levels, also showed a spontaneous phenotype secondary to PrPC overexpression, as described by Westaway and coworkers [39], which needs to be taken into account when evaluating the results of any given experiment. Similar changes have been observed previously in other useful transgenic models overexpressing PrPc [40,41]. In this phenotype, between 300 and 400 days, the majority (over 80%) of hemizygous mice (5x-6x PrPc expression compared to normal levels; S2 Fig) developed gait abnormalities in the hindquarters that progressed slowly to complete hind-limb paralysis and atrophy of muscles (S3 Fig). The animals were able to feed, drink and groom normally and when it was not the case, as with any infected animal that reached the end point criteria, they were humanely euthanatized. See death time points for control groups in Fig 1. The same clinical presentation, but with enhanced severity, appeared in mice homozygous (10x-12x PrPc expression levels) for the transgene as early as 60 days of age.
Microscopically, the skeletal muscle tissue showed irregular diameter of the muscle fibers along with the presence of anguloid fibers, centralization of nuclei and substitution by adipose tissue proliferation in the endomysium (S3E Fig and Fig 3). These changes are compatible with neurogenic atrophy. Lesions were observed also in the central nervous system and consisted of an intense spongiform change in the white matter, particularly in the corpus callosum and internal capsule (S4 Fig). The remaining brain parenchyma also showed diffuse moderate spongiosis, which was more evident in the diencephalon and brainstem and particularly intense in mice euthanized at older ages.
Even though no PrPd was detected by western blotting or ELISA in any of the control animals, upon immunohistochemistry an intense PrPC background immunolabelling was present throughout the brain in agreement with the known overexpression of PrPC. Additionally, a more intense labelling was observed, that could be mistaken for PrPd signaling, which consisted of punctuate labelling around and within the cytoplasm of neurons, mainly located in the cortices but occasionally in the diencephalon and brain stem. Also, in the white matter, a punctate immunolabelling pattern was observed. Certain regions consistently showed strong immunolabelling of PrPC including the cochlear nucleus in the medulla oblongata and the cerebellar cortex where a diffuse labelling was observed in the molecular layer and intense labelling in the granular layer, depicting the synaptic glomeruli (S4 Fig). Despite most of the animals displaying an overt overexpression phenotype, characterization allowed clear discrimination of this from bona fide prion infection in this model.
Materials and Methods
Inocula preparation for in vivo prion propagation studies
Brain homogenates (10−1 in PBS) for use as seeds for PMCA or direct intracerebral inocula were prepared manually using a glazed mortar and pestle from brains of animals clinically affected by various TSE: BSE-C and BSE-L field cases supplied by the Laboratorio Central de Veterinaria (Algete, Madrid, Spain), SSBP/1 and ME7 supplied by Animal Heath and Veterinary Laboratory Agency (New Haw, Addlestone, Surrey, UK), CWD from the thalamus area of the brain of a female Mule deer, genotype 225SS, infected with CWD (04–22412 WSV2 EJW/JEJ), supplied by Department of Veterinary Sciences (Laramie, WY, USA), RML supplied by Rocky Mountain Laboratories (Hamilton, MT, USA) and Sheep BSE supplied by Ecole Nationale Vétérinaire (Toulouse, France). The atypical scrapie isolate was obtained from a field case diagnosed in PRIOCAT laboratory, CReSA-IRTA (Barcelona, Spain). Rabbit spontaneous prions were those obtained in the rabbit bioassays conducted in the Moredun Research Institute, Scotland [23].
Generation of in vitro PrPres by serial automated PMCA (saPMCA)
The in vitro prion replication and PrPres detection of amplified samples was performed as described previously with minor modifications [23,42]. Briefly, rabbit brains used for substrate were perfused using PBS + 5 mM EDTA and the blood-depleted brains were frozen immediately until required for preparing the 10% rabbit brain homogenates (PBS + NaCl 1% + 1% Triton X-100). 50–60 μl of 10% rabbit brain homogenate, either unseeded or seeded with the corresponding prion strain were loaded onto 0.2-ml PCR tubes and placed into a sonicating water bath at 37–38°C without shaking. Tubes were positioned on an adaptor placed on the plate holder of the sonicator (model S-700MPX, QSonica, Newtown, CT, USA) and subjected to incubation cycles of 30 min followed by a 20 s pulse of 150–220 watts sonication at 70–90% of amplitude. Serial rounds of PMCA consisted of 24-48h of standard PMCA followed by serial in vitro 1:10 passages in fresh 10% rabbit brain homogenate substrate. An equivalent number of unseeded (4–6 duplicates) tubes containing the corresponding brain substrate were subjected to the same number of rounds of saPMCA in order to control cross-contamination and/or the generation of spontaneous PrPres. The detailed protocol for PMCA, including reagents, solutions and troubleshooting, has been published elsewhere [43].
Biochemical characterization of in vitro- and in vivo-generated prion strains
PMCA treated samples were incubated with 85–200 μg/ml of protease K (PK) for 1 h at 42°C with shaking (450 rpm) as described previously [44]. Digestion was stopped by adding electrophoresis Laemmli loading buffer and the samples were analyzed by Western blotting.
Generation of TgRab mice
After isolation by PCR amplification using 5’ CCGCCGTACGTCATCATGGCGCACCTCGGCTAC 3’ and 5’ GGGGCCGGCCTCATCCCACGATCAGGAAG 3’ as primers, the open reading frame (ORF) of the rabbit PRNP gene was cloned into the pGEM-T vector (Promega). The rabbit-PrP ORF was excised from the cloning vector by using the restriction enzymes BsiWI (Thermo Fisher Scientific Inc.) and FseI (New England Biolabs Ltd.) and then inserted into a modified version of MoPrP.Xho vector [45] as described previously [46], which was also digested with BstWI and FseI. This vector contains the murine PrP promoter and exon-1, intron-1, exon-2 and 3’ untranslated sequences. The transgene was excised using NotI and purified with Invisorb Spin DNA Extraction Kit (Inviteck) according to the manufacturer recommendations.
Transgenic mouse founders were generated by microinjection of DNA into pronuclei following standard procedures [40]. DNA extracted from tail biopsies was analyzed by PCR using specific primers for the mouse exon 2 and 3’ untranslated sequences (5’ GAACTGAACCATTTCAACCGAG 3’ and 5’ AGAGCTACAGGTGGATAACC 3’). Those which tested positive were bred to mice null for the mouse Prnp gene in order to avoid endogenous expression of mouse prion protein. Absence of the mouse endogenous Prnp was assessed using the following primers: 5’ ATGGCGAACCTTGGCTACTGGC 3’ and 5’ GATTATGGGTACCCCCTCCTTGG 3’. The rabbit PrP expression levels of brain homogenates from transgenic mouse founders were determined by western blot using anti-PrP MAb L42 antibody (RIDA-Biopharm, Darmstadt, Germany) and compared with the PrP expression levels from NZW rabbit brain homogenates.
Animals homozygous for the transgene showed a spontaneous clinical phenotype as early as 60 days old, resembling the one described in the results section, but more severe, requiring euthanasia at 60–120 days old. Due to this, hemizygous mice were maintained for subsequent studies. The international code to identify this transgenic mouse line is STOCK-Prnptm2Edin Tg(moPrpn rabPrP)58Bps although throughout the paper they are referred to as TgRab mice.
TgRab mice inoculation
Mice of 42–56 days of age were intracerebrally inoculated under gaseous anesthesia (Isoflurane) through the right parietal bone. A 50 μl SGC precision syringe was used with a 25 G gauge needle and coupled to a repeatability adaptor fixed at 20 μl. A dose of buprenorphine was subcutaneously injected before recovery to consciousness to reduce post-inoculation pain.
Mice were kept in a controlled environment at a room temperature of 22°C, 12 h light-darkness cycle and 60% relative humidity in HEPA filtered cages (both air inflow and extraction) in ventilated racks. The mice were fed ad libitum, observed daily and their clinical status assessed twice a week. The presence of ten different TSE-associated clinical signs [47] was scored.
The experimental groups are listed in Table 2. As the hemizygous mice had a slight spontaneous phenotype due to PrPC overexpression (see Results), involving gait abnormalities, animals were euthanized following the end-point criteria (body weight, measurable clinical signs, physical appearance, unprovoked behavior and response to external stimuli). Positive TSE diagnosis relied principally on the detection of PrPd (either by immunohistochemistry and/or western blotting or ELISA) and associated spongiform change in the brain parenchyma.
Ethics statement
All experiments involving animals were approved by the animal experimentation ethics committee of the Autonomous University of Barcelona (Reference number: 585–3487) in agreement with Article 28, sections a), b), c) and d) of the “Real Decreto 214/1997 de 30 de Julio” and the European Directive 86/609/CEE and the European council Guidelines included in the European Convention for the Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes.
Sample processing and general procedures
When the clinical end-point criteria were reached, mice were euthanized by an overdose of pentobarbital administered intraperitoneally followed by decapitation. The brain was immediately extracted and placed into 10% phosphate buffered formalin. From each mouse a rostral section of the brain (including olfactory bulbs and frontal cortex), a caudal fraction of the medulla oblongata and the whole spinal cord were kept frozen (for biochemical studies and second passage). Transversal sections of the remaining brain tissue were performed at the level of the piriform cortex, optic chiasm and medulla oblongata. Samples were embedded in paraffin-wax after dehydration through increasing alcohol concentrations and xylene. Four micrometer sections were mounted on glass microscope slides which were stained with hematoxylin and eosin for morphological evaluation. Additional sections were mounted in 3-trietoxysilil-propilamine-coated glass microscope slides for immunohistochemistry.
Detection of PrPd
A pool of all frozen central nervous tissues samples was homogenized 1:10 (W/V) in PBS using closed tubes containing ceramic beads, placed in a ribolyzer (Precess, Bio-Rad) and subsequently analyzed either by western blotting, as described above, or by ELISA (IDEXX, Herdcheck). The latter is a commercial ELISA based on the affinity of misfolded prions to an anionic substrate (termed Seprion). A new threshold was defined to adapt to the higher densitometry readings obtained when working with samples with PrPc overexpression: only samples with a ratio spectrophotometry reading/cutoff over 5 were considered positive.
Immunohistochemistry
Immunohistochemistry (IHC) against PrPd was performed as described previously [48]. Briefly, deparaffinized sections were subjected to epitope unmasking treatments: immersed in formic acid and boiled at low pH (6.15) in a pressure cooker and pre-treated with proteinase K. Endogenous peroxidases were blocked by immersion in a 3% H2O2 in methanol. Then, the sections were incubated overnight with anti-PrP MAb 6H4 primary antibody (1:2000, Prionics AG) and subsequently visualised using the DAKO Goat anti-mouse EnVision system (Ref. K400111/0) and 3,3’diaminobenzidine as the chromagen substrate. As a background control, incubation with the primary antibody was omitted.
Semi-quantification and data analysis
Histological lesions (i.e. spongiform change) and PrPd immunolabelling were evaluated under a light microscope by a pathologist. A semi-quantitative approach was used to obtain comparable data from the different prions used to challenge mice. Spongiform lesion and PrPd immunolabelling were scored separately. A total of 15 different brain regions were chosen: piriform cortex (Pfc), hippocampus (H), frontal cortex (Fc), parietal cortex (Pc), temporal cortex (Tc), occipital cortex (Oc), thalamus (T), hypothalamus (HT), mesencephalon (M), medulla oblongata (Mobl), cerebellar nuclei (Cm), cerebellar vermis (Cv) and cerebellar cortex (Cc). Scores ranging from (0) absence of spongiosis or immunolabelling: (1) mild, (2) moderate, (3) intense and (4) maximum intensity of lesion or immunolabelling were assigned to each brain area studied (Fig 3). Each area was investigated globally as region for the scoring. Brain profiles were plotted as a function of the anatomical areas which were ordered along the X axis in an attempt to represent the caudo-rostral axis of the brain. This methodology was adapted from a previous study performed on BSE-infected cattle [49]. Graphs were plotted using Microsoft Office 2007 Excel software.
Discussion
This is the first report of in vivo evidence suggesting that TgRab mice are susceptible to cross species transmission of prion strains. This not only reinforces that rabbits can no longer be considered TSE resistant, but also that there is a possibility they could act as a reservoir for other prion strains. As such, rabbits must be taken into account when determining the epidemiology of several TSE both in relation to the species of origin, especially sympatric ones, but also to potential zoonotic transmission.
In previous studies we demonstrated that rabbits were able to propagate abnormal prions and that these were transmissible to other rabbits. However, this was only one prion strain which was generated de novo in an in vitro PMCA assay in rabbit brain homogenate (a spontaneous rabbit prion strain) and on first passage it had only a very limited attack rate [23]. This new mouse model, which responded in a comparable manner to rabbits when challenged with the same in vitro generated rabbit derived inoculum, has allowed us to evaluate the TgRab’s susceptibility to a number of actual field prions strains from a variety of different species. Although the use of rabbits would have been the most appropriate model there are strong, particularly budgetary, limitations due to the longer lifespan of rabbits and the need to use level 3 biosafety facilities. Thus, a transgenic mouse model overexpressing rabbit PrPC was designed to overcome these limitations and allow us to determine its susceptibility to different prion strains.
No polymorphisms have been described in the PRNP rabbit gene, therefore several mouse transgenic lines were generated expressing rabbit PrPC at different expression levels. The line with the highest possible PrPC expression levels was selected to allow for easier prion propagation capacity but the overexpression was not so high as to generate a spontaneous phenotype at an early age which would preclude the attainment of infectivity/susceptibility data. The hemizygous TgRab line met these criteria with levels of PrPC between 5 to 6 times those present in rabbits. The use of transgenic mice overexpressing ovine PrPC to obtain the infectivity titer of specific prion isolates has been shown to be equivalent to titrations obtained through bioassay in the natural host [50]. Phenotyping of the newly developed prion transgenic model was essential, especially as the levels of PrPc expression present have not been shown to be problematic in other models [41,46]. Eighty percent of the TgRab mice presented with a late onset spontaneous neurological disease phenotype (S3 Fig and S4 Fig) which, fortunately, did not interfere in the interpretation of prion susceptibility results. This allowed us to work with a model that faithfully reproduced the behavior in rabbits with respect to their capability to propagate different prion strains. One cannot exclude the possibility that the presence of spontaneous disease might create a toxic environment in the brain which artificially enhances the transmission of certain strains. Therefore a thorough knowledge of the PrPC overexpression-related changes in uninfected controls was essential to identify the true prion disease status and validity of susceptibility.
Lesion morphology and profiling within the brain and identification of specific PrPd deposition-types allowed unequivocal identification of infected animals (either spontaneous or as a result of an inoculation). Further biochemical detection of the presence of PrPres by western blotting confirmed the ability of morphological techniques to identify an infected animal. Additionally, as PrPC overexpression may mask an incipient infection, second passages are required to confirm if rabbits are totally resistant to those prion isolates to which they initially appeared to be, such as SSBP/1, atypical scrapie or CWD, and these experiments are ongoing.
Once validated the TgRab model was used to evaluate which TSE strains were able to infect the model (Table 2). Previous attempts in rabbits had concluded they were resistant, probably due to incomplete studies and the strong barrier of rabbits to propagate prions [34]. Initially classical cattle BSE, the most relevant field strain, was tested and found to be infectious on first passage with a low attack rate (4/9) and relatively long incubation period (551dpi±10). The strain properties observed in the infected TgRab mice (western blotting, brain lesion and PrPd deposition profiles) were typical of BSE and indistinguishable from those obtained in other BSE murine models [36]. Parallel bioassay studies were conducted with the BSE isolate previously amplified in vitro using rabbit normal brain homogenate as a substrate (BSE-RaPrPres, this inoculum was characterised previously in a TgBov mouse model by our group [36]). These animals showed a 100% (12/12) attack rate and a shortened incubation period (396dpi ±12 vs 551dpi ±10) compared to the cattle BSE inoculated TgRab mice. This reduction already indicated that a transmission barrier between species had been overcome thanks to the in vitro adaptation of the cattle BSE-C to rabbit PrPC, a second passage was performed from that isolate which also showed a 100% attack rate (3/3). Its incubation period was reduced to 322dpi ±12 (mean ± s.e.m.) indicating further host adaptation (S5 Fig).
SheepBSE, derived from BSE-C, infected TgRab mice with a 100% attack rate (9/9), a relatively short incubation time (368±10 dpi) and with lesion and PrPd brain profiles identical to those of BSE-C inoculated mice, suggesting that the same strain was being propagated through both isolates. This enhanced virulence of sheepBSE compared to BSE-C has been previously demonstrated in other experimental scenarios [29,51]. The results obtained with sheep scrapie differed completely as, in agreement with early experiments in rabbits [34], none of the TgRab mice inoculated with SSBP/1 showed any evidence of a prion disease on first passage. However, this result does not preclude that, if further in vivo SSBP/1 passages were to be performed, the transmission barrier would be crossed. As in the case of BSE in the bank vole (Myodes glareolus), where after an initial resistance a bank vole adapted BSE strain was obtained which was highly transmissible [52,53]. Conversely, ME7 and RML scrapie, both murine adapted sheep scrapie strains, infected TgRab mice on first passage and their incubation times, PrPres biochemical profiles, lesion profiles and PrPd deposition patterns were clearly distinguishable from cattle derived strains. Together these data are the first evidence that TgRab mice are not only able to propagate prions but they do it maintaining clearly the different distinguishing strain features (Figs 1, 3 and 4) which strongly suggests that rabbits may also.
It is noteworthy that both ME7 and RML, which originated from serial passages of SSBP/1 in different rodents [54,55], directly propagated in TgRab mice on first passage. Conversely, SSBP/1 did not infect TgRab mice on first passage. The murine adapted prion strains behaved differently to their parent strain, possibly because passage through rodents had selected for a strain capable of crossing the rodent species barriers. The situation is analogous to CWD which will infect hamsters after initial passage through ferrets [9]. In the present work, previous adaptation of scrapie to rodents, possibly resulting in a higher sequence identity in some specific and crucial PrP regions with rabbits compared to sheep, allowed rodent adapted scrapie prions to misfold rabbit PrPC. In previous studies ME7 did not infect rabbits after 4–5 years of incubation, with the exception of a single inconclusive case [23,34]. This result is difficult to extrapolate since we are discussing different species, of differing lifespans and with a species barrier between them. The PrPC overexpression in TgRab may have allowed ME7 to propagate more efficiently than in rabbits which suggests that if the original rabbit experiments had been performed over the maximum lifespan of rabbits ME7 may have propagated on first passage also.
Once BSE in cattle has been virtually controlled, CWD in cervids is the animal prion disease with the most repercussions, at least in the North American continent. The uncertainty of its transmissibility to humans [56] and its unique ability to spread through the free ranging cervid population make its study highly relevant with respect to transmissibility to other species. Moreover CWD prions are known to be shed and are highly persistent in the environment. Rabbits are a sympatric species with cervids. Even though CWD has been shown to transmit on first passage to many species including sheep, cattle [57], squirrel monkeys [58], cats [59], hamsters [60], ferrets [9], mink [61], bank voles and deer mice (Genus Peromyscus) [62] its transmissibility efficiency is relatively low with very long incubation periods and low attack rates. For instance, wild type mice could not be readily infected, so tga20 mice overexpressing murine PrPC were required to prove susceptibility to CWD [63] or required a second passage [64]. Another example is the transmission of CWD to cats, which required an incubation period of longer than 4 years [59]. The present study showed CWD was not able to infect TgRab on first passage (0/12). Further experiments are required to confirm the resistance of rabbits to CWD including a blind second passage and bioassays with CWD previously passaged in other species, especially rodents [9]. This will rule out an analogous situation as the one observed in this paper with sheep scrapie whereby SSBP/1 does not transmit to TgRab but murine passaged counterparts, ME7 and RML, do.
With respect to the atypical prion strains of purported spontaneous origin [18,65,66], BSE-L infected TgRab mice on first passage and, although the attack rate was low (3/11), they had the shortest incubation period observed in this model so far (221dpi for the first animal to die, mean 280±26dpi). The lesion and PrPd deposition brain profiles differed considerably from those of BSE-C. None of the TgRab mice inoculated with atypical scrapie showed evidence of a TSE with the exception of one animal, euthanized at 742 dpi which, even though no histological lesions nor PrPd deposits were present suggestive of infection, it was positive by PrPd ELISA. This result could not be confirmed by western blotting. However, this ELISA detects PrPd through its affinity to an anionic ligand not due to its resistance to protease K so we cannot rule out this single mouse was positive. A second passage is ongoing which will determine the result.
Initial in vitro experiments predicted that BSE as well as SSBP/1 and CWD isolates were able to missfold rabbit PrPC. However, a discrepancy was found with the bioassay results since neither SSBP/1 nor CWD infected TgRab mice on first passage. Several saPMCA rounds were needed in order to amplify the different isolates, varying in number depending of each strain. Thus, it is not surprising that on first passage some of the isolates do not transmit.
Besides the PRNP sequence, another component of the transmission barrier is the genetic background in which each PrPC is contained. This has been demonstrated by infectivity studies showing BSE propagated more efficiently in RIII mice than C57/Black mice, two mice strains of the same species with the same PRNP gene [67]. Or when the genetic background (i.e. passage through different inbred mouse lines) determined not only the incubation period but also the propagation of two biochemically different BSE-derived strains [68]. For these reasons the belief that rabbits were resistant to prion infection was not only attributed to the rabbit PrPC sequence but also to its genetic background. To study whether the genetic background of rabbits was responsible for the apparent prion resistance, Houdebine’s group generated transgenic rabbits expressing an ovine PrPC which was known to easily misfold. Upon inoculation with ovine prion strains these rabbits succumbed to prion disease further proving that rabbits are not resistant to prions (results published paired with this article) and that the genetic background is not a limiting factor [37].
The differential susceptibility observed between actual rabbits and the transgenic model presented here can be explained by the higher PrPC expression levels of TgRab mice. Lower expression mouse lines would probably only be susceptible on first passage to strains previously adapted to rabbit PrPC as occurs with rabbits. It has taken more than three decades to finally dismiss the rabbit as a prion resistant species. We believe that the studies presented here confirm that in vitro studies are of great help in interpreting in vivo results, leave no room for misinterpretation, and that it can be ascertained that rabbits, and probably all other mammal species [21], are susceptible to infection by specific prion strains. The prion strain and its species of origin determine the extent of susceptibility, but neither rabbit PRNP nor their genetic background suggest they are resistant to prion propagation. Unfortunately, as with other mammals, the exact molecular mechanisms governing the capricious choice of strains that can be propagated in a certain species is still unknown.
In light of our results, especially susceptibility to spontaneous cattle prions (BSE-L), the restrictions on rabbits being fed ruminant protein should be maintained sine die to minimize the chances of any prion strain transmitting to rabbits.
Supporting Information
Rabbit brain homogenates seeded with different prion strains (cattle: BSE-C, sheep: SSPB/1 and sheep BSE-C, deer: CWD) or unseeded (de novo) were subjected to saPMCA. Seeded samples from round 10 and the unseeded sample from round 20 from in vivo isolates were digested with 100 μg/ml of proteinase K (PK) and analyzed by western blot using monoclonal antibody 6H4 to compare their differential electrophoretic migration and glycosylation patterns. Control: Normal rabbit brain homogenate. MW: Molecular weight.
(TIF)
Samples were diluted as described in figure and monoclonal antibody 6H4 was used at 1:10,000. The level of PrPC expression observed in TgRab brain was 5–6 times higher than NZW rabbit brain and 10–12 times higher than mouse brain. MW: Molecular weight.
(TIF)
A and B: Atrophy and paralysis of the hindquarters. C: Hindquarter footprints (black ink) of a healthy normal mouse. D: Footprints of a 500 days old non-inoculated mouse showing severe gait alteration. E: Histological image of the muscular tissue of a TgRab mouse with the spontaneous phenotype showing remarkable loss of muscular fibers (substituted by adipose tissue proliferation in the endomysium), irregular diameter and nuclear centralization in the remaining muscular fibers. F: normal muscular tissue of an unaffected mouse.
(TIF)
All images were taken at the same magnification. Bar 50 μm. A: In some mice a severe spongiform change was observed in the corpus callosum (cc) and deeper layers of the neocotex, the image corresponds to parietal cortex (Pc). This lesion could also be observed in the internal capsule. B: Immunolabelling of PrPd in the white matter showed punctiform morphology (arrowheads). C: Spongiosis of variable intensity was seen throughout the remaining brain areas which increased with the age of the mouse. In the image a section of thalamus with mild age-related spongiosis is seen. D: Parietal cortex. Punctifom immunolabelling around neurons was frequently observed in TgRab mice. Its distribution varied between mice but was present frequently in the cortices, striatum and thalamus. The fact that these cells are overexpressing rabbit PrPC could be an explanation of this signaling. All these animals yielded a negative result to PrPd by western blotting. E: The cochlear nucleus of the medulla oblongata consistently showed intense labelling of the neuropil in all negative controls. F: Cerebellar cortex. Intense, diffuse labelling of the neuropil was typically observed in the molecular (mol) and the granular (gra) layers and co-localized with the synaptic glomeruli.
(TIF)
A: TgRab survival times after inoculation. Each dot corresponds to a mouse. Dots with a red margin represent TSE positive animals. B: Brain lesion and PrPd deposit distribution of the first passage of BSE-RaPrPres in TgRab mice. C: 2nd passage of BSE-RaPrPres in TgRab mice. Lesion brain profiles and PrPd deposition profiles represent the mean semi-quantitative scoring (0–4, vertical axis) of the spongiform lesions (black) and the immunohistochemical labelling of PrPd deposits (red) against 14 brain regions (Pfc: piriform cortex, H: hippocampus, Oc: occipital cortex, Tc: temporal cortex, Pc: parietal cortex, Fc: frontal cortex, S: striatum, T: thalamus, HT: hypothalamus, M: mesencephalon, Mob: medulla oblongata, Cm: cerebellar nuclei, Cv: cerebellar vermis, Cc: cerebellar cortex). D: Histopathological characterization of BSE-RaPrPres in TgRab mice. Lesion and PrPd deposition patterns are remarkably similar to those observed in BSE C-derived strains. PrPd plaques are readily conspicuous in haematoxylin and eosin (H&E) stained sections (arrowheads). Upon immunohistochemical (IHC) labelling with 6H4 antibody against prion protein, the predominant pattern also consists of intensely labelled round-shaped plaques which can coalesce and form large aggregates. All images were taken at the same magnification. Bar 50 μm. E: Brain schematic mapping summary of the distribution of spongiform lesions and PrPd deposits in the brains of TgRab mice. The red dots depict the areas where spongiosis and/or PrPd deposits were mostly found in each group of infected mice. The image is consistent with that found in BSE-C derived strains strongly involving the medulla oblongata, ventral mesencephalon, thalamus and deep parietal cortex while sparing the remaining cortices and the hypothalamus.
(TIF)
Acknowledgments
The authors thank the support from IKERBasque foundation, vivarium and maintenance from CIC bioGUNE, Sierra Espinar, Marta Valle, Mariano Moreno, Paola Marco and IRTA-CReSA’s biocontainment unit staff for care and maintenance of the animals, Tomás Mayoral, Jean Jewell for the BSE, CWD and BASE (BSE-L) brain tissue samples, respectively, and G. Telling and Jifeng Bian for the plasmid phggPrP-MCS.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was financially supported by 3 national grants from Spain [AGL2009-11553-C02-01 (JC), AGL2012-37988-C04-01 (JC) and AGL2008-05296-C02 (EV)], a Basque government grant (PI2010-18) (JC), two CTP grants (CTP11-P04 and CTP2013-P05) (JC), 3 InterReg grants [EFA205/11 and EFA218/11) (JC); EFA282/13—Transprion (MP, DF, EV)], Etortek Research Programs 2011/2013 (JC) and by Agència de Salut pública de Catalunya, Departament de Salut, Generalitat de Catalunya (EV). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science (80-). 1982;216: 136–144. [DOI] [PubMed] [Google Scholar]
- 2. Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95: 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bruce ME, Will RG, Ironside JW, McConnell I, Drummond D, Suttie A, et al. Transmissions to mice indicate that “new variant” CJD is caused by the BSE agent. Nature. 1997;389: 498–501. [DOI] [PubMed] [Google Scholar]
- 4. Eloit M, Adjou K, Coulpier M, Fontaine JJ, Hamel R, Lilin T, et al. BSE agent signatures in a goat. Vet Rec. 2005;156: 523–524. [DOI] [PubMed] [Google Scholar]
- 5. Pearson GR, Gruffydd-Jones TJ, Wyatt JM, Hope J, Chong A, Scott AC, et al. Feline spongiform encephalopathy. Vet Rec. 1991;128: 532. [DOI] [PubMed] [Google Scholar]
- 6. Stack MJ, Chaplin MJ, Clark J. Differentiation of prion protein glycoforms from naturally occurring sheep scrapie, sheep-passaged scrapie strains (CH1641 and SSBP1), bovine spongiform encephalopathy (BSE) cases and Romney and Cheviot breed sheep experimentally inoculated with BSE using. Acta Neuropathol. 2002;104: 279–286. [DOI] [PubMed] [Google Scholar]
- 7. Bellworthy SJ, Dexter G, Stack M, Chaplin M, Hawkins SA, Simmons MM, et al. Oral transmission of BSE to VRQ/VRQ sheep in an experimental flock. Vet Rec. 2008;162: 130–131. [DOI] [PubMed] [Google Scholar]
- 8. Pearson GR, Wyatt JM, Gruffydd-Jones TJ, Hope J, Chong A, Higgins RJ, et al. Feline spongiform encephalopathy: fibril and PrP studies. Vet Rec. 1992;131: 307–310. [DOI] [PubMed] [Google Scholar]
- 9. Bartz JC, Marsh RF, McKenzie DI, Aiken JM. The host range of chronic wasting disease is altered on passage in ferrets. Virology. 1998;251: 297–301. 10.1006/viro.1998.9427 [DOI] [PubMed] [Google Scholar]
- 10. Lezmi S, Bencsik A, Monks E, Petit T, Baron T. First case of feline spongiform encephalopathy in a captive cheetah born in France: PrP(sc) analysis in various tissues revealed unexpected targeting of kidney and adrenal gland. Histochem Cell Biol. 2003;119: 415–422. [DOI] [PubMed] [Google Scholar]
- 11. Poser CM. Notes on the history of the prion diseases. Part I. Clin Neurol Neurosurg. 2002;104: 1–9. [DOI] [PubMed] [Google Scholar]
- 12. Wells GA, Wilesmith JW. The neuropathology and epidemiology of bovine spongiform encephalopathy. Brain Pathol. 1995;5: 91–103. [DOI] [PubMed] [Google Scholar]
- 13. Wyatt JM, Pearson GR, Smerdon TN, Gruffydd-Jones TJ, Wells GA, Wilesmith JW. Naturally occurring scrapie-like spongiform encephalopathy in five domestic cats. Vet Rec. 1991;129: 233–236. [DOI] [PubMed] [Google Scholar]
- 14. Barlow RM. Transmissible mink encephalopathy: pathogenesis and nature of the aetiological agent. J Clin Pathol Suppl (R Coll Pathol). 1972;6: 102–109. [PMC free article] [PubMed] [Google Scholar]
- 15. Gajdusek DC. Unconventional viruses and the origin and disappearance of kuru. Science (80-). 1977;197: 943–960. [DOI] [PubMed] [Google Scholar]
- 16. Williams ES. Chronic wasting disease. Vet Pathol. 2005;42: 530–549. [DOI] [PubMed] [Google Scholar]
- 17. Biacabe A-G, Laplanche J-L, Ryder S, Baron T. Distinct molecular phenotypes in bovine prion diseases. EMBO Rep. 2004;5: 110–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Casalone C, Zanusso G, Acutis P, Ferrari S, Capucci L, Tagliavini F, et al. Identification of a second bovine amyloidotic spongiform encephalopathy: Molecular similarities with sporadic Creutzfeldt-Jakob disease. PNAS. 2004;101:3065–3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Le Dur A, Beringue V, Andreoletti O, Reine F, Lai TL, Baron T, et al. A newly identified type of scrapie agent can naturally infect sheep with resistant PrP genotypes. Proc Natl Acad Sci U S A. 2005;102: 16031–16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Benestad SL, Sarradin P, Thu B, Schönheit J, Tranulis M a, Bratberg B. Cases of scrapie with unusual features in Norway and designation of a new type, Nor98. Vet Rec. 2003;153: 202–8. [DOI] [PubMed] [Google Scholar]
- 21. Fernández-Borges N, Chianini F, Eraña H, Vidal E, Eaton SL, Pintado B, et al. Naturally prion resistant mammals: a utopia? Prion. 2012;6: 425–9. 10.4161/pri.22057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chianini F, Fernández-Borges N, Eraña H, Pang Y, Vidal E, Eaton SL, et al. Prion-resistant or prion-susceptible species, this is the question. Virulence. Landes Bioscience; 2013;4: 333–334. 10.4161/viru.24456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chianini F, Fernandez-Borges N, Vidal E, Gibbard L, Pintado B, de Castro J, et al. Rabbits are not resistant to prion infection. Proceedings of the National Academy of Sciences. 2012;109;5080–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Di Bari MA, Chianini F, Vaccari G, Esposito E, Conte M, Eaton SL, et al. The bank vole (Myodes glareolus) as a sensitive bioassay for sheep scrapie. J Gen Virol. 2008;89: 2975–2985. 10.1099/vir.0.2008/005520-0 [DOI] [PubMed] [Google Scholar]
- 25. Espinosa JC, Herva ME, Andréoletti O, Padilla D, Lacroux C, Cassard H, et al. Transgenic mice expressing porcine prion protein resistant to classical scrapie but susceptible to sheep bovine spongiform encephalopathy and atypical scrapie. Emerg Infect Dis. 2009;15: 1214–21. 10.3201/eid1508.081218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fernández-Borges N, de Castro J, Castilla J. In vitro studies of the transmission barrier. Prion. 2009;3: 220–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kurt TD, Seelig DM, Schneider JR, Johnson CJ, Telling GC, Heisey DM, et al. Alteration of the chronic wasting disease species barrier by in vitro prion amplification. J Virol. 2011;85: 8528–37. 10.1128/JVI.00809-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Le Dur A, Béringue V, Andréoletti O, Reine F, Laï TL, Baron T, et al. A newly identified type of scrapie agent can naturally infect sheep with resistant PrP genotypes. Proc Natl Acad Sci U S A. 2005;102: 16031–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Espinosa JC, Andreoletti O, Castilla J, Herva ME, Morales M, Alamillo E, et al. Sheep-Passaged Bovine Spongiform Encephalopathy Agent Exhibits Altered Pathobiological Properties in Bovine-PrP Transgenic Mice. J Virol. 2007;81: 835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Padilla D, Béringue V, Espinosa JC, Andreoletti O, Jaumain E, Reine F, et al. Sheep and goat BSE propagate more efficiently than cattle BSE in human PrP transgenic mice. PLoS Pathog. 2011;7: e1001319 10.1371/journal.ppat.1001319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stewart P, Campbell L, Skogtvedt S, Griffin K a, Arnemo JM, Tryland M, et al. Genetic predictions of prion disease susceptibility in carnivore species based on variability of the prion gene coding region. PLoS One. 2012;7: e50623 10.1371/journal.pone.0050623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vorberg I, Groschup MH, Pfaff E, Priola SA. Multiple amino acid residues within the rabbit prion protein inhibit formation of its abnormal isoform. J Virol. 2003;77: 2003–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang J. The Structural Stability of Wild-type Horse Prion Protein. J Biomol Struct Dyn. 2011;29: 369–377. [DOI] [PubMed] [Google Scholar]
- 34. Barlow RM, Rennie JC. The fate of ME7 scrapie infection in rats, guinea-pigs and rabbits. Res Vet Sci. 1976;21: 110–111. [PubMed] [Google Scholar]
- 35. Gibbs CJ, Gajdusek DC. Experimental subacute spongiform virus encephalopathies in primates and other laboratory animals. Science. 1973;182: 67–68. [DOI] [PubMed] [Google Scholar]
- 36. Vidal E, Fernández-Borges N, Pintado B, Ordóñez M, Márquez M, Fondevila D, et al. Bovine spongiform encephalopathy induces misfolding of alleged prion-resistant species cellular prion protein without altering its pathobiological features. J Neurosci. 2013;33: 7778–86. 10.1523/JNEUROSCI.0244-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sarradin P, Viglietta C, Limouzin C, Andreoletti O, Daniel-Carlier N, Barc C, et al. Transgenic rabbits expressing ovine PrP are susceptible to scrapie. PLoS Pathog. 2015;11:e1005077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lombardi G, Casalone C, A DA, Gelmetti D, Torcoli G, Barbieri I, et al. Intraspecies transmission of BASE induces clinical dullness and amyotrophic changes. PLoS Pathog. 2008;4: e1000075 10.1371/journal.ppat.1000075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Westaway D, DeArmond SJ, Cayetano-Canlas J, Groth D, Foster D, Yang SL, et al. Degeneration of skeletal muscle, peripheral nerves, and the central nervous system in transgenic mice overexpressing wild-type prion proteins. Cell. 1994;76: 117–29. [DOI] [PubMed] [Google Scholar]
- 40. Castilla J, Gutierrez Adan A, Brun A, Pintado B, Ramirez MA, Parra B, et al. Early detection of PrPres in BSE-infected bovine PrP transgenic mice. Arch Virol. 2003;148: 677–691. [DOI] [PubMed] [Google Scholar]
- 41. Fischer M, Rülicke T, Raeber A, Sailer A, Moser M, Oesch B, et al. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 1996;15: 1255–64f. [PMC free article] [PubMed] [Google Scholar]
- 42. Castilla J, Saa P, Morales R, Abid K, Maundrell K, Soto C. Protein misfolding cyclic amplification for diagnosis and prion propagation studies. Methods Enzym. 2006;412: 3–21. [DOI] [PubMed] [Google Scholar]
- 43. Saa P, Castilla J, Soto C. Cyclic amplification of protein misfolding and aggregation. Methods Mol Biol. 2005;299: 53–65. [DOI] [PubMed] [Google Scholar]
- 44. Castilla J, Saá P, Hetz C, Soto C. In vitro generation of infectious scrapie prions. Cell. 2005;121: 195–206. [DOI] [PubMed] [Google Scholar]
- 45. Borchelt DR, Davis J, Fischer M, Lee MK, Slunt HH, Ratovitsky T, et al. A vector for expressing foreign genes in the brains and hearts of transgenic mice. Genet Anal. 1996;13: 159–163. [DOI] [PubMed] [Google Scholar]
- 46. Castilla J, Gutiérrez Adán a, Brun a, Pintado B, Ramírez M a, Parra B, et al. Early detection of PrPres in BSE-infected bovine PrP transgenic mice. Arch Virol. 2003;148: 677–91. [DOI] [PubMed] [Google Scholar]
- 47. Scott M, Foster D, Mirenda C, Serban D, Coufal F, Walchli M, et al. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell. 1989;59: 847–857. [DOI] [PubMed] [Google Scholar]
- 48. Siso S, Ordoñez M, Cordón I, Vidal E, Pumarola M. Distribution of PrPres in the brains of BSE-affected cows detected by active surveillance in Catalonia, Spain. Vet Rec. 2004;155: 524–525. [DOI] [PubMed] [Google Scholar]
- 49. Vidal E, Marquez M, Ordonez M, Raeber AJ, Struckmeyer T, Oesch B, et al. Comparative study of the PrPBSE distribution in brains from BSE field cases using rapid tests. J Virol Methods. 2005;127: 24–32. [DOI] [PubMed] [Google Scholar]
- 50. Douet J-Y, Lacroux C, Corbière F, Litaise C, Simmons H, Lugan S, et al. PrP expression level and sensitivity to Prion infection. J Virol. 2014; 88:5870–5872. 10.1128/JVI.00369-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Padilla D, Béringue V, Espinosa JC, Andreoletti O, Jaumain E, Reine F, et al. Sheep and goat BSE propagate more efficiently than cattle BSE in human PrP transgenic mice. PLoS Pathog. 2011;7: e1001319 10.1371/journal.ppat.1001319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pirisinu L, Marcon S, Bari M, D’Agostino C, Agrimi U, Nonno R. Biochemical Characterization of Prion Strains in Bank Voles. Pathogens. Multidisciplinary Digital Publishing Institute; 2013;2: 446–456. 10.3390/pathogens2030446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nonno R, Di Bari MA, Cardone F, Vaccari G, Fazzi P, Dell’Omo G, et al. Efficient transmission and characterization of Creutzfeldt-Jakob disease strains in bank voles. Westaway D, editor. PLoS Pathog. Public Library of Science; 2006;2: e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. CHANDLER RL. Encephalopathy in mice produced by inoculation with scrapie brain material. Lancet. 1961;1: 1378–9. [DOI] [PubMed] [Google Scholar]
- 55. Dickinson AG, Meikle VM. A comparison of some biological characteristics of the mouse-passaged scrapie agents, 22A and ME7. Genet Res. 1969;13: 213–25. [DOI] [PubMed] [Google Scholar]
- 56. Saunders SE, Bartelt-Hunt SL, Bartz JC. Occurrence, transmission, and zoonotic potential of chronic wasting disease. Emerg Infect Dis. 2012;18: 369–76. 10.3201/eid1803.110685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hamir AN, Kehrli ME, Kunkle RA, Greenlee JJ, Nicholson EM, Richt JA, et al. Experimental interspecies transmission studies of the transmissible spongiform encephalopathies to cattle: comparison to bovine spongiform encephalopathy in cattle. J Vet Diagn Invest. 2011;23: 407–20. 10.1177/1040638711403404 [DOI] [PubMed] [Google Scholar]
- 58. Race B, Meade-White KD, Miller MW, Barbian KD, Rubenstein R, LaFauci G, et al. Susceptibilities of nonhuman primates to chronic wasting disease. Emerg Infect Dis. 2009;15: 1366–76. 10.3201/eid1509.090253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mathiason CK, Nalls A V, Seelig DM, Kraft SL, Carnes K, Anderson KR, et al. Susceptibility of domestic cats to chronic wasting disease. J Virol. 2013;87: 1947–56. 10.1128/JVI.02592-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Raymond GJ, Raymond LD, Meade-White KD, Hughson AG, Favara C, Gardner D, et al. Transmission and adaptation of chronic wasting disease to hamsters and transgenic mice: evidence for strains. J Virol. 2007;81: 4305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Harrington RD, Baszler T V, O’Rourke KI, Schneider DA, Spraker TR, Liggitt HD, et al. A species barrier limits transmission of chronic wasting disease to mink (Mustela vison). J Gen Virol. 2008;89: 1086–96. 10.1099/vir.0.83422-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Heisey DM, Mickelsen NA, Schneider JR, Johnson CJ, Johnson CJ, Langenberg JA, et al. Chronic wasting disease (CWD) susceptibility of several North American rodents that are sympatric with cervid CWD epidemics. J Virol. 2010;84: 210–5. 10.1128/JVI.00560-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sigurdson CJ, Manco G, Schwarz P, Liberski P, Hoover EA, Hornemann S, et al. Strain fidelity of chronic wasting disease upon murine adaptation. J Virol. 2006;80: 12303–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lee Y-H, Sohn H-J, Kim M-J, Kim H-J, Park K-J, Lee W-Y, et al. Experimental chronic wasting disease in wild type VM mice. J Vet Med Sci. 2013;75: 1107–10. [DOI] [PubMed] [Google Scholar]
- 65. Baron T, Biacabe AG, Arsac JN, Benestad S, Groschup MH. Atypical transmissible spongiform encephalopathies (TSEs) in ruminants. Vaccine. 2006; 22(6):823–42. [DOI] [PubMed] [Google Scholar]
- 66. Benestad SL, Sarradin P, Thu B, Schonheit J, Tranulis MA, Bratberg B. Cases of scrapie with unusual features in Norway and designation of a new type, Nor98. Vet Rec. 2003;153: 202–208. [DOI] [PubMed] [Google Scholar]
- 67. Green R, Horrocks C, Wilkinson A, Hawkins SA, Ryder SJ. Primary Isolation of the Bovine Spongiform Encephalopathy Agent in Mice: Agent Definition based on a Review of 150 Transmissions. J Comp Pathol. 2005;132: 117–131. [DOI] [PubMed] [Google Scholar]
- 68. Lloyd SE, Linehan JM, Desbruslais M, Joiner S, Buckell J, Brandner S, et al. Characterization of two distinct prion strains derived from bovine spongiform encephalopathy transmissions to inbred mice. J Gen Virol. 2004;85: 2471–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rabbit brain homogenates seeded with different prion strains (cattle: BSE-C, sheep: SSPB/1 and sheep BSE-C, deer: CWD) or unseeded (de novo) were subjected to saPMCA. Seeded samples from round 10 and the unseeded sample from round 20 from in vivo isolates were digested with 100 μg/ml of proteinase K (PK) and analyzed by western blot using monoclonal antibody 6H4 to compare their differential electrophoretic migration and glycosylation patterns. Control: Normal rabbit brain homogenate. MW: Molecular weight.
(TIF)
Samples were diluted as described in figure and monoclonal antibody 6H4 was used at 1:10,000. The level of PrPC expression observed in TgRab brain was 5–6 times higher than NZW rabbit brain and 10–12 times higher than mouse brain. MW: Molecular weight.
(TIF)
A and B: Atrophy and paralysis of the hindquarters. C: Hindquarter footprints (black ink) of a healthy normal mouse. D: Footprints of a 500 days old non-inoculated mouse showing severe gait alteration. E: Histological image of the muscular tissue of a TgRab mouse with the spontaneous phenotype showing remarkable loss of muscular fibers (substituted by adipose tissue proliferation in the endomysium), irregular diameter and nuclear centralization in the remaining muscular fibers. F: normal muscular tissue of an unaffected mouse.
(TIF)
All images were taken at the same magnification. Bar 50 μm. A: In some mice a severe spongiform change was observed in the corpus callosum (cc) and deeper layers of the neocotex, the image corresponds to parietal cortex (Pc). This lesion could also be observed in the internal capsule. B: Immunolabelling of PrPd in the white matter showed punctiform morphology (arrowheads). C: Spongiosis of variable intensity was seen throughout the remaining brain areas which increased with the age of the mouse. In the image a section of thalamus with mild age-related spongiosis is seen. D: Parietal cortex. Punctifom immunolabelling around neurons was frequently observed in TgRab mice. Its distribution varied between mice but was present frequently in the cortices, striatum and thalamus. The fact that these cells are overexpressing rabbit PrPC could be an explanation of this signaling. All these animals yielded a negative result to PrPd by western blotting. E: The cochlear nucleus of the medulla oblongata consistently showed intense labelling of the neuropil in all negative controls. F: Cerebellar cortex. Intense, diffuse labelling of the neuropil was typically observed in the molecular (mol) and the granular (gra) layers and co-localized with the synaptic glomeruli.
(TIF)
A: TgRab survival times after inoculation. Each dot corresponds to a mouse. Dots with a red margin represent TSE positive animals. B: Brain lesion and PrPd deposit distribution of the first passage of BSE-RaPrPres in TgRab mice. C: 2nd passage of BSE-RaPrPres in TgRab mice. Lesion brain profiles and PrPd deposition profiles represent the mean semi-quantitative scoring (0–4, vertical axis) of the spongiform lesions (black) and the immunohistochemical labelling of PrPd deposits (red) against 14 brain regions (Pfc: piriform cortex, H: hippocampus, Oc: occipital cortex, Tc: temporal cortex, Pc: parietal cortex, Fc: frontal cortex, S: striatum, T: thalamus, HT: hypothalamus, M: mesencephalon, Mob: medulla oblongata, Cm: cerebellar nuclei, Cv: cerebellar vermis, Cc: cerebellar cortex). D: Histopathological characterization of BSE-RaPrPres in TgRab mice. Lesion and PrPd deposition patterns are remarkably similar to those observed in BSE C-derived strains. PrPd plaques are readily conspicuous in haematoxylin and eosin (H&E) stained sections (arrowheads). Upon immunohistochemical (IHC) labelling with 6H4 antibody against prion protein, the predominant pattern also consists of intensely labelled round-shaped plaques which can coalesce and form large aggregates. All images were taken at the same magnification. Bar 50 μm. E: Brain schematic mapping summary of the distribution of spongiform lesions and PrPd deposits in the brains of TgRab mice. The red dots depict the areas where spongiosis and/or PrPd deposits were mostly found in each group of infected mice. The image is consistent with that found in BSE-C derived strains strongly involving the medulla oblongata, ventral mesencephalon, thalamus and deep parietal cortex while sparing the remaining cortices and the hypothalamus.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.