Abstract
Transmissible spongiform encephalopathies (TSEs) are a group of neurodegenerative diseases affecting a wide range of mammalian species. They are caused by prions, a proteinaceous pathogen essentially composed of PrPSc, an abnormal isoform of the host encoded cellular prion protein PrPC. Constrained steric interactions between PrPSc and PrPC are thought to provide prions with species specificity, and to control cross-species transmission into other host populations, including humans. Transgenetic expression of foreign PrP genes has been successfully and widely used to overcome the recognized resistance of mouse to foreign TSE sources. Rabbit is one of the species that exhibit a pronounced resistance to TSEs. Most attempts to infect experimentally rabbit have failed, except after inoculation with cell-free generated rabbit prions. To gain insights on the molecular determinants of the relative resistance of rabbits to prions, we generated transgenic rabbits expressing the susceptible V136R154Q171 allele of the ovine PRNP gene on a rabbit wild type PRNP New Zealand background and assessed their experimental susceptibility to scrapie prions. All transgenic animals developed a typical TSE 6–8 months after intracerebral inoculation, whereas wild type rabbits remained healthy more than 700 days after inoculation. Despite the endogenous presence of rabbit PrPC, only ovine PrPSc was detectable in the brains of diseased animals. Collectively these data indicate that the low susceptibility of rabbits to prion infection is not enciphered within their non-PrP genetic background.
Author Summary
Prions are infectious pathogens causing irremediably fatal neurodegenerative diseases in human and in farmed or wild animals. They are formed from abnormally folded assemblies (PrPSc) of the host-encoded prion protein (PrPC). Different PrPSc conformational variants or ‘strains’ can propagate in the same host, giving distinct biological phenotypes. Like other pathogens, prions can transmit from one species to another, representing a zoonotic risk. A barrier, commonly referred to as “species barrier”, limits prion cross-species transmission. This barrier is supposed to reflect the steric incompatibility between invading prion PrPSc and PrPC of the infected host. Rabbit is one of the species that exhibit a pronounced resistance to prions. To gain insights on the molecular determinants of the relative resistance of this species to prions, we generated transgenic rabbits expressing sheep PrPC and assessed their experimental susceptibility to sheep scrapie prions, as routinely done with transgenic mouse models of prion disease. All transgenic rabbits developed a typical prion disease within 200 days, whereas wild type rabbits remained healthy more than 700 days after inoculation. These data indicate that the low susceptibility of rabbits to prion infection is not enciphered within their non-PrP genetic background.
Introduction
Scrapie in small ruminants, bovine spongiform encephalopathy (BSE) in cattle, chronic wasting disease (CWD) in cervids, transmissible mink encephalopathy (TME) or Creutzfeldt-Jakob disease (CJD) in humans belong to a group of fatal neurodegenerative diseases referred to as transmissible spongiform encephalopathies (TSEs) or prion diseases [1]. TSEs are characterized by the accumulation in the central nervous system (CNS) of a ß-sheet enriched, protease-resistant and aggregated isoform (PrPSc) of the host encoded cellular prion protein (PrPC). During TSE pathogenesis, PrPSc seeds, acquired through infection or arising from spontaneous conversion of wild type or mutant PrPC, are believed to template the conformational change of host PrPC to nascent PrPSc forms. This autocatalytic polymerization process leads to deposition of injurious deposits into the brain. PrPSc particles are thought to be the major if not the sole component of TSEs infectious agent or prion [2]. Distinct strains of prions are recognized phenotypically in a given host species. They cause TSEs with specific phenotypic traits, including time course to disease, neuropathological features and PrPSc biochemical properties. There is compelling evidence that prion strain diversity reflects stable differences in PrPSc conformations, at the level of the tertiary and/or quaternary structure [3–5].
A wide range of mammals like ruminants, pigs, rodents, carnivores or primates can be naturally and/or experimentally infected with prions. Prions are usually easy to transmit between individuals of the same species. Prions can also transmit between species, as exemplified by the emergence of variant CJD, following dietary exposure of humans to BSE prions. However, such events are restricted by a so-called ‘species’ or ‘transmission’ barrier, the strength of which depends essentially on interactions between host PrPC and the infecting prion strain type(s) [3, 4]. The force of the transmission barrier is classically gauged by the appearance of disease-specific, clinical signs and/or PrPSc in the brain and, sometimes, extraneural tissues of the new host. Their concomitant absence would usually suggest a resistance to infection or a disease incubation time exceeding that of the exposed host life span [6–9]. Rabbits have long been recognized as one of the best examples of a species refractory to TSEs agents as attempts to transmit Kuru, CJD, sheep scrapie, TME isolates and mouse adapted scrapie strains repeatedly failed [6, 10]. In marked contrast, cell-free PrPSc-templated conversion of PrPC by assays such as the protein misfolding cyclic amplification (PMCA, [11]) revealed that rabbit PrPC was fully convertible into rabbit PrPSc by seeds from scrapie infected, BSE and mouse scrapie brain sources ([12–14]). Yet, the amplified agents propagated with limited success in rabbits [12], leading support for the hypothesis that rabbit may barely develop clinical TSEs.
Numerous transgenetic studies in mice have demonstrated that mouse resistance to foreign prions can be abrogated by introducing in the murine genome the corresponding PRNP gene (the gene encoding PrPC). Transgenes can be introduced in animals knocked out for their own Prnp gene or on a murine wild-type background (reviews: [15, 16]). Vidal et al. demonstrates that transgenic mice expressing rabbit PrP are permissive to a broad panel of TSE sources from different species [14], strongly advocating for the full convertibility of rabbit PrPC into disease-associated isoforms. While transgenesis on a murine Prnp knockout background usually abolishes the transmission barrier, co-expression of the transgene with the wild-type Prnp gene can interfere with prion replication [17–19]. In sheep, different PRNP alleles tightly modulate the incidence and pathogenesis of classical scrapie. Three codons (for amino acids at positions 136, 154, and 171) act as major determinants, the V136R154Q171 allele conferring the highest susceptibility [20–22]. Experimental transmission of scrapie prions was markedly improved in transgenic mice overexpressing this allele [3, 23].
In this study, we report that rabbits engineered to express transgenetically the V136R154Q171 ovine PRNP allele develop a TSE syndrome upon experimental inoculation with scrapie. Disease occurred at full attack rate, with no apparent interfering effect of the rabbit wild-type PrPC.
Material and Methods
Ethics statement
All the experiments involving animals were done in strict accordance with the European Community Council Directive 86/609/EEC. The French committee for GMO,-formerly the governmental "Commission de Génie Génétique" that belonged to the French Ministry for Education, Research and Technology-, has approved the creation and experimental infection with prions of the rabbit transgenic animal model. The agreement number is 2992-II. It was issued the 12th of November 1998.
Generation of transgenic rabbits
A fragment of the sheep genome containing the V136R154Q171 allele of the Prnp gene was cloned in a bacterial artificial chromosome (BAC) vector previously used to generate the tg338 mouse line [23, 24]. This 125 kb insert, including 40 kb upstream and 60 kb downstream of the PRNP gene, was purified following a Not1 digestion and microinjected into New Zealand White rabbit embryos. Approximately 700 hundred embryos (from 41 donor females) were microinjected (mostly in the male pronuclei). 600 embryos were implanted into 30 pseudo-pregnant females. There were 28 live births, of which one was transgenic, as identified by PCR. Rabbit DNA was extracted by digesting ear fragments using proteinase K-SDS method. Digestion was followed by a DNA precipitation by ethanol. The two primers used to identify the sheep PRNP gene were TAGGCAGTTGGATCCTGGTT and CCCTATCCTACTATGAGAAA. The primers used to identify the control endogenous αS1-casein gene were CACTCCCTTGTTGAAAACTCTCCTCAG and ATTTTGTGGTTTCAGATCAACCAATAGG.
RT-qPCR analyses
To estimate the concentration of ovine and rabbit PRNP RNA in ovine PrP transgenic rabbits (TgOv), RT-qPCR analyses were performed on 3 animals per line, as previously described [25]. Two micrograms of total RNA (extracted from brain tissue) were reverse-transcribed with random adapters, following the manufacturer’s instructions [25]. Quantification was achieved by SYBR Green quantitative PCR (Applied Biosystems) using sets of primers specific to the PRNP ovine and rabbit sequences, resulting in the amplification of 100 bp long fragments, respectively. Two sets of primers were used for rabbit PRNP, owing the existence of two variants (with or without exon 2). The sequences of the specific primers were as follows: forward primer: 5’- TCATGGTGAAAAGCCACATAGG-3’, reverse primer: 5’- CCTCCGCCAGGTTTTGGT-3’ for ovine PRNP; forward primer: 5’-TCCTCTCGGCAGCTGTCAT-3’, reverse primer: 5’-GCTTCGGCCGCTTCTTG-3’ for Rabbit PRNP without exon 2; forward primer: 5’-AGAGGCCCCAGTCCAGTGTA-3’; reverse primer: 5’-CACTCCACGTGGCCACAA-3’ for Rabbit PRNP with exon 2. These primers were chosen so that they are located in different exons. The primers for β-actin were as follows: forward: 5’- CCGCATGCAGAAGGAGATCA-3’; reverse: 5’-AGAGCGAGGCCAGGATGGA-3’. For each sample, RNA concentration normalization was achieved using RT-qPCR on β-actin, as previously described [24]. It is thus given by the formula 2(CtPRNP-Ctβ-actin).
Scrapie inoculation of transgenic rabbits
Eighteen tgOv rabbits (9 males and 9 females) and 6 wild type (WT) rabbits (3 males and 3 females) aged 103 to 132 days (all bred at INRA Nouzilly) were selected for the experiment. Among them, twelve individually identified tgOv animals and six WT rabbits were injected with 50 μL of a 1% (wt/vol in 5% glucose) brain homogenate from tg338 mice infected with the LA21K fast strain. As controls, six tgOv animals were injected similarly with 50 μL of a 10% brain homogenate from a healthy tg338 mouse.
The inocula were prepared in a class II microbiological cabinet using disposable equipment, with strict safety rules, and immediately inoculated to the animals by the intracerebral route, at the level of the right parietal cortex (depth: 1 cm). Inoculations were performed under general anesthesia by injecting a mixture of xylazine (Rompun, Bayer, France) and ketamine (Imalgene, Merial, France) by the intramuscular route. All inoculations were carried out in compliance with ethics and animal welfare according to regulation requirements. The LA21K fast strain has been obtained through serial transmission and biological cloning by limiting dilutions of the Langlade field scrapie isolate (INRA Toulouse [26]) to tg338 mice. The LA21K fast infectious titer is 109.4 50% lethal doses (LD50)/g of tg338 brain [26].
Rabbits were housed in individual cages, in a dedicated biosafety level-3 facility. They were monitored using a video system and monitored daily for clinical signs and food consumption, by different investigators. Any death arising during the experiment was recorded and animals were necropsied. At clinical stage, rabbits were sacrificed by carbon dioxide suffocation and autopsied for brain and spleen collection. One half of each brain and spleen was stored at –20°C for immunoblotting and biochemical analyses, while the other half of organs was fixed in neutral-buffered 4% formalin for 1 week before paraffin-embedding for immunohistochemistry and histology.
Immunoblotting
Brains and spleens were analyzed for the presence of either PrPC or PrPSc by Western blotting as previously described [27]. Briefly, homogenates were prepared with a tissue homogenizer (Precellys, Bertin Technologies, France) in a 5% glucose solution. PrP was purified and concentrated using the Bio-Rad TeSeE purification kit. When needed, suspensions were treated with 100 μg/mL of proteinase K (Roche diagnostics, Germany) for 30 min at 37°C and the final pellet was suspended in Laemmli buffer. Samples were denatured at 100°C for 10 minutes, centrifuged at 20 000 g for 5 minutes and the supernatants were run on 12% SDS-PAGE gels. When two-dimensional gel electrophoresis was performed, as previously described [28], the second dimension was run on a 6–16% linear gradient SDS-PAGE. After transfer onto nitrocellulose membranes, samples were probed with Sha31 anti-PrP monoclonal antibody (generous gift of J. Grassi, CEA, Saclay, France, [29]), which binds to the YEDRYYRE amino acid sequence of the PrP protein (amino acid (aa) residues 146 to 153 of the rabbit PrP, Fig 1). This step was followed by the addition of a horseradish peroxidase-conjugate. Peroxidase activity was revealed using a chemiluminescent substrate (SuperSignal West Dura, Pierce, USA), and the signals were captured with a digital imager (Fluorchem 8900, Alpha Innotech, USA) or GeneGnome digital imager (Syngene, Frederick, Maryland, United States). The PrP levels and glycoforms ratios were quantified with the GeneTools software.
Fig 1. PrP amino acid sequences from New Zealand White rabbit and the sheep (VRQ allele).

Fragments identified by mass spectrometry in the brains of scrapie-diseased tgOv rabbit and specific of the sheep sequence are colored. Bold: Sha31 anti-PrP epitope.
Expression levels of PrPC in brains and spleens of rabbits and sheep were compared. Immunoblots revealed with the mAb Sha31 were quantified using the Alpha-Ease software (Alpha-Innotech, USA). Brain samples were diluted and PrP amounts were calculated according to a dilution curve of full-length recombinant ovine PrP (a generous gift of D. Marc, INRA, Tours, France), used as a reference.
Immunohistochemistry
Rabbit brain sections (5 μm thick) were treated as described previously [30]. Briefly, after being deparaffinised and rehydrated, tissue sections were incubated in 98% formic acid (MERCK) for 30 min at room temperature, then autoclaved for 15 min at 121°C in 10 mM citrate buffer (pH 6.1) and allowed to cool for 20 min. Sections were then subjected to a 15 min proteolysis at 37°C with 20 μg/mL of proteinase K. Endogenous peroxidase was inhibited using 0.3% hydrogen peroxide in methyl alcohol for 30 min at room temperature. Immunostaining was performed on a DAKO Autostainer according to the manufacturer’s instructions using the mouse monoclonal anti-PrP antibody 2G11 (gift from J. Grosclaude, INRA, Jouy-en-Josas, France, [30]) as primary antibody, followed by the DAKO EnVision+ System labelled polymer-HRP anti-mouse with 3, 3’-diaminobenzidine (DAB) as chromogen. After immunostaining, sections were counterstained with Mayer’s haematoxylin and cover-slipped. The 2G11 antibody was selected for optimal results with TSE-infected-tgOv rabbit brains without any background staining on tissue sections from both uninfected-tgOv and–WT rabbits.
Paraffin-embedded tissue blot (PET blot)
PET blot were performed using a method previously described [31]. Immunodetection was performed with Sha31 monoclonal antibody (4 μg/mL), followed by application of an alkaline phosphatase labeled secondary antibody (Dako, 1/500 final dilution). Enzymatic activity was revealed using NBT/BCIP substrate chromogen.
Histology
Paraffin-embedded sections were mounted on glass microscope slides and stained with hematoxylin and eosin.
Mass spectrometry
The equivalent of 6 mL of 20% brain homogenate from tgOv and WT rabbit, treated or not with proteinase K and denatured (see above) were resuspended in 100 mM NaCl, 10 mM EDTA, 10 mM TrisHCl pH 7.8, 0.5% DOC, 0.5% Igepal (Sigma). Monoclonal antibody Sha31 coupled to magnetic beads (Dynabeads M-280 Tosylactivated, Dynal) was added at the rate of 100 μL beads/240 mg tissue equivalent and reacted for 2 hours at 37°C. Beads were collected with a magnet, washed twice with PBS buffer and denatured in Laemmli buffer (10 min at 99°C). Samples were loaded on a 12% acrylamide gel, before electrophoresis and either immunoblotted (60 mg of tissue equivalent) or silver stained (1140 mg of tissue equivalent). The silver stained gels were compared to the western blot and protein bands that were at the same molecular masses than the PrPSc reactive bands were cut, rinsed and then reduced with dithiothreitol and alkylated with iodoacetamide. Samples were incubated overnight at 37°C with 12.5ng/μl trypsin (sequencing grade, Roche, Meylan, France) in 25 mM NH4HCO3 [28]. Tryptic peptides were analyzed by nanoLC-MS/MS with Q-q-TOF and Linear Ion trap.
For CapLC system coupled to Q-TOF Ultima Global (Waters Micromass, Manchester), the digested peptides were loaded on a precolumn (300μm i.d x 5mm, packed with C18 PepMap, LC Packings, Dionex) and desalted. Peptide separations were conducted on a C18 column (Atlantis dC18, 75mm I.D x 150 mm Nano Ease, Waters). Peptides were eluted with a 5–60% linear gradient with water/acetonitrile 98/2 (v/v) containing 0.1% formic acid in buffer A and water/acetonitrile 20/80 (v/v) containing 0.1% formic acid inbuffer B. Mass data were acquired using one MS survey followed by MS/MS scans on the 3 most intense ions detected. Data were processed using ProteinLynx Global server 2.2. The peptide and fragment masses were matched in database (nrNCBI) using MASCOT software (http://www.matrixscience.com). The mass tolerance was 0.2 Da for both precursor and fragment ions.
For Ettan MDLC system (GE Healthcare, Germany) coupled to LTQ Linear Ion Trap Mass Spectrometer (Thermo Electron, US), each sample was desalted using Zorbax 300-SB C18 trap column, 300μm i.d x 5 mm (Agilent Technologies, Germany). Peptide separations were conducted on a Zorbax 300-SB C18 column, 75 μm i.d x 150 mm (Agilent Technologies, Germany). Buffer A consisted of water with 0.1% formic acid while buffer B was 84% acetonitrile with 0.1% formic acid. Separation was performed by applying gradient of 15–55% B for 60 minutes at a flow rate of 400 nL/min. Mass data were acquired using one MS survey (m/z 500–2000) followed by MS/MS scans on the 3 most intense ions detected using Collision Induced Dissociation fragmentation mode. Identification was then performed with Bioworks 3.2 (Thermo Finnigan, San Jose, CA) software. MS/MS spectra were searched against the non-redundant Uniprot database (2006_12) and analysed using TurboSEQUEST (Thermo Finnigan, San Jose, CA). Search parameters included differential amino acid mass shifts for oxidized methionine (+16 Da) and carbamidomethylation on cystein (+57 Da). The output data were evaluated in term of Xcorr magnitude up to 1.7, 2.2 and 3.5 for charge states 1+, 2+ and 3+, respectively.
Cell culture
Confluent Rov cells (P2FJ6 clone, [26, 32]) were grown for 2 days in single wells of 12-well plates. Rov cells were incubated in culture medium containing 10 μL of 20% brain homogenate from tgOv rabbits, WT rabbits and tg338 mice challenged with LA21K fast prions, and from uninfected tgOv rabbits. After 2 days, the medium was removed; the cells were rinsed in phosphate-buffered saline (PBS) and split into 25 cm2 flasks. Each week, one flask was used for subpassaging, whereas another was used to prepare a cell lysate for PrP content analysis (see above). The total protein content was estimated by using a protein assay kit (bicinchoninic acid assay (BCA); Pierce).
Accession number
The Swiss-Prot accession numbers for the proteins mentioned in the text are sheep (P23907) and rabbit PrP (Q95211).
Results
Production and characterization of transgenic rabbits
One transgenic rabbit founder animal was obtained following microinjection of an ovine BAC DNA insert encompassing the entire PRNP transcription unit. This insert has already been used to produce various mouse transgenic lines that express the PrPVRQ allele [23]. For animal production, the tgOv transgenic founder was mated with a WT rabbit. Transmission of the transgene was of about 50% indicating that the rabbit founder was not a mosaic. The transgenic F1 rabbits were mated with WT rabbits giving birth to 39 offspring including 50% of heterozygous transgenic rabbits and 50% of non-transgenic control rabbits. Health of the rabbits did not appear to be affected by the presence of the ovine PRNP transgene (period of observation > 700 days).
The concentration of ovine PRNP RNA relative to that of rabbit PRNP RNA was estimated by RT-qPCR analyses on brain tissue extracts of tgOv rabbits and WT rabbits. In WT and tgOv rabbits, two rabbit PRNP RNA variants were found, as expected, resulting from the splicing (or not) of exon 2. The values were cumulated to obtain the total concentration of rabbit PRNP transcripts. In tgOv rabbits, ovine PRNP RNA levels were 1.5–2 fold higher than those of rabbit PRNP RNA (n = 3 rabbits analyzed). Rabbit PRNP RNA levels were similar between WT and TgOv rabbits, suggesting that expression of ovine PRNP has no interfering effects on the transcription of rabbit PRNP.
In the absence of rabbit-specific anti-PrP antibody, it was not possible to quantify the relative expression levels of ovine versus endogenous rabbit PrPC in tgOv rabbits. Immunoblots analyses indicate that the total levels of PrPC in brain of tgOv rabbits were approx. 1.5–2 fold higher than those found in the brain of their WT counterparts or sheep carrying the VRQ allele (Fig 2A), in agreement with the transcriptional analysis. TgOv rabbits expressed about 50 fold more PrPC in the brain than in the spleen, which compares with the ratio of about 60 found in WT rabbit.
Fig 2. Imunoblot analyses of PrP in the brains of wild-type and ovine PrP transgenic rabbits.
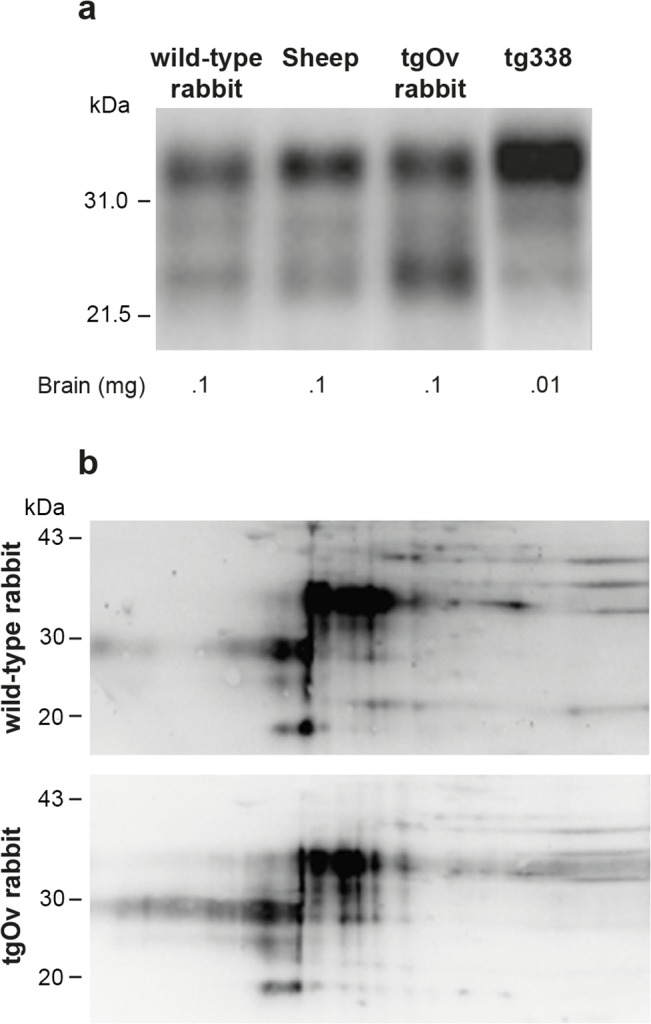
(a) PrPC electrophoretic pattern and level of expression in the brain of wild-type, sheep (VRQ allele), tgOv rabbit and tg338 mice. The amounts of material loaded are indicated. (b) Two-dimensional electrophoretic gel analysis of PrPC from wild-type and tgOv rabbit. The equivalent of 1mg of brain extract was used for comparison (Acidic side at left). Blots were probed with Sha31 anti-PrP antibody.
Two-dimensional gel electrophoresis was performed to determine whether any change in the isoforms pattern of PrPC was visible between WT and tgOv rabbits. Equivalent amounts of brain extracts were separated and transferred to nitrocellulose and probed with the Sha31 antibody (Fig 2B). Both extracts gave similar 2D patterns and none of the isoforms appeared specific or quantitatively different between the two types of rabbits.
Transgenic rabbits expressing ovine PrP are susceptible to scrapie
TgOv and WT rabbits were intracerebrally inoculated with LA21K fast scrapie strain, a fast strain that kills tg338 mice in less than 2 months [26].This agent induced a neurological disease in all inoculated tgOv rabbits (n = 12). The behavioral and clinical signs were invariant from animal to animal. The first behavioral signs were a drop in food consumption and restlessness. Early neurological signs (referred to as moderate in the S1 Video) were characterized by amaurosis and decreased time of random exploration in the cage. With disease progression, more severe neurological and behavioral signs were progressively observed, including loss of balance, disordered gait, paparesis, drop in food consumption and more severe amaurosis, (S2 Video). Animals were euthanized as soon as at least 3 of these signs were recorded. Neither pruritus nor tremor was observed. The clinical phase lasted less than 2 weeks. Animals were euthanized between 161 and 239 days post-inoculation (mean ± SEM incubation time: 192 ± 8 days; Fig 3). None of the mock-infected tgOv rabbits (n = 6) or LA21K fast-challenged WT rabbits (n = 6) presented any clinical signs during the time course of the experiment (Fig 3, S3 Video). They were euthanized healthy at 701 days post-inoculation, except two animals from each control group that were sacrificed, for comparison purpose with scrapie-sick animals.
Fig 3. Survival time of ovine PrP transgenic rabbits infected with scrapie.
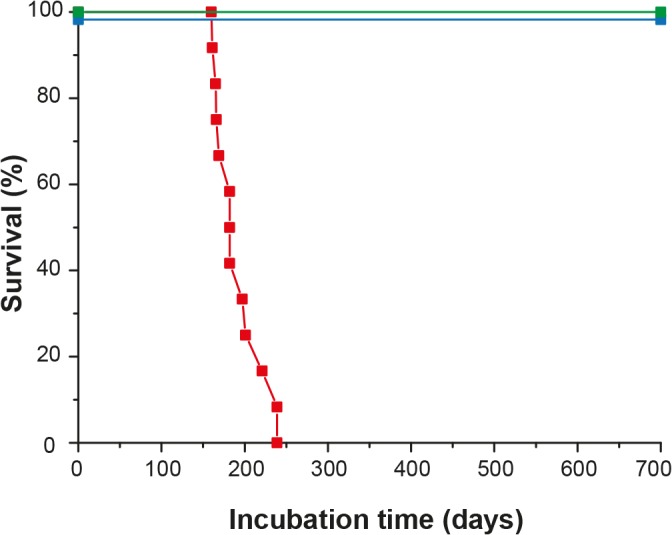
Kaplan-Meier survival plots of wild-type and tgOv rabbits inoculated intracerebrally with LA21K fast scrapie prions or with healthy brain homogenates. Animals were monitored for clinical signs and analyzed for the presence of PrPSc either by WB or immunohistochemistry. LA21K fast-inoculated tgOv rabbits: red symbol; LA21K fast-inoculated-WT rabbits: blue symbol; mock-inoculated tgOv rabbits: green symbol.
The brains of diseased tgOv animals were analyzed by immunoblotting and immunohistochemistry for the presence of PrPSc. Proteinase-K resistant PrPSc (PrPres) was readily detected in all infected tgOv rabbits (Figs 4 and 5), consistent with the efficient transmission. All controls remained PrPres negative (Fig 4A and 4B). In the absence of PK-treatment, PrPSc was essentially detected as full-length PrPSc (Fig 4A) as in tg338 brain [33], suggesting absence of endogenous cleavage generating the so-called C2 fragment [33]. Remarkably, LA21K fast electrophoretic pattern was conserved in tgOv rabbits with regards to apparent molecular mass and relative proportions of glycoforms (Fig 4B and 4C). None of the animals of the experiment, including positive transgenic rabbits, showed any detectable PrPres deposits in the spleen (Fig 4B). LA21K fast, as other fast ovine strains is lymphotropic in tg338 mice ([34], Fig 4B). It is likely that disease after intracerebral inoculation has occurred too rapidly in the rabbits to allow centrifugal spreading and replication of LA21K fast scrapie prions in the spleen.
Fig 4. Brain PrPSc in ovine PrP transgenic rabbit infected with LA21K fast scrapie prions.
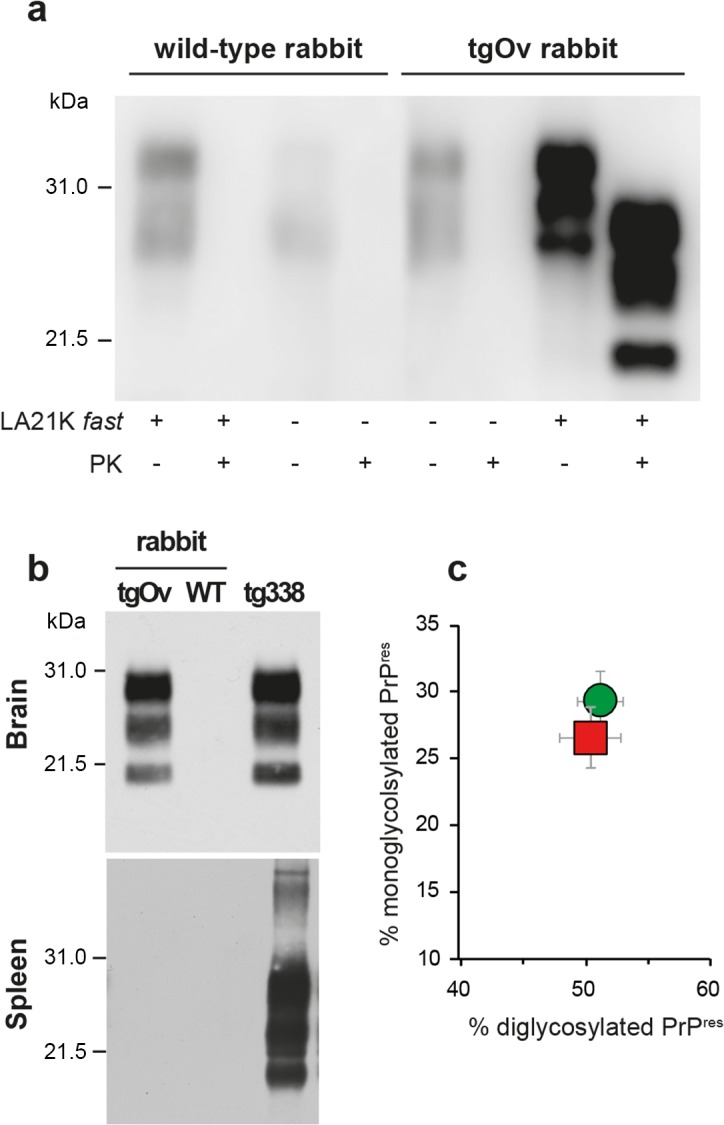
(a) Western blot analyses of the brain from wild-type and tgOv rabbits mock-infected or inoculated with LA21K fast prions for the presence of proteinase K (PK)-resistant PrPSc. The equivalent of 2 mg brain tissue were loaded in lane 1–6, 3 mg in lanes 7 and 8. (b) Electrophoretic pattern and (c) glycoform ratios (plotted as means ± SEM(c)) of PrPres in the brains and spleens of tgOv rabbit and tg338 mice infected with LA21K fast prions. The same amount of brain (0.5 mg) and spleen (3 mg) tissue equivalent was loaded on the gel.
Fig 5. Pathological PrP deposition and vacuolation in the brains of wild-type and tgOv rabbits inoculated with LA21K fast scrapie prions.
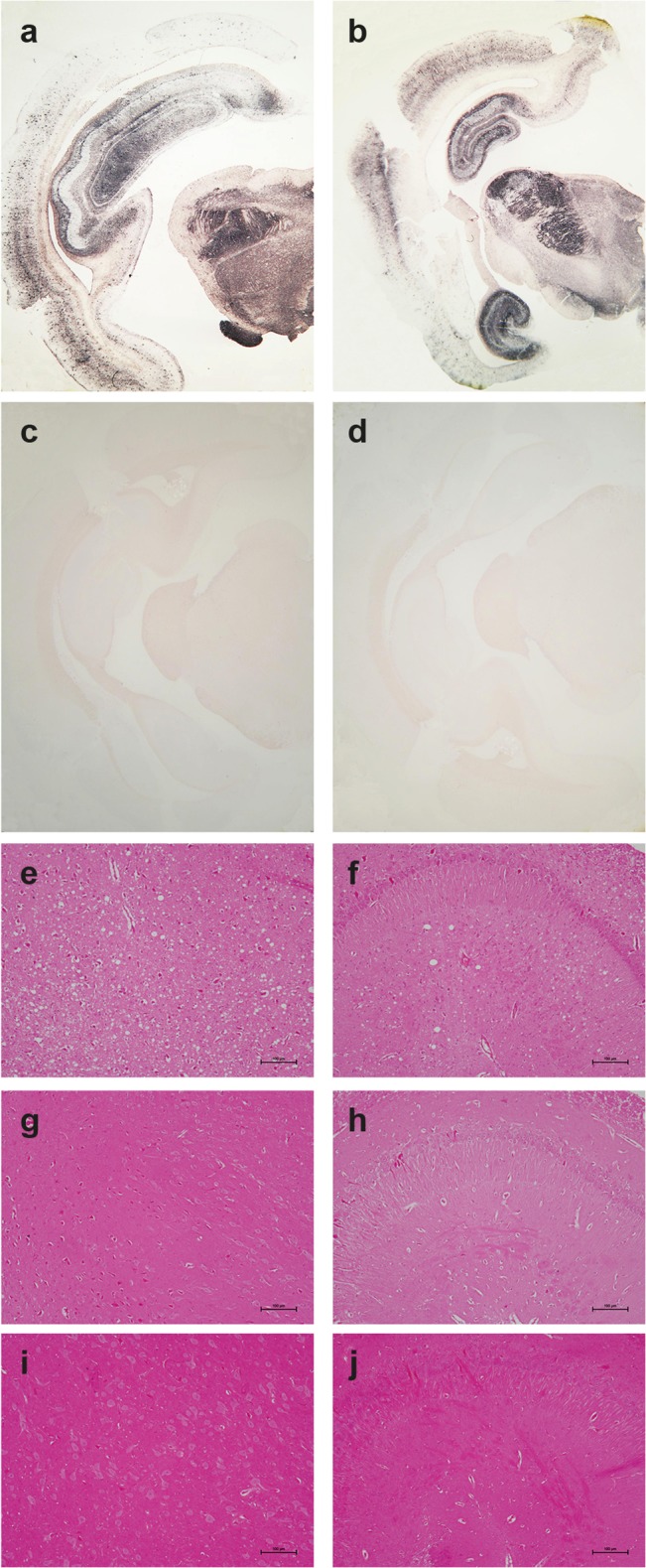
Midbrain sections from tgOv (a-b, e-f) and wild-type (c, g-i) rabbits challenged with LA21K fast scrapie prions and from a mock-infected tgOv rabbit (d, i-j). (a-d) PET blot analyses using monoclonal antibody Sha31 showed PrPres accumulation solely in LA21K fast challenged tgOv rabbits. (e-j) Hematoxylin and eosin-stained section at the level of the thalamus (e, g, i) and hippocampus (f, h, j) showing vacuolation, predominantly in the thalamus (e) and to a lesser extend in the hippocampus (f) of the LA21K fast challenged tgOv rabbits, but none in the control animals (g-j). Scale bar: 100 μm.
PrPres distribution in the brain was examined by PET blot analyses and immunohistochemistry. The greatest levels of PrPres deposits were observed in the thalamus, hippocampus and frontal cortex of scrapie-sick TgOv rabbits (Fig 5A and 5B). Sparse or moderate PrPSc deposition was seen in the cerebellum, obex and medulla oblongata (Table 1, analysis of 9 animals). Aged, uninfected tgOv rabbits and LA21K fast-challenged WT rabbits remained PrPres negative (Fig 5C and 5D).
Table 1. Regional distribution of PrPres and vacuolation in the brain of transgenic rabbits expressing ovine PrP challenged with LA21K fast prions.
| Brain Regions | PrPres deposits | Spongiosis |
|---|---|---|
| Cortex | +++ | - |
| Hippocampus | +++ | + |
| Thalamus | +++ | ++ |
| Cerebellum | +/- | +/- |
| Obex | + | +/- |
| Medulla oblongata | + | + |
-: no staining, no vacuolation; +/-: low; +: moderate; ++: pronounced; +++, intense staining or spongiosis.
Examination of histopathologic lesions in several brain areas of scrapie-sick TgOv rabbits (5 animals analyzed) indicated that spongiosis was prominent in the thalamus (Fig 5E). Mild spongiosis was also observed in the hippocampus (Fig 5F). Sparse spongiosis was observed in the medulla oblongata, obex and cerebellum (Table 1). The cortex was not vacuolated. There was no evidence of vacuolation in aged, uninfected tgOv rabbits and LA21K fast-challenged WT rabbits (Fig 5H–5J).
Collectively, these data indicate that scrapie infected tgOv rabbits exhibited the major clinical, biochemical and neuropathological hallmarks of TSEs.
Diseased transgenic rabbits accumulate only ovine PrPSc
To determine which of the rabbit or the ovine PrPC had been converted in scrapie-sick tgOv rabbits, immunoprecipitated brain extracts of healthy and infected tgOv rabbits,—treated or not with PK-, were analyzed by two techniques of mass spectrometry. Both analyses allowed detecting PrP fragments in the different gel bands corresponding to the western blot immunoreactive bands. In PK-treated, LA21K fast infected tgOv rabbit brains, five fragments were identified: ESQAYYQR; GENFTETDIK; VVEQMCITQYQR; GENFTETDIKIMER; EHTVTTTTKGENFTETDIK. Only one fragment VVEQMCITQYQR was identified in mock-infected tgOv rabbit, while no fragments were obtained from WT rabbits. All these fragments were located within the C-terminal part of the PrP protein and two could be assigned without ambiguities to the sheep PrP sequence (ESQAYYQR; VVEQMCITQYQR; Fig 1). Thus, PrPres molecules that accumulate in the brain of scrapie-sick tgOv rabbits was essentially of ovine origin, suggesting limited conversion, if any of endogenous rabbit PrPC during disease pathogenesis.
Prions from diseased transgenic rabbits readily infect Rov cells
LA21K fast scrapie prions can be efficiently passaged in Rov cells expressing ovine PrP [26, 32, 35]. We examined whether the prions produced in the brain of LA21K fast-sick tgOv rabbits would infect Rov cells with similar efficacy. Rov cells were exposed to similar amounts of brain homogenate from LA21K fast-infected tgOv rabbit and tg338 mice, and grown for up to 4 passages. At each passage, PrPres accumulation was monitored to assess the success of the infection and compare the levels of protein produced. In parallel, cells were exposed to brain extracts from aged, uninfected tgOv rabbits and from LA21K fast-inoculated WT rabbits. While immunoblots analyses of PK-digested cell lysates failed to detect PrPres in these controls, cells exposed to LA21K fast prions from either tgOv rabbit or tg338 mouse origin accumulated similar levels of PrPres at each passage (Fig 6). LA21K fast prions derived from tgOv rabbits and tg338 mice exhibit therefore similar efficacy to infect Rov cells. These data would further sustain the view that the prions produced in the brains of tgOv rabbits are of ovine origin.
Fig 6. Infection of Rov cells with TgOv- and tg338-passaged LA21K fast prions.
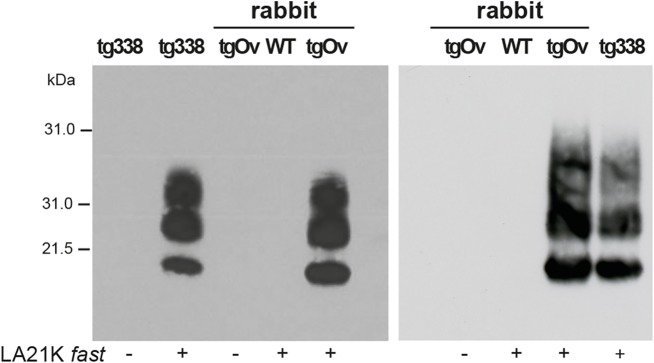
Western blot analyses for the presence of PrPres in lysates from Rov cells exposed to brain homogenates from WT, tgOv rabbits and tg338 mice that were mock infected or challenged with LA21K fast prions. Analysis was performed two (left panel) and four passages (right panel) after initial exposure of the cells. The loaded samples were normalized relative to total protein concentration.
Discussion
The limited number of prion-permissive cell models, the prolonged incubation time in farm species and the low susceptibility of conventional mouse lines to TSEs agents have favored development of mouse transgenesis in the TSE field. Some of the Prnp 0/0 mouse lines used to demonstrate the key role of PrPC in susceptibility to prions [36] were eventually engineered to (over)express PRNP genes from a wide range of mammalian species. These models considerably improved our knowledge on prion diversity, and most particularly on the molecular determinants of the transmission barrier during interspecies prion transmission [3]. A side effect of these studies was that transmission barriers that were considered as strong or ‘absolute’ were essentially abrogated [19, 23, 37, 38]. Here, we applied the same strategy to the rabbit species to clarify the respective roles of rabbit PrPC and non-PrP host factors in their pronounced, albeit not absolute [12], resistance to TSEs. We show that transgenic rabbits expressing a scrapie-susceptible ovine PRNP allele develop, at full attack rate, classical hallmarks of TSEs upon inoculation with scrapie prions, including fatal neurological diseases, clinical signs, PrPres deposition and vacuolation in the brain. This demonstrates that rabbits do not bear non-PrP factors that make them intrinsically resistant to prions.
The strategy used to generate the tg338 mouse line, a model highly susceptible to sheep scrapie sources [3, 23, 26, 34], was transposed to rabbit. At variance with the tg338 mouse line, transgenesis was performed on a wild type rabbit background, as Prnp 0/0 rabbits are not available, leading to the likely expression of both rabbit and sheep PrP proteins. Analyses of the PRNP transcripts indicated that ovine PRNP transcripts were present in tgOv rabbit brain at levels 1.5–2 fold higher than those of rabbit PRNP. Consistently, expression level of total PrPC in tgOv rabbit brains was 1.5–2 fold higher than that in WT rabbit and sheep. Co-expression of two different prion proteins can have a strong inhibitory effect on the conversion into prions of the transgenic PrPC protein, resulting in either no transmission, or a marked increase of the incubation time or no clinical disease [17–19, 39]. Here, scrapie developed at full attack rate in tgOv rabbits. The molecular, LA21K fast strain-specific [3] signature was conserved upon passage to another species expressing the same transgene in a different genetic background. The potential of LA21K fast prions to infect Rov cells [26, 32] was unaltered by the intermediate passage onto TgOv rabbits. Collectively, these data support the view that transgenic expression of the ovine PrPVRQ allele abrogated the rabbit species barrier to LA21K fast scrapie prions. Demonstrating formally that the species barrier has been fully abrogated and/or that co-expression of rabbit and sheep PrPC had no major interfering effects on scrapie pathogenesis would necessitate further subpassaging on tgOv rabbits to measure a potential reduction in incubation time, if any [3]. This experiment has not been done. However, it can be noticed that the mean incubation time observed at primary passage in tgOv rabbits (<200 days) is within the range of incubation time observed in sheep and transgenic mice expressing physiological levels of ovine PrP upon intracerebral infection, at the same dose, of fast scrapie prions ([23, 30]; 140 ± 5 days (6/6) in tg335 mice [23]).
While scrapie-infected tgOv rabbits showed characteristic signs of prion diseases, their WT counterparts remained healthy for more than 700 days, with neither detectable signs nor lesions or PrPSc deposits. This confirmed with another natural TSEs source the pronounced resistance of the rabbit species to foreign prions [6, 10]. We also showed by mass spectrometry that most if not all of the converted PrPC molecules in the brains of scrapie-sick tgOv rabbits were of ovine origin, indicating that conversion of rabbit PrPC was not favored by the conversion of ovine PrPC at vicinity. Collectively, these data suggest that rabbit PrPC is poorly convertible into LA21K fast PrPSc in vivo. Consistently, cell-free conversion of rabbit PrPC by SSBP/1 prions,-which exhibit a LA21K fast phenotype in tg338 mice (mean survival time of 63 ± 1 days in 6/6 tg338 mice on primary passage)-, necessitated a relatively high number of PMCA rounds as compared to other prion sources [12]. To further address the issue of the presence of rabbit PrPSc in the brain of diseased tgOv rabbits, secondary passage to transgenic mice expressing rabbit PrPC are planned ([12] and the accompanying paper by Vidal et al.).
The common shared view that rabbits were resistant to prion infection was not only attributed to rabbit PrPC sequence but also to its genetic background [12, 40–42]. Vidal and Castilla’s groups demonstrate by using transgenic modeling that rabbit PrPC, as many other mammalian PrPC, is fully convertible into disease-specific isoforms after infection with a broad panel of TSE sources [14]. Thus, taken separately, rabbit genetic background and rabbit PrPC cannot explain the apparently low susceptibility of rabbits to prion infection. What makes the rabbit species comparatively resistant to prion disease remains to be clarified. On the one hand, the diversity of prion sources inoculated to this species has remained too limited [6, 10] to definitely conclude that rabbits are poor acceptor for prions. The difficulty in identifying TSEs agents able to replicate on certain PrP sequence has been recently exemplified by studies on scrapie prions zoonotic potential. Such evidence (contrarily to a common- shared view) was provided because a panel of diverse scrapie sources was inoculated to human PrP transgenic mice [43]. On the other hand, it is possible that prion disease in rabbit would develop too slowly to be observed, because of a low conversion rate of rabbit PrPC. Transgenic modeling with mice expressing PrP at variable levels may help to verify this hypothesis [23, 44].
To summarize, we found that rabbit expressing ovine PrP at near physiological levels can develop a bona fide TSE upon infection with scrapie prions. The low susceptibility of rabbits to prion infection is not enciphered within the rabbit genetic background. Owing to its sensitivity and intermediate size, this model may be a valuable tool for studying TSE pathogenesis, most notably prionemia.
Supporting Information
(MOV)
The animals were euthanized at that stage.
(MOV)
(MOV)
Acknowledgments
We thank A. Le Dur (INRA-Jouy) for expert technical help and the P. Vincent and J. Auger of the PFIE facility (INRA-Nouzilly) for their expertise in rabbit breeding and in vivo experimentation.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1. Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–50. Epub 2001/04/03. 10.1146/annurev.neuro.24.1.519 24/1/519 [pii]. . [DOI] [PubMed] [Google Scholar]
- 2. Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95(23):13363–83. Epub 1998/11/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beringue V, Vilotte JL, Laude H. Prion agent diversity and species barrier. Vet Res. 2008;39(4):47 Epub 2008/06/04. 10.1051/vetres:2008024 v08241 [pii]. . [DOI] [PubMed] [Google Scholar]
- 4. Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318(5852):930–6. Epub 2007/11/10. doi: 318/5852/930 [pii] 10.1126/science.1138718 . [DOI] [PubMed] [Google Scholar]
- 5. Diaz-Espinoza R, Soto C. High-resolution structure of infectious prion protein: the final frontier. Nature structural & molecular biology. 2012;19(4):370–7. 10.1038/nsmb.2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gibbs CJ Jr., Gajdusek DC. Experimental subacute spongiform virus encephalopathies in primates and other laboratory animals. Science. 1973;182(107):67–8. Epub 1973/10/05. . [DOI] [PubMed] [Google Scholar]
- 7. Nonno R, Di Bari MA, Cardone F, Vaccari G, Fazzi P, Dell'Omo G, et al. Efficient transmission and characterization of Creutzfeldt-Jakob disease strains in bank voles. PLoS Pathog. 2006;2(2):e12 Epub 2006/03/07. 10.1371/journal.ppat.0020012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wadsworth JD, Asante EA, Desbruslais M, Linehan JM, Joiner S, Gowland I, et al. Human prion protein with valine 129 prevents expression of variant CJD phenotype. Science. 2004;306(5702):1793–6. Epub 2004/11/13. doi: 1103932 [pii] 10.1126/science.1103932 . [DOI] [PubMed] [Google Scholar]
- 9. Beringue V, Herzog L, Jaumain E, Reine F, Sibille P, Le Dur A, et al. Facilitated cross-species transmission of prions in extraneural tissue. Science. 2012;335(6067):472–5. 10.1126/science.1215659 . [DOI] [PubMed] [Google Scholar]
- 10. Barlow RM, Rennie JC. The fate of ME7 scrapie infection in rats, guinea-pigs and rabbits. Research in veterinary science. 1976;21(1):110–1. . [PubMed] [Google Scholar]
- 11. Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411(6839):810–3. 10.1038/35081095 . [DOI] [PubMed] [Google Scholar]
- 12. Chianini F, Fernandez-Borges N, Vidal E, Gibbard L, Pintado B, de Castro J, et al. Rabbits are not resistant to prion infection. Proc Natl Acad Sci U S A. 2012;109(13):5080–5. 10.1073/pnas.1120076109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vidal E, Fernandez-Borges N, Pintado B, Ordonez M, Marquez M, Fondevila D, et al. Bovine spongiform encephalopathy induces misfolding of alleged prion-resistant species cellular prion protein without altering its pathobiological features. J Neurosci. 2013;33(18):7778–86. 10.1523/JNEUROSCI.0244-13.2013 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vidal E, Fernández-Borges N, Pintado B, Eraña R, Ordoñez M, Márquez M, et al. Transgenic mouse bioassay: evidence that rabbits are susceptible to a variety of prion isolates. PLoS Pathog. 2015(8): ppat.1004977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Groschup MH, Buschmann A. Rodent models for prion diseases. Vet Res. 2008;39(4):32 10.1051/vetres:2008008 . [DOI] [PubMed] [Google Scholar]
- 16. Wadsworth JD, Asante EA, Collinge J. Review: contribution of transgenic models to understanding human prion disease. Neuropathol Appl Neurobiol. 2010;36(7):576–97. 10.1111/j.1365-2990.2010.01129.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Buschmann A, Pfaff E, Reifenberg K, Muller HM, Groschup MH. Detection of cattle-derived BSE prions using transgenic mice overexpressing bovine PrP(C). Archives of virology Supplementum. 2000;(16):75–86. . [DOI] [PubMed] [Google Scholar]
- 18. Telling GC, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen FE, et al. Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell. 1995;83(1):79–90. . [DOI] [PubMed] [Google Scholar]
- 19. Windl O, Buchholz M, Neubauer A, Schulz-Schaeffer W, Groschup M, Walter S, et al. Breaking an absolute species barrier: transgenic mice expressing the mink PrP gene are susceptible to transmissible mink encephalopathy. J Virol. 2005;79(23):14971–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bossers A, Schreuder BE, Muileman IH, Belt PB, Smits MA. PrP genotype contributes to determining survival times of sheep with natural scrapie. J Gen Virol. 1996;77 (Pt 10):2669–73. . [DOI] [PubMed] [Google Scholar]
- 21. Elsen JM, Amigues Y, Schelcher F, Ducrocq V, Andreoletti O, Eychenne F, et al. Genetic susceptibility and transmission factors in scrapie: detailed analysis of an epidemic in a closed flock of Romanov. Archives of virology. 1999;144(3):431–45. . [DOI] [PubMed] [Google Scholar]
- 22. Goldmann W, Hunter N, Smith G, Foster J, Hope J. PrP genotype and agent effects in scrapie: change in allelic interaction with different isolates of agent in sheep, a natural host of scrapie. J Gen Virol. 1994;75 (Pt 5):989–95. . [DOI] [PubMed] [Google Scholar]
- 23. Vilotte JL, Soulier S, Essalmani R, Stinnakre MG, Vaiman D, Lepourry L, et al. Markedly increased susceptibility to natural sheep scrapie of transgenic mice expressing ovine prp. J Virol. 2001;75(13):5977–84. Epub 2001/06/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vaiman D, Billault A, Tabet-Aoul K, Schibler L, Vilette D, Oustry-Vaiman A, et al. Construction and characterization of a sheep BAC library of three genome equivalents. Mamm Genome. 1999;10(6):585–7. . [DOI] [PubMed] [Google Scholar]
- 25. Daniel-Carlier N, Sawafta A, Passet B, Thepot D, Leroux-Coyau M, Lefevre F, et al. Viral infection resistance conferred on mice by siRNA transgenesis. Transgenic Res. 2013;22(3):489–500. 10.1007/s11248-012-9649-4 . [DOI] [PubMed] [Google Scholar]
- 26. Tixador P, Herzog L, Reine F, Jaumain E, Chapuis J, Le Dur A, et al. The physical relationship between infectivity and prion protein aggregates is strain-dependent. PLoS Pathog. 2010;6(4):e1000859 Epub 2010/04/27. 10.1371/journal.ppat.1000859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benestad SL, Sarradin P, Thu B, Schonheit J, Tranulis MA, Bratberg B. Cases of scrapie with unusual features in Norway and designation of a new type, Nor98. Vet Rec. 2003;153(7):202–8. . [DOI] [PubMed] [Google Scholar]
- 28. Syntin P, Dacheux F, Druart X, Gatti JL, Okamura N, Dacheux JL. Characterization and identification of proteins secreted in the various regions of the adult boar epididymis. Biology of reproduction. 1996;55(5):956–74. . [DOI] [PubMed] [Google Scholar]
- 29. Feraudet C, Morel N, Simon S, Volland H, Frobert Y, Creminon C, et al. Screening of 145 anti-PrP monoclonal antibodies for their capacity to inhibit PrPSc replication in infected cells. J Biol Chem. 2005;280(12):11247–58. 10.1074/jbc.M407006200 . [DOI] [PubMed] [Google Scholar]
- 30. Andreoletti O, Berthon P, Marc D, Sarradin P, Grosclaude J, van Keulen L, et al. Early accumulation of PrP(Sc) in gut-associated lymphoid and nervous tissues of susceptible sheep from a Romanov flock with natural scrapie. J Gen Virol. 2000;81(Pt 12):3115–26. . [DOI] [PubMed] [Google Scholar]
- 31. Andreoletti O, Orge L, Benestad SL, Beringue V, Litaise C, Simon S, et al. Atypical/Nor98 scrapie infectivity in sheep peripheral tissues. PLoS Pathog. 2011;7(2):e1001285 10.1371/journal.ppat.1001285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laferriere F, Tixador P, Moudjou M, Chapuis J, Sibille P, Herzog L, et al. Quaternary structure of pathological prion protein as a determining factor of strain-specific prion replication dynamics. PLoS Pathog. 2013;9(10):e1003702 10.1371/journal.ppat.1003702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dron M, Moudjou M, Chapuis J, Salamat MK, Bernard J, Cronier S, et al. Endogenous proteolytic cleavage of disease-associated prion protein to produce C2 fragments is strongly cell- and tissue-dependent. J Biol Chem. 2010;285(14):10252–64. 10.1074/jbc.M109.083857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Langevin C, Andreoletti O, Le Dur A, Laude H, Beringue V. Marked influence of the route of infection on prion strain apparent phenotype in a scrapie transgenic mouse model. Neurobiol Dis. 2011;41(1):219–25. Epub 2010/09/30. doi: S0969-9961(10)00311-6 [pii] 10.1016/j.nbd.2010.09.010 . [DOI] [PubMed] [Google Scholar]
- 35. Vilette D, Andreoletti O, Archer F, Madelaine MF, Vilotte JL, Lehmann S, et al. Ex vivo propagation of infectious sheep scrapie agent in heterologous epithelial cells expressing ovine prion protein. Proc Natl Acad Sci U S A. 2001;98(7):4055–9. 10.1073/pnas.061337998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bueler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, et al. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73(7):1339–47. . [DOI] [PubMed] [Google Scholar]
- 37. Scott M, Foster D, Mirenda C, Serban D, Coufal F, Walchli M, et al. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell. 1989;59(5):847–57. . [DOI] [PubMed] [Google Scholar]
- 38. Telling GC, Scott M, Hsiao KK, Foster D, Yang SL, Torchia M, et al. Transmission of Creutzfeldt-Jakob disease from humans to transgenic mice expressing chimeric human-mouse prion protein. Proc Natl Acad Sci U S A. 1994;91(21):9936–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perrier V, Kaneko K, Safar J, Vergara J, Tremblay P, DeArmond SJ, et al. Dominant-negative inhibition of prion replication in transgenic mice. Proc Natl Acad Sci U S A. 2002;99(20):13079–84. 10.1073/pnas.182425299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Loftus B, Rogers M. Characterization of a prion protein (PrP) gene from rabbit; a species with apparent resistance to infection by prions. Gene. 1997;184(2):215–9. . [DOI] [PubMed] [Google Scholar]
- 41. Qing LL, Zhao H, Liu LL. Progress on low susceptibility mechanisms of transmissible spongiform encephalopathies. Dongwuxue Yanjiu. 2014;35(5):436–45. 10.13918/j.issn.2095-8137.2014.5.436 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vorberg I, Groschup MH, Pfaff E, Priola SA. Multiple amino acid residues within the rabbit prion protein inhibit formation of its abnormal isoform. J Virol. 2003;77(3):2003–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cassard H, Torres JM, Lacroux C, Douet JY, Benestad SL, Lantier F, et al. Evidence for zoonotic potential of ovine scrapie prions. Nature communications. 2014;5:5821 10.1038/ncomms6821 . [DOI] [PubMed] [Google Scholar]
- 44. Fischer M, Rulicke T, Raeber A, Sailer A, Moser M, Oesch B, et al. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 1996;15(6):1255–64. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(MOV)
The animals were euthanized at that stage.
(MOV)
(MOV)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.


